Abstract
Microsatellite DNA sequences display allele length alterations or microsatellite instability (MSI) in tumor tissues, and MSI is used diagnostically for tumor detection and classification. We discuss the known types of tumor-specific MSI patterns and the relevant mechanisms underlying each pattern. Mutation rates of individual microsatellites vary greatly, and the intrinsic DNA features of motif size, sequence, and length contribute to this variation. MSI is used for detecting mismatch repair (MMR)-deficient tumors, which display an MSI-High phenotype due to genome-wide microsatellite destabilization. Because several pathways maintain microsatellite stability, tumors that have undergone other events associated with moderate genome instability may display diagnostic MSI only at specific mono-, di- or tetranucleotide markers. We summarize evidence for such alternative MSI forms (A-MSI) in sporadic cancers, also referred to as MSI-Low and EMAST. While the existence of A-MSI is not disputed, there is disagreement about the origin and pathologic significance of this phenomenon. Although ambiguities due to PCR methods may be a source, evidence exists for other mechanisms to explain tumor-specific A-MSI. Some portion of A-MSI tumors may result from random mutational events arising during neoplastic cell evolution. However, this mechanism fails to explain the specificity of A-MSI for di- and tetranucleotide instability. We present evidence supporting the alternative argument that some A-MSI tumors arise by a distinct genetic pathway, and give examples of DNA metabolic pathways that, when altered, may be responsible for instability at specific microsatellite motifs. Finally, we suggest that A-MSI in tumors could be molecular signatures of environmental influences and DNA damage. Importantly, A-MSI occurs in several preneoplastic inflammatory states, including inflammatory bowel diseases, consistent with a role of oxidative stress in A-MSI. Understanding the biochemical basis of A-MSI tumor phenotypes will advance the development of new diagnostic tools and positively impact the clinical management of individual cancers.
1. Introduction
In 1993, cancer geneticists were introduced to a new paradigm of tumor pathogenesis, namely that of microsatellite instability due to defects in the postreplication mismatch repair (MMR) pathway [1–3]. In these studies, comparisons of microsatellite DNA markers in normal versus colorectal tumor tissues revealed alterations in microsatellite allele lengths, a phenomenon later termed microsatellite instability (MSI). Tumor-specific MSI has become a widely used diagnostic assay for loss of MMR function in tumors. In this review, we discuss the various types of tumor-specific MSI patterns that have been described in the literature over the ensuing 20 years, and the relevant mechanisms that underlie each pattern. We focus primarily on two types of tumor-specific MSI patterns, MSI-low (MSI-L) and elevated microsatellite alterations at selected tetranucleotides (EMAST). We have designated these MSI patterns as alternative MSI (A-MSI). A-MSI has been detected to varying extents in a wide range of sporadic cancers, including colon, rectal, endometrial, ovarian, lung, melanoma, pancreatic, gastric and bladder. We will not discuss MSI that is associated with Lynch Syndrome (formerly known as hereditary non-polyposis colorectal carcinoma), a familial cancer predisposition syndrome caused by MMR defects (see [4] for a review of Lynch Syndrome).
2. Microsatellite Genetics and Mismatch Repair
Microsatellite sequences are tandem repeats of short (1 – 6 bp) DNA motifs and are ubiquitous in eukaryotic genomes, making up ~3% of the total DNA in the human genome [5]. Microsatellites display high levels of polymorphism among individuals, which has resulted in their widespread use as markers in association studies, population genetics, forensics, and cancer diagnostics [6, 7]. DNA features intrinsic to the microsatellite itself, such as motif size (mononucleotide, dinucleotide, etc), sequence composition, and repeat number (tract length), are the strongest predictors of mutability in a genome-wide analysis [8]. Consequently, the germline mutation rates of individual microsatellites vary greatly from locus to locus [6].
Over the past 12 years, our laboratory has extensively studied the mechanisms of somatic microsatellite mutagenesis in human cells. We have analyzed how DNA features affect mutagenesis, using both ex vivo (non-tumorigenic cell lines) [9–11] and in vitro (DNA polymerase fidelity) assays [12–16]. Our direct experimental analyses demonstrated that the mutability of each microsatellite is dependent upon features intrinsic to the repeated DNA sequence itself (reviewed in [17]). Examples of the variations in microsatellite mutation rates are given in Table 1. In non-tumorigenic, MMR-proficient human lymphoblastoid cells, microsatellite mutation rates vary over two orders of magnitude, from a low of ~ 2 × 10−7 mutations/cell/generation for a [GT/CA]10 allele to a high of ~ 5 × 10−5 mutations/cell/generation for a [TCTA/AGAT]9 allele (Table 1). Recently, we have used a combination of computational, mathematical and experimental approaches to determine at what length a short tandem repeat sequence becomes a microsatellite [18, 19]. In both studies, a critical threshold length was identified where tandem repeats change their mutational behavior. This threshold value is 8 units for mononucleotides, 5 units for dinucleotides, and 4 units for tetranucleotide repeats [18, 19].
Table 1.
Comparison of Human Cell and In Vitro Polymerase Mutation Rates at Selected Mono-, Di-, and Tetranucleotide Microsatellites
| Microsatellite Mutation Rate × 10−5 | |||||
|---|---|---|---|---|---|
| Ex Vivo Shuttle Vectora (lymphoblastoid cell lines) |
In Vitro Polymeraseb | ||||
| Motif | Sequence | MMR+ c | PMS2 −/−d | Pol αe | Pol βf |
| Mononucleotide | [A/T]8 | n.d. | n.d. | 990 | 7300 |
| [G/C]7 | 0.03 | 1.4 (47X)g | n.d. | n.d. | |
| [G/C]10 | 5.5 | n.d. | n.d. | 8300 | |
| Dinucleotide | [GT/CA]10 | 0.021 | 3.9 (190X) | n.d. | 500 |
| [GT/CA]19 | 0.62 | 720 (1160X) | n.d. | 1700 | |
| [TC/AG]11 | 0.33 | 28 (85X) | 730 | 1100 | |
| Tetranucleotide | [TTCC/AAGG]9 | 0.56 | 11 (20X) | 690 | 920 |
| [TTTC/AAAG]9 | 3.5 | 13 (3.7X) | n.d. | n.d. | |
| [TCTA/AGAT]9 | 4.8 | n.d. | n.d. | n.d. | |
Median microsatellite mutation frequencies per locus per generation are shown.
Polymerase error frequency per round of synthesis. Polymerase error rates for a microsatellite pair were calculated by adding the polymerase error rates of each complementary strand together.
Data from [9, 11], and unpublished (see footnote 1)
Data from [11] and unpublished (see footnote 1)
Data from [14] and unpublished
Data from [13] and unpublished (see footnote 1)
Fold increase in mutation rate upon loss of PMS2 (PMS2−/PMS2+)
n.d., not determined
The favored model to explain microsatellite mutagenesis is the slipped strand mispairing model [6, 20]. During DNA replication, a polymerase encountering unusual DNA secondary structures or DNA damage may pause and/or dissociate from the primer-template, allowing the DNA strands to transiently denature. Because of the repetitive nature of microsatellites, bases comprising the repeat motifs may renature in an incorrect alignment, forming a looped DNA structure that includes one or more units of the repeat. If the DNA polymerase rebinds to such misaligned, slipped strand intermediates and continues DNA synthesis, and if the loop is not repaired (see below), the slipped strand intermediate will be fixed into either a unit-based insertion (loop in the nascent strand) or a unit-based deletion (loop in the template strand) during the next round of DNA replication. For the majority of DNA polymerases studied, including the replicative Pols α, δ and ε, the error rates for unit-based insertions/deletions (Indels) within microsatellite sequences are 10- to 100- fold higher than those for traditional frameshifts within a gene coding sequence [12, 14, 16]. One very notable exception to this rule is human DNA Pol κ, which produces few unit-based Indel errors during synthesis of di- and tetranucleotide microsatellites [16, 21]. DNA polymerases also vary as to the effect of intrinsic DNA features on error rates. As shown in Table 1, Pol α-primase error rates during synthesis of mono-, di- and tetranucleotide repeats is fairly constant, whereas Pol β error rates are much higher during synthesis of mononucleotide than of di- or tetranucelotide repeats of similar length. The exact biochemistry and thermodynamics involved in the simplified slipped strand mispairing model described above must still be elucidated. However, our data indicate that the probability of strand slippage errors arising within microsatellites is highly dependent upon the exact interaction of a specific polymerase with a specific microsatellite motif (Table 1).
The insertion/deletion loops (IDLs) created during microsatellite DNA replication are efficiently corrected by MMR [22, 23]. In human cells, recognition of polymerase errors in the newly synthesized strand is carried out by the MutS homologues MutSα (hMSH2 and hMSH6) and MutS β hMSH2 and hMSH3). Upon recognition of the mismatch, MutL heterodimers, either MutLα (hMLH1 and hPMS2) or MutLγ (hMLH1 and hMLH3), are recruited and coordinate the excision of the nascent strand, followed by DNA resynthesis. Using our ex vivo assay, we have demonstrated that hPMS2 correction efficiency within microsatellite sequences depends on both the microsatellite motif size and sequence ([11] and unpublished results1). In hPMS2-deficient lymphoblastoid cells, dinucleotides are destabilized to the greatest extent, followed by mononucleotides; tetranucleotide microsatellites are destabilized the least of any motifs tested (Table 1). Using a reporter assay and comparing hMLH1-deficient and proficient human colorectal carcinoma cell lines, Campregher et al. demonstrated that hMLH1was more effective for a mononucleotide [G/C]16 allele than for either a mononucleotide [A/T]10 or [CA/GT]13 allele [24]. Thus, the resulting microsatellite mutation frequencies in MMR-deficient cells varies at least two orders of magnitude for alleles of varying motif size, sequence composition, and lengths.
3. Microsatellite Instability and Cancer Diagnostics: The MSI-High Phenotype
Microsatellites have been used extensively as diagnostic tools for the detection and classification of tumors. In these assays, specific microsatellite loci in genomic DNA from both normal and tumor tissues are amplified, and the resulting fragment lengths are analyzed. Loci that show alterations in fragment length between normal and tumor tissue are considered to be unstable, whereas loci that exhibit no length variations between normal and tumor are considered stable. Tumors that exhibit no length variation for any of the marker loci analyzed are termed MSS, for microsatellite stable.
