Abstract
Interstitial collagen mechanical and biological properties are altered by proteases that catalyze the hydrolysis of the collagen triple-helical structure. Collagenolysis is critical in development and homeostasis but also contributes to numerous pathologies. Mammalian collagenolytic enzymes include matrix metalloproteinases, cathepsin K, and neutrophil elastase, and a variety of invertebrates and pathogens possess collagenolytic enzymes. Components of the mechanism of action for the collagenolytic enzyme MMP-1 have been defined experimentally, and insights into other collagenolytic mechanisms have been provided. Ancillary biomolecules may modulate the action of collagenolytic enzymes.
Keywords: Arthritis, Cancer, Collagen, Matrix Metalloproteinase (MMP), Proteolytic Enzymes
Enzymes That Catalyze Interstitial Collagen Catabolism
Collagens are composed of three α chains of primarily repeating Gly-Xaa-Yaa triplets, which induce each α chain to adopt a left-handed poly-Pro II helix. Three chains then intertwine, staggered by one residue and coiled, to form a right-handed triple helix. Triple helices assemble to form semicrystalline aggregates referred to as fibrils, and bundles of fibrils form fibers. The proteolysis of interstitial collagen (types I–III) is integral for numerous physiological functions, including morphogenesis, tissue remodeling, and wound healing, and has been recognized as a contributing factor to multiple pathologies, including tumor cell spreading (metastasis), arthritis, glomerulonephritis, periodontal disease, tissue ulcerations, cardiovascular disease, and neurodegenerative diseases. Identifying proteases capable of processing triple helices provides a starting point for defining the roles of collagen catabolism in health and disease.
Members of the matrix metalloproteinase (MMP)2 family of zinc-dependent endopeptidases possess collagenolytic activity (1). Interstitial collagens are hydrolyzed by the “classic” collagenases, MMP-1, MMP-8, and MMP-13 (Fig. 1), into 1/4 and 3/4 length fragments (Table 1) (1, 2). MMP-2 (Fig. 1) cleaves type I collagen (3), although how robust the collagenolytic activity is has been brought into question (4). MMP-9 (Fig. 1) cleaves type I and III collagens (5). Hydrolysis of type I collagen was monitored at 37 °C, at which some denatured triple helices might exist. For MMP-2 and MMP-9, the cleavage site is the same as for the classic collagenases (Table 1).
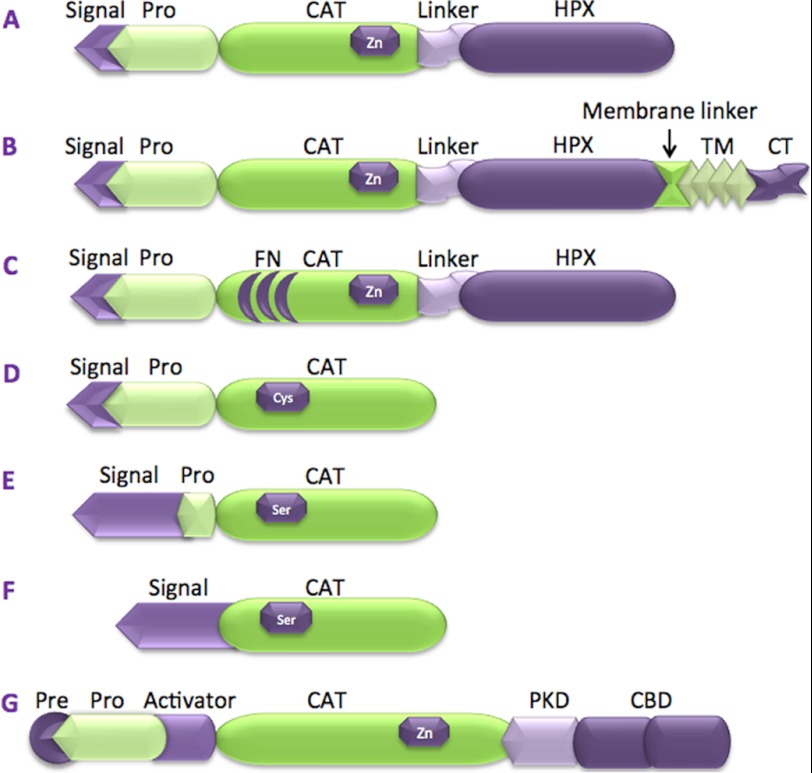
FIGURE 1.
Domain structures of collagenolytic proteases. A, MMP-1, MMP-8, and MMP-13. B, MT1-MMP. C, MMP-2 and MMP-9. D, cathepsin K. E, neutrophil elastase. F, fiddler crab collagenase. G, C. histolyticum class I collagenase (ColG). Signal, secretory signaling peptide; Pro, prodomain; TM, transmembrane domain; CT, cytoplasmic tail; FN, fibronectin domain; Pre, predomain; PKD, polycystic kidney disease-like domain.
TABLE 1.
Representative protease cleavage sites within interstitial collagen triple-helical domains
| Enzyme | Collagen chain | Sequencea |
|---|---|---|
| MMP-1/2/8/9/12/13 and MT1-MMP | α1(I) | Pro-Gln-Gly775∼Ile776-Ala-Gly |
| MMP-1/2/8/9/12/13 and MT1-MMP | α2(I) | Pro-Gln-Gly775∼Leu776-Leu-Gly |
| MMP-1/8/13 and MT1-MMP | α1(II) | Pