Abstract
The SMC5/6 protein complex consists of the Smc5, Smc6 and Non-Smc-Element (Nse) proteins and is important for genome stability in many species. To identify novel components in the DNA repair pathway, we carried out a genetic screen to identify mutations that confer reduced resistance to the genotoxic effects of caffeine, which inhibits the ATM and ATR DNA damage response proteins. This approach identified inactivating mutations in CG5524 and MAGE, homologs of genes encoding Smc6 and Nse3 in yeasts. The fact that Smc5 mutants are also caffeine-sensitive and that Mage physically interacts with Drosophila homologs of Nse proteins suggests that the structure of the Smc5/6 complex is conserved in Drosophila. Although Smc5/6 proteins are required for viability in S. cerevisiae, they are not essential under normal circumstances in Drosophila. However, flies carrying mutations in Smc5, Smc6 and MAGE are hypersensitive to genotoxic agents such as ionizing radiation, camptothecin, hydroxyurea and MMS, consistent with the Smc5/6 complex serving a conserved role in genome stability. We also show that mutant flies are not compromised for pre-mitotic cell cycle checkpoint responses. Rather, caffeine-induced apoptosis in these mutants is exacerbated by inhibition of ATM or ATR checkpoint kinases but suppressed by Rad51 depletion, suggesting a functional interaction involving homologous DNA repair pathways that deserves further scrutiny. Our insights into the SMC5/6 complex provide new challenges for understanding the role of this enigmatic chromatin factor in multi-cellular organisms.
Introduction
The evolutionarily conserved Structural Maintenance of Chromosomes proteins are essential for the organization, segregation, and stability of the genome [1], [2], [3]. Three functionally distinct SMC complexes have been defined in eukaryotes: cohesin (Smc1/3), condensin (Smc2/4), and the otherwise unnamed Smc5/6 complex, each accompanied by a unique set of regulatory subunits. Cohesin holds sister chromatids together after DNA replication and plays important roles in regulation of gene expression and DNA repair [4], while condensin is essential for mitotic chromosome organization and segregation [5]. The Smc5/6 complex is less well characterized but is required for homologous DNA recombination-based processes, including repair of DNA double strand breaks, restart of stalled replication forks, ribosomal DNA maintenance, telomere elongation, and chromosome dynamics during meiosis [6], [7], [8], [9], [10].
The Smc5/6 complex in the yeasts is made up of eight subunits that form three sub-complexes: Smc6-Smc5-Nse2, Nse1-Nse3-Nse4, and Nse5-Nse6 [11]. Smc5 and Smc6 dimerize through their hinge regions to form the core. The Sumo ligase Nse2 associates with the Smc5-Smc6 heterodimer through a direct interaction with Smc5 [12], [13], [14]. Nse1, a RING finger protein with E3 ubiquitin ligase activity, Nse4, the kleisin component of the complex, and Nse3, a MAGE homolog, interact with each other to form the sub-complex that bridges the head domain of the Smc5-Smc6 heterodimer [7], [14], [15], [16], [17]. Nse5 and Nse6 form the third sub-complex in yeasts, but these proteins have no counterparts in higher eukaryotes [11].
In humans, the Nse3 gene is represented by an expanded family of “MAGE” (melanoma antigen gene) genes with over 50 members, classified into two types. Type I MAGE genes are frequently over-expressed in human primary cancers and cancer cell lines, and may play a role in resistance to chemotherapeutic agents [18]. In fact, 85% of cancer cell lines over-express at least one Type I MAGE gene [19]. In contrast, Type II MAGE genes, such as NDN, MAGEL2 and MAGED1 are expressed in normal tissues and have important roles in mammalian development [20], [21], [22]. MAGEG1 was identified as a component of the human Smc5/6 complex [23]. The crystal structure of MAGEG1 revealed its interaction with RING protein Nse1, and this interaction stimulates the ubiquitin ligase activity of Nse1 [17], [23]. Other MAGE proteins interact with the mammalian homologs of Nse1 and Nse4, suggesting a conserved role of MAGE proteins as part of distinct Smc5/6 complexes [15], [17], [23], [24], [25].
All components of the Smc5/6 complex are essential in S. cerevisiae [13], and, except for Nse5 and Nse6, also in S. pombe [11]. Many hypomorphic Smc5/6 mutants are hypersensitive to genotoxic agents such as ionizing radiation (IR), the alkylating agent methyl methanesulfonate (MMS), hydroxyurea (HU) and UV light in yeasts [26]. Epistasis experiments in yeasts and vertebrate cells have placed Smc5/6 genes in the homologous recombination-based DNA repair pathway that involves Rad51 nucleofilament proteins [8]. In Drosophila, Smc5/6 plays a role in maintaining genome stability in heterochromatin regions by repressing non-sister chromosome recombination events [9], [27]. Drosophila Smc5/6 also serves a conserved molecular role in blocking Rad51 loading during this process and compromising Smc6 activity in S2 cells caused chromosome defects, suggesting Smc5/6 functions are essential [27]. Regulation of homologous recombination-mediated repair relies largely on two kinases, ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR). ATM and ATR are phosphoinositide 3-kinase-like kinases (PIKK) that are activated by double strand breaks, turning on a network of DNA damage response signaling pathways that coordinate cell cycle progression and DNA repair [28]. Caffeine is a PIKK inhibitor commonly used to inhibit ATM and ATR [29], [30]. We sought to identify novel genes functioning in DNA damage response pathways that are redundant with ATM and ATR, by screening for conditional eye phenotypes in adult flies that were fed caffeine throughout larval development. We found unexpectedly that three Drosophila genes, Smc5, Smc6 and MAGE, are not essential under normal growth conditions, but are essential for resistance to caffeine exposure throughout development. Interestingly, these mutants are also hypersensitive to genotoxic agents, suggesting a conserved role for the Smc5/6 in DNA damage repair. Caffeine induces apoptosis in the mutant flies in a process mediated by ATM and ATR that does not involve conventional cell cycle checkpoints. We have thus identified a novel caffeine-sensitive mechanism that prevents apoptosis in cells exposed to genotoxic stress.
Results
A Screen for Caffeine-sensitive Eye Mutants Reveals Three Loci on Chromosome 3R
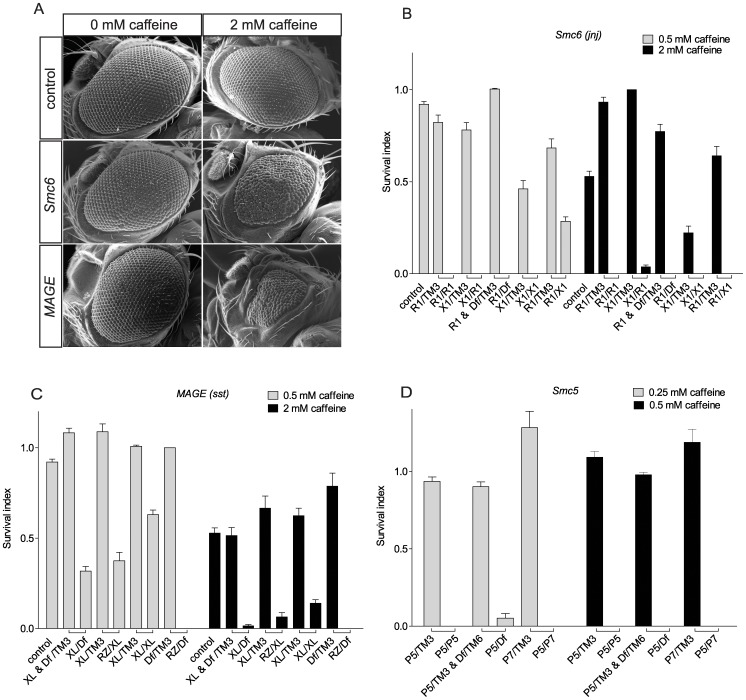
The compound eyes of Drosophila are ideal tissues to detect defects in proliferation and apoptosis as they are not essential for survival, but they are sensitive to developmental perturbations and easy to score for mutant phenotypes. To identify novel genes functioning in DNA damage response pathways that are redundant with ATM and ATR, we previously performed a genetic screen to identify conditional eye phenotypes in adult flies fed 2 mM caffeine and 3 mM hydroxyurea (HU) throughout larval development [31]. While caffeine inhibits ATM and ATR, HU stalls replication forks through inhibition of dNTP production, eventually generating single strand or double strand DNA breaks, thereby activating DNA damage responses regulated by ATM and ATR. At the drug concentrations used, there were no phenotypic effects in wildtype flies. In this screen, we used the “EGUF, GMR-hid” (EGUF) system to produce homozygous mutant clonal cells in the entire adult eye of an otherwise heterozygous fly [32]. This screen identified a single caffeine-sensitive locus (huc95E) on chromosome arm 3R, here renamed java no jive (jnj), which we mapped to cytological region 95E by complementation testing with chromosomal deficiencies [31]. Flies that were mosaic hemizygous for jnj in the eye exhibit caffeine-dependent small, rough eyes associated with increased apoptosis. To identify novel DNA damage pathway components, we have now carried out a new screen of chromosome arm 3R for conditional caffeine-sensitive eye phenotypes. By screening 9098 males, we identified three loci on chromosome arm 3R including six additional alleles of jnj, two mutant alleles of a locus called sleepless in seattle (sst), and one allele of a novel locus called double double trouble (ddt), that has not yet been linked to a specific gene (Fig. 1A, Fig. S1). All hemizygous jnj, sst and ddt mutants exhibit caffeine-dependent pupal lethality (Fig. 1B–D and data not shown).
Figure 1. Eye phenotypes in caffeine-sensitive mutant flies.
(A) Caffeine-dependent eye phenotype of Smc6 (jnj) and MAGE (sst) mutants. Fly genotypes are as follows. Control: EGUF/+; FRT82B +/FRT82B GMR-hid. Smc6 (loss of Smc6 in eye cells): EGUF/+; FRT82B jnjR1/FRT82B GMR-hid. MAGE (loss of MAGE in eye cells): EGUF/+; FRT82B sstRZ/FRT82B GMR-hid. (B-D) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when raised in media with caffeine. Bars represent the survival index (p) and error bars represent SEM. “□” indicates flies eclosed from the same cross. Absence of a bar indicates no surviving flies. Wildtype control flies are w1118. (B) Smc6 mutants are sensitive to caffeine. R1 (jnjR1) is an allele from the caffeine screen, X1 (jnjX1) was generated by an imprecise excision of a P-element adjacent to the 5′UTR of Smc6, and Df (Df(3R)Exel6198) is a deficiency chromosome uncovering the Smc6 locus. (C) MAGE mutants are sensitive to caffeine. RZ (sstRZ) is an allele from the caffeine screen, XL (sstXL) is a targeted knockout, and Df (Df(3R)Antp1) is a deficiency chromosome uncovering the MAGE locus. (D) Smc5 mutants are sensitive to caffeine. Both P5 (Smc5P{GSV1}GS3245) and P7 (Smc5P{GSV6}GS14577) contain P-element insertions in a coding exon of Smc5, and Df (Df(3L)BSC418) is a deficiency chromosome uncovering the Smc5 locus.