Microsatellite markers are perhaps best known for their use in the diagnosis of tumors that have lost MMR activity. Efforts to discover markers specific for loss of MMR led to the adoption of the Bethesda panel. Proposed in 1998 by a National Cancer Institute workshop, this internationally standardized diagnostic test consists of two [A/T]n mononucleotide and three [GT/CA]n dinucleotide microsatellite markers [25] (Table 2). The mononucleotide markers are considered quasimonomorphic, in that most individuals have the same number of repeat units at that allele (Figure 1A). The dinucleotide markers are polymorphic, such that >1% of the population show heterozygosity in the number of repeats [26] (Figure 1B). MMR-deficient tumors generally display instability at ≥ 2 loci when using the Bethesda panel, or instability at ≥ 30 – 40% loci if more than five loci are analyzed [25]. This phenotype is defined as MSI-High (MSI-H). Because mononucleotide markers have proven to be more sensitive and specific for detecting MSI-H [27, 28], the revised Bethesda guidelines [29] suggest using a panel of five [A/T]n mononucleotide markers (Table 2). Screening tumors for an MSI-H phenotype is important clinically, because colorectal cancers exhibiting MSI-H present with distinct clinicopathological features, including poor tumor differentiation, lymphocytic infiltration, mucinous character, lower stage, less invasiveness, and better prognosis [30]. Genetically, the MSI-H phenotype in sporadic cancers results from silencing of the hMLH1 gene by promoter hypermethylation. For a more detailed review of MSI-H tumors, the interested reader is referred to reference [26].
Table 2.
Representative Markers Analyzed to Distinguish Microsatellite Instability Phenotypes in Cancer Diagnostics
| Type | Marker | Motif a | Sequence b | Reference |
|---|---|---|---|---|
| MSI-H | BAT25c | Interrupted Mono | gag [T]4GTG[T]4G[T]7GA[T]25 gag | [25] |
| BAT26c | Pure Mono | ggt [A]27 ggg | [25] | |
| D2S123c | Interrupted Di | gat [AC]21[AT]2 ttta[taga]3 | [25] | |
| D5S346c | Interrupted Di | ttt [AC]20[AG]2 taa | [25] | |
| D17S250c | Pure Di (2) | gtt [TA]7[CA]2TACA….CAAA[CA]19 ctt | [25] | |
| NR-21d | Pure Mono | gcc [T]21 agc | [29] | |
| NR-24d | Pure Mono | cta [T]23 gtg | [29] | |
| NR-27d | Pure Mono | ggt [A]26 gcc | [29] | |
| MSI-Le | D2S136 | Pure Di | att [CA]19 cca | [37, 90] |
| D3S1611 | Interrupted Di | gtt [AC]3A[AC]14 tag | [32] | |
| D5S107 | Compound Di | atg [CA]9AA[CA]20[GA]7 cat | [25, 34,37,48,90,92] | |
| D6S87 | Complex (Di) | aac [A]10..[AT]6 [AC]2[AT]3[GT]3AT[AC]17[AG]2 taa | [37, 90] | |
| D10S197 | Interrupted Di | cat [CA]3GA[CA]21 aga | [25,27,33,37,46,59] | |
| D17S261 | Pure Di | ttt [AC]17 ggc | [28,34,37,90,92] | |
| D18S34 | Interrupted Di | ttt AG[AC]26 att | [32,37,48,62,90] | |
| D6S344f | Complex Di | cta AG[AC]17[TC]9[TA]6[CA]8[TA]2TGTA ttt | [91] | |
| EMASTg | D9S242 | Complex (Tetra) | caa [AAAG]3AAG[AAAG]15GG[AG]5A [AAGG]6AGAAG[AAAAG]3AAAAAG cca |
[42,43,70,71] |
| MYCL1h | Compound (Tetra) | aag GAAAA[GAAA]2TAAA[A/G]16[GAAA]13 [GAAAA]2G[A]8[GAAAA]3G[A]6[GAAAA]2 |
[42,43,70,71] | |
| UT5320 | Complex (Tetra) | caa [AAT]9GAAA[GA]7[GGAA]5AGG[AGGG]2 [A/G]7[AAAG]14[AAGG]8AAG[AAAG]5AAAA taa |
[71] | |
A pure microsatellite is defined as an array containing a single tandem repeat motif sequence. Compound microsatellites contain two types of microsatellites. Complex microsatellites contain three or more types of microsatellites. For complex alleles, the motif size of the longest microsatellite is indicated in parentheses. Interrupted microsatellites contain one or more non-repeated basepairs internal to the array.
Sequences within the amplicon were obtained using the GRCh37/hg19 February 2009 Human Genome Assembly found on the University of California Santa Cruz (UCSC) Genome Browser (http://www.genome.ucsc.edu). Underlines indicate tandem repeats above the microsatellite threshold length [18, 19].
Bethesda consensus panel of microsatellite markers. The MSI-H phenotype is defined as instability at ≥ 2 markers, and the MSI-L phenotype is defined as instability at one marker, using this panel.
Revised Bethesda consensus panel of microsatellite markers. The MSI-H phenotype is defined as instability at 2 or more markers in this panel.
Examples of MSI-L markers that have been used in addition to the Bethesda panel.
This marker used for MSI analysis in COPD patients.
The EMAST phenotype is tested using a panel of 5–7 markers, only some of which are listed. EMAST is defined as instability at one marker or at 2 markers of the panel, depending on the study.
The MYCL1 marker is also used to detect the MSI-L phenotype.
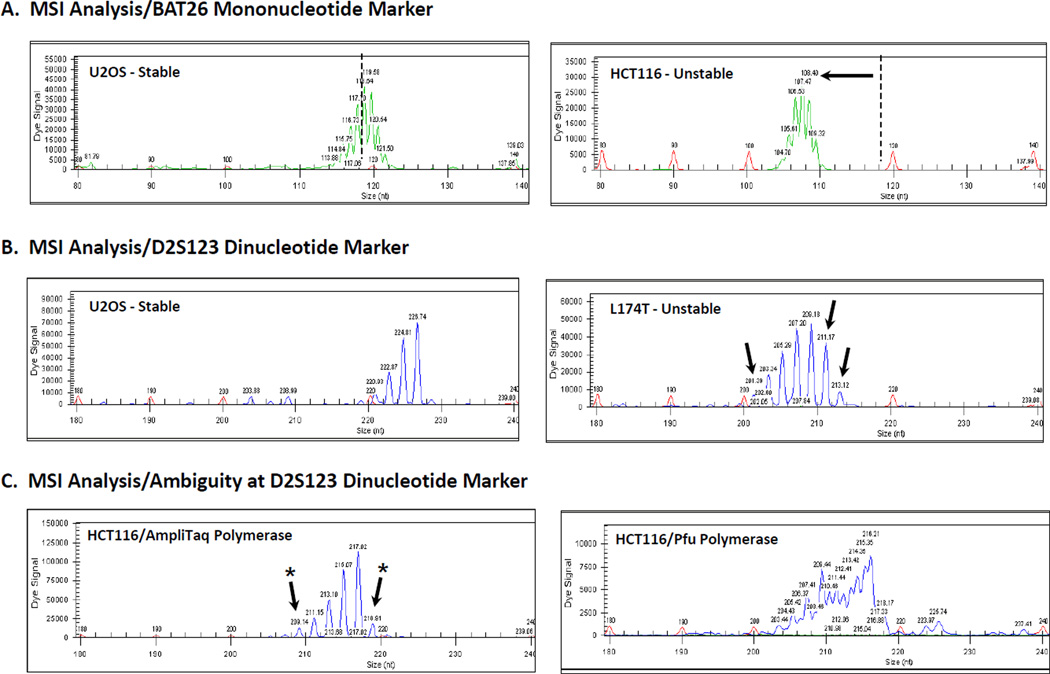
Figure 1. MSI Analysis of MMR+ and MMR- cell lines at the BAT26 and D2S123 loci.
PCR reactions were performed using genomic DNA isolated from MMR+ U2OS and MMR-HCT116 or L174T cell lines and primer sets in which one primer was labeled with WellRED D3 dye (BAT26) or WellRED D4 dye (D2S123). The amount of input DNA, MgCl2 concentration, and annealing temperature were optimized for each primer set. PCR products and size standards were mixed with formamide, run on a Beckman Coulter CEQ 8000 system, and analyzed using the Fragment Analysis software. A. MSI analysis using the BAT26 marker. DNA from U2OS cells shows stability at the quasimonomorphic locus with a fragment of 119nt (PCR stutter bands are also evident). DNA from MMR-deficient HCT116 cells display instability with a shift to a smaller allele size. B. MSI analysis using the D2S123 marker. DNA from U2OS cells is homozygous and stable for this allele, but DNA from MMR− L174T show extra alleles as indicated by the arrows. C. Ambiguity at the D2S123 marker. Amplification of DNA from HCT116 cells showed minor new alleles (arrows with asterisk) that were not resolved upon utilizing a proofreading-proficient DNA polymerase in the PCR reaction.
4. Alternative Forms Of MSI Observed In Cancers
Microsatellite mutation rates vary greatly from locus to locus, and the intrinsic DNA features of motif size and sequence, as well as tract length, greatly influence microsatellite mutagenesis. Mechanistically, this variation is due to differences in both DNA polymerase error rates and in MMR correction efficiency within repeats of varying size and sequence. Diagnostically, then, we argue that instability at mono-, di- and tetranucleotide markers will arise by distinct pathways, since several genome maintenance pathways are involved in maintaining microsatellite stability (Figure 2). Tumors that have lost hMSH2 or hMLH1 expression are likely to be deficient in all MMR functions, including polymerase error correction as well as DNA damage signaling. Therefore, these tumors display the MSI-H phenotype, due to widespread destabilization of all sequences, including microsatellites, throughout the genome (Figure 2). On the other hand, tumors that have undergone other molecular events associated with moderate genome instability may display MSI only at selected microsatellite markers. In this section, we summarize evidence for such alternative forms of tumor-specific MSI phenotypes (A-MSI) at di- and tetranucleotide microsatellites.
