Summary
Polyglutamine-repeat diseases are neurodegenerative ailments elicited by glutamine-encoding CAG nucleotide expansions within endogenous human genes. Despite efforts to understand the basis of these diseases, the precise mechanism of cell death remains stubbornly unclear. Much of the data seems consistent with a model in which toxicity is an inherent property of the polyglutamine repeat, whereas host protein sequences surrounding the polyQ expansion modulate severity, age of onset, and cell specificity. Recently, a gene, pqn-41encoding a glutamine-rich protein was found to promote normally-occurring non-apoptotic cell death in C. elegans. Here we review evidence for toxic and modulatory roles for polyQ repeats and their host proteins, respectively, and suggest similarities with pqn-41 function. We explore the hypothesis that toxicity mediated by glutamine-rich motifs may be important not only in pathology, but also in normal development.
Glutamine repeats in disease and development
Of the human nucleotide repeat diseases, ten are caused by CAG expansions with the capacity to encode expanded glutamine stretches within endogenous proteins. These diseases, referred to as polyglutamine or polyQ repeat diseases, include Huntington’s disease (HD), six of the spinocerebellar ataxias (SCA 1–3, 6, 7, 17), spino-bulbar muscular atrophy (SBMA), and dentatorubral-pallidoluysian atrophy (DRPLA) [1–7] (Table 1). Huntington’s disease-like 2 (HDL2) was also recently suggested to derive from a polyQ expansion encoded by an antisense mRNA from the Junctophilin-3 locus [8]. In addition to expanded polyQ tracts, these human diseases share other characteristics. Except for SBMA, all are dominant gain-of-function disorders [9]. Neural tissue is the principal site of pathology, and in both human patients and animal models, insoluble protein aggregates containing the mutant proteins are found within affected neurons- a pathological hallmark of disease. However, differences between these diseases are equally striking. The polyQ expansions affect genes encoding proteins with little apparent functional similarities, aside from generally broad expression patterns; and the diseases affect different regions of the brain and different neuronal subtypes (with some overlap) [9]
Table 1.
Polyglutamine expansion disease proteins
| Disease/ Process | Protein | Wild type Q length | Pathogenic Q length |
|---|---|---|---|
| Huntington’s disease (HD) | Huntington | 6–34 | 36–121 [1] |
| Spinocerebellar ataxia (SCA1) | Ataxin-1 | 6–38, 39–44 CAT interrupteda | 39–44 CAGs uninterrupted; 45–91 [3] |
| SCA2 | Ataxin-2 | <32 | 32–500 [3] |
| SCA3 | Ataxin-3 | 11–44 | 45–86 [3,5] |
| SCA6 | CACNA1A | 4–18 | 19–33 [3] |
| SCA7 | Ataxin-7 | 4–19 | 34–460 [3] |
| SCA17 | TATA-box binding protein (TBP) | 25–40 | 42–66 [3,4] |
| SBMA | Androgen receptor | 9–34 | 38–62 [2,6] |
| DRPLA | Atrophin-1 | 6–35 | 49–93 [3] |
| HDL2 | Unknownb | 6–28 | 40–59 [8] |
CAG tract may be interrupted by 1–4 CAT sequences which affect pathogenicity of tract length
CAG expanded transcript was found to be derived antisense to junctophillin-3 (JPH3) [10].
Polyglutamine sequences have generally been discussed in the context of human pathology. However, a recent study in C. elegans suggests a role for a glutamine-rich protein, PQN-41, in naturally occurring non-apoptotic developmental cell death [10]. The linker cell dies in the normal course of C. elegans development, during gonadal morphogenesis of the male [11–13] (Figure 1). Linker cell death is independent of caspases a nd all other known apoptotic and necrotic C. elegans cell death genes [13, 14]. Mutations in the pqn-41 gene block linker cell death, suggesting an important role in linker cell demise. Furthermore, transcription of pqn-41 is induced just prior to the onset of cell death, suggesting that this locus may be intimately connected with the killing process [10]. pqn-41 encodes multiple alternative transcripts, most of which can encode a domain of 427 amino-acids, of which 35% are glutamines, arranged in tracts of 1–8 residues in length [10].
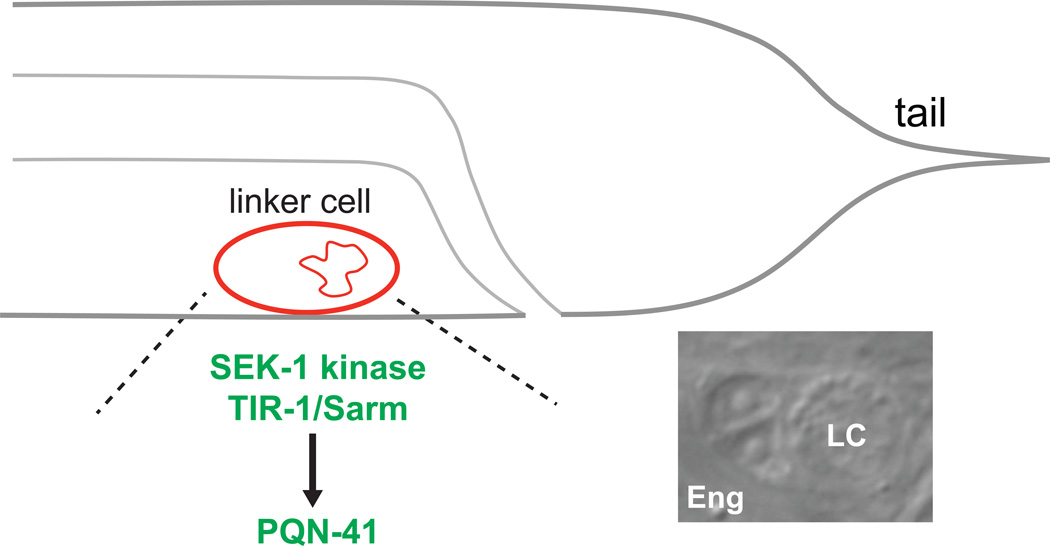
Figure 1. Linker cell death is controlled by PQN-41.