Mutations in Smc6 Cause Caffeine-dependent Defects in java no jive Mutant Flies
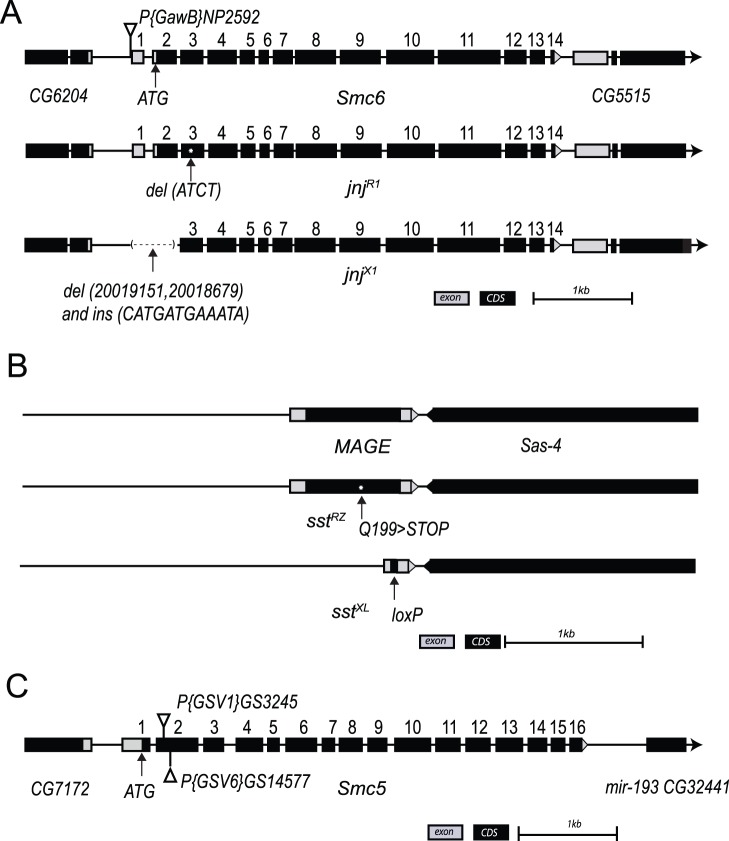
Deletion mapping indicated that all of the caffeine-sensitive jnj alleles were viable in hemizygous combinations with deletions uncovering region 95E, indicating that the homozygous lethality of most jnj alleles was caused by second site mutation(s). Homozygotes for one allele, jnjR1, were viable on regular media, but died at the pupal stage when raised in media containing caffeine (Fig. 1B). Sequencing of candidate genes in the jnj region identified a four base pair deletion in exon two of the FlyBase annotated gene CG5524 (del_ATCT at position 334–337 bp from the presumptive start codon), creating a frameshift resulting in a stop codon at position 133 of the presumptive 1122 amino acid protein (Fig. 2A). The predicted CG5524 protein has highest amino acid identity with SMC6 (Structural Maintenance of Chromosomes 6) in other species. SMC6 regulates chromosome stability in yeasts [7], [8], [9], and is implicated in heterochromatic DNA repair in Drosophila [27]. We tested CG5524 (hereafter called Smc6) and four neighboring genes for levels of expression by quantitative RT-PCR of RNA from whole flies. Levels of Smc6 RNA were greatly reduced with all seven alleles of jnj, ranging from 9% to 24% of control levels (Fig. S2A) whereas nearby genes showed little change in expression. Despite extensive sequencing efforts, we were not able to identify the nature of jnj alleles other than jnjR1, suggesting that these unmapped mutations reside in as yet unidentified regulatory regions of Smc6. To be certain that our jnj alleles corresponded to Smc6, we generated additional Smc6 lines by imprecise excision of the P-element present in line NP2592, including the new line jnjX1 that lacks exon 1 and sequences up- and downstream of this exon (Fig. 2A). We tested caffeine sensitivity in all of the jnj allelic combinations and found that raising larvae on 0.5 mM caffeine resulted in almost complete lethality (Fig. 1B). Using RNAi to deplete Smc6 expression in developing eye discs also resulted in a caffeine-dependent rough eye phenotype (Fig. S2B). Collectively, the presence of a frame shift mutation in Smc6 in jnjR1, the reduced expression levels of Smc6 in all seven alleles of jnj, the caffeine-dependent lethality of the deletion allele jnjX1, and caffeine-dependent eye phenotypes induced by Smc6 RNAi all implicate CG5524/Smc6 as the relevant gene in jnj mutants.
Figure 2. Overview of Smc6, MAGE, and Smc5 gene location, structural organization and mutant alleles.
(A) Smc6 is a 14 exon gene located on 3R:95E8–95F1. jnjR1 contains a 4 bp deletion in the 2nd coding exon. jnjX1 contains a 473 bp deletion of sequences upstream of exon 1 (196 bp), the entire exon 1 (252 bp), and a portion of intron 1 (25 bp), with a 12 bp vestige of the original P element remaining. Smc6 genomic locus (3R:20,014,770.20,019,145 [−]) is shown. (B) MAGE is a single exon gene located on the right arm of the 3rd chromosome at position 84C7–84C7. sstRZ has a point mutation that converts a glutamine at position 109 to a stop codon. sstXL carries a targeted deletion of the entire coding sequence of MAGE. MAGE genomic locus (3R:2,979,960.2,980,898 [−]) is shown. (C) Smc5 is a 16 exon gene located in 78D6–78D7 of the left arm of the 3rd chromosome. Exons encoding the longest transcripts are shown. Both P{GSV1}GS3245 and P{GSV6}GS14577 are inserted in the second coding exon. The Smc5 genomic locus (3L:21,562,309.21,566,623 [+]) is shown. CDS, coding sequence.
Caffeine-sensitivity in sleepless in seattle Mutants is Due to Mutations in the MAGE Gene
The sstRZ mutation exhibits caffeine-dependent pupal lethality in combination with a chromosomal deficiency (Df(3R)Antp1, Fig. 1C) but sstRZ homozygotes are not viable on regular media, presumably because of a second site mutation. Further deletion mapping refined the position of the caffeine-sensitive sst locus to a region containing seven candidate genes, each of which were sequenced. We identified a glutamine to stop mutation affecting the MAGE gene [33] in sstRZ, at position 109 of the 232 amino acid Mage protein (Fig. 2B). In previous studies, depletion of MAGE mRNA using double strand RNA injection suggested that MAGE was essential for viability during early embryogenesis, whereas conditional knockdown at later developmental stages suggested a role in postembryonic neuronal survival and proliferation [34]. Moreover, DNA fibers connecting mitotic cells were observed after RNAi-mediated depletion of Smc5 or Smc6 in S2 cells, suggesting that the Smc5/6 complex could be essential for mitosis in Drosophila [27]. We therefore initially reasoned that sstRZ was a partial loss-of-function allele, since hemizygous sstRZ flies were viable. To test this idea we synthesized a knockout allele by homologous recombination [35]. In this new allele (sstXL) the complete coding sequence of MAGE was deleted (Fig. 2B). Surprisingly, homozygous sstXL flies displayed no increased lethality or obvious mutant phenotype when raised on media without caffeine. As with sstRZ hemizygotes, sstXL flies reared in caffeine media were inviable, but they were less sensitive to a lower dose of caffeine (0.5 mM) than jnj mutants (Fig. 1C). About 15% of predicted sstXL homozygous flies survived 2 mM caffeine exposure and the surviving flies often had small or rough eyes, similar to sstRZ mutants (Fig. 1A). Transheterozygous sstRZ/sstXL progeny were also viable on normal media, but only 6% survived on 2 mM caffeine (Fig. 1C). Using polyclonal antibodies directed against Mage [36] we found that Mage was absent from protein lysates derived from sst adult flies (Fig. S3). In addition, caffeine-dependent lethality of sstXL can be complemented by a genomic MAGE transgene (Table S1) that includes the full coding region of MAGE and 3 kb sequence upstream and expresses Mage protein at normal levels (Fig. S3). Collectively, the identification of a stop mutation in the MAGE gene (sstRZ), the caffeine-sensitivity of a MAGE knockout allele sstXL, the loss of Mage protein in sst flies and the rescue of caffeine sensitivity by a MAGE transgene all implicate MAGE as the mutated gene in sst flies.
Smc5 Mutant Flies are also Caffeine Sensitive
In yeasts and mammalian cells, all known SMC6 functions involve SMC5 [23], [37], so we predicted that loss of Smc5 activity would also cause caffeine sensitivity in flies (Fig. 3A). We tested two P insertion alleles predicted to affect Smc5 for caffeine sensitivity, namely Smc5P{GSV1}GS3245, referred to as Smc5P5, and Smc5P{GSV6}GS14577, referred to as Smc5P7 [38]. As predicted, both Smc5 mutants were sensitive to caffeine (Fig. 1D). Both of these alleles have P-element insertions within the second exon of Smc5 and the insertion sites are very close to the putative start codon (Fig. 2C). Therefore, they are very likely to be null alleles. To rule out the possibility that caffeine-sensitivity of Smc5 flies was caused by second site mutations, we generated fly lines in which the P-elements in both alleles were excised by a transposase, either restoring the wild-type sequence or resulting in an insertion or deletion of the original P element insertion in the coding exon of Smc5. We therefore predicted that some excision lines would no longer be caffeine-sensitive while others would retain the mutant phenotype. As expected, of 13 independent fly lines produced by the excision of P7, seven lines were no longer caffeine sensitive (Table S2A). Similar results were obtained from the excision of P5 (Table S2B). In conclusion, as with Smc6 and MAGE, loss of Smc5 function results in caffeine-dependent lethality.
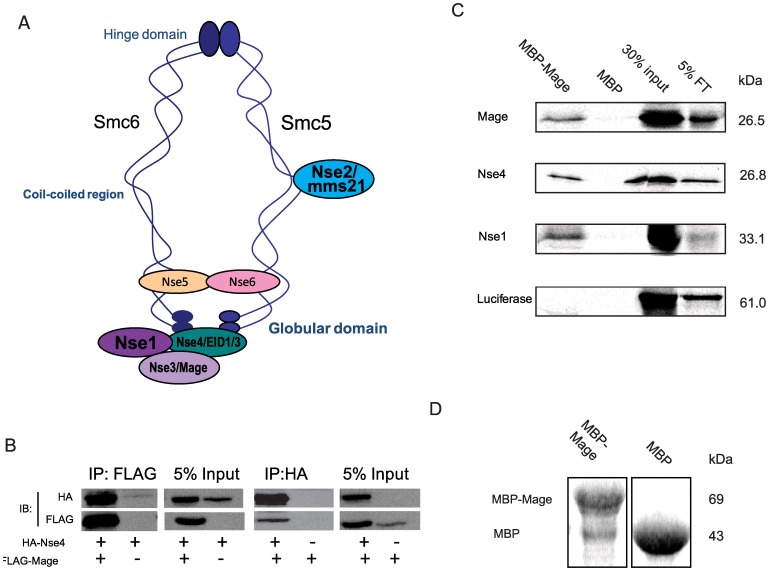
Figure 3. Mage is part of the Drosophila Smc5/6 complex.
(A) Diagram of a generic Smc5/6 complex in S. pombe (adapted from [70]). The structure in S. cerevisiae is different in that Nse5/6 were found to bind at the hinge. (B) Mage interacts with Nse4 when both proteins are co-expressed in S2 cells. HA-Nse4 co-immunoprecipitated (co-IP) with FLAG-Mage from an S2 cell lysate when two proteins were co-expressed; FLAG-Mage co-IPed with HA-Nse4 from the S2 cell lysate when two proteins were co-expressed. (C) Recombinant Mage interacts with Nse4 and Nse1 directly. Immobilized maltose binding protein (MBP)-fused MAGE or MBP were incubated with 35S-methionine labeled Mage, Nse4, Nse1, or luciferase (as a negative control), respectively. Proteins that were associated with immobilized MBP-Mage or MBP were resolved with SDS-PAGE and visualized by autoradiography. Results show that Mage, Nse4, and Nse1 each interact with MBP-Mage but not with MBP and luciferase does not interact with either of these proteins. (D) Coomassie staining of protein immobilized on 10 µl of amylose beads showed that approximately equal amounts of MBP-Mage and MBP proteins were immobilized on resin beads.
Caffeine Sensitivity is Mediated through Smc5/6
At the whole organism level, a higher proportion of MAGE mutants were able to survive exposure to 0.5 mM caffeine throughout larval development than Smc6 and Smc5 mutants. Indeed all genetic combinations of MAGE mutant flies had some survivors on media containing 2 mM caffeine, while there were essentially no survivors among the Smc5 or Smc6 mutants raised on 2 mM caffeine (Fig. 1B–D). This suggests that the Mage protein is less important for caffeine resistance than the Smc5 and Smc6 proteins. To further test this hypothesis, we measured the viability of flies carrying mutations in two different components of the protein complex when raised on media containing caffeine. Flies deficient for both Mage and Smc6 were more sensitive to caffeine than flies deficient for Mage alone, but were similar in sensitivity to flies deficient for Smc6 alone (Table S3). This suggests that the Smc5/6 heterodimer has a more critical role in caffeine resistance than does the sub-complex containing Nse1-Mage, consistent with observations in yeasts [1].