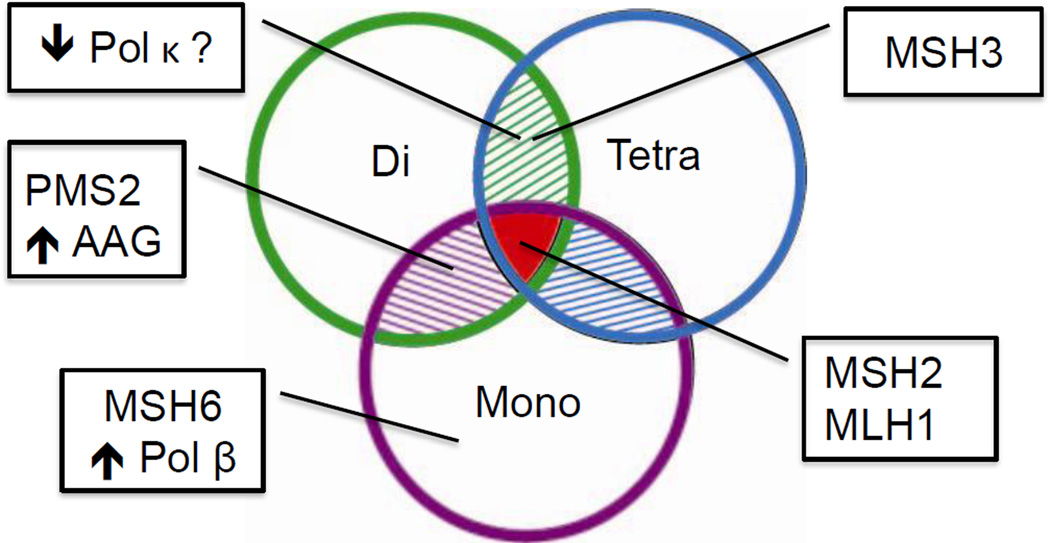
Figure 2. MSI specificity in tumor cells reflects underlying genetic alterations.
The microsatellite markers used to detect tumor-specific MSI are very diverse at the DNA sequence level (see Table 2 for details). In diagnostics, the primary microsatellite markers used to diagnose Lynch Syndrome are mononucleotide repeats (purple circle). Dinucleotide repeats (green circle) and tetranucleotide repeats (blue circle) have been used to detect the MSI-L and EMAST phenotypes, respectively. Loss of the major mismatch repair proteins MSH2 and MLH1 results in widespread destabilized of all microsatellite motifs, and the MSI-H phenotype (red intersect). Alternative MSI (A-MSI) phenotypes arise when other genome maintenance pathways are disrupted. Loss of MSH6 disrupts only MutSα-mediated repair, and destabilizes only mononucleotides, whereas loss of MSH3 disrupts only MutSβ-mediated repair and destabilizes both di- and tetranucleotides. Loss of PMS2 results in a higher rate of instability at dinucleotide repeats. Increased expression of alkyladenine glycosylase (AAG) results in MSI at mono- and dinucleotides. Based on in vitro characterization of DNA polymerase fidelity at various microsatellites, tumors with increased expression of Pol β are expected to display A-MSI restricted to mononucleotides, while tumors with decreased expression of Pol κ are expected to display A-MSI restricted to di- and tetranucleotide repeats. See text for details and references.
4.1. Dinucleotide microsatellite instability
A form of MSI that differs from MSI-H has been routinely observed during the course of applying the Bethesda panel to cancer diagnostics. This phenotype, termed MSI-low (MSI-L), was defined as instability at one microsatellite marker when using the Bethesda consensus panel, or instability at < 30 – 40% of loci if greater than five markers are analyzed [25]. MSI-L tumors display microsatellite allele length changes primarily at dinucleotide microsatellites [27, 28, 31]. As shown in Table 2, the precise sequence compositions of markers used to detect MSI-L varies greatly, and includes complex microsatellites comprised of several repeated motifs. In one study of MSI-L in melanoma, instability was restricted to di- and trinucleotide microsatellites, and no MSI was observed at several mononucleotide markers [32]. MSI-L tumors have been consistently identified over the past two decades [28, 33–36]. The phenotype occurs in 3 – 15% of all colorectal cancers [26], and has been detected in sporadic endometrial and ovarian carcinomas [33], melanoma [32] and pancreatic cancer [37].
4.2. Tetranucleotide microsatellite instability
A set of microsatellites distinct from the Bethesda panel also has been used in cancer diagnostics. This panel includes complex microsatellites containing long tetranucleotide repeats (Table 2), and allele length variation that is detected using this panel is termed EMAST, for elevated microsatellite alterations at selected tetranucleotides [38]. Although this type of MSI has been less intensively studied than MSI at mono- and dinucleotide markers, EMAST has been found to varying extents in non-small cell lung, bladder, ovarian, colon, rectal and skin cancers [38–44]. EMAST is common in bladder cancer, with up to 40% of samples displaying this instability. Conversely, bladder cancers rarely exhibit MSI-H, and EMAST-positive primary bladder tumors are not correlated with absence of either MSH2 or MLH1 [45]. Similarly, EMAST is common, but MSI-H is rare, in rectal cancer [42]. Primary lung cancers displaying MSI at the EMAST markers also do not display a phenotype consistent with MMR defects [39]. A higher EMAST frequency is associated with advanced stage and an ulcerated pathologic phenotype in adenocarcinomas [43].
5. The A-MSI Debate
While the existence of A-MSI is not disputed, there is a wide-ranging disagreement particularly about the origin and pathologic significance of MSI-L and whether it should be recognized as a distinct genetic subgroup in cancer [36, 46, 47]. A bimodal distribution of MSI frequency (number of unstable microsatellite loci/total number of loci examined) is observed using mono- and dinucleotide markers, with a breakpoint near 30% [27, 48, 49]. One interpretation of this distribution is that the breakpoint distinguishes MSI-H from MSI-L cancers, with MSS tumors being those that show 0% MSI. An alternative interpretation is that the breakpoint distinguishes MSI-H from MSS cancers, and that tumors displaying MSI frequencies in the 1 – 30% range are actually the same as MSS tumors, but with differing degrees of spontaneous microsatellite mutation events. The EMAST phenotype has not been as intensely debated in the literature; however, we feel that that similar arguments can apply to this panel of markers as well. In the following sections, we discuss four arguments that have been proposed to explain the phenomenon of A-MSI in tumors: 1). A-MSI is an experimental artifact; 2). A-MSI reflects spontaneous mutation events at microsatellite markers; 3). A-MSI is a distinct genetic subgroup with a moderate defect in repair or replication and 4). A-MSI reflects a damage-induced phenotype. We note that A-MSI tumors most likely represent a heterogeneous grouping of tumors, and that these arguments are not mutually exclusive.
5.1. Argument #1: A-MSI is an Experimental Artifact
The diagnostic sensitivity of a given microsatellite marker is highly dependent upon its sequence [27, 48]. Using a 31 microsatellite marker panel of varying motif sizes, sequences and complexities, Dietmaier et al. demonstrated that the intrinsic DNA features of the marker determined the frequency of MSI, as well as the reliability of PCR interpretation [27]. In this study, pure or complex dinucleotide markers showed the highest ambiguity scores, defined by assessment of PCR patterns by three independent observers. The D5S346 dinucleotide from the Bethesda consensus panel is one such highly ambiguous marker. This finding is significant, as many of the markers used to identify MSI-L are pure or complex dinucleotides (Table 2). Two multicenter studies were performed to evaluate the reliability of MSI testing [50, 51]. Different groups were able to stratify the tumors according to MSI-H and MSS with good agreement. However, there was much variability between different laboratories regarding interpretation of MSI at certain microsatellite loci, one of which was D5S346. Variability in the sensitivity of MSI-L detection was verified in three separate studies, which showed that different tumor sets were identified as exhibiting MSI-L when different microsatellite marker panels were used [35, 48, 52]. Based on these results, one could argue that the assignment of the MSI-L phenotype can be a technical artifact resulting from peak ambiguity generated during PCR amplication of various microsatellite motifs and sequences. Thermostabile DNA polymerases contribute directly to the stutter bands seen in analyses of microsatellite markers, as such artifacts are not observed during PCR amplification using mesophilic DNA polymerases [53]. Figure 1C shows an example of such stutter peaks generated during Amplitaq and Pfu polymerase PCR using the D2S123 marker and genomic DNA isolated from colorectal cancer cell lines. In addition to the enzyme choice, a highly significant relationship has been demonstrated between PCR artifacts at MSI loci and the quality of input DNA [54]. With regards to EMAST, different groups have obtained frequencies of EMAST-positive tumors in colorectal cancer cohorts ranging from 14% to 60%, depending on the tetranucleotide markers analyzed and the number of markers defining positivity [42, 43, 55–58]. In summary, some variation in the assignment of A-MSI tumors can be attributed to differences in the number and sequences of microsatellites analyzed, cut-off levels for defining A-MSI, and thermostable PCR amplification errors.
5.2. Argument #2: A-MSI is Due to Spontaneous Microsatellite Mutation
Because some microsatellites can display a relatively high mutation rate (Table 1), A-MSI may be due to spontaneous mutations arising within the tumor genome during clonal evolution. Two studies examined MSI using an extensive panel of microsatellite markers, with the notion that if MSI-L represents spontaneous “background” mutations arising in the tumor, nearly all tumors will exhibit MSI-L if enough microsatellite loci are analyzed. One study analyzed non-MSI-H colorectal tumors for MSI at 377 markers, and observed some degree of MSI in 79% of the samples [35]. A direct relationship was measured between age of the patient (at the time of tumor removal) and MSI frequency, with a statistically estimated mutation rate of 4 × 10−7 per cell division. This value is within the mutation rate range of shorter mono- and dinucleotide alleles measured in nontumorigenic cells using our experimental system (Table 1), consistent with the interpretation that the observed MSI-L phenotypes represent stochastic mutation events. A second study analyzed MSI using 44 microsatellite markers and found that 68% of the samples displayed some degree of MSI-L [52]. These authors also concluded that MSI-L reflects spontaneous mutational events and the evolutionary history of the tumor. Importantly, both studies were unable to define a distinction between the MSI-L and MSS phenotypes, based on the observed frequency of tumor-specific MSI.