The C. elegans male-specific linker cell (red) dies at the L4-adult transition at the posterior of the animal next to the cloacal tube (grey) [11–13]. A DIC image of the dying linker cell (LC) shows its engulfment by a binucleate neighboring cell. Linker cell death is controlled by PQN-41 downstream of MAPKK SEK-1 and TIR-1 [10].
The involvement of glutamines in neurodegenerative human disease and in programmed cell death in C. elegans begs the question of whether these processes are related. Dying linker cells in C. elegans fail to display classic apoptotic features. Rather, cell death is accompanied by lack of chromatin condensation, nuclear envelope crenellation, and swelling of cytoplasmic organelles. Intriguingly, similar ultrastructural features are found in normally dying cells during development of the vertebrate nervous system [15–17], as well as in biopsy samples from polyQ disease patients, and in mice and cell culture models of polyQ disease [18–21]. These observations raise the possibility that linker cell death and polyQ degeneration may have common molecular features, and a number of observations support this possibility.
Below we examine what is known about the mechanism of polyQ-induced degeneration, specifically as relevant to assessing similarities and differences with PQN-41 function in C. elegans. This paper is therefore not intended as a comprehensive description of the polyQ disease field, and the reader is referred to other excellent recent reviews for a more general discussion [9, 22].
Are glutamines the business end?
The pqn-41 locus produces multiple alternative mRNAs. One mRNA variant, pqn-41Cencodes only the C-terminal glutamine-rich domain of the protein and is sufficient to restore linker cell death to pqn-41 mutants. A sequence just upstream of the region encoding this mRNA is sufficient to induce expression of a reporter protein in the linker cell just prior to onset of cell death [10]. These results suggest that the glutamine-rich domain of PQN-41 is a key effector of linker cell death.
The observation that proteins of apparently disparate functions promote similar forms of neurodegeneration when imbued with polyQ expansions suggests that the toxic element in polyglutamine diseases may be the polyglutamine peptide itself. Indeed, the longer the repeat, the more severe is the disease, and the earlier the age of onset [7]. Several other observations support this notion:
Overexpression
Overexpression of polyglutamine fragments outside the context of the endogenous protein promotes length-dependent toxicity in many model systems. For example, overexpression of human huntingtin exon 1 containing 115 to 150 glutamines in mice induces a neurodegenerative phenotype similar to Huntington’s Disease [23]. Similarly, expression of polyQ-only peptides throughout the nervous system of C. elegans promotes neurotoxicity, and toxicity correlates with increased length of the polyQ repeat [24]. In Drosophilaexpression of a human huntingtin fragment derived from exon 1 and containing an expanded polyQ tract also promotes length-dependent cellular dysfunction [25, 26]. These studies are all consistent with an inherent toxicity of polyQ peptides.
Proteolysis
A number of studies suggest that sequences surrounding a polyQ stretch reduce its toxicity. These observations suggest the possibility that in neurodegeneration, polyQ stretches might escape host protein protective sequences through selective proteolysis. While in some polyQ diseases proteolysis has not been described, in others, such protein scission has indeed been reported [27]. The contribution of huntingtin (Htt) proteolysis in promoting pathogenesis has been studied extensively. A variety of N-terminal fragments housing the polyQ expansion are formed by proteolytic cleavage of full-length polyQ-expanded Htt [28]. These fragments can form toxic soluble oligomers in vitro [29]. Furthermore, biochemical studies of polyQ-expanded and wild-type Htt suggest that the expanded protein is more susceptible to cleavage, presumably due to a conformational difference resulting from the longer polyQ region [30]. A particularly important cleavage event may be one generating a 586 amino-acid N-terminal fragment, housing the polyglutamine repeat. This fragment can be detected in mice expressing the polyQ-expanded protein, and mice expressing this fragment show neurodegeneration [31, 32]. This N-terminal fragment may itself be cleaved to form smaller fragments in vivo. Mutations preventing at least some of these cleavage events reduce toxicity in cultured neurons [31–34].
The cleavage at amino acid 586 has been proposed to be mediated by caspase-6. Mice in which this cleavage site is mutated show reduced degeneration and fewer behavioral abnormalities compared to mice expressing the unmodified expanded protein [35]. However, the 586 amino-acid fragment can still be detected in caspase-6 knockout mice [36], raising the possibility that other proteases promote cleavage together with or independently of caspase-6. Indeed, Htt cleavage by many different proteases has been described, and cleavage at amino acid 402 by Matrix Metallopeptidase 10 (MMP-10) promotes toxicity of mutant Htt in cultured neurons and in mouse models [34].