Drosophila Smc5/6 Components Form a Protein Complex
In yeasts, the Smc5/6 complex consists of Smc5, Smc6 and six Nse (non-Smc element) subunits [14], four of which were also identified in humans [15], [23]. In searches of Drosophila genome databases, we uncovered a set of putative transcription units that appear to correspond to SMC5/6 complex subunits in yeasts (Table S4). Of these, MAGE has previously been described as a homolog of yeast Nse3 and human MAGEG1 [23]. In Drosophila, Mage protein was shown to interact with Drosophila Nse4 (Nse4) using a yeast two-hybrid system [39]. When we examined the Gene Expression Omnibus (GEO, [40]) to compare gene expression profiles, we found that these two genes have very similar expression patterns across different tissues, supporting the idea that the encoded proteins function in a complex (Fig. S4). Fission yeast Nse1 has been detected in the same sub-complex as Nse3 and Nse4, as part of the larger Smc5/6 complex (Fig. 3A) [11]. We first tested for a physical interaction between Drosophila Mage and Nse4 in cell culture, by generating epitope-tagged plasmid constructs that produce HA-tagged Nse4 or FLAG-tagged Mage, and co-transfecting them into Drosophila Schneider 2 (S2) cells. We were able to co-immunoprecipitate HA-Nse4 and FLAG-Mage from S2 cell lysates (Fig. 3B). We then performed in vitro pull down experiments to show that this interaction is likely direct, and that Mage also interacts with Nse1 directly (Fig. 3C). These results indicate that the three Drosophila proteins (Nse1, Mage and Nse4) form a sub-complex analogous to that found in yeast, consistent with conservation of structure across species.
Loss of Function for Smc6 or MAGE Sensitizes Imaginal Cells to Caffeine-induced Apoptosis
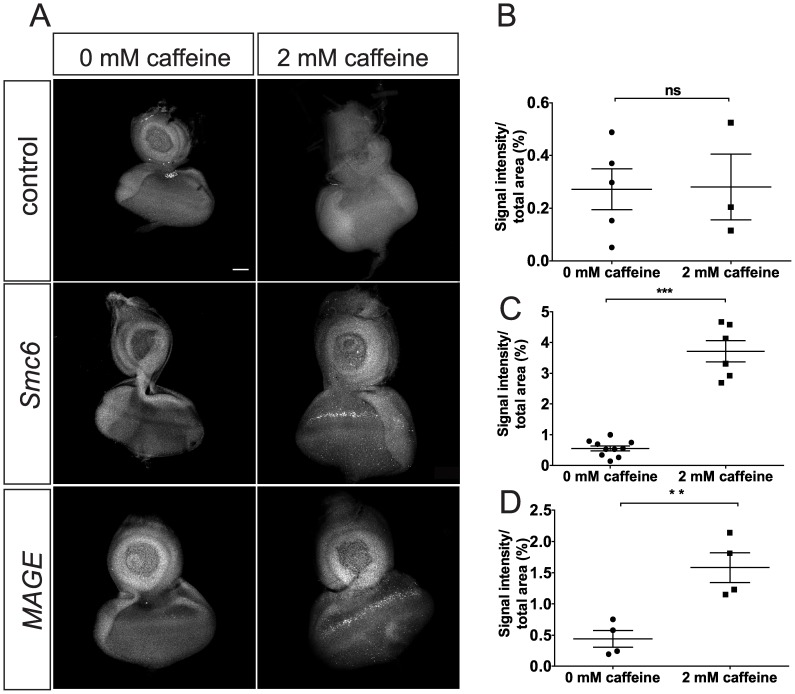
Previous examinations of jnjhuc95E hemizygous mutants were based on the EGUF eye mosaic system [31]. In this experiment, we observed caffeine-dependent defects in ommatidial patterning and increased apoptosis in the eye discs. Larvae mutant for Smc6 or MAGE die at the pupal stage when raised long term on caffeine-containing media. Remarkably, upon dissection of these larvae we noticed that the imaginal discs were severely damaged or altogether absent, suggesting increased cell death as the cause of this defect. To test this hypothesis, we dissected eye imaginal discs from late third instar larvae and labeled them with antibodies against activated caspase 3 to mark apoptotic cells. We detected minimal labeling of apoptotic foci in eye discs of control larvae, regardless of caffeine exposure (Fig. 4). In contrast, dramatically increased labeling of apoptotic foci were seen in the eye discs of Smc6 or MAGE mutant third instar larvae after short term (12 hours) caffeine exposure. Apoptotic labeling was markedly enhanced in a band of cells immediately anterior to the morphogenetic furrow, where cells become synchronized in G1 phase [41]. These results suggest that caffeine-induced apoptosis in developing imaginal discs likely underlies caffeine-dependent pupal lethality in MAGE and Smc6 mutant flies.
Figure 4. Caffeine exposure results in apoptosis in eye discs of MAGE and Smc6 mutants.
(A) Anti-cleaved-caspase-3 antibody staining of eye discs from third instar larvae of control (WT, FRT82B), MAGE (sstRZ/sstXL), and Smc6 (jnjX1/jnjR1) genotypes raised in either standard media (0 mM caffeine) or media supplemented with 2 mM caffeine for 12 hours before dissection. Images are single stacks of confocal images. More cleaved-caspase-3 foci in eye discs of sstRZ/sstXL and jnjX1/jnjR1 larvae were observed after caffeine exposure. A narrow band of apoptotic cells (white arrow heads) anterior to the presumptive morphogenetic furrow are most noticeable. Scale bar represents 50 µM. (B-D) Quantification and comparison of cleaved caspase-3 staining levels in WT (B), MAGE (C) or Smc6 (D) eye discs, comparing the no caffeine and 2 mM caffeine groups. Data represent mean area stained from multiple eye discs for each genotype per treatment. A maximum projection of all stacks of a confocal image was used to quantify the signal intensity of staining. This value was divided by the area of each eye disc to obtain a ratio representing the relative amount of immunostaining. Error bars represent SEM. A non-paired two-tailed t-test was used to determine statistical significance. **, P = 0.006, ***, P<0.0001.
Smc5/6 Mutant Flies are Hypersensitive to Genotoxic Stress
The DNA damage response is a multi-step process that involves sensing of damage, cell cycle arrest, and repair of the damaged DNA. Yeast with hypomorphic mutations affecting Smc6, Nse1, Nse2, Nse3 or Nse4 are hypersensitive to gamma irradiation, UV light, MMS, camptothecin (a topoisomerase I inhibitor), and inhibition of DNA replication by HU [26]. All of these genotoxic stresses directly or indirectly generate DNA single-stranded or double-stranded breaks. To explore whether Drosophila Smc5/6 provides similar responses to genotoxic stress, we analyzed the effects of ionizing radiation, camptothecin, HU or MMS on viability. Exposure to 40 Gy ionizing radiation caused increased lethality in MAGE, Smc6 and Smc5 mutants compared to controls (Fig. 5). Moreover, all three mutants were hypersensitive to camptothecin, HU and MMS, compared to controls (Figs. S5 and data not shown).
Figure 5. Smc5/6 mutants are hypersensitive to ionizing radiation.
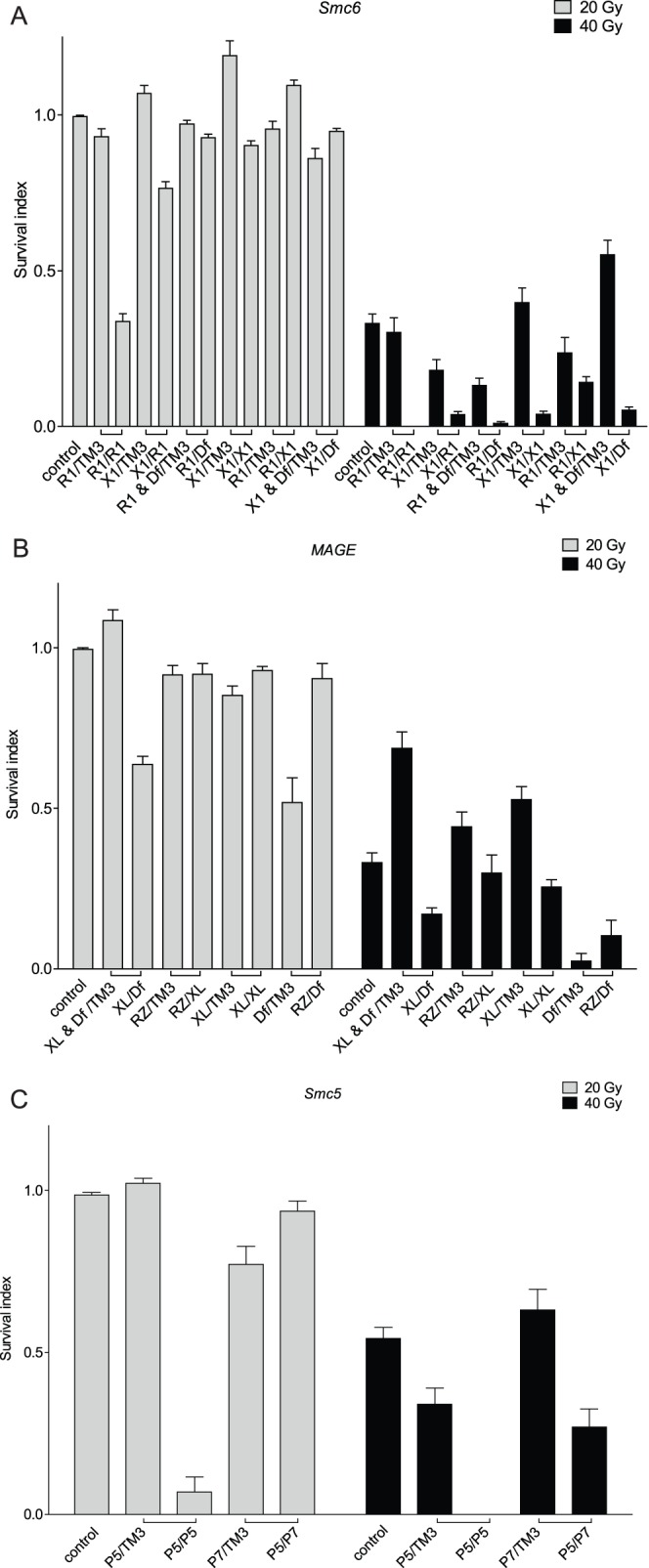
(A–C) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when exposed to 40 Gy of IR. Bars represent the survival index (p) ± SEM. “□” indicates flies eclosed from the same cross. Absence of a bar indicates that no flies survived at that IR dose. (A) Smc6 mutants are hypersensitive to IR. R1 (jnjR1) and X1 (jnjX1) are Smc6 alleles. Df (Df(3R)Exel6198) is a deficiency chromosome uncovering the Smc6 locus. (B) MAGE mutants are hypersensitive to IR. RZ (sstRZ) and XL (sstXL) are MAGE alleles. Df (Df(3R)Antp1) is a deficiency chromosome uncovering the MAGE locus. (C) Smc5 mutants are hypersensitive to IR. P5 (Smc5P{GSV1}GS3245) and P7 (Smc5P{GSV6}GS14577) are Smc5 alleles. Df (Df(3L)BSC418) is a deficiency chromosome uncovering the Smc5 locus.