In 2008, a statistical modeling analysis of MSI was performed to test the null hypothesis that the frequency of MSI-L tumors was not different from that expected by spontaneous microsatellite mutations arising during growth of the tumor cell population [59]. Importantly, acceptance versus rejection of the null hypothesis was dependent upon only a five-fold variation in the microsatellite mutation rate (10−5 to 5 × 10−5 per locus per division). This study analyzed MSI at nine dinucleotide microsatellite markers. As a point of comparison, we measured a mutation rate of ~6 × 10−6 mutations/cell/generation after replication of a [GT/CA]19 allele in MMR-proficient, non-tumorigenic cells (Table 1), below the range used in the above study. Two possible explanations exist to reconcile our experimental observations with the statistical model: 1). MSI-L results from random mutagenesis at specific microsatellite sequences that display an increased rate of spontaneous mutation due to their sequence composition, or 2). MSI-L arises in cells with a mild mutator phenotype or arises as a consequence of a DNA damaging microenvironment (see Sections 5.3.1 – 3 and 5.4). Because intrinsic DNA features drive microsatellite mutagenesis, explanation #1 will remain a possibility until MSI tests are conducted that control for the effect of marker sequence. In this regard, the marker used most often for detection of EMAST, MYCL1 (Table 2), has a high rate of instability in MMR-proficient cells [60]. We obtained similar results, as the [TTTC/AAAG]9 motif showed one of the highest mutation rates in MMR-proficient cells (Table 1). Thus, the data available at this time support the argument that at least a portion of A-MSI observances could result from random mutational events arising during neoplastic cell evolution. However, this mechanism fails to fully explain the specificity of microsatellite instability within A-MSI tumors (Table 2). Direct experimental studies in MMR-proficient mitotic human cell model systems have shown that mononucleotide alleles have a higher mutation rate than di- and tetranucleotide alleles, for a given length and for the specific sequences analyzed (Table 1 and reviewed in [17]). Therefore, if MSI-L was only due to spontaneous strand slippage mutagenesis, then mononucleotide markers should be as unstable, if not more unstable, than dinucleotide or tetranucleotide markers. In actuality, the microsatellite mutation rates within a developing tumor cell population will depend not only upon the sequence of the microsatellite, but also upon the genetic landscape of the individual tumor and the tumor microenvironment, all of which are expected to impact mutation rates and specificity.
5.3. Argument #3: A-MSI is a Discrete Genetic Subgroup
If the A-MSI phenotype represents tumors arising by a distinct genetic pathway, then we might expect a correlation between A-MSI and other tumor phenotypes. Consistent with this, MSI-L colorectal tumors are associated with an alternative histological pathway, namely that involving serrated or hyperplastic polyps [61, 62]. Furthermore, MSI-L tumors have an increased frequency of K-RAS mutations and a decreased frequency of LOH at 5q (APC), compared to MSS tumors [63]. The MSI-L phenotype also is associated with LOH at 1p32 and 8p12–22, both of which are associated with poor prognosis [34]. Even in cases where MSI-L and MSS tumors exhibit similar pathologic features, poorer survival was associated with MSI-L tumors, indicating that an underlying genetic difference might exist between some MSI-L and MSS tumors [64, 65]. EMAST-positive colorectal tumors showed a higher infiltration of CD8+ T-cells than EMAST-negative tumors [66]. However, EMAST in non-small cell lung cancers were associated with lymph node metastasis [67]. Interestingly, when EMAST and MSI-L stage II and III colorectal tumors are grouped together, Kaplan-Meier analyses indicate a significantly lower recurrence-free survival for these cases compared to MSI-H and MSS cases [55].
Perhaps the most convincing data suggesting that MSI-L is a distinct subgroup originates from a cDNA microarray study that was interpreted using principal component analysis [68]. Complex gene expression data from different tumor types were reduced to sets of independent hierarchal components, arranged according to the variance in the data (e.g., component 1 contained data with the most variability, then component 2, etc.) This analysis identified a component (“component 3”) with 7% variance that clearly distinguished MSI-H from non MSI-H, and another distinct component (“component 10”) with 2.5% variance that significantly differentiated MSI-L from both MSI-H and MSS tumors. Genes showing a greater relative impact for component 3 include those previously shown to be altered in MSI-H, such as MLH1 and BAX. Selenoprotein P, an antioxidant gene, showed a high relative impact for component 10, supporting a role for oxidative stress in MSI-L (see section 5.4). The component 10 finding strongly indicates that MSI-L tumors display a distinct global molecular phenotype. Similar studies with EMAST have not been performed. Given that A-MSI may be a distinct pathological group, we next discuss the DNA metabolic pathways that directly or indirectly affect A-MSI and play a role in generating the instability observed at specific microsatellite motifs.
5.3.1. Mismatch repair
Genetic and biochemical studies have demonstrated that the cellular phenotype resulting from inactivation of the MMR pathway depends upon the precise gene that is inactivated [22, 23]. The tumor tissue specificity and incidence phenotypes of knockout mouse models for MMR-deficiencies vary, depending on the precise genetic defect [69], consistent with differing biochemical specificities of the MMR proteins [70]. Microsatellite mutation frequency and specificity also differs among the MMR-knockout mouse models [71]. For example, the mutation frequencies for mononucleotide [G/C] Indels within a supFG1 reporter in various MMR-deficient mouse strains followed the order: Mlh1−/− > Msh2−/− > Pms2−/− > Msh6−/− > Msh3−/−. Thus, we expect that the MSI phenotypes of MMR-deficient cells at specific microsatellite alleles (mono, di, etc) will depend on the precise gene deficiency (Figure 2). The MutSα heterodimer (MSH2•MSH6) shows biochemical specificity for single base mismatches and one base loops [23], and correspondingly, MSH6-deficient tumors can be detected by the use of a pentaplex mononucleotide panel [72]. The MutSβ heterodimer (MSH2•MSH3) primarily binds to and repairs 2- to 12 base IDLs [73]. In vitro, MutSβ is more efficient at repair of dinucleotide substrates than is MutSα [74]. An important discovery in understanding the di- and tetranucleotide specificity of A-MSI was the finding that knockdown of MSH3, or cells deficient in MSH3, results in EMAST and MSI-L phenotypes [56]. Interestingly, tetranucleotide EMAST loci were five times as unstable as dinucleotide loci when MSH3 was absent. EMAST was shown to be common in sporadic colorectal cancer [58]. EMAST is associated with heterogeneous expression of MSH3 [43, 56], and a portion of MSI-L tumors showed LOH and decreased expression of MSH3 [75]. Our examination of MSI frequency data at mono-, di-, and tetranucleotide loci (presented by Haugen et al. 2008, ref. [56]) revealed that a discrete group of tumors with moderate MSI frequencies of 7–36% displayed the highest percentages (22–50%) of MSH3-negative cells. The increased extent of MSH3-negative cells in the MSI-L/EMAST tumors, relative to the MSS tumors, provides a clear distinction between the two groups.
While the MutLα heterodimer, MLH1•PMS2, repairs a wide range of IDL substrates [23], repair attributable to the MutLγ heterodimer (MLH1•MLH3) has been less well described. Consistent with a role of MLH3 in microsatellite stability, genetic evidence exists for residual MMR in PMS2-deficient cells. PMS2−/− knockout mice display a lower frequency of instability at [A/T]n mononucleotide alleles than do MLH1−/− mice, but the frequency of dinucleotide [CA/GT]n instability did not differ between PMS2−/− and MLH1−/− mice [76]. Double knockout MLH3−/−, PMS2−/− mice display microsatellite instability patterns indistinguishable from MLH1−/− mice, demonstrating a role for both PMS2 and MLH3 in MMR [77]. These genetic and biochemical differences have been suggested to contribute, at least in part, to the differential tissue-specificities of PMS2−/−, MLH3−/−, and MLH1−/− mice [70]. The effects of hMLH3 loss on MSI in human tumors has not been rigorously examined using a comprehensive microsatellite panel.
Deficiencies of proteins that functionally interact with MMR proteins also might produce a mild MMR-deficiency and an A-MSI phenotype. This mechanism has been documented for hMRE11, a member of the MRN complex. In a human cell line model, knockdown of hMRE11 lead to microsatellite instability and mismatch repair-deficiency, as measured using reporter assays [78]. Since MMR components form complexes with numerous proteins involved in genome maintenance, genetic mutation or deficiencies in other genes could result in A-MSI by this mechanism.
5.3.2. Base excision repair
Several genes involved in DNA repair and DNA damage response pathways are differentially expressed in sporadic colon cancers, relative to normal tissue, and the extent of aberrant expression varies widely among tumor samples [79]. Thus, it is possible that an individual tumor’s gene expression profile will affect MSI status. Alkyladenine glycosylase (AAG), a component of the base excision repair pathway, has been shown directly to be involved in MSI. Overexpression of AAG in human cultured cells leads to MSI, most often at dinucleotide alleles [80, 81]. Purified AAG binds 1–2 base IDLs in vitro; thus, AAG overexpression may allow for increased binding to IDLs, shielding the IDLs from repair by MMR. Interestingly, overexpression of AAG induced frameshifts in the absence of MMR, suggesting that an MMR-independent mechanism for IDL repair exists [80].
5.3.3. DNA Polymerases
Numerous DNA polymerases are required for the completion of DNA repair, including Pol δ, Pol β, and Pol κ [82, 83]. Our in vitro studies have revealed that the relative accuracy of DNA polymerases within microsatellite sequences is distinct from that in non-repetitive sequences, such that a distinct hierarchy of DNA polymerase fidelity is observed within microsatellites [16]. The replicative Pol δ and Pol ε holoenzymes display a high rate of indel errors within [GT/CA] microsatellites, and the polymerase proofreading activity contributes little to fidelity at microsatellites [12]. The identity of the polymerase also contributes to the specificity of microsatellite mutations. For example, both Pol β and Pol κ display high error frequencies at mononucleotide repeats, but lower frequencies at dinucleotide repeats [16]. Pol κ displays an additional signature of low Indel error rates at tetranucleotide repeats [21]. Overexpression of both Pols β and κ has been demonstrated in several types of cancer [82, 84, 85], and reduced expression of Pol κ has been observed in colorectal cancer [86]. Overexpression of Pol β in MMR-proficient cells can increase MSI at mononucleotides by up to three-fold [87]. In addition, tumor-specific mutant forms of Pol β have been detected in various cancers [88], with POLB gene coding mutations detected in 40% of colon cancers [89]. We have shown that, in vitro, the gastric cancer-associated D160N and prostate cancer-associated I260M Pol β variants display altered fidelity during dinucleotide microsatellite DNA synthesis, as compared to wild-type Pol β [90, 91].