In addition to huntingtin, there is evidence that other polyQ disease promoting proteins are susceptible to proteolysis, which may contribute to toxicity. For example, mutation of a caspase cleavage site in polyQ-expanded Atrophin-1 (ATN1) reduces toxicity in vitro [37]. Furthermore, ATN1 cleavage products are found in patient brain tissue [38]; however, at least in this case, a causative link between these fragments and degeneration was not investigated. Similarly, proteolysis of Ataxin-3 (ATXN3) has been reported in mouse models of spinocerebellar ataxia 3 (SCA3) and in human disease tissue [39, 40]. In a Drosophila model of SCA3, mutation of proposed caspase cleavage sites in ATXN3 dramatically reduces production of polyQ-enriched fragments [41] and ameliorates degeneration caused by polyQ-expanded ATXN3. Ataxin-7 (ATXN7) cleavage by caspase 7 at amino acid 266 also enhances its toxicity in vitro [42], and cytotoxicity of the expanded androgen receptor is enhanced by caspase cleavage at amino acid 146 [43].
Alternative splicing
Alternative splicing could theoretically also produce proteins specifically enriched in the polyQ moiety, and has been documented for several polyQ disease genes, including Atrophin-1 [44], the alpha1A voltage-dependent calcium-channel [45], and ATXN3 [46, 47]. However, roles for transcript variants in disease have not been extensively explored.
Together, the studies reviewed here suggest that protein fragments enriched in glutamines can promote cell death in the context of linker cell death in C. elegans and at least in some polyQ diseases.
Continuous or interrupted polyQ tracts?
In C. elegansfusion of PQN-41C, composed of interrupted runs of glutamines, to green fluorescent protein (GFP), promotes aggregate formation in the linker cell. Secondary structure prediction algorithms suggest that PQN-41C has a high propensity for forming coiled-coils [10], supersecondary helical structures that mediate protein-protein interactions and oligomerization. Importantly, mutations predicted to disrupt the coiled-coil motifs partially or fully abrogate the ability of PQN-41C to rescue pqn-41 mutants [10, 48]. Additional experiments are required to determine whether aggregation is an intrinsic property of PQN-41C (and not influenced by the GFP tag or interacting proteins) and whether this aggregation is required for PQN-41C toxicity.
Exactly how expansion of the polyQ tract in human disease leads to toxicity remains unclear. One hypothesis is that the length-dependence of toxicity is tied to structural transitions of the protein. Supporting this idea, expansion of the polyQ tract increases the propensity for protein aggregation and inclusion body formation. Thus, aggregation may be crudely used to monitor structural transitions in the protein. It is important to note that the link between aggregation and toxicity remains highly debated. It has been suggested that, at least in the case of huntingtin, a soluble oligomeric form may be the toxic species, and that the large insoluble aggregates are non-toxic, and perhaps even protective [49, 50]. Some studies suggest that long polyQ repeats have a propensity to form β sheets stabilized by intermolecular hydrogen bonds between main chain and side chain amides [51]. It is the formation of these beta-sheets, or “polar zippers”, that might promote aggregation. Other studies suggest that the polyQ tracts cause the host protein to partially unfold, resulting in solvent exposure of hydrophobic residues and the amide-backbone, thus increasing the propensity to aggregate [52–54].
Recently, Fiumara et al. [55] proposed that aggregation of polyQ disease proteins and their interacting partners, as well as oligomerization/aggregation of some glutamine/asparagine (Q/N) rich protein domains, may be mediated by their propensity to form coiled coils [55]. Supporting this assertion, circular dichroism measurements of polyQ peptides reveals helical structure. Furthermore, disruption of coiled-coil domain formation in the Htt exon 1 sequence containing an expansion of 72 glutamine residues decreases its propensity to aggregate and its toxicity in HEK293 cells [55]. This study, therefore, supports the possibility of a structural relatedness between pure polyQ sequences and glutamine-rich motifs.
Indeed, although a large body of evidence implicates uninterrupted polyQ peptides in toxicity in culture and in vivo [56], a strictly homogenous peptide sequence may not be required to induce a structural transition and/or toxicity. For example, a glutamine-alanine repeat peptide derived from the huntingtin-interacting protein CA150 spontaneously aggregates with similar kinetics to that of an uninterrupted polyQ peptide of the same length [57]. Furthermore, the same study showed that the glutamine-alanine aggregates can seed polyQ peptide elongation [57]. In another study, the insertion of two alanines in the center of a glutamine peptide was also found to have a minimal effect on aggregration [58]. These experiments suggest that interrupted polyglutamine peptides can aggregate readily, however, it remains to be tested whether any of these sequences can promote toxicity in vivo. Indeed, studies of ATXN1 indicate that interruption of the homogenous polyQ domain by histidine residues reduces SCA1 pathogenesis even above the typical polyQ length threshold required for disease [59, 60].