Loss of Smc5/6 Function does not Compromise G2/M and S Phase Checkpoints Induced by Genotoxic Agents
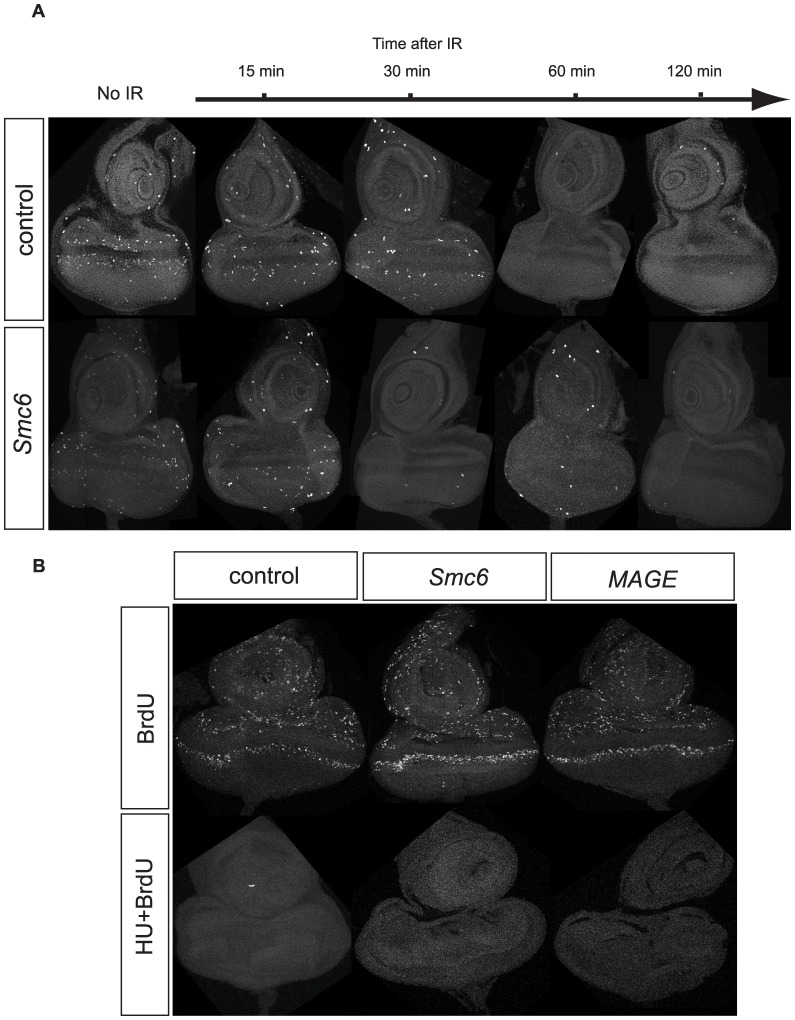
Studies in Drosophila have proven to be valuable for the study of proteins and pathways controlling DNA repair and checkpoint responses, which are remarkably well conserved among flies and other organisms [27], [42]. In S. pombe, nse3-1 hypomorphic mutants activate a DNA damage checkpoint that arrests cells in late S phase/G2 [7], and Smc6 (Rad18) is required for maintenance but not activation of the G2 checkpoint [43], [44]. We therefore tested whether cell cycle checkpoints important for DNA damage response pathways were perturbed in caffeine-sensitive MAGE or Smc6 mutant flies. To assess G2/M checkpoint function we used ionizing radiation (IR) to determine if IR exposure decreased the number of mitotic cells [45]. We dissected eye imaginal discs from late third instar larvae and labeled them with anti-phospho histone H3 antibodies to mark mitotic cells. The number of mitotic cells in un-irradiated eye imaginal discs of jnjR1 (Smc6) or sstXL (MAGE) larvae was comparable to that of control eye discs (Fig. 6A). Larvae were exposed to 40 Gy of IR and dissected eye discs were examined from 15 to 120 min. after exposure. Phospho-histone H3 foci disappeared after 30 or 60 min in wild-type (Iso) controls, jnjR1/X1 (Smc6) and sstXL/RZ (MAGE) eye discs (Fig. 6A), demonstrating that neither Mage nor Smc6 is required for activation of the G2/M checkpoint.
Figure 6. Smc5/6 genes are not required for G2/M and S phase checkpoints induced by genotoxic agents.
(A) Wandering third instar larvae were irradiated with 40 Gy of ionizing radiation and the eye-antenna discs were dissected and fixed 15 minutes, 30 minutes, 1 hour or two hours after radiation, with discs from unirradiated larvae serving as controls. Representative images of PH3 staining for mitotic cells in eye-antenna discs from control (WT, FRT82B) and Smc6, (jnjR1/jnjX1) transheterozygous larvae are shown. (B) Eye-antenna discs from wandering third instar larvae were incubated with or without HU before adding BrdU to the incubation solution. Representative images of BrdU staining for cells in S phase in eye-antenna discs from control (WT, FRT82B), transheterozygous Smc6 (jnjR1/jnjX1) or transheterozygous MAGE (sstRZ/sstXL) eye-antenna discs are shown.
The caffeine sensitive ATM/ATR kinases are important mediators of DNA damage checkpoints [28]. In S. pombe, the SMC5/6 complex is recruited to and stabilizes stalled replication forks after Rad3 (ATR homolog) activation [46]. To investigate whether the S phase checkpoint was intact in jnjR1/X1 (Smc6) and sstXL/RZ (MAGE) mutant flies, we monitored BrdU incorporation pattern in eye imaginal discs before and after treatment with HU, which induces the S phase checkpoint [47]. We observed many S-phase cells incorporating BrdU in control untreated eye discs, however incorporation was abolished upon exposure to HU. BrdU incorporation was also abolished by HU treatment in jnjR1/X1 and sstXL/RZ mutant discs (Fig. 6B), demonstrating that Mage and Smc6 are also not essential for S phase checkpoint activity in Drosophila.
Smc6 and MAGE Genetically Interact with Proteins Required for DNA Damage Responses
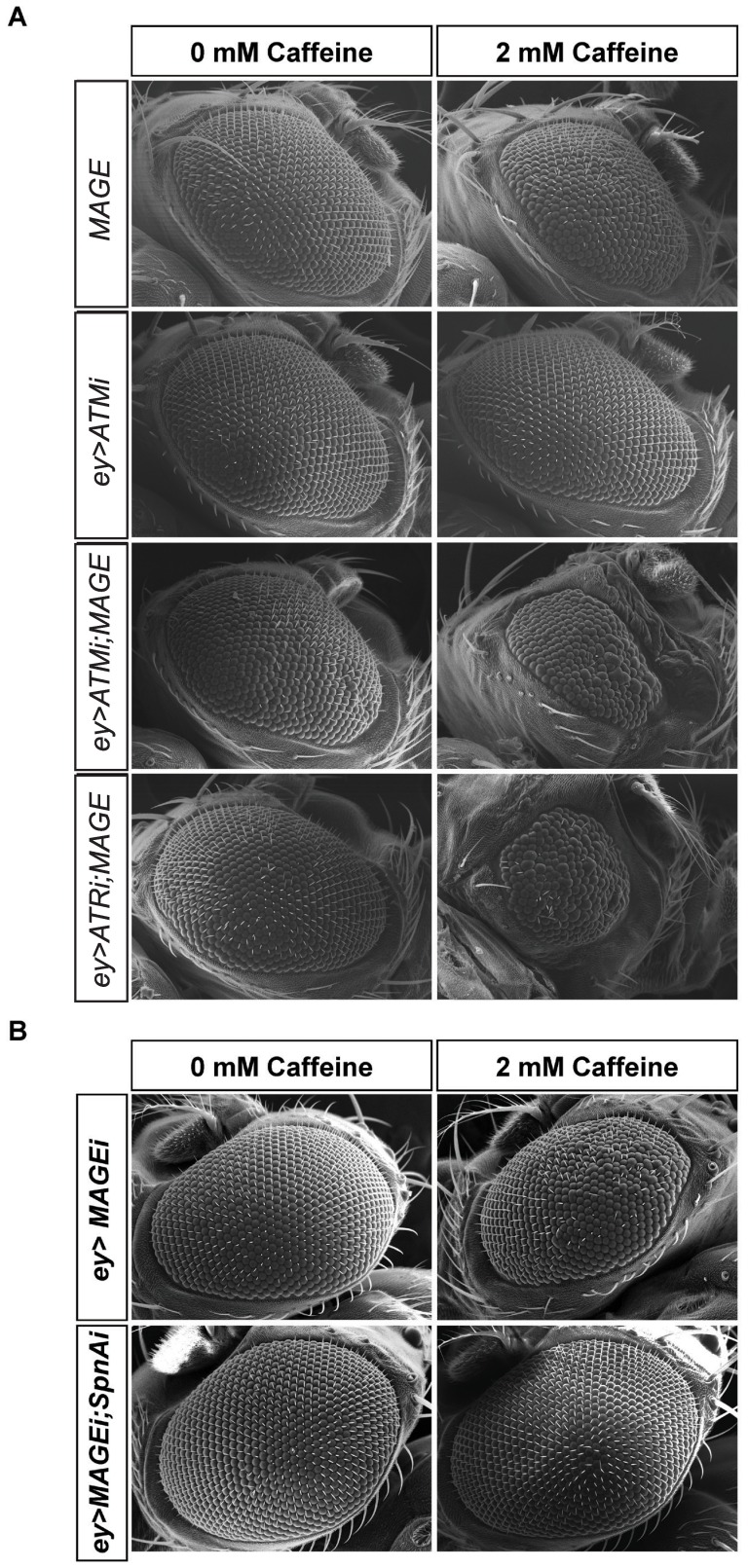
Caffeine inhibits ATR and ATM kinase activity [29], [30], raising the possibility that partial loss of ATM or ATR function could be contributing to the caffeine-induced defects that we observed in Smc5/6 mutant flies. We therefore examined whether genetically reducing ATM or ATR function in an Smc6 mutant background would cause synthetic lethality. The Drosophila homolog of ATR is Mei-41 [48] and mei-41 mutants are homozygous viable but not caffeine-sensitive on their own [31]. To test for genetic interactions between mei-41 and Smc6, we generated double mutants and measured the proportion that survived to adulthood when raised on caffeine-free media. There was no increased lethality associated with mei-41;Smc6 double mutants (Table S5), implying that the inhibition of ATR alone by caffeine was not the main cause of caffeine-dependent lethality of Smc6 homozygotes. To further examine genetic interactions between ATR and MAGE or Smc6, we used the EGUF system as a more sensitive system for detecting mutant phenotypes than lethality. Raised on standard media, adult flies with homozygous MAGE mutant eyes were indistinguishable from control flies (Fig. 7). Raised on 2 mM caffeine, however, MAGE mutant eyes were moderately rough relative to control eyes. ATR RNAi alone caused no observable roughness in the eye but when ATR RNAi was expressed in MAGE-deficient eyes, moderate to severely rough caffeine-dependent eye defects were observed that were not seen on caffeine-free media (Fig. 7, quantification in Fig. S6). We then tested whether ATM plays a role in caffeine sensitivity. Drosophila ATM (tefu) null mutants are non-conditional pupal lethal [49], so we used the EGUF system to examine these interactions as well. ATM-RNAi knockdown alone produced a normal looking eye, either in the absence or presence of caffeine. When MAGE mutant eyes were combined with ATM-RNAi, however, we observed a range of caffeine-dependent rough eye phenotypes, similar to eye defects caused by ATR-RNAi in MAGE-deficient eyes (Fig. 7, S6). ATR-RNAi knockdown alone produced a normal looking eye, either in the absence or presence of caffeine. We noted differences in expressivity between the MAGE-deficient eyes (compare Figs. 1A and 7A) that could be caused by slight differences in the genetic background (the genetic interaction study used CyO balancers while the original screen had wild type chromosomes) or the accumulation of genetic modifiers. We propose that the caffeine-induced partial loss of function of both ATM and ATR causes the rough eye phenotype in the MAGE-deficient background, and that further loss of either ATM or ATR increases the severity of this phenotype, We also examined interactions with NBS1, a component of the MRN (Mre11, Rad50 and Nbs1) complex that collaborates with ATM in DNA repair and telomere maintenance [50]. While NBS1-knockdown alone produced no effect, a dramatic caffeine-dependent enhancement of the rough eye phenotype was observed when NBS1-RNAi was combined with eye-specific MAGE mutants (Fig. S7). These striking caffeine-dependent genetic interactions between MAGE and ATR, ATM, and NBS1 suggest that these proteins act together in maintaining genome stability. Similar genetic interactions were observed between ATR and ATM in Smc6 eye-specific mutants, supporting this conclusion (data not shown).
Figure 7. Caffeine-dependent genetic interaction of MAGE with ATM, ATR and Rad51(SpnA).