As summarized in Figure 2, published studies clearly show that hMSH3 efficiency is associated with both of the A-MSI phenotypes, while hMSH6 deficiency is primarily associated with instability at mononucleotides. Basic experimental studies suggest that hPMS2 deficiency may not result in tetranucleotide instability, and may present as an A-MSI phenotype. Based on the studies described, we expect that A-MSI at specific microsatellite markers will be caused by alterations in the relative levels and forms of AAG, Pol β and Pol κ present in tumor cells.
5.4. Argument #4: A-MSI Reflects a Damage-Induced Phenotype
The environmental influences on microsatellite instability in cancer have been understudied, but elucidating the role of damage-induced microsatellite mutagenesis is important to resolving whether A-MSI phenotypes represent stochastic mutational events during tumor growth or a discrete genetic subgroup of tumors. Various types of MSI have been reported in several inflammatory states, including inflammatory diseases of the colon (ulcerative colitis (UC) and Crohn’s disease) [92], pancreas (pancreatitis) [37], and lung (Chronic Pulmonary Disease) [93]. If the definition of MSI-L is used (1–40% of markers unstable), then a portion of UC specimens exhibiting high grade dysplasia and a portion with no evidence of dysplasia display an MSI-L phenotype [92, 94]. In a study of DNA from the pancreatic juice of patients with pancreatitis, 60% exhibit an A-MSI phenotype consistent with the MSI-L definition [37]. Finally, inflammation was suggested to be an etiologic agent in the generation of EMAST positive colon and rectal neoplasias [42, 43].
Indirect evidence from mouse models support a mechanism of damage-induced microsatellite mutagenesis. Expanded short tandem repeats (ESTR) loci in mice closely resemble microsatellites, in that they display a replication-dependent mechanism, and both germline and somatic instability [95]. Examples include the Ms6-hm locus (CAGGG pentanucleotide with repeat units from 200 to >1000) and the Hm-2 locus (GGCA tetranucleotide with up to 5000 repeats) [96, 97]. The frequency of germline ESTR loci mutagenesis was significantly increased in mice exposed to mainstream and sidestream tobacco smoke [98], and in the offspring of mice exposed to chemotherapeutic agents [99]. Epidemiologic studies also support a damage-induced MSI mechanism. Two reports found a significant association of MSI cancers with cigarette smoking [100, 101]. Dietary mutagens play an important role in human carcinogenesis (reviewed in [102]), and a significant association was found between patients with MSI-H colon cancers and cooking practices related to the production of heterocyclic amines [101]. In a Dutch population study, red meat intake was positively associated with sporadic MSI-L/MSS colorectal carcinomas, but inversely associated with MSI-H tumors [103].
5.4.1. DNA damage and repair
MMR proteins are intimately involved in DNA damage recognition and repair [104, 105]. Downregulation of O6-methylguanine DNA alkyltransferase (MGMT) by promoter hypermethylation is significantly associated with MSI [106, 107]. Since alkylating agent exposure induces the rapid recruitment of MMR proteins to chromatin [108], one mechanism to explain this result is that in the absence of MGMT, the MMR system becomes the primary pathway for alkylation lesion repair, resulting in saturation of cellular MMR capacity and onset of MSI. Alternatively, exposure to DNA damaging agents may alter the expression of DNA repair genes, thus leading to MSI. For example, injection of an oxidative stress-inducing agent into mice led to increased DNA damage and up-regulation of DNA polymerase beta [109]. Moreover, oxidative stress leads to decreased expression of MMR proteins, possibly by causing their denaturation [110].
The expression of DNA repair genes and proteins is altered in various inflammatory conditions, which can be characterized by areas of oxidative stress as well as necrosis and hypoxia. The expression of several MMR genes and MGMT is downregulated during Heliobactor pylori-induced gastritis (reviewed in [111, 112]). Epigenetic silencing of MLH1 has been observed in mouse models of inflammatory bowel disease [113], and epigenetic downregulation of hMLH1 by hypoxia causes increased mutation at a [GT/CA]29 microsatellite in a reporter assay [114]. Interestingly, the BER enzymes AAG and APE1 are overexpressed in inflamed tissue in UC patients, and overexpression is associated with MSI-H and MSI-L [81]. EMAST is associated with inflammation [42, 43] and recent studies have shown that the hypoxia marker, glucose transporter 1, is associated with EMAST and with down-regulation of MSH3, suggesting that hypoxia can reduce MSH3 and thus cause EMAST [115]. This association of hypoxia with reduced expression of MSH3 may explain the heterogeneity of MSH3 expression in colorectal tumor tissue, such that cells experiencing hypoxic conditions may exhibit transient reduction of MSH3 [43, 56]. Therefore, MSI associated with inflammatory states is expected to present as either an MSI-H or A-MSI phenotype, depending upon the exact DNA repair genes affected.
5.4.2. Direct DNA damage and MSI specificity
The tumor-specific mutational events observed in MSI-L and EMAST tumors could be molecular signatures of direct DNA damage. The question thus arises, does DNA damage induce MSI at specific microsatellite sequences? Our laboratory has shown that H2O2 treatment of MMR-proficient human lymphoblastoid cells leads to increased Indel mutation rates at an [AAGG/TTCC]9 microsatellite, along with increased point mutations within non-repetitive sequences [116]. Other laboratories have reported increased frameshift mutations at a [GT/CA]13 microsatellite sequence upon H2O2 treatment of MMR-deficient and MMR-proficient colorectal carcinoma cell lines [117]. Mutagenesis at tetranucleotide microsatellite reporters can be induced by alkylating and oxidatizing agents, in a sequence-dependent manner [118]. Keeping in mind that MSI-L is often detected by instability at dinucleotide [GT/CA] motifs and EMAST is detected at tetranucleotide [AAAG/TTTC], while the Revised Bethesda panel is exclusively [A/T] mononucleotides (Table 2), the available specificity studies support a role of DNA damage in the etiology of specific forms of MSI. Guanine residues, particularly repeated guanine bases, are very susceptible to oxidative and alkylation damage, and an increased frequency of [G]n microsatellite mutation is induced by oxidative damage [119]. In support of this idea, oxidative damage, specifically at G residues within the hairpin loop formed by CAG/CTG trinucleotide microsatellites, is thought to initiate a toxic oxidation cycle leading to sequential microsatellite expansions [120–122].
6. Perspective
Currently published data indicate that numerous tumor types exhibit an A-MSI phenotype. Whether-or-not these alternative forms of MSI represent a discrete genetic group of tumors that differ mechanistically may be debated for many years. Resolution of the debate may be attained by future analysis of MSI-L or EMAST versus MSS tumors using a whole genome approach, similar to that recently performed for MSI-H and MSS tumors [123]. Nevertheless, differences in the histopathological characteristics of MSI-L tumors versus MSS tumors have been described, and MSI-L tumors are associated with a poor prognosis and more advanced disease, compared to MSS tumors. To our knowledge, no published studies have examined adjuvant therapy responsiveness of MSI-L or EMAST versus MSS tumors. However, MMR-deficient cells are resistant to 5-flurouracil (5-FU) cytotoxicity [124, 125], and patients with tumors exhibiting an MSI-H phenotype do not benefit from the widely used 5-FU – based adjuvant chemotherapy [126–128]. Importantly, a recent study revealed that MutSβ, in addition to MutSα, can bind to 5-FU modified DNA and can contribute to 5-FU cytotoxicity [129]. Therefore, patients with tumors of the A-MSI phenotype attributable to decreased MSH3 expression also may show decreased responsiveness to 5-FU, compared to patients with MSS tumors. MSH3 is also involved in the repair of intrastrand cross-links that are induced by chemotherapeutic drugs such as cisplatin and oxaliplatin [130]. MSH3-deficient colorectal cells are sensitive to these drugs, particularly in combination with poly (ADP-ribose) polymerase (PARP) inhibitors [130]. Thus, patients with MSH3-defective tumors may benefit from such treatments. Based on experimental evidence, an argument can be made that altered expression of DNA repair genes (other than MMR) in tumor cells may give rise to an A-MSI phenotype. Synthetic lethality approaches for DNA repair defective tumors have been effectively used in recent years to increase the efficacy of anti-cancer therapies [131]. Therefore, if tumors displaying the A-MSI phenotype do, in fact, harbor defects in DNA repair or DNA damage response pathways, this knowledge could directly impact the clinical management of patients.
MSI-L and EMAST tumors generally display a very specific microsatellite instability, primarily at dinucleotide and tetranucleotide microsatellites, respectively, rather than mononucleotide microsatellites. Revising the Bethesda consensus panel of microsatellite markers to include only mononucleotide markers has increased the specificity and sensitivity of the MSI diagnostic assay to detect MSI-H tumors that are primarily hMSH2 or hMLH1-deficient, and to diagnose Lynch syndrome patients. Unfortunately, by doing so, we may have done a disservice to a large proportion of cancer patients who could potentially benefit from being correctly classified as MSI-L or EMAST. To detect cancers in which other mechanisms may be operating, we must develop a microsatellite panel that is less restrictive and contains markers that include dinucleotide and tetranucleotide microsatellites. Compelling evidence exists that the A-MSI phenotype exists in cells under inflammatory conditions, consistent with a mechanistic role for DNA damage in MSI. Notwithstanding the key role played by MMR in maintaining microsatellite stability, we suggest that the current paradigm of diagnostic MSI could be expanded, to include the use of specific microsatellites as biomarkers of oxidative DNA damage. Understanding the biochemical basis of the MSI-L and EMAST tumor phenotypes will advance the development of new diagnostic tools and positively impact the clinical management of cancers, allowing improved personalized care for all patients with tumors presenting with MSI.
Highlights.
Alternative forms of microsatellite instability at di- and tetranucleotide markers (A-MSI) are observed in tumors.
Some A-MSI may be due to a higher spontaneous mutation rate within a subset of microsatellite markers.
A-MSI can result from defects in, or aberrant expression of, specific DNA repair proteins.
DNA damage, in particular oxidative stress, may contribute to A-MSI in tumors.
Understanding the biochemical basis of A-MSI will positively impact the clinical management of individual cancers.
Acknowledgements
We thank members of the Eckert laboratory for in-depth discussions of this topic and critical reading of the manuscript. Our research was supported by grants from the National Institutes of Health CA100060 and GM087472 to K.A.E., and by generous donations to the Gittlen Memorial Cancer Research Foundation.