The genetic instabilities that lead to CAG genomic expansions are unlikely to result in impure polyQ repeats, as repeat formation likely occurs through DNA homology seeking mechanisms. Thus, even if interrupted glutamine repeats could produce human disease, identifying patients with such lesions is exceedingly unlikely. However, evidence for the possible role of interrupted glutamine-rich domains in human neurodegeneration has been obtained from studies of amyotrophic lateral sclerosis (ALS), another neurodegenerative disease. Mutations in the gene TDP-43 promote ALS, and TDP-43 protein aggregates are found in ALS patients [61, 62]. Most TDP-43 mutations in patients affect the C-terminal glycine-rich domain. Fuentealba et al. [63] argue that the portion of the TDP-43 protein, which is critical for its sequestration in the cytosol, is more accurately characterized as a glutamine/asparagine (Q/N) rich domain [62, 63]. Similar domains are found in yeast prions that are prone to aggregation [64–66], and as described above, tend to form coiled-coils. The C-terminal region of TDP-43 seems to allow co-aggregation with polyQ disease proteins including Htt [63]. Furthermore, intermediate length polyQ expansions in ATXN2 are a risk factor for ALS [67].
Taken together, the results described here, although not definitive, raise the possibility that runs of glutamines in PQN-41C or polyQ disease peptides may not be the essential determinant of toxicity. Rather, it may be a specific protein secondary structure (perhaps a coiled coil) that is the relevant effector of cell death.
No polyQ peptide is an island
mRNAs encoding the glutamine-rich region of PQN-41 fused to upstream sequences can lead to enhanced linker cell survival in wild-type animals [10]; and GFP fusion to these larger proteins are not found in aggregates. Thus, sequences surrounding the glutamine-rich domain may modulate toxicity.
Similarly, sequences outside the polyQ tract can influence the kinetics of aggregation of polyQ-expanded proteins [68]. For example, using β-lactamase as a model protein scaffold, it was observed that internal insertion of successively longer polyQ tracts promoted aggregation into amyloid-fibrils as the structural integrity of the flanking β-lactamase moiety was compromised [69]. This suggests that the inherent propensity for polyQ tracts to aggregate can be countered by conformational constraints of the host protein up to a critical threshold repeat size. Similar host protein constraints have been observed in huntingtin, in which the flanking polyproline sequence can suppress the aggregation of the polyQ-expanded protein [70].
The map kinase SEK-1 was shown to promote linker cell death [10] suggesting that substrate phosphorylation likely plays a role in this process. Whether PQN-41 is the relevant target for SEK-1 is, however, not yet known. Post-translational modification of polyQ disease proteins can have well-defined effects on protein function. Huntingtin, for example, is extensively modified. Acetylation at lysine 444 is observed in human HD patients and has been shown to promote clearance of the mutant protein by targeting it to autophagosomes for degradation [71]. This modification is largely unique to the expanded form of the protein, and mutation of the modification site results in increased aggregation and toxicity in cultured cells and in a mouse model [71]. Toxicity of polyQ-expanded ATXN7 may also be regulated by acetylation. Cleavage of this protein at amino acid 266 enhances its toxicity in culture [42] (see above), however, the polyQ-containing cleavage product is cleared by macroautophagy. Acetylation of lysine 257 blocks degradation of the toxic ATXN7 peptide, enhancing toxicity [72].
The association between pathogenesis and phosphorylation of residues in the conserved 17 N-terminal amino acids of Htt has recently come into focus. Phosphate addition has been described at threonine 3 (T3) and at serines 13 and 16 (S13 and S16) of huntingtin, with all three modifications apparently reducing toxicity of polyQ-expanded Htt [32, 73, 74]. Similarly, a phospho-mimetic T3D mutation reduces toxicity of polyQ-expanded Htt in a Drosophila model; although in this case increased aggregation is found [73]. Similarly, phospho-mimetic mutations at S13 and S16 reduce polyQ-expanded Htt neurotoxicity (and aggregation) in the mouse [74]; and phospho-resistant S13A and S16A mutations do not affect toxicity [74]. A possible mechanism explaining reduced toxicity of the phosphorylated protein may be that these modifications promote protein degradation by the lysosome and proteasome [75].
And neither is its host protein
While the role of C. elegans pqn-41 outside the linker cell has not been studied, at least some transcripts are expressed in most cells in the animal [10]. Furthermore, animals carrying pqn-41 mutations are slow growing and egg-laying defective. Thus, pqn-41 may be important at some level for the basic function of all cells [10]. It is therefore possible that pqn-41-derived death-promoting transcripts expressed in the linker cell interfere with normal pqn-41 function. Supporting this idea, expression of an N-terminal domain of PQN-41 without the glutamine-rich region seems to prevent linker cell death in otherwise wild-type animals [10].
Several recent studies suggest that polyQ expansions may also promote disease by altering the native functions of their host proteins. A striking example of this idea is described by Duvick et al., studying ATXN1 [76]. The authors demonstrated that substitution of a phospho-mimicking aspartic acid for serine 776 in wild-type ATXN1 induces Purkinje cell disease sharing many features with SCA1. These observations suggest that altering wild-type ATXN1 functions may contribute to polyQ disease. Indeed, ATXN1 normally binds the RNA splicing factor RBM-17 and the transcriptional repressor Capicua, and the relative strengths of these interactions is affected by the length of the polyQ repeat as well as by phosphorylation of serine 776 [77]. However, Duvick et al. report that unlike SCA1, neuronal cell death is not observed in the serine 776 mutant, supporting the idea that the inherent toxicity of the polyQ moiety still plays an important role in disease pathology.