(A) Representative eye phenotypes of MAGE (EGUF/+; FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE in eye cells), ey>ATMi (knockdown of ATM in eye cells), ey>ATMi;MAGE (EGUF/UAS-ATM-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATM in eye cells) and ey>ATRi;MAGE (EGUF/UAS-ATR-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATR in eye cells) flies that were reared on either standard media or media containing 2 mM caffeine. The EGUF system carrying the eyeless-Gal4 driver was used to drive the UAS-RNAi transgenes in the eye and also makes the eye homozygous for MAGE (sstRZ). Controls for the effects of each eyeless-driven RNAi alone were carried out for ATM and ATR resulting in wild type appearing eyes, but only the results of ATM RNAi are shown here as an example. (B) Representative eye phenotypes of MAGE knockdown (eyeless-Gal4/+;UAS-MAGE-RNAi/UAS-Dicer2, knockdown of MAGE in eye cells) and MAGE Rad51 double knockdown (eyeless-Gal4/UAS-SpnA-RNAi;UAS-MAGE-RNAi/UAS-Dicer2, knockdown of MAGE and Rad51 in eye cells) flies that were reared on either standard media or media containing 2 mM caffeine.
Drosophila MAGE RNAi Caffeine Sensitive Phenotype is Rescued by Rad 51 Knockdown
In Drosophila and other organisms, Smc5/6 functions in the homologous recombination repair pathway in DNA double strand break repair [26], [51], [52]. Rad51 is a key component of the homologous recombination pathway, regulating the rate-limiting step of homology searching and strand invasion. In Drosophila, Smc5/6 prevents precocious Rad51 loading onto irradiation damaged heterochromatin region before it moves outside of the HP1a domain for proper repair [27]. In yeast, Smc5/6 mutants accumulate unresolved DNA structures, and Smc5/6 actively resolves DNA mediated sister chromatin linkages [53], [54], [55]. We therefore tested whether the caffeine-dependent rough eye phenotype of Smc5/6 mutants is related to deregulated Rad51 activity. Knocking down Rad51 in the MAGE-RNAi background rescued the rough eye phenotype of MAGE-RNAi flies in 80% of the double RNAi flies raised on 2 mM caffeine (Figs. 7B, S8). Taken together, these data indicate that the caffeine sensitivity of the Smc5/6 complex or at least of MAGE mutants is largely attributable to improper Rad51 activity. It is also possible that Rad51 action is normal during HR, but the Smc5/6 complex mutants are unable to complete HR repair or resolve HR intermediates.
Discussion
In a genetic screen for mutations conferring caffeine sensitivity in flies, we identified viable alleles of Drosophila Smc6 (jnj; CG5524) and MAGE (sst; CG10059) as well as an unknown gene (ddt). Additional loss-of-function alleles created by imprecise P-element excision of Smc6 (jnjX1) or targeted knockout of MAGE (sstXL) were also viable under normal conditions, but exhibited caffeine-sensitive lethality. Although no molecular lesions were identified for most jnj (Smc6) alleles, transcript levels were dramatically reduced in all these mutants when hemizygous, implying that either mutations in regulatory regions affected expression, or that, like jnjR1, transcripts were subjected to nonsense-mediated decay. There was no detectable MAGE expression in homozygous, transheterozygous, or hemizygous sst mutants. Furthermore, a genomic MAGE transgene restored expression and rescued the caffeine-dependent lethality of sst mutants. Loss of Smc5 by P-element insertion also resulted in caffeine sensitivity. These genetic results as well as biochemical data showing physical interactions among SMC6, MAGE, Nse1 and Nse4 indicate that the Drosophila Smc5/6 complex is structurally and functionally conserved between yeast and flies. Our screen only covered one chromosome arm (3R) to obtain seven alleles of Smc6 and two alleles of MAGE, representing ∼20% of the genome. Homologs of the remaining SMC5/6 components reside on chromosome arms 2L and 3L (Table S4) and were thus not discovered in our screen. As there are no known Smc5/6 homologs mapping to the ddt locus, identifying this gene and screening remaining chromosome arms for mutations conferring caffeine sensitivity may lead to novel Smc5/6 components or other pathways in which Smc5/6 is involved.
The SMC5/6 complex has been intensively studied in yeasts and human cells for its roles in chromosome replication, segregation and repair of DNA double strand breaks by homologous recombination [10]. Depletion of Smc5 or Smc6 in Drosophila tissue culture cells resulted in heterochromatin bridges in 50% of mitotic cells [27], suggesting that the Smc5 or Smc6 genes would be essential for viability. On the contrary, we found that the loss of Smc5, Smc6, or MAGE did not result in lethality in vivo, and indeed homozygous mutant flies have been maintained for generations (data not shown). There was a slight reduction in hatching rates among null eggs from null mothers in some of the mutant lines, so we cannot rule out a contribution of the maternal RNA to viability in early development. We also did not observe DNA links between sister chromatids, excess aneuploidy, or translocations in mitotic chromosomes of neuroblast squashes from Smc5/6 mutant flies (data not shown). Homologs of Smc5 and Smc6 in Caenorhabditis elegans are also dispensable for viability, however the homozygous mutant strains were prone to sterility and germ cell defects because of compromised inter-sister chromatid recombinational repair and excessive germ cell apoptosis [56].
In both S. cerevisiae and S. pombe, genes encoding SMC5/6 and Nse1–4 are essential and hypomorphic mutants are sensitive to genotoxic agents [7]. In C. elegans, smc-5 and smc-6 mutant germ cells are also hypersensitive to IR and exhibit increased germ cell apoptosis even without IR exposure [56]. In vertebrates, Smc5-deficient chicken DT40 cells are sensitive to MMS and IR [52]. Interfering with the function of human NSE2 by RNAi sensitizes HeLa cells to MMS-induced DNA damage [57]. The Smc5, Smc6 and MAGE mutants described here are also sensitive to IR (40 Gy), HU (4 mM to 8 mM), camptothecin (0.025 mM) and MMS (0.05–0.015%), consistent with an evolutionarily conserved role in resistance to genotoxic agents. Components of the Smc5/6 complex may be responsible for existing Drosophila mutagen sensitive (mus) mutants (e.g. [58]) or may not yet be represented among these collections so constitute novel genes important for mutagen resistance.
Our experiments suggested that cells located just before the morphogenetic furrow in the imaginal eye discs of larvae lacking Smc5/6 components were most sensitive to caffeine (Fig. 4). Many of these cells normally become synchronized in G1 phase by being forced through mitosis through induction of the Cdc25stg gene suggesting that the Smc5/6 and MAGE mutants described here are particularly sensitive to mitotic kinase Cdk1 activity when treated with caffeine [41]. G2/M checkpoint responses to DNA damage and the S-phase checkpoint induced by stalled replication forks were both intact in Drosophila Smc6 or MAGE mutants, however. These results may be explained by accumulating evidence that yeast Smc5/6 mutants undergo normal initiation of the checkpoint response but then fail to complete repair before entering mitosis leading to the formation of DNA bridges and aberrant mitosis [9], [43], [59], [60]. Consistent with this explanation, Drosophila MAGE and Smc6 mutants genetically interact with ATM and ATR to increase the severity of the caffeine-induced rough eye phenotypes (Fig. 7). Similar dependencies were also recently reported for S. cerevisiae, where Nse2 mutants deficient in SUMO ligase activity were viable but needed Mec1 kinase (ATR) to survive, even in the absence of genotoxic stress [61].
Studies of protein complexes that are critical for cellular responses to genotoxic stress are also highly relevant to cancer therapy in humans. It is increasingly apparent that the gene expression signature of each tumor dictates in part the success or failure of chemotherapeutic treatment or radiotherapy [62]. The expression of human Type I MAGE genes is commonly dysregulated in cancer cells. Moreover, many studies have correlated the levels of expression of particular MAGE genes with therapeutic response, prognosis and probability of metastasis [18]. The unexpected synergy between caffeine and loss of SMC5/6 activity could potentially be exploited for new therapeutic strategies where one could preferentially sensitize checkpoint-compromised cancer cells to apoptosis. Although the therapeutic potential of caffeine for causing premature chromosome condensation in G1 checkpoint-compromised cancer cells has long been recognized, the concentrations needed to fully inhibit ATR kinases are toxic [63]. In cells exposed to UV-light, caffeine inhibits rescue of stalled replication forks by translesion DNA synthesis, causing a switch to homologous recombination that can result in chromosomal aberrations [64], [65]. Further studies are needed to elucidate the relationships among MAGE proteins, Smc5/6 components, and proteins such as ATM and ATR that are also important for resistance to genotoxic agents in normal and cancer cells. In turn, mechanistic understanding of how cells respond to genotoxic stress will aid in the selection and dose of chemotherapeutic agents that target specific disruptions to DNA damage response pathways, in order to improve cancer prognosis and survival.
Materials and Methods
Drosophila Stocks and Husbandry
All crosses were carried out at 25°C, and flies were maintained on media formulated at the Bloomington Drosophila Stock Center at Indiana University (BDSC) with p-Hydroxy-benzoic acid methyl ester or propionic acid as the fungicide. Stocks were obtained from the BDSC, the Vienna Drosophila RNAi Center (VDRC), or the Drosophila Genetic Resource Center at Kyoto (DGRC) or generated in our laboratories where specified. Fly stocks used were:
y1 w*; P{70FLP}11 P{70I-SceI}2B snaSco/CyO, S2.
w1118; P{70FLP}10; Sb 1/TM6, Ubx.
y1 w67c23 P{Crey}1b; D*/TM3, Sb1.
P{GawB}NP2592.
w*; Dr1/TMS, P{Delta2-3}99B.
P{GSV1}GS3245.
P{GSV6}GS14577.
P{ey3.5-GAL4.Exel}2.
C(1)DX, y [1] f [1]/w [1] mei-41[D3].
UAS-ATR-RNAi.
UAS-ATM-RNAi.
UAS-NBS1-RNAi.
UAS-SpnA-RNAi.
UAS-MAGE-RNAi/CyO (TRiP).
Ethyl Methanesulfonate (EMS) Screen for Caffeine-sensitive Mutants on Chromosome 3R
The isogenized fly stock FRT82B carries a transgenic Flippase Recognition Target (FRT) site inserted at polytene segment 82B on chromosome 3R and was used to screen for caffeine sensitivity. Adult male flies were mutagenized by feeding with 15 mM EMS dissolved in 1% sucrose for 12 h. After a one day recovery period, mutagenized males were crossed to EGUF; FRT82B GMR-hid, CL/TM3, Sb virgin females. Three to five F1 progeny EGUF/+; FRT82B/FRT82B GMR-hid, CL males with normal eye morphology were crossed to EGUF; FRT82B GMR-hid, CL/TM3, Sb virgin females. The F2 progeny were raised in media with 2 mM caffeine. Individual male non-balancer F2 flies displaying abnormal eye morphology in both eyes were backcrossed to EGUF; FRT82B GMR-hid, CL/TM3, Sb virgin females, and the F3 progeny were raised in media without caffeine to identify any flies with caffeine-independent eye defects. Once the caffeine-dependence of the eye phenotype was confirmed, each mutation was mapped by complementation with the original jnjhuc95E allele [31] or using the Drosophila 3R deficiency kit (BDSC). Both the jnjR1 and sstRZ lines emerged from this screen.
Sequencing of Candidate Genes
Targeted re-sequencing of mapped caffeine-sensitive loci was used to identify mutations in candidate genes. Genomic DNA from 50 adult flies was extracted using DNAzol reagent (Invitrogen, Burlington, ON, Canada). Overlapping PCR fragments about 10 kb in size were amplified using a Long Range PCR kit (Invitrogen). These fragments covered each region predicted to contain a mutation and 10 kb on either side. The PCR products were sequenced using Illumina technology and data was analyzed with Bowtie software (Illumina Inc., San Diego, CA) [66]. Mutations were confirmed by Sanger sequencing with BigDye v3.1 (Applied Biosystems, Carlsbad, CA). Restriction digestion (BpmI) of a genomic PCR fragment was used to confirm the mutation in jnjR1.