Abbreviations used
- EMAST
elevated microsatellite alterations at selected tetranucleotides
- ESTR
expanded short tandem repeat
- Indel
insertion/deletion
- IDL
insertion/deletion loop
- MMR
mismatch repair
- MSI
microsatellite instability
- MSI-H
microsatellite instability-high
- MSI-L
microsatellite instability-low
- MSS
microsatellite stable
- Pol
DNA polymerase
- UC
Ulcerative colitis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
B. Baptiste, G. Ananda, N. Strubczewski, A. Lutzkanin, S. Khoo, A. Srikanth, N. Kim, K. Makova, M. Krasilnikova and K. Eckert, manuscript submitted.
Conflict of Interest Statement:
Each author declares no competing or conflicts of interest.
References
- 1.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 2.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 3.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 4.Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 6.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 7.Sidransky D. Nucleic Acid-based methods for the detection of cancer. Science. 1997;278:1054–1058. doi: 10.1126/science.278.5340.1054. [DOI] [PubMed] [Google Scholar]
- 8.Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 2008;18:30–38. doi: 10.1101/gr.7113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckert KA, Yan G, Hile SE. Mutation Rate and Specificity Analysis of Tetranucleotide Microsatellite DNA Alleles in Somatic Human Cells. Molecular Carcinogenesis. 2002;34:140–150. doi: 10.1002/mc.10058. [DOI] [PubMed] [Google Scholar]
- 10.Hile SE, Yan G, Eckert KA. Somatic mutation rates and specificities at TC/AG and GT/CA microsatellite sequences in nontumorigenic human lymphoblastoid cells. Cancer Res. 2000;60:1698–1703. [PubMed] [Google Scholar]
- 11.Shah SN, Eckert KA. Human postmeiotic segregation 2 exhibits biased repair at tetranucleotide microsatellite sequences. Cancer Res. 2009;69:1143–1149. doi: 10.1158/0008-5472.CAN-08-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdulovic AL, Hile SE, Kunkel TA, Eckert KA. The in vitro fidelity of yeast DNA polymerase delta and polymerase varepsilon holoenzymes during dinucleotide microsatellite DNA synthesis. DNA Repair (Amst) 2011;10:497–505. doi: 10.1016/j.dnarep.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckert KA, Mowery A, Hile SE. Misalignment-mediated DNA polymerase beta mutations: comparison of microsatellite and frame-shift error rates using a forward mutation assay. Biochemistry. 2002;41:10490–10498. doi: 10.1021/bi025918c. [DOI] [PubMed] [Google Scholar]
- 14.Hile SE, Eckert KA. Positive correlation between DNA polymerase alpha-primase pausing and mutagenesis within polypyrimidine/polypurine microsatellite sequences. Journal of Molecular Biology. 2004;335:745–759. doi: 10.1016/j.jmb.2003.10.075. [DOI] [PubMed] [Google Scholar]
- 15.Hile SE, Eckert KA. DNA polymerase kappa produces interrupted mutations and displays polar pausing within mononucleotide microsatellite sequences. Nucleic Acids Res. 2008;36:688–696. doi: 10.1093/nar/gkm1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hile SE, Wang X, Lee MY, Eckert KA. Beyond translesion synthesis: polymerase kappa fidelity as a potential determinant of microsatellite stability. Nucleic Acids Res. 2012;40:1636–1647. doi: 10.1093/nar/gkr889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckert KA, Hile SE. Every microsatellite is different: Intrinsic DNA features dictate mutagenesis of common microsatellites present in the human genome. Mol Carcinog. 2009;48:379–388. doi: 10.1002/mc.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelkar YD, Strubczewski N, Hile SE, Chiaromonte F, Eckert KA, Makova KD. What is a microsatellite: a computational and experimental definition based upon repeat mutational behavior at A/T and GT/AC repeats. Genome Biol Evol. 2010;2:620–635. doi: 10.1093/gbe/evq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ananda G, Walsh E, T.G.P.P.S.I.A. Consortium. Jacob KD, Krasilnikova M, Eckert KA, Chiaromonte F, Makova K. Distinct mutational behaviors distinguish simple tandem repeats from microsatellites in the human genome. Genome Biology and Evolution. 2012 doi: 10.1093/gbe/evs116. in publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levinson G, Gutman GA. Slipped strand mispairing: a major mechanism for DNA sequence evolution. Molecular Biology Evolution. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 21.Baptiste BA, Eckert KA. DNA Polymerase kappa microsatellite synthesis: Two distinct mechanisms of slippage-mediated errors. Environmental and Molecular Mutagenesis. 2012 doi: 10.1002/em.21721. in press. [DOI] [PubMed] [Google Scholar]
- 22.Buermeyer A, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA Mismatch Repair. Annual Review of Genetics. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 23.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 24.Campregher C, Scharl T, Nemeth M, Honeder C, Jascur T, Boland CR, Gasche C. The nucleotide composition of microsatellites impacts both replication fidelity and mismatch repair in human colorectal cells. Hum Mol Genet. 2010;19:2648–2657. doi: 10.1093/hmg/ddq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 26.de la Chapelle A, Hampel H. Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol. 2010;28:3380–3387. doi: 10.1200/JCO.2009.27.0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 28.Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, Dowell JM, Wasserman L, Compton C, Mayer RJ, Bertagnolli MM, Boland CR. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res. 2003;63:1608–1614. [PubMed] [Google Scholar]
- 29.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. e2073. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatch SB, Lightfoot HM, Jr, Garwacki CP, Moore DT, Calvo BF, Woosley JT, Sciarrotta J, Funkhouser WK, Farber RA. Microsatellite instability testing in colorectal carcinoma: choice of markers affects sensitivity of detection of mismatch repair-deficient tumors. Clin Cancer Res. 2005;11:2180–2187. doi: 10.1158/1078-0432.CCR-04-0234. [DOI] [PubMed] [Google Scholar]
- 32.Richetta A, Ottini L, Falchetti M, Innocenzi D, Bottoni U, Faiola R, Mariani-Costantini R, Calvieri S. Instability at sequence repeats in melanocytic tumours. Melanoma Res. 2001;11:283–289. doi: 10.1097/00008390-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 33.Halford SE, Sawyer EJ, Lambros MB, Gorman P, Macdonald ND, Talbot IC, Foulkes WD, Gillett CE, Barnes DM, Akslen LA, Lee K, Jacobs IJ, Hanby AM, Ganesan TS, Salvesen HB, Bodmer WF, Tomlinson IP, Roylance RR. MSI-low, a real phenomenon which varies in frequency among cancer types. J Pathol. 2003;201:389–394. doi: 10.1002/path.1453. [DOI] [PubMed] [Google Scholar]
- 34.Kambara T, Matsubara N, Nakagawa H, Notohara K, Nagasaka T, Yoshino T, Isozaki H, Sharp GB, Shimizu K, Jass J, Tanaka N. High frequency of low-level microsatellite instability in early colorectal cancer. Cancer Res. 2001;61:7743–7746. [PubMed] [Google Scholar]
- 35.Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, Mecklin JP, Jarvinen H, Sistonen P, Kim KM, Shibata D, Houlston RS, Aaltonen LA. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–1170. [PubMed] [Google Scholar]
- 36.Tomlinson I, Halford S, Aaltonen L, Hawkins N, Ward R. Does MSI-low exist? J Pathol. 2002;197:6–13. doi: 10.1002/path.1071. [DOI] [PubMed] [Google Scholar]
- 37.Brentnall TA, Chen R, Lee JG, Kimmey MB, Bronner MP, Haggitt RC, Kowdley KV, Hecker LM, Byrd DR. Microsatellite instability and K-ras mutations associated with pancreatic adenocarcinoma and pancreatitis. Cancer Res. 1995;55:4264–4267. [PubMed] [Google Scholar]
- 38.Xu L, Chow J, Bonacum J, Eisenberger C, Ahrendt SA, Spafford M, Wu L, Lee SM, Piantadosi S, Tockman MS, Sidransky D, Jen J. Microsatellite instability at AAAG repeat sequences in respiratory tract cancers. Int J Cancer. 2001;91:200–204. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1031>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Ahrendt SA, Decker PA, Doffek K, Wang B, Xu L, Demeure MJ, Jen J, Sidransky D. Microsatellite instability at selected tetranucleotide repeats is associated with p53 mutations in non-small cell lung cancer. Cancer Res. 2000;60:2488–2491. [PubMed] [Google Scholar]
- 40.Mao L, Schoenberg MP, Scicchitano M, Erozan YS, Merlo A, Schwab D, Sidransky D. Molecular detection of primary bladder cancer by microsatellite analysis. Science. 1996;271:659–662. doi: 10.1126/science.271.5249.659. [DOI] [PubMed] [Google Scholar]
- 41.Danaee H, Nelson HH, Karagas MR, Schned AR, Ashok TD, Hirao T, Perry AE, Kelsey KT. Microsatellite instability at tetranucleotide repeats in skin and bladder cancer. Oncogene. 2002;21:4894–4899. doi: 10.1038/sj.onc.1205619. [DOI] [PubMed] [Google Scholar]
- 42.Devaraj B, Lee A, Cabrera BL, Miyai K, Luo L, Ramamoorthy S, Keku T, Sandler RS, McGuire KL, Carethers JM. Relationship of EMAST and microsatellite instability among patients with rectal cancer. J Gastrointest Surg. 2010;14:1521–1528. doi: 10.1007/s11605-010-1340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SY, Chung H, Devaraj B, Iwaizumi M, Han HS, Hwang DY, Seong MK, Jung BH, Carethers JM. Microsatellite alterations at selected tetranucleotide repeats are associated with morphologies of colorectal neoplasias. Gastroenterology. 2010;139:1519–1525. doi: 10.1053/j.gastro.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singer G, Kallinowski T, Hartmann A, Dietmaier W, Wild PJ, Schraml P, Sauter G, Mihatsch MJ, Moch H. Different types of microsatellite instability in ovarian carcinoma. Int J Cancer. 2004;112:643–646. doi: 10.1002/ijc.20455. [DOI] [PubMed] [Google Scholar]