Models of SBMA also suggest that disease progression may depend on native androgen receptor (AR) functions. Binding of polyQ-expanded AR to its normal ligand, testosterone is required for toxicity [78, 79]. Furthermore, upon ligand binding, wild-type AR accumulates in the nucleus and binds DNA. While expression of a polyQ-expanded AR promotes cell degeneration in the Drosophila eye, expression of a similar protein with a defective DNA binding domain fails to induce degeneration even in the presence of ligand [80]. In addition, the AR interacts with transcription factors through its AF1 and AF2 protein domains. Disruption of the AF2 interaction surface can also rescue polyQ-expanded AR toxicity [80]. These results, together with the observation that overexpression of wild-type AR can promote toxicity similar to that observed with polyQ-expanded AR in both Drosophila and mice [80, 81], suggest that pathogenesis may arise, at least in part, from amplification of AR activity in the nucleus.
Thus, death-independent functions of both PQN-41 and polyQ disease proteins may modulate cellular toxicity.
Cell- and age-specific toxicity
PolyQ-mediated pathogenesis is markedly specific, at least at early stages of disease, affecting well-defined subsets of cells, despite broad expression of the proteins. Although several studies have shown that there is relative uniform expression of the disease proteins [82–84], it is possible that even small changes in protein concentration, which may be below the limits of experimental detection, could have a large impact on aggregation kinetics [85]. Indeed, broadly expressed promoters driving disease genes can cause disease in atypical cell populations for the given disease [32].
Another plausible determinant of cell specificity is that proteins that modify or interact with polyQ-expanded proteins may function or be expressed in a cell-specific manner. Rhes, a small G-protein specifically expressed in striatum, binds to polyQ-expanded Htt promoting its SUMOylation and increased toxicity [86, 87]. Havel et al. (2011) showed that phosphorylation of serine 16 of Htt N-terminal fragments results in increased nuclear Htt, an important step for pathogenesis [88, 89]. In vitro studies suggest that S16 is highly phosphorylated when Htt is incubated with striatal lysates but not with cortical or cerebellar lysates [88], suggesting cell-specific kinases or kinase levels in action. However, this kinase specificity is at odds with the seemingly protective roles of S16 phsophorylation (see above), and further studies are required to determine the full significance of these results.
Cell-specific cleavage of polyQ proteins may also account for cytotoxic specificity. A recent paper [90] showed that calcium-dependent calpain cleavage of mutant ATXN3 is promoted by excitation-induced rise in calcium levels in neurons. This effect could explain the restriction of SCA3 pathology to neurons with specific activity profiles, although the ability of cleaved ATXN3 to mediate neurodegeneration in vivo has not yet been conclusively shown [90].
The problem of cell-specificity may also relate to that of late onset of polyQ diseases. Indeed, even early onset patients must express disease proteins for years before clinical signs are evident. While damage could be cumulative, manifesting clinically only after a substantial number of cells have degenerated, another possibility is that polyQ peptides are not sufficient on their own to induce cellular degeneration. Other cellular defects, which may be cell-specific, could then contribute to disease. A possible effector of such a two-hit mechanism is the proteasome. Indeed, proteasome function has been shown to decline with age [91], and the proteasome has been implicated in pathogenesis of polyQ-expansion diseases in vertebrates (see above) and in C. elegans models [92, 93].
A one-two punch hypothesis is particularly appealing in the case of linker cell death in C. elegans. Although pqn-41 can promote cell death, mutations in the gene do not fully block linker cell death [10]. Furthermore, overexpression of PQN-41C in other cells in the animal or in the linker cell, well before the cell normally dies, does not promote ectopic cell death [10]. Thus, pqn-41 alone seems not sufficient to induce death, and may require a specific cellular setting to express its effects. Thus, as with polyQ disease proteins, PQN-41 function in promoting cell death is highly dependent on cellular and temporal context.
Concluding Remarks
The morphological and molecular similarities between polyQ-induced neurodegeneration and linker cell death in C. elegans are intriguing, and suggest the highly speculative, but exciting possibility of a shared mechanism of toxicity. These results also raise the possibility that polyQ disease may reflect, in part, inappropriate activation of an endogenous developmental cell death program. Ultrastructural similarities among linker cell death, polyQ-induced neurodegeneration, and developmental cell death in the vertebrate nervous system support this idea [13, 15–21]. Furthermore, the kinase scaffold protein TIR-1 is important for linker cell death in C. elegansand its Drosophila and mouse homologs, dSarm and Sarm, respectively, were recently implicated in the progression of Wallerian degeneration, a form of neuronal process degeneration suppressed by the Wlds fusion protein [10, 94]. Thus, a molecular link between linker cell death and neurodegeneration is already established.
The vertebrate proteins most similar in motif structure to PQN-41 are MED12 and p400, which have glutamine-rich C-termini and have been implicated in tumor formation [95, 96], a process requiring evasion of cell death. Interestingly, p400 was identified as a potentially conserved interactor (interlog) in the ataxia network generated by Lim and colleagues [97]. These results hint at similarities between linker cell death and known vertebrate neurodegenerative processes.