Generation of the MAGE Allele sstXL Using Gene Targeting
The “ends-out” method [35] was used to produce a targeted deletion of MAGE. Specifically, 3 kb genomic regions upstream and downstream of the MAGE genomic locus were amplified by PCR from a Drosophila BAC clone (BACPAC Resources Center, RP98-3E11), using the following PCR primers 5′-ATTCATGCGGCCGCCGAAACTCAAACGCAGCGAA and 5′-ATTCTAGGTACCGAGAAGTGCTAGCCATTTCGAG or 5′-ATTCTAGGCGCGCCGGAGTAAACGCGGAGTAGAATACC and 5′-ATTCATCGTACGGGAAGGGGATCAGGATTGAA. The two PCR fragments were subcloned into the NotI-KpnI (Acc65I) or AscI-BsiWI sites of the ends-out vector P[w25.2] to produce a donor construct P[w25.2]_NK_AB. Seven transgenic lines were generated by P element transformation of a w1118 strain using P[w25.2]_NK_AB (BestGene Inc, Chino Hills. CA). The three lines in which the P[w25.2]_NK_AB was located on chromosome 2 were tested for efficient excision by crossing to a line carrying the FLP recombinase (w1118; P{ry+t7.2 70FLP}10; Sb1/TM6, Ubx). One of the three transgenic lines (6030-1-6M) with the highest excision efficiency was chosen as the donor line, and crossed to y1 w*; P{70FLP}11 P{70I-SceI}2B snaSco/CyO, S2 (BDSC #6934). The parents were allowed to lay eggs for two days in a vial, and on the third day the larvae were heat-shocked for 1 h in a 38°C water bath. F1 virgin females were collected and crossed to w1118; P{70FLP}10; Sb 1/TM6, Ubx (BDSC #6938) males. About 100 F2 progeny were selected by screening for nonwhite flies from about 1000 independent crosses. Each of these progeny was crossed to w 1118; P{70FLP}10; Sb 1/TM6, Ubx to make stocks. Twenty five independent lines were identified that exhibited correct targeting as detected by PCR of genomic DNA and loss of Mage protein expression by immunoblotting with a guinea-pig anti-Mage antibody [36]. The white marker of these lines was removed by crossing to a line carrying a Cre recombinase (y1 w67c23 P{Crey}1b; D*/TM3, Sb1 (BDSC #851). The resulting lines were tested for heterozygote and homozygote viability under normal conditions, yielding the line named sstXL.
Generation of a Genomic Rescue Construct for MAGE on Chromosome 2
Genomic DNA was isolated from the isogenized strain P{ry[+t7.2] = neoFRT}82B to PCR amplify (Sequal Prep Long PCR Kit, Invitrogen) a 4 kb fragment spanning from 3 kb upstream of the MAGE gene (genomic locus 3R:2983898, based on the predicted transcription start site), to 206 bp downstream MAGE stop codon (genomic locus 3R: 2979891). The PCR product was digested with the restriction enzyme XbaI and cloned into the pCasper-hs vector. Transgenic flies were generated by BestGene Inc.
Generation of Additional Smc6 Alleles by P-element Mediated Excision
The Smc6 deletion allele jnjX1 was generated by imprecise excision of a P element in P{GawB}NP2592 (DGRC #104251). This insertion, hereafter referred to as NP2592, is located 7 bp upstream of the putative transcriptional initiation site of CG5524 (Smc6) (3R:20,014,770.20,019,145). Its location was confirmed by genomic PCR using primers flanking the NP2592 locus. To excise out NP2592, NP2592 virgin females were crossed to w*; Dr1/TMS, P{Delta2-3}99B (BDSC #1610) males carrying a Δ2–3 transposase. Single virgin F1 females of genotype ΔNP2592/TMS,{Δ2–3}99B were crossed to Ly/TM3, Sb males. Single F2 males of genotype ΔNP2592/TM3, Sb were crossed to virgin Ly/TM3, Sb virgin females to establish balanced lines. About 200 candidate lines were produced and subsequently tested for sensitivity to 2 mM caffeine. Six lines were found to be homozygous viable but caffeine-dependent lethal. Genomic PCR was used to confirm that there were deletions around the original P insertion sites in these stocks. One of the resulting lines was renamed jnjX1.
Molecular Characterization of Smc5 Alleles
The location of P{GSV1}GS3245 (BDSC #200582) and P{GSV6}GS14577 (BDSC #205862) within coding exon 2 of the Smc5 gene was confirmed by genomic PCR using primers 5′-CGTTTCCACGATTTGTTACTGACA and 5′-CGTTTTTGCTTCTTAACCAGATCAC. These lines were renamed Smc5P5 and Smc5P7, respectively. Df(3L)BSC418 (BDSC #24922) is a sequence mapped chromosome deletion (78C9;78E1) that includes the Smc5 locus and nearby genes.
Embryo Collection, Drug Administration and Ionizing Radiation (IR) Treatment
Parental flies were allowed to lay eggs in collection cages on apple juice agar plates with yeast paste for 20 h. The eggs were gently removed from the agar plates using distilled water and a brush and collected using a small cloth-bottomed basket, and then arrayed on new apple juice agar plates. For each drug or radiation treatment, at least 100 embryos were transferred with a thin layer of agar underneath into each of 3 vials containing medium. Drug stocks were pre-added into the media to the appropriate working concentration, with the exception of methyl methanesulfonate, which was added into the medium 48 hours after transferring the embryos. For drugs dissolved in DMSO, an equal amount of DMSO alone was added into medium fed to control flies. The following drugs were used: caffeine (Sigma-Aldrich, St. Louis, MO, stock 1 M in water, final concentration 0.25–2 mM); camptothecin (Sigma-Aldrich, stock 25 mM in DMSO, final concentration 0.025 mM), methyl methanesulfonate (Sigma-Aldrich, stock 99%, final concentration 0.005–0.015%) and HU (Sigma-Aldrich, stock 1 M, final concentration 4–8 mM). For IR, third instar larvae were irradiated at doses of 20 and 40 Gray using an irradiator (Gammacell 220–Cobalt-60, Atomic Energy of Canada, 1979). The survival index (p) of a given genotype was calculated by dividing the number of adult survivors of the genotype resulting from media with a given reagent concentration or treatment (n) by the number of adult survivors of the same genotype resulting from media without that reagent or treatment (N).
Immunoblotting
For each sample, ten 3–4 day-old adult flies were collected, frozen in liquid nitrogen and ground using a pestle in a 1.5 ml eppendorf tube. Mild lysis buffer (50 mM Tris, 150 mM NaCl, and 1% Triton X-100, pH 8.0) was then added (10 µl per fly) to solubilize the tissue. The suspension was centrifuged at 20,000g for 10 min. at 4°C and the supernatant was mixed and boiled with 2X Laemmli Buffer. Proteins were resolved by SDS-PAGE and transferred onto PVDF membranes for immunoblotting. A 1∶2500 dilution of guinea pig anti-Mage serum was used to detect Mage protein [36].
Genetic Interactions of ATM, ATR, NBS1 and RAD51 Loss-of-function with MAGE and Smc6
Double mutants of ATR and Smc6 used mei-41D3 [48] and Smc6 alleles jnjX1 and jnjDf(3R)Exel6198. Knockdown of ATM, ATR or NBS1 function in MAGE or Smc6 homozygous mutant eye clones was achieved using the EGUF system, which uses the eyeless-Gal4 driver to express transgenes throughout eye development [32]. The EGUF system also ensures that all ommatidia of the adult eye are homozygous for either Smc6 or MAGE mutant alleles, because of an eye-specific GMR-hid transgene that eliminates non-mutant ommatidia. RNAi knockdown of MAGE alone or double RNAi of MAGE and Rad51 ortholog SpnA in the eye was achieved by crossing appropriate RNAi constructs containing males to UAS-Dcr2/CyO; ey-Gal4/TM3,Ser virgin females. For each genotype, five to nine specimens were photographed, and representative phenotypes are shown.
cDNA Clones, Cell Culture, Transfections, and Co-immunoprecipitation
Full-length cDNA clones for Nse1 (GM14348) and Nse4 (IP09347) were obtained from the Canadian Drosophila Microarray Centre, the MAGE (RE25453) clone was obtained from the Drosophila Genomics Resource Center (DGRC, Indiana University). Drosophila S2 cells (from the DGRC) were grown at 25°C in TNM-FH medium (SH30280.02, Thermo Scientific, Waltham, MA) supplemented with 10% fetal bovine serum. Expression constructs for transfection of S2 cells were created by inserting relevant full-length coding sequences into the Drosophila Gateway destination vectors (obtained from the DGRC). S2 cells were transfected with relevant expression constructs using dimethyldioctadecyl-ammonium [67]. Cells were harvested 24 h after transfection, washed once in phosphate buffered saline, pH 7.2, and re-suspended in the mild lysis buffer supplemented with a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The lysate was centrifuged for 10 min. at 20,000g at 4°C, and the supernatant transferred to a fresh tube. 200 µl of supernatant was mixed with 20 µl of protein G agarose beads (GE Healthcare Life Sciences, Piscataway, NJ) pre-bound with 5 µg of antibody in 800 µl mild lysis buffer. The agarose beads were then incubated for 1 h at 4°C with rocking, washed six times using mild lysis buffer and the bound proteins analyzed on immunoblots.
In vitro Pulldown Assays
pMBP-Mage was previously described [36] and the control pMBP construct was supplied with a Maltose binding protein (MBP) purification kit (New England Biolabs, Ipswich, MA). Expression constructs were produced by inserting relevant full-length coding sequences into a Gateway pDEST-14 expression vector. MBP fused Mage (MBP-Mage) was expressed in Escherichia coli (ER2523, New England Biolabs) and immobilized onto amylose resin (E8200S) according to the manufacturer’s directions. 35S labeled probe proteins were expressed from Gateway pDest14 vectors using the TNT-coupled in vitro transcription-translation system (Promega, Madison, WI). For the in vitro binding assay, 35S-labeled probe proteins were incubated with immobilized MBP-Mage proteins in 500 µl of buffer (20 mM Tris, 100 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 1% Tween-20, pH 7.6) containing 0.25% bovine serum albumin (BSA) and protease inhibitor cocktail [68] overnight at 4°C with end-over-end mixing. The resin was washed six times in 500 µl of the same buffer, and the bound proteins were resolved by SDS-PAGE and detected by autoradiography.
Scanning Electron Microscopy (SEM) and Immunohistochemistry
Adult heads were prepared for SEM according to the HMDS method described in Drosophila Protocols [69] and imaged using a Scanning Electron Microscope (FEI (XL30), Philips, Hillsboro, OR). Dissection, fixation, BrdU labeling, and antibody staining of third larval instar eye-antennal discs were also carried out as described in Drosophila Protocols. Antibodies for immunohistochemistry included anti-cleaved caspase 3 (1∶1600 dilution, Cell Signaling Technologies, Beverly, MA), anti-BrdU (1∶200 dilution, Pharmingen San Jose, CA), and anti-phospho-histone H3 (Cell Signaling, 1∶1000 dilution). Secondary antibodies were used at a dilution of 1∶1000 (Alexa Fluor 488 and 586, Invitrogen). For the detection of apoptosis in third instar imaginal discs with an anti-cleaved caspase 3 antibody, embryos were collected at one hour intervals on grape juice plates and larvae were reared on yeast paste plates until the L3 molt. They were then transferred to 2 mM caffeine medium 32 h after the L3 molt and allowed to develop for a further 12 h before dissection. Images of the dissected discs were acquired using a LSM 700 confocal microscope (Carl Zeiss Inc., Thornwood, NY) and processed using Zen (Carl Zeiss). A maximum projection of all stacks of a confocal image was used to quantify the signal intensity of staining using a lower threshold to eliminate background staining. This value was divided by the area of each eye disc to obtain a ratio representing the relative amount of immunostaining. Data represent at least 7 eye discs per genotype per treatment.
Supporting Information
An ethyl methanesulfonate (EMS) screen for caffeine-sensitive mutants on chromosome 3R. Ethylmethane sulfonate (EMS) mutagenized male flies carrying transgenic FRT82B sites were crossed en masse to y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in standard media. Non-TM3, Sb progeny males containing normal looking eyes were then collected and crossed in pools of 3–5 males to 3–5 y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in molasses and cornmeal media containing 2 mM caffeine. Non-TM3, Sb progeny males containing developmental defects in both eyes were selected and individually tested with y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in normal media to eliminate any false positive caffeine-independent mutations that might have arisen in the male germline. Once a caffeine-dependent phenotype was confirmed, the mutant was then crossed to y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females to establish balanced stocks. “*” indicates a putative mutation.