- 45.Catto JW, Azzouzi AR, Amira N, Rehman I, Feeley KM, Cross SS, Fromont G, Sibony M, Hamdy FC, Cussenot O, Meuth M. Distinct patterns of microsatellite instability are seen in tumours of the urinary tract. Oncogene. 2003;22:8699–8706. doi: 10.1038/sj.onc.1206964. [DOI] [PubMed] [Google Scholar]
- 46.Jass J, Whitehall V, Young J, Leggett B, Meltzer SJ, Matsubara N, Fishel R. Correspondance re: Laiho et al., Low level microsatellite instability in most colorectal carcinomas. Cancer Res., 62: 1166–1170, 2002. Cancer Res. 2002;62:5988–5990. [PubMed] [Google Scholar]
- 47.Jass JR, Young J, Leggett BA. Biological significance of microsatellite instability-low (MSI-L) status in colorectal tumors. Am J Pathol. 2001;158:779–781. doi: 10.1016/s0002-9440(10)64020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thibodeau SN, French AJ, Cunningham JM, Tester D, Burgart LJ, Roche PC, McDonnell SK, Schaid DJ, Vockley CW, Michels VV, Farr GH, Jr, O'Connell MJ. Microsatellite instability in colorectal cancer: different mutator phenotypes and the principal involvement of hMLH1. Cancer Res. 1998;58:1713–1718. [PubMed] [Google Scholar]
- 49.Gonzalez-Garcia I, Moreno V, Navarro M, Marti-Rague J, Marcuello E, Benasco C, Campos O, Capella G, Peinado MA. Standardized approach for microsatellite instability detection in colorectal carcinomas. J Natl Cancer Inst. 2000;92:544–549. doi: 10.1093/jnci/92.7.544. [DOI] [PubMed] [Google Scholar]
- 50.Bocker T, Diermann J, Friedl W, Gebert J, Holinski-Feder E, Karner-Hanusch J, von Knebel-Doeberitz M, Koelble K, Moeslein G, Schackert HK, Wirtz HC, Fishel R, Ruschoff J. Microsatellite instability analysis: a multicenter study for reliability and quality control. Cancer Res. 1997;57:4739–4743. [PubMed] [Google Scholar]
- 51.Lindor NM, Smalley R, Barker M, Bigler J, Krumroy LM, Lum-Jones A, Plummer SJ, Selander T, Thomas S, Youash M, Seminara D, Casey G, Bapat B, Thibodeau SN. Ascending the learning curve--MSI testing experience of a six-laboratory consortium. Cancer Biomark. 2006;2:5–9. doi: 10.3233/cbm-2006-21-202. [DOI] [PubMed] [Google Scholar]
- 52.Halford S, Sasieni P, Rowan A, Wasan H, Bodmer W, Talbot I, Hawkins N, Ward R, Tomlinson I. Low-level microsatellite instability occurs in most colorectal cancers and is a nonrandomly distributed quantitative trait. Cancer Res. 2002;62:53–57. [PubMed] [Google Scholar]
- 53.Hite JM, Eckert KA, Cheng KC. Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n.d(G-T)n microsatellite repeats. Nucleic Acids Res. 1996;24:2429–2434. doi: 10.1093/nar/24.12.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sieben NL, ter Haar NT, Cornelisse CJ, Fleuren GJ, Cleton-Jansen AM. PCR artifacts in LOH and MSI analysis of microdissected tumor cells. Hum Pathol. 2000;31:1414–1419. [PubMed] [Google Scholar]
- 55.Garcia M, Choi C, Kim HR, Daoud Y, Toiyama Y, Takahashi M, Goel A, Boland CR, Koi M. Association Between Recurrent Metastasis From Stage II and III Primary Colorectal Tumors and Moderate Microsatellite Instability. Gastroenterology. 2012;143:48–50. e41. doi: 10.1053/j.gastro.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haugen AC, Goel A, Yamada K, Marra G, Nguyen T-P, Nagasaka T, Kanazawa S, Koike J, Kikuchi Y, Zhong X, Arita M, Shibuya K, Oshimura M, Hemmi H, Boland CR, Koi M. Genetic Instability Caused by Loss of MutS Homologue 3 in Human Colorectal Cancer. Cancer Res. 2008;68:8465–8472. doi: 10.1158/0008-5472.CAN-08-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, Leppert MF, Slattery ML. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada K, Kanazawa S, Koike J, Sugiyama H, Xu C, Funahashi K, Boland CR, Koi M, Hemmi H. Microsatellite instability at tetranucleotide repeats in sporadic colorectal cancer in Japan. Oncol Rep. 2010;23:551–561. [PMC free article] [PubMed] [Google Scholar]
- 59.Graham T, Halford S, Page KM, Tomlinson IP. Most low-level microsatellite instability in colorectal cancers can be explained without an elevated slippage rate. J Pathol. 2008;215:204–210. doi: 10.1002/path.2351. [DOI] [PubMed] [Google Scholar]
- 60.Hatch SB, Farber RA. Mutation rates in the complex microsatellite MYCL1 and related simple repeats in cultured human cells. Mutat Res. 2004;545:117–126. doi: 10.1016/j.mrfmmm.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 61.Jass JR. Serrated adenoma and colorectal cancer. J Pathol. 1999;187:499–502. doi: 10.1002/(SICI)1096-9896(199904)187:5<499::AID-PATH309>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 62.Iino H, Jass JR, Simms LA, Young J, Leggett B, Ajioka Y, Watanabe H. DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol. 1999;52:5–9. doi: 10.1136/jcp.52.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jass JR, Biden KG, Cummings MC, Simms LA, Walsh M, Schoch E, Meltzer SJ, Wright C, Searle J, Young J, Leggett BA. Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol. 1999;52:455–460. doi: 10.1136/jcp.52.6.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wright CM, Dent OF, Newland RC, Barker M, Chapuis PH, Bokey EL, Young JP, Leggett BA, Jass JR, Macdonald GA. Low level microsatellite instability may be associated with reduced cancer specific survival in sporadic stage C colorectal carcinoma. Gut. 2005;54:103–108. doi: 10.1136/gut.2003.034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gafa R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025–2037. [PubMed] [Google Scholar]
- 66.Lee SY, Miyai K, Han HS, Hwang DY, Seong MK, Chung H, Jung BH, Devaraj B, McGuire KL, Carethers JM. Microsatellite instability, EMAST, and morphology associations with T cell infiltration in colorectal neoplasia. Digestive diseases and sciences. 2012;57:72–78. doi: 10.1007/s10620-011-1825-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woenckhaus M, Stoehr R, Dietmaier W, Wild PJ, Zieglmeier U, Foerster J, Merk J, Blaszyk H, Pfeifer M, Hofstaedter F, Hartmann A. Microsatellite instability at chromosome 8p in non-small cell lung cancer is associated with lymph node metastasis and squamous differentiation. International journal of oncology. 2003;23:1357–1363. [PubMed] [Google Scholar]
- 68.Mori Y, Selaru FM, Sato F, Yin J, Simms LA, Xu Y, Olaru A, Deacu E, Wang S, Taylor JM, Young J, Leggett B, Jass JR, Abraham JM, Shibata D, Meltzer SJ. The impact of microsatellite instability on the molecular phenotype of colorectal tumors. Cancer Res. 2003;63:4577–4582. [PubMed] [Google Scholar]
- 69.Edelmann L, Edelmann W. Loss of DNA mismatch repair function and cancer predisposition in the mouse: Animal models for human hereditary nonpolyposis colorectal cancer. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2004;129C:91–99. doi: 10.1002/ajmg.c.30021. [DOI] [PubMed] [Google Scholar]
- 70.Chao EC, Lipkin SM. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res. 2006;34:840–852. doi: 10.1093/nar/gkj489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–2408. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.You JF, Buhard O, Ligtenberg MJ, Kets CM, Niessen RC, Hofstra RM, Wagner A, Dinjens WN, Colas C, Lascols O, Collura A, Flejou JF, Duval A, Hamelin R. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer. 2010;103:1840–1845. doi: 10.1038/sj.bjc.6605988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutS� from human cells and comparison of the mismatch repair specificities of MutS� and MutSalpha. Journal of Biological Chemistry. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 74.Kantelinen J, Kansikas M, Korhonen MK, Ollila S, Heinimann K, Kariola R, Nystrom M. MutSbeta exceeds MutSalpha in dinucleotide loop repair. Br J Cancer. 2010;102:1068–1073. doi: 10.1038/sj.bjc.6605531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plaschke J, Preussler M, Ziegler A, Schackert HK. Aberrant protein expression and frequent allelic loss of MSH3 in colorectal cancer with low-level microsatellite instability. Int J Colorectal Dis. 2012;27:911–919. doi: 10.1007/s00384-011-1408-0. [DOI] [PubMed] [Google Scholar]
- 76.Yao X, Buermeyer AB, Narayanan L, Tran D, Baker SM, Prolla TA, Glazer PM, Liskay RM, Arnheim N. Different mutator phenotypes in Mlh1- versus Pms2-deficient mice. Proc Natl Acad Sci U S A. 1999;96:6850–6855. doi: 10.1073/pnas.96.12.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen PC, Dudley S, Hagen W, Dizon D, Paxton L, Reichow D, Yoon SR, Yang K, Arnheim N, Liskay RM, Lipkin SM. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65:8662–8670. doi: 10.1158/0008-5472.CAN-05-0742. [DOI] [PubMed] [Google Scholar]
- 78.Vo AT, Zhu F, Wu X, Yuan F, Gao Y, Gu L, Li GM, Lee TH, Her C. hMRE11 deficiency leads to microsatellite instability and defective DNA mismatch repair. EMBO Rep. 2005;6:438–444. doi: 10.1038/sj.embor.7400392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu J, Mallon MA, Zhang W, Freimuth RR, Marsh S, Watson MA, Goodfellow PJ, McLeod HL. DNA repair pathway profiling and microsatellite instability in colorectal cancer. Clin Cancer Res. 2006;12:5104–5111. doi: 10.1158/1078-0432.CCR-06-0547. [DOI] [PubMed] [Google Scholar]
- 80.Klapacz J, Lingaraju GM, Guo HH, Shah D, Moar-Shoshani A, Loeb LA, Samson LD. Frameshift mutagenesis and microsatellite instability induced by human alkyladenine DNA glycosylase. Mol Cell. 2010;37:843–853. doi: 10.1016/j.molcel.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hofseth LJ, Khan MA, Ambrose M, Nikolayeva O, Xu-Welliver M, Kartalou M, Hussain SP, Roth RB, Zhou X, Mechanic LE, Zurer I, Rotter V, Samson LD, Harris CC. The adaptive imbalance in base excision-repair enzymes generates microsatellite instability in chronic inflammation. J Clin Invest. 2003;112:1887–1894. doi: 10.1172/JCI19757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- 83.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 84.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffmann JS, Cazaux C. Aberrant expression of alternative DNA polymerases: a source of mutator phenotype as well as replicative stress in cancer. Semin Cancer Biol. 2010;20:312–319. doi: 10.1016/j.semcancer.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 86.Pillaire MJ, Selves J, Gordien K, Gourraud PA, Gentil C, Danjoux M, Do C, Negre V, Bieth A, Guimbaud R, Trouche D, Pasero P, Mechali M, Hoffmann JS, Cazaux C. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene. 2010;29:876–887. doi: 10.1038/onc.2009.378. [DOI] [PubMed] [Google Scholar]