Nonetheless, several questions must be addressed to solidify or refute the notion of similarity between these cell death processes. For example, the requirement for uninterrupted glutamine stretches in disease contrasts with the interspersed glutamine-rich domain of PQN-41. Are these differences significant? Besides glutamine-rich motifs, are there other common molecular regulators of these cell death processes? Are glutamine-rich proteins important in vertebrate developmental cell death? What is the lethal blow to the cell, and is this event the same in both paradigms?
Linker cell death research is at an early stage, and the answers to the questions raised here, as well as many others, are not yet known. Nonetheless, if future studies do suggest a link between linker cell death and polyQ disease, the implications could be important. C. elegans is an excellent organism for gene discovery, and would offer a promising setting for characterizing the genetics and cell biology of key genes involved in disease pathogenesis. Furthermore, mechanistic similarities between linker cell death and polyQ disease would allow linker cell death to model the diseases in both research and therapy applications.
Acknowledgements
We thank Dr. Chris Link for helpful comments on the manuscript. Funding was provided by NIH grant NS081490 to S.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington“s disease chromosomes. The Huntington”s Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.La Spada AR, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 3.Sequeiros J, et al. Consensus and controversies in best practices for molecular genetic testing of spinocerebellar ataxias. Eur. J. Hum. Genet. 2010;18:1188–1195. doi: 10.1038/ejhg.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nolte D, et al. Spinocerebellar ataxia type 17 associated with an expansion of 42 glutamine residues in TATA-box binding protein gene. J. Neurol. Neurosurg. Psychiatr. 2010;81:1396–1399. doi: 10.1136/jnnp.2009.180711. [DOI] [PubMed] [Google Scholar]
- 5.Padiath QS, et al. Identification of a novel 45 repeat unstable allele associated with a disease phenotype at the MJD1/SCA3 locus. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;133:124–126. doi: 10.1002/ajmg.b.30088. [DOI] [PubMed] [Google Scholar]
- 6.La Spada A. Spinal and Bulbar Muscular Atrophy. In: Pagon RA, Bird TD, Dolan CR, et al., editors. GeneReviews. Seattle: University of Washington; 2011. (Internet) Available from: http://www.ncbi.nlm.nih.gov/books/NBK1333/ [PubMed] [Google Scholar]
- 7.Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 8.Wilburn B, et al. An antisense CAG repeat transcript at JPH3 locus mediates expanded polyglutamine protein toxicity in Huntington's disease-like 2 mice. Neuron. 2011;70:427–440. doi: 10.1016/j.neuron.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orr HT, Zoghbi HY. Trinucleotide Repeat Disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 10.Blum ES, et al. Control of Nonapoptotic Developmental Cell Death in Caenorhabditis elegans by a Polyglutamine-Repeat Protein. Science. 2012;335:970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- 12.Sulston JE, et al. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- 13.Abraham MC, et al. A Morphologically Conserved Nonapoptotic Program Promotes Linker Cell Death in Caenorhabditis elegans. Developmental Cell. 2007;12:73–86. doi: 10.1016/j.devcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 15.Pilar G, Landmesser L. Ultrastructural differences during embryonic cell death in normal and peripherally deprived ciliary ganglia. The Journal of Cell Biology. 1976;68:339–356. doi: 10.1083/jcb.68.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu-Wang IW, Oppenheim RW. Cell death of motoneurons in the chick embryo spinal cordIA light and electron microscopic study of naturally occurring and induced cell loss during development. J. Comp. Neurol. 1978;177:33–57. doi: 10.1002/cne.901770105. [DOI] [PubMed] [Google Scholar]
- 17.Oppenheim RW, et al. Programmed cell death of developing mammalian neurons after genetic deletion of caspases. Journal of Neuroscience. 2001;21:4752–4760. doi: 10.1523/JNEUROSCI.21-13-04752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bots GT, Bruyn GW. Neuropathological changes of the nucleus accumbens in Huntington's chorea. Acta Neuropathol. 1981;55:21–22. doi: 10.1007/BF00691525. [DOI] [PubMed] [Google Scholar]
- 19.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi H, et al. Neuronal nuclear alterations in dentatorubral-pallidoluysian atrophy: ultrastructural and morphometric studies of the cerebellar granule cells. Brain Res. 2001;919:12–19. doi: 10.1016/s0006-8993(01)02986-9. [DOI] [PubMed] [Google Scholar]
- 21.Zander C, et al. Similarities between spinocerebellar ataxia type 7 (SCA7) cell models and human brain: proteins recruited in inclusions and activation of caspase-3. Human Molecular Genetics. 2001;10:2569–2579. doi: 10.1093/hmg/10.22.2569. [DOI] [PubMed] [Google Scholar]
- 22.Orr HT. The cell biology of disease: Cell biology of spinocerebellar ataxia. The Journal of Cell Biology. 2012;197:167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 24.Brignull HR, et al. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. Journal of Neuroscience. 2006;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson GR, et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 26.Warrick JM, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 27.Wellington CL, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J. Biol. Chem. 1998;273:9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 28.Landles C, et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. Journal of Biological Chemistry. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nucifora LG, et al. Identification of Novel Potentially Toxic Oligomers Formed in Vitro from Mammalian-derived Expanded huntingtin Exon-1 Protein. Journal of Biological Chemistry. 