(PDF)
Caffeine sensitivity of jnj alleles is caused by loss of Smc6 . (A) mRNA transcript levels of Smc6 and its neighboring genes CHORD, CG5515 and CG6204 in control and jnj mutant flies were measured by quantitative RT-PCR. All seven jnj alleles tested had reduced Smc6 transcript levels ranging from 7% to 24% of the control level, while the transcript levels of the neighboring genes comparable to the control level. The caffeine screen starting stock “Iso” carrying the transgenic FRT82B site crossed to Df to normalize the Smc6 level was used to generate control flies. “Df” is the deficiency chromosome Df(3R)Exel6198. (B) Knocking-down Smc6 expression using RNAi in developing eye discs resulted in a caffeine-dependent adult rough eye phenotype. Control, Eyeless-Gal4/+ was from a cross of Eyeless-Gal4/Eyeless-Gal4 X w1118 and Smc6-RNAi, Eyeless-Gal4/+; UAS-Smc6-RNAi/+ resulted from the cross Eyeless-Gal4/Eyeless-Gal4 X UAS-Smc6-RNAi/+. UAS-Smc6-RNAi was obtained from VDRC (#107055).
(PDF)
Immunoblot for Mage. Levels of endogenous Mage were measured in protein lysates from whole flies derived from various lines, immunoblotted with anti-Mage antibody. Genotypes were as follows: Lane 1: sstXL/TM3,Sb, 2: sstRZ/TM3,Ser,ActGFP, 3: sstXL/sstRZ, 4: Df(3R)Antp1/TM3,Sb, 5: Df(3R)Antp1/sstRZ, 6: Df(3R)Antp1/sstXL, 7. w1118, 8: S2 cells, 9: S2 cells dMAGE RNAi, 10: sstXL/TM3,Ser,ActGFP, 11: sstXL/sstXL, 12∶3Kb+MAGE transgene/CyO; sstXL/sstXL.
(PDF)
Expression profiles of genes encoding Smc5/6 complex proteins. The expression profile figure for each gene was obtained from GEO Profiles database at NCBI (GDS2784) from the original data of Chintapalli et al. [40].
(PDF)
Smc6, MAGE and Smc5 mutants are sensitive to camptothecin, HU and MMS. Flies eclosed from the same cross are indicated with a ‘□’. Embryos (n = 360, expected to be half homozygous or transheterozygous mutants and half heterozygous mutants) were collected from a given cross for each drug concentration and allowed to develop in media without or with each drug. Bars represent the survival index (p) ± SEM. Absence of a bar indicates that no flies survived at that drug concentration. The survival index was calculated by normalizing the number of eclosed adults from each drug treatment against the number of eclosed adults from the no treatment control. (A–C) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when raised in media supplemented with 0.025 mM camptothecin; (D–F) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when raised in media supplemented with hydroxyurea (HU); (G) MAGE mutants are sensitive to MMS; (H) Smc5 mutants are sensitive to MMS. Smc6 mutants are also sensitive to MMS (data not shown). Smc6: R1 (jnjR1) and X1 (jnjX1) are Smc6 alleles. Df (Df(3R)Exel6198) is a deficiency chromosome uncovering the Smc6 locus; MAGE: RZ (sstRZ) and XL (sstXL) are MAGE alleles. Df (Df(3R)Antp1) is a deficiency chromosome uncovering the MAGE locus. Smc5: P5 (Smc5P{GSV1}GS3245) and P7 (Smc5P{GSV6}GS14577) are Smc5 alleles. Df (Df(3L)BSC418) is a deficiency chromosome uncovering the Smc5 locus.
(PDF)
Quantification the area of the adult eye as a measure of the genetic interaction of MAGE with ATM , ATR or NBS1 . MAGE (EGUF/+; FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE in eye cells), ey>ATM-RNAi (knockdown of ATM in eye cells), ey>ATR-RNAi (knockdown of ATR in eye cells), ey>NBS1-RNAi (knockdown of NBS1 in eye cells), ey>ATM-RNAi;MAGE (EGUF/UAS-ATM-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATM in eye cells), ey>ATR-RNAi;MAGE (EGUF/UAS-ATR-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATR in eye cells), and ey>NBS1-RNAi;MAGE (EGUF/UAS-NBS1-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of NBS1 in eye cells) flies were reared on either standard media or media containing 2 mM caffeine. A Student two-tailed t-test was performed to compare between genotypes.
(PDF)
NBS1 interacts with MAGE . Representative eye phenotypes of MAGE (EGUF/+; FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE in eye cells) and ey>NBS1i (knockdown of NBS1 in eye cells) and ey>NBS1i;MAGE (EGUF/UAS-NBS1-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of NBS1 in eye cells) flies that were reared on either standard media or media containing 2 mM caffeine. The EGUF system carrying the eyeless-Gal4 driver was used to drive the UAS-RNAi transgene in the eye and was also made the eyes homozygous for sstRZ.
(PDF)
Rad51 (SpnA-RNAi) depletion rescues the MAGE-RNAi caffeine-sensitive eye phenotype. Bars represent the percentage of flies with wildtype eye phenotypes among MAGE knockdown (UAS-Drc2/+; UAS-MAGE-RNAi/+) and MAGE Rad51 double knockdown (Drc2/+; UAS-MAGE-RNAi/UAS-SpnA-RNAi) flies that were reared on either standard media or media containing 2 mM caffeine. Data were collected from 4 replicates of each cross. Absence of error bar indicates flies of this genotype had consistent phenotypes.
(PDF)
sst caffeine sensitivity can be rescued by a MAGE transgene.
(PDF)
P-element excision of P{GSV1}GS3245 and P{GSV6}GS14577 produce both caffeine-sensitive and -insensitive lines.
(PDF)
Caffeine sensitivity of MAGE and Smc6 double mutants is similar to sensitivity of flies mutant for Smc6 alone.
(PDF)
Genes encoding Smc5/6 complexes in different model organisms.
(PDF)
mei-41/ATM and jnj/Smc6 double mutants have normal viability.
(PDF)
Supporting Methods.
(PDF)
Acknowledgments
We thank Dr. K. Yoshikawa (Osaka University) for the MBP plasmid and the anti-Mage antibody. We thank Dr. A. Simmonds and members of his laboratory, in particular Dr. Hua Deng, for helpful discussions and Gina Catena for validation of the sstRZ allele.
Funding Statement
This study was funded by an operating grant from the Cancer Research Society (Canada) to RW and SCH, a Canadian Institutes of Health Research Operating Grant to KK-J (MOP 93761), and an NSERC Discovery Grant (G121210462) to SDC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lehmann AR (2005) The role of SMC proteins in the responses to DNA damage. DNA Repair (Amst) 4: 309–314. [DOI] [PubMed] [Google Scholar]
- 2. Wu N, Yu H (2012) The Smc complexes in DNA damage response. Cell Biosci 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7: 311–322. [DOI] [PubMed] [Google Scholar]
- 4. Dorsett D, Strom L (2012) The ancient and evolving roles of cohesin in gene expression and DNA repair. Curr Biol 22: R240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuylen S, Haering CH (2011) Deciphering condensin action during chromosome segregation. Trends Cell Biol 21: 552–559. [DOI] [PubMed] [Google Scholar]
- 6. Potts PR, Yu H (2007) The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat Struct Mol Biol 14: 581–590. [DOI] [PubMed] [Google Scholar]
- 7. Pebernard S, McDonald WH, Pavlova Y, Yates JR 3rd, Boddy MN (2004) Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell 15: 4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murray JM, Carr AM (2008) Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol 9: 177–182. [DOI] [PubMed] [Google Scholar]
- 9. Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, et al. (2005) SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol 7: 412–419. [DOI] [PubMed] [Google Scholar]
- 10. Kegel A, Sjogren C (2010) The Smc5/6 complex: more than repair? Cold Spring Harb Symp Quant Biol 75: 179–187. [DOI] [PubMed] [Google Scholar]
- 11. Pebernard S, Wohlschlegel J, McDonald WH, Yates JR 3rd, Boddy MN (2006) The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol 26: 1617–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, et al. (2009) Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5–6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J Biol Chem 284: 8507–8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao X, Blobel G (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews EA, et al. (2005) Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol Cell Biol 25: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hudson JJ, Bednarova K, Kozakova L, Liao C, Guerineau M, et al. (2011) Interactions between the Nse3 and Nse4 components of the SMC5–6 complex identify evolutionarily conserved interactions between MAGE and EID Families. PLoS One 6: e17270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR (2006) The Smc5-Smc6 DNA repair complex. bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J Biol Chem 281: 36952–36959. [DOI] [PubMed] [Google Scholar]
- 17. Doyle JM, Gao J, Wang J, Yang M, Potts PR (2010) MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 39: 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Miranda EI (2010) MAGE, biological functions and potential clinical applications. Leuk Res 34: 1121–1122. [DOI] [PubMed] [Google Scholar]
- 19. Sang M, Wang L, Ding C, Zhou X, Wang B, et al. (2011) Melanoma-associated antigen genes - an update. Cancer Lett 302: 85–90. [DOI] [PubMed] [Google Scholar]
- 20. Lee S, Kozlov S, Hernandez L, Chamberlain SJ, Brannan CI, et al. (2000) Expression and imprinting of MAGEL2 suggest a role in Prader-willi syndrome and the homologous murine imprinting phenotype. Hum Mol Genet 9: 1813–1819. [DOI] [PubMed] [Google Scholar]
- 21. Lee S, Walker CL, Karten B, Kuny SL, Tennese AA, et al. (2005) Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum Mol Genet 14: 627–637. [DOI] [PubMed] [Google Scholar]
- 22. Bertrand MJ, Kenchappa RS, Andrieu D, Leclercq-Smekens M, Nguyen HN, et al. (2008) NRAGE, a p75NTR adaptor protein, is required for developmental apoptosis in vivo. Cell Death Differ 15: 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR (2008) Identification of the proteins, including MAGEG1, that make up the human SMC5–6 protein complex. Mol Cell Biol 28: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guerineau M, Kriz Z, Kozakova L, Bednarova K, Janos P, et al. (2012) Analysis of the Nse3/MAGE-binding domain of the Nse4/EID family proteins. PLoS One 7: e35813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bush JR, Wevrick R (2008) The Prader-Willi syndrome protein necdin interacts with the E1A-like inhibitor of differentiation EID-1 and promotes myoblast differentiation. Differentiation 76: 994–1005. [DOI] [PubMed] [Google Scholar]
- 26. De Piccoli G, Torres-Rosell J, Aragon L (2009) The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res 17: 251–263. [DOI] [PubMed] [Google Scholar]
- 27. Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, et al. (2011) Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, et al. (1999) Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res 59: 4375–4382. [PubMed] [Google Scholar]
- 30. Blasina A, Price BD, Turenne GA, McGowan CH (1999) Caffeine inhibits the checkpoint kinase ATM. Curr Biol 9: 1135–1138. [DOI] [PubMed] [Google Scholar]
- 31. Silva EA, Lee BJ, Caceres LS, Renouf D, Vilay BR, et al. (2006) A novel strategy for identifying mutations that sensitize Drosophila eye development to caffeine and hydroxyurea. Genome 49: 1416–1427. [DOI] [PubMed] [Google Scholar]
- 32. Stowers RS, Schwarz TL (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pold M, Pold A, Ma HJ, Sjak-Shieb NN, Vescio RA, et al. (2000) Cloning of the first invertebrate MAGE paralogue: an epitope that activates T-cells in humans is highly conserved in evolution. Dev Comp Immunol 24: 719–731. [DOI] [PubMed] [Google Scholar]
- 34. Nishimura I, Sakoda JY, Yoshikawa K (2008) Drosophila MAGE controls neural precursor proliferation in postembryonic neurogenesis. Neuroscience 154: 572–581. [DOI] [PubMed] [Google Scholar]
- 35. Maggert KA, Gong WJ, Golic KG (2008) Methods for homologous recombination in Drosophila. Methods Mol Biol 420: 155–174. [DOI] [PubMed] [Google Scholar]
- 36. Nishimura I, Shimizu S, Sakoda JY, Yoshikawa K (2007) Expression of Drosophila MAGE gene encoding a necdin homologous protein in postembryonic neurogenesis. Gene Expr Patterns 7: 244–251. [DOI] [PubMed] [Google Scholar]
- 37. Fujioka Y, Kimata Y, Nomaguchi K, Watanabe K, Kohno K (2002) Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J Biol Chem 277: 21585–21591. [DOI] [PubMed] [Google Scholar]