- 87.Yamada NA, Farber RA. Induction of a low level of microsatellite instability by overexpression of DNA polymerase beta. Cancer Res. 2002;62:6061–6064. [PubMed] [Google Scholar]
- 88.Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001. [PubMed] [Google Scholar]
- 89.Donigan KA, Sun KW, Nemec AA, Murphy DL, Cong X, Northrup V, Zelterman D, Sweasy JB. The Human POLB Gene is Mutated in a High Percentage of Colorectal Tumors. J Biol Chem. 2012 doi: 10.1074/jbc.M111.324947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dalal S, Hile S, Eckert KA, Sun KW, Starcevic D, Sweasy JB. Prostate-cancer-associated I260M variant of DNA polymerase beta is a sequence-specific mutator. Biochemistry. 2005;44:15664–15673. doi: 10.1021/bi051179z. [DOI] [PubMed] [Google Scholar]
- 91.Donigan KA, Hile SE, Eckert KA, Sweasy JB. The human gastric cancer-associated DNA polymerase beta variant D160N is a mutator that induces cellular transformation. DNA Repair (Amst) 2012;11:381–390. doi: 10.1016/j.dnarep.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brentnall TA, Crispin DA, Bronner MP, Cherian SP, Hueffed M, Rabinovitch PS, Rubin CE, Haggitt RC, Boland CR. Microsatellite instability in nonneoplastic mucosa from patients with chronic ulcerative colitis. Cancer Res. 1996;56:1237–1240. [PubMed] [Google Scholar]
- 93.Tzortzaki EG, Dimakou K, Neofytou E, Tsikritsaki K, Samara K, Avgousti M, Amargianitakis V, Gousiou A, Menikou S, Siafakas NM. Oxidative DNA damage and somatic mutations: a link to the molecular pathogenesis of chronic inflammatory airway diseases. Chest. 2012;141:1243–1250. doi: 10.1378/chest.11-1653. [DOI] [PubMed] [Google Scholar]
- 94.Ozaki K, Nagasaka T, Notohara K, Kambara T, Takeda M, Sasamoto H, Jass JR, Tanaka N, Matsubara N. Heterogeneous microsatellite instability observed within epithelium of ulcerative colitis. Int J Cancer. 2006;119:2513–2519. doi: 10.1002/ijc.22095. [DOI] [PubMed] [Google Scholar]
- 95.Hardwick RJ, Tretyakov MV, Dubrova YE. Age-related accumulation of mutations supports a replication-dependent mechanism of spontaneous mutation at tandem repeat DNA Loci in mice. Mol Biol Evol. 2009;26:2647–2654. doi: 10.1093/molbev/msp182. [DOI] [PubMed] [Google Scholar]
- 96.Weitzmann MN, Woodford KJ, Usdin K. The mouse Ms6-hm hypervariable microsatellite forms a hairpin and two unusual tetraplexes. The Journal of biological chemistry. 1998;273:30742–30749. doi: 10.1074/jbc.273.46.30742. [DOI] [PubMed] [Google Scholar]
- 97.Gibbs M, Collick A, Kelly RG, Jeffreys AJ. A tetranucleotide repeat mouse minisatellite displaying substantial somatic instability during early preimplantation development. Genomics. 1993;17:121–128. doi: 10.1006/geno.1993.1292. [DOI] [PubMed] [Google Scholar]
- 98.Marchetti F, Rowan-Carroll A, Williams A, Polyzos A, Berndt-Weis ML, Yauk CL. Sidestream tobacco smoke is a male germ cell mutagen. Proc Natl Acad Sci U S A. 2011;108:12811–12814. doi: 10.1073/pnas.1106896108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Glen CD, Dubrova YE. Exposure to anticancer drugs can result in transgenerational genomic instability in mice. Proc Natl Acad Sci U S A. 2012;109:2984–2988. doi: 10.1073/pnas.1119396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Slattery ML, Curtin K, Anderson K, Ma KN, Ballard L, Edwards S, Schaffer D, Potter J, Leppert M, Samowitz WS. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92:1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 101.Wu AH, Shibata D, Yu MC, Lai MY, Ross RK. Dietary heterocyclic amines and microsatellite instability in colon adenocarcinomas. Carcinogenesis. 2001;22:1681–1684. doi: 10.1093/carcin/22.10.1681. [DOI] [PubMed] [Google Scholar]
- 102.Ferguson LR, Philpott M. Nutrition and mutagenesis. Annu Rev Nutr. 2008;28:313–329. doi: 10.1146/annurev.nutr.28.061807.155449. [DOI] [PubMed] [Google Scholar]
- 103.Diergaarde B, Braam H, van Muijen GN, Ligtenberg MJ, Kok FJ, Kampman E. Dietary factors and microsatellite instability in sporadic colon carcinomas. Cancer Epidemiol Biomarkers Prev. 2003;12:1130–1136. [PubMed] [Google Scholar]
- 104.Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–444. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- 105.Duckett DR, Drummond JT, Murchie AIH, Reardon JT, Sancar A, Lilley DMJ, Modrich P. Human MutSa recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proceedings of the National Academy of Science U.S.A. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, Seruca R, Gaub MP, Legrain M, Collura A, Lascols O, Tiret E, Flejou JF, Duval A. Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers. Gut. 2010;59:1516–1526. doi: 10.1136/gut.2009.194787. [DOI] [PubMed] [Google Scholar]
- 107.Whitehall VL, Walsh MD, Young J, Leggett BA, Jass JR. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 108.Schroering AG, Williams KJ. Rapid induction of chromatin-associated DNA mismatch repair proteins after MNNG treatment. DNA Repair (Amst) 2008;7:951–969. doi: 10.1016/j.dnarep.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cabelof DC, Raffoul JJ, Yanamadala S, Guo Z, Heydari AR. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002;23:1419–1425. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- 110.Chang CL, Marra G, Chauhan DP, Ha HT, Chang DK, Ricciardiello L, Randolph A, Carethers JM, Boland CR. Oxidative stress inactivates the human DNA mismatch repair system. Am J Physiol Cell Physiol. 2002;283:C148–C154. doi: 10.1152/ajpcell.00422.2001. [DOI] [PubMed] [Google Scholar]
- 111.Machado AM, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection generates genetic instability in gastric cells. Biochim Biophys Acta. 2010;1806:58–65. doi: 10.1016/j.bbcan.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 112.Touati E. When bacteria become mutagenic and carcinogenic: lessons from H. pylori. Mutat Res. 2010;703:66–70. doi: 10.1016/j.mrgentox.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 113.Edwards RA, Witherspoon M, Wang K, Afrasiabi K, Pham T, Birnbaumer L, Lipkin SM. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69:6423–6429. doi: 10.1158/0008-5472.CAN-09-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li J, Koike J, Kugoh H, Arita M, Ohhira T, Kikuchi Y, Funahashi K, Takamatsu K, Boland CR, Koi M, Hemmi H. Down-regulation of MutS homolog 3 by hypoxia in human colorectal cancer. Biochim Biophys Acta. 2012;1823:889–899. doi: 10.1016/j.bbamcr.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Maneval ML, Eckert KA. Effects of oxidative and alkylating damage on microsatellite instability in nontumorigenic human cells. Mutat Res. 2004;546:29–38. doi: 10.1016/j.mrfmmm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 117.Gasche C, Chang CL, Rhees J, Goel A, Boland CR. Oxidative stress increases frameshift mutations in human colorectal cancer cells. Cancer Res. 2001;61:7444–7448. [PubMed] [Google Scholar]
- 118.Slebos R, Oh D, Umbach D, Taylor J. Mutations in tetranucleotide repeats following DNA damage depend on repeat sequence and carcinogenic agents. Cancer Res. 2002;62:6052–6060. [PubMed] [Google Scholar]
- 119.Jackson AL, Loeb LA. Microsatellite instability induced by hydrogen peroxide in Escherichia coli. Mutat Res. 2000;447:187–198. doi: 10.1016/s0027-5107(99)00206-7. [DOI] [PubMed] [Google Scholar]
- 120.Jarem DA, Wilson NR, Delaney S. Structure-dependent DNA damage and repair in a trinucleotide repeat sequence. Biochemistry. 2009;48:6655–6663. doi: 10.1021/bi9007403. [DOI] [PubMed] [Google Scholar]
- 121.Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jarem DA, Wilson NR, Schermerhorn KM, Delaney S. Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion. DNA repair. 2011;10:887–896. doi: 10.1016/j.dnarep.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Timmermann B, Kerick M, Roehr C, Fischer A, Isau M, Boerno ST, Wunderlich A, Barmeyer C, Seemann P, Koenig J, Lappe M, Kuss AW, Garshasbi M, Bertram L, Trappe K, Werber M, Herrmann BG, Zatloukal K, Lehrach H, Schweiger MR. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One. 2011;5:e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch repair proficiency and in vitro response to 5-fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA mismatch repair-dependent response to fluoropyrimidine-generated damage. J Biol Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 126.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 127.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tajima A, Iwaizumi M, Tseng-Rogenski S, Cabrera BL, Carethers JM. Both hMutSalpha and hMutSss DNA mismatch repair complexes participate in 5-fluorouracil cytotoxicity. PLoS One. 2011;6:e28117. doi: 10.1371/journal.pone.0028117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Takahashi M, Koi M, Balaguer F, Boland CR, Goel A. MSH3 mediates sensitization of colorectal cancer cells to cisplatin, oxaliplatin, and a poly(ADP-ribose) polymerase inhibitor. The Journal of biological chemistry. 2011;286:12157–12165. doi: 10.1074/jbc.M110.198804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]