2012;287:16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Legleiter J, et al. Mutant Huntingtin Fragments Form Oligomers in a Polyglutamine Length-dependent Manner in Vitro and in Vivo. Journal of Biological Chemistry. 2010;285:14777–14790. doi: 10.1074/jbc.M109.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldron-Roby E, et al. Transgenic Mouse Model Expressing the Caspase 6 Fragment of Mutant Huntingtin. Journal of Neuroscience. 2012;32:183–193. doi: 10.1523/JNEUROSCI.1305-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tebbenkamp ATN, et al. Transgenic mice expressing caspase-6-derived N-terminal fragments of mutant huntingtin develop neurologic abnormalities with predominant cytoplasmic inclusion pathology composed largely of a smaller proteolytic derivative. Human Molecular Genetics. 2011;20:2770–2782. doi: 10.1093/hmg/ddr176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juenemann K, et al. Modulation of mutant huntingtin N-terminal cleavage and its effect on aggregation and cell death. Neurotox Res. 2011;20:120–133. doi: 10.1007/s12640-010-9227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JP, et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington's disease. Neuron. 2010;67:199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham RK, et al. Cleavage at the Caspase-6 Site Is Required for Neuronal Dysfunction and Degeneration Due to Mutant Huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 36.Gafni J, et al. Caspase-6 Activity in a BACHD Mouse Modulates Steady-State Levels of Mutant Huntingtin Protein But Is Not Necessary for Production of a 586 Amino Acid Proteolytic Fragment. J. Neurosci. 2012;32:7454–7465. doi: 10.1523/JNEUROSCI.6379-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellerby LM, et al. Cleavage of atrophin-1 at caspase site aspartic acid 109 modulates cytotoxicity. J. Biol. Chem. 1999;274:8730–8736. doi: 10.1074/jbc.274.13.8730. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki Y, et al. Proteolytic processing regulates pathological accumulation in dentatorubral-pallidoluysian atrophy. FEBS Journal. 2010;277:4873–4887. doi: 10.1111/j.1742-4658.2010.07893.x. [DOI] [PubMed] [Google Scholar]
- 39.Goti D, et al. A mutant ataxin-3 putative-cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. Journal of Neuroscience. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colomer Gould VF, et al. A mutant ataxin-3 fragment results from processing at a site N-terminal to amino acid 190 in brain of Machado-Joseph disease-like transgenic mice. Neurobiol. Dis. 2007;27:362–369. doi: 10.1016/j.nbd.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jung J, et al. Preventing Ataxin-3 protein cleavage mitigates degeneration in a Drosophila model of SCA3. Human Molecular Genetics. 2009;18:4843–4852. doi: 10.1093/hmg/ddp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young JE, et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J. Biol. Chem. 2007;282:30150–30160. doi: 10.1074/jbc.M705265200. [DOI] [PubMed] [Google Scholar]
- 43.Ellerby LM, et al. Kennedy's disease: caspase cleavage of the androgen receptor is a crucial event in cytotoxicity. Journal of Neurochemistry. 1999;72:185–195. doi: 10.1046/j.1471-4159.1999.0720185.x. [DOI] [PubMed] [Google Scholar]
- 44.Tadokoro K, et al. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: the case of Gln in DRPLA affects subcellular localization of the products. J. Hum. Genet. 2005;50:382–394. doi: 10.1007/s10038-005-0261-9. [DOI] [PubMed] [Google Scholar]
- 45.Tsunemi T, et al. Cell-type-specific alternative splicing in spinocerebellar ataxia type 6. Neurosci. Lett. 2008;447:78–81. doi: 10.1016/j.neulet.2008.09.065. [DOI] [PubMed] [Google Scholar]
- 46.Harris GM, et al. Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS ONE. 2010;5:e13695. doi: 10.1371/journal.pone.0013695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bettencourt C, et al. Increased transcript diversity: novel splicing variants of Machado-Joseph disease gene (ATXN3) Neurogenetics. 2010;11:193–202. doi: 10.1007/s10048-009-0216-y. [DOI] [PubMed] [Google Scholar]
- 48.Parry DAD, et al. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. Journal of structural biology. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi T, et al. Soluble polyglutamine oligomers formed prior to inclusion body formation are cytotoxic. Human Molecular Genetics. 2007;17:345–356. doi: 10.1093/hmg/ddm311. [DOI] [PubMed] [Google Scholar]
- 50.Arrasate M, et al. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 51.Perutz MF, et al. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. SciU.SA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ignatova Z, et al. In-cell aggregation of a polyglutamine-containing chimera is a multistep process initiated by the flanking sequence. J. Biol. Chem. 2007;282:36736–36743. doi: 10.1074/jbc.M703682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka M, et al. Intra- and intermolecular beta-pleated sheet formation in glutamine-repeat inserted myoglobin as a model for polyglutamine diseases. J. Biol. Chem. 2001;276:45470–45475. doi: 10.1074/jbc.M107502200. [DOI] [PubMed] [Google Scholar]
- 54.Thakur AK, et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nature Structural & Molecular Biology. 2009;16:380–389. doi: 10.1038/nsmb.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiumara F, et al. Essential Role of Coiled Coils for Aggregation and Activity of Q/N-Rich Prions and PolyQ Proteins. Cell. 2010;143:1121–1135. doi: 10.1016/j.cell.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi T, et al. Polyglutamine diseases: where does toxicity come from? what is toxicity? where are we going? Journal of molecular cell biology. 2010;2:180–191. doi: 10.1093/jmcb/mjq005. [DOI] [PubMed] [Google Scholar]