- 38. Toba G, Ohsako T, Miyata N, Ohtsuka T, Seong KH, et al. (1999) The gene search system. A method for efficient detection and rapid molecular identification of genes in Drosophila melanogaster. Genetics 151: 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- 40. Chintapalli VR, Wang J, Dow JA (2007) Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet 39: 715–720. [DOI] [PubMed] [Google Scholar]
- 41. Thomas BJ, Zipursky SL (1994) Early pattern formation in the developing Drosophila eye. Trends Cell Biol 4: 389–394. [DOI] [PubMed] [Google Scholar]
- 42. Sekelsky JJ, Brodsky MH, Burtis KC (2000) DNA repair in Drosophila: insights from the Drosophila genome sequence. J Cell Biol 150: F31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Verkade HM, Bugg SJ, Lindsay HD, Carr AM, O’Connell MJ (1999) Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol Biol Cell 10: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miyabe I, Morishita T, Hishida T, Yonei S, Shinagawa H (2006) Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol Cell Biol 26: 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, Rubin GM (2000) mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev 14: 666–678. [PMC free article] [PubMed] [Google Scholar]
- 46. Irmisch A, Ampatzidou E, Mizuno K, O’Connell MJ, Murray JM (2009) Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J 28: 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klovstad M, Abdu U, Schupbach T (2008) Drosophila brca2 is required for mitotic and meiotic DNA repair and efficient activation of the meiotic recombination checkpoint. PLoS Genet 4: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laurencon A, Purdy A, Sekelsky J, Hawley RS, Su TT (2003) Phenotypic analysis of separation-of-function alleles of MEI-41, Drosophila ATM/ATR. Genetics 164: 589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silva E, Tiong S, Pedersen M, Homola E, Royou A, et al. (2004) ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr Biol 14: 1341–1347. [DOI] [PubMed] [Google Scholar]
- 50. Czornak K, Chughtai S, Chrzanowska KH (2008) Mystery of DNA repair: the role of the MRN complex and ATM kinase in DNA damage repair. J Appl Genet 49: 383–396. [DOI] [PubMed] [Google Scholar]
- 51. Watanabe K, Pacher M, Dukowic S, Schubert V, Puchta H, et al. (2009) The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 21: 2688–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stephan AK, Kliszczak M, Dodson H, Cooley C, Morrison CG (2011) Roles of vertebrate Smc5 in sister chromatid cohesion and homologous recombinational repair. Mol Cell Biol 31: 1369–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, et al. (2006) Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509–522. [DOI] [PubMed] [Google Scholar]
- 54. Ampatzidou E, Irmisch A, O’Connell MJ, Murray JM (2006) Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol 26: 9387–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, et al. (2009) The Saccharomyces cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction-mediated intra-S repair. Mol Biol Cell 20: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bickel JS, Chen L, Hayward J, Yeap SL, Alkers AE, et al. (2010) Structural maintenance of chromosomes (SMC) proteins promote homolog-independent recombination repair in meiosis crucial for germ cell genomic stability. PLoS Genet 6: e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Potts PR, Yu H (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Boyd JB, Golino MD, Nguyen TD, Green MM (1976) Isolation and characterization of X-linked mutants of Drosophila melanogaster which are sensitive to mutagens. Genetics 84: 485–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Harvey SH, Sheedy DM, Cuddihy AR, O’Connell MJ (2004) Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol Cell Biol 24: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bermudez-Lopez M, Ceschia A, de Piccoli G, Colomina N, Pasero P, et al. (2010) The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res 38: 6502–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rai R, Varma SP, Shinde N, Ghosh S, Kumaran SP, et al. (2011) Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J Biol Chem 286: 14516–14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wong KM, Hudson TJ, McPherson JD (2011) Unraveling the genetics of cancer: genome sequencing and beyond. Annu Rev Genomics Hum Genet 12: 407–430. [DOI] [PubMed] [Google Scholar]
- 63. Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL (2001) ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A 98: 9092–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Johansson F, Lagerqvist A, Filippi S, Palitti F, Erixon K, et al. (2006) Caffeine delays replication fork progression and enhances UV-induced homologous recombination in Chinese hamster cell lines. DNA Repair (Amst) 5: 1449–1458. [DOI] [PubMed] [Google Scholar]
- 65. Han W, Ming M, He YY (2011) Caffeine promotes ultraviolet B-induced apoptosis in human keratinocytes without complete DNA repair. J Biol Chem 286: 22825–22832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Han K (1996) An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res 24: 4362–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Deng H, Hughes SC, Bell JB, Simmonds AJ (2009) Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster. Mol Biol Cell 20: 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan W, Ashburner M, Hawley RS (2000) Drosophila protocols. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. xiv, 697 p.
- 70. Stephan AK, Kliszczak M, Morrison CG (2011) The Nse2/Mms21 SUMO ligase of the Smc5/6 complex in the maintenance of genome stability. FEBS Lett 585: 2907–2913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
An ethyl methanesulfonate (EMS) screen for caffeine-sensitive mutants on chromosome 3R. Ethylmethane sulfonate (EMS) mutagenized male flies carrying transgenic FRT82B sites were crossed en masse to y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in standard media. Non-TM3, Sb progeny males containing normal looking eyes were then collected and crossed in pools of 3–5 males to 3–5 y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in molasses and cornmeal media containing 2 mM caffeine. Non-TM3, Sb progeny males containing developmental defects in both eyes were selected and individually tested with y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females in normal media to eliminate any false positive caffeine-independent mutations that might have arisen in the male germline. Once a caffeine-dependent phenotype was confirmed, the mutant was then crossed to y,w; EGUF; FRT82B GMR-hid/TM3, Sb virgin females to establish balanced stocks. “*” indicates a putative mutation.
(PDF)
Caffeine sensitivity of jnj alleles is caused by loss of Smc6 . (A) mRNA transcript levels of Smc6 and its neighboring genes CHORD, CG5515 and CG6204 in control and jnj mutant flies were measured by quantitative RT-PCR. All seven jnj alleles tested had reduced Smc6 transcript levels ranging from 7% to 24% of the control level, while the transcript levels of the neighboring genes comparable to the control level. The caffeine screen starting stock “Iso” carrying the transgenic FRT82B site crossed to Df to normalize the Smc6 level was used to generate control flies. “Df” is the deficiency chromosome Df(3R)Exel6198. (B) Knocking-down Smc6 expression using RNAi in developing eye discs resulted in a caffeine-dependent adult rough eye phenotype. Control, Eyeless-Gal4/+ was from a cross of Eyeless-Gal4/Eyeless-Gal4 X w1118 and Smc6-RNAi, Eyeless-Gal4/+; UAS-Smc6-RNAi/+ resulted from the cross Eyeless-Gal4/Eyeless-Gal4 X UAS-Smc6-RNAi/+. UAS-Smc6-RNAi was obtained from VDRC (#107055).
(PDF)
Immunoblot for Mage. Levels of endogenous Mage were measured in protein lysates from whole flies derived from various lines, immunoblotted with anti-Mage antibody. Genotypes were as follows: Lane 1: sstXL/TM3,Sb, 2: sstRZ/TM3,Ser,ActGFP, 3: sstXL/sstRZ, 4: Df(3R)Antp1/TM3,Sb, 5: Df(3R)Antp1/sstRZ, 6: Df(3R)Antp1/sstXL, 7. w1118, 8: S2 cells, 9: S2 cells dMAGE RNAi, 10: sstXL/TM3,Ser,ActGFP, 11: sstXL/sstXL, 12∶3Kb+MAGE transgene/CyO; sstXL/sstXL.
(PDF)
Expression profiles of genes encoding Smc5/6 complex proteins. The expression profile figure for each gene was obtained from GEO Profiles database at NCBI (GDS2784) from the original data of Chintapalli et al. [40].
(PDF)
Smc6, MAGE and Smc5 mutants are sensitive to camptothecin, HU and MMS. Flies eclosed from the same cross are indicated with a ‘□’. Embryos (n = 360, expected to be half homozygous or transheterozygous mutants and half heterozygous mutants) were collected from a given cross for each drug concentration and allowed to develop in media without or with each drug. Bars represent the survival index (p) ± SEM. Absence of a bar indicates that no flies survived at that drug concentration. The survival index was calculated by normalizing the number of eclosed adults from each drug treatment against the number of eclosed adults from the no treatment control. (A–C) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when raised in media supplemented with 0.025 mM camptothecin; (D–F) Smc6, MAGE or Smc5 homozygous, trans-heterozygous or hemizygous mutants have reduced survival when raised in media supplemented with hydroxyurea (HU); (G) MAGE mutants are sensitive to MMS; (H) Smc5 mutants are sensitive to MMS. Smc6 mutants are also sensitive to MMS (data not shown). Smc6: R1 (jnjR1) and X1 (jnjX1) are Smc6 alleles. Df (Df(3R)Exel6198) is a deficiency chromosome uncovering the Smc6 locus; MAGE: RZ (sstRZ) and XL (sstXL) are MAGE alleles. Df (Df(3R)Antp1) is a deficiency chromosome uncovering the MAGE locus. Smc5: P5 (Smc5P{GSV1}GS3245) and P7 (Smc5P{GSV6}GS14577) are Smc5 alleles. Df (Df(3L)BSC418) is a deficiency chromosome uncovering the Smc5 locus.
(PDF)
Quantification the area of the adult eye as a measure of the genetic interaction of MAGE with ATM , ATR or NBS1 . MAGE (EGUF/+; FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE in eye cells), ey>ATM-RNAi (knockdown of ATM in eye cells), ey>ATR-RNAi (knockdown of ATR in eye cells), ey>NBS1-RNAi (knockdown of NBS1 in eye cells), ey>ATM-RNAi;MAGE (EGUF/UAS-ATM-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATM in eye cells), ey>ATR-RNAi;MAGE (EGUF/UAS-ATR-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of ATR in eye cells), and ey>NBS1-RNAi;MAGE (EGUF/UAS-NBS1-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of NBS1 in eye cells) flies were reared on either standard media or media containing 2 mM caffeine. A Student two-tailed t-test was performed to compare between genotypes.
(PDF)
NBS1 interacts with MAGE . Representative eye phenotypes of MAGE (EGUF/+; FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE in eye cells) and ey>NBS1i (knockdown of NBS1 in eye cells) and ey>NBS1i;MAGE (EGUF/UAS-NBS1-RNAi;FRT82B sstRZ/FRT82B GMR-hid, loss of MAGE and knockdown of NBS1 in eye cells) flies that were reared on either standard media or media containing 2 mM caffeine. The EGUF system carrying the eyeless-Gal4 driver was used to drive the UAS-RNAi transgene in the eye and was also made the eyes homozygous for sstRZ.
(PDF)
Rad51 (SpnA-RNAi) depletion rescues the MAGE-RNAi caffeine-sensitive eye phenotype. Bars represent the percentage of flies with wildtype eye phenotypes among MAGE knockdown (UAS-Drc2/+; UAS-MAGE-RNAi/+) and MAGE Rad51 double knockdown (Drc2/+; UAS-MAGE-RNAi/UAS-SpnA-RNAi) flies that were reared on either standard media or media containing 2 mM caffeine. Data were collected from 4 replicates of each cross. Absence of error bar indicates flies of this genotype had consistent phenotypes.
(PDF)
sst caffeine sensitivity can be rescued by a MAGE transgene.
(PDF)
P-element excision of P{GSV1}GS3245 and P{GSV6}GS14577 produce both caffeine-sensitive and -insensitive lines.
(PDF)
Caffeine sensitivity of MAGE and Smc6 double mutants is similar to sensitivity of flies mutant for Smc6 alone.
(PDF)
Genes encoding Smc5/6 complexes in different model organisms.
(PDF)
mei-41/ATM and jnj/Smc6 double mutants have normal viability.
(PDF)
Supporting Methods.
(PDF)