- 57.Arango M, et al. CA150 expression delays striatal cell death in overexpression and knock-in conditions for mutant huntingtin neurotoxicity. J. Neurosci. 2006;26:4649–4659. doi: 10.1523/JNEUROSCI.5409-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walters RH, Murphy RM. Aggregation kinetics of interrupted polyglutamine peptides. J. Mol. Biol. 2011;412:505–519. doi: 10.1016/j.jmb.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sen S, et al. Role of histidine interruption in mitigating the pathological effects of long polyglutamine stretches in SCA1: A molecular approach. Protein Sci. 2003;12:953–962. doi: 10.1110/ps.0224403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jayaraman M, et al. The impact of ataxin-1-like histidine insertions on polyglutamine aggregation. Protein Eng. Des. Sel. 2009;22:469–478. doi: 10.1093/protein/gzp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 62.Pesiridis GS, et al. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Human Molecular Genetics. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fuentealba RA, et al. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. Journal of Biological Chemistry. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santoso A, et al. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 65.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 66.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl. Acad. SciU.SA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robertson AL, Bottomley SP. Towards the treatment of polyglutamine diseases: the modulatory role of protein context. Current medicinal chemistry. 2010;17:3058–3068. doi: 10.2174/092986710791959800. [DOI] [PubMed] [Google Scholar]
- 69.Scarafone N, et al. Amyloid-like fibril formation by polyQ proteins: a critical balance between the polyQ length and the constraints imposed by the host protein. PLoS ONE. 2012;7:e31253. doi: 10.1371/journal.pone.0031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhattacharyya A, et al. Oligoproline effects on polyglutamine conformation and aggregation. J. Mol. Biol. 2006;355:524–535. doi: 10.1016/j.jmb.2005.10.053. [DOI] [PubMed] [Google Scholar]
- 71.Jeong H, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mookerjee S, et al. Posttranslational Modification of Ataxin-7 at Lysine 257 Prevents Autophagy-Mediated Turnover of an N-Terminal Caspase-7 Cleavage Fragment. Journal of Neuroscience. 2009;29:15134–15144. doi: 10.1523/JNEUROSCI.4720-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aiken CT, et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. Journal of Biological Chemistry. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gu X, et al. Serines 13 and 16 Are Critical Determinants of Full-Length Human Mutant Huntingtin Induced Disease Pathogenesis in HD Mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson LM, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. The Journal of Cell Biology. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–935. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Katsuno M, et al. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–854. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 79.Takeyama K-I, et al. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- 80.Nedelsky NB, et al. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinobulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Monks DA, et al. Overexpression of wild-type androgen receptor in muscle recapitulates polyglutamine disease. Proc. Natl. Acad. SciU.SA. 2007;104:18259–18264. doi: 10.1073/pnas.0705501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aronin N, et al. CAG expansion affects the expression of mutant Huntingtin in the Huntington's disease brain. Neuron. 1995;15:1193–1201. doi: 10.1016/0896-6273(95)90106-x. [DOI] [PubMed] [Google Scholar]
- 83.Servadio A, et al. Expression analysis of the ataxin-1 protein in tissues from normal and spinocerebellar ataxia type 1 individuals. Nat Genet. 1995;10:94–98. doi: 10.1038/ng0595-94. [DOI] [PubMed] [Google Scholar]
- 84.Imbert G, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 85.Colby DW, et al. Stochastic kinetics of intracellular huntingtin aggregate formation. Nature Chemical Biology. 2006;2:319–323. doi: 10.1038/nchembio792. [DOI] [PubMed] [Google Scholar]
- 86.Subramaniam S, et al. Rhes, a Striatal Specific Protein, Mediates Mutant-Huntingtin Cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Falk JD, et al. Rhes: A striatal-specific Ras homolog related to Dexras1. J. Neurosci. Res. 1999;57:782–788. [PubMed] [Google Scholar]
- 88.Havel LS, et al. Preferential accumulation of N-terminal mutant huntingtin in the nuclei of striatal neurons is regulated by phosphorylation. Human Molecular Genetics. 2011;20:1424–1437. doi: 10.1093/hmg/ddr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Raamsdonk JM, et al. Selective degeneration and nuclear localization of mutant huntingtin in the YAC128 mouse model of Huntington disease. Human Molecular Genetics. 2005;14:3823–3835. doi: 10.1093/hmg/ddi407. [DOI] [PubMed] [Google Scholar]
- 90.Koch P, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 91.Vernace VA, et al. Aging and regulated protein degradation: who has the UPPer hand? Aging cell. 2007;6:599–606. doi: 10.1111/j.1474-9726.2007.00329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Morley JF, et al. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. SciU.SA. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ben-Zvi A, et al. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. SciU.SA. 2009;106:14914–14919. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Osterloh JM, et al. dSarm/Sarm1 Is Required for Activation of an Injury-Induced Axon Death Pathway. Science. 2012 doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fuchs M, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 96.Makinen N, et al. MED12, the Mediator Complex Subunit 12 Gene, Is Mutated at High Frequency in Uterine Leiomyomas. Science. 2011;334:252–255. doi: 10.1126/science.1208930. [DOI] [PubMed] [Google Scholar]
- 97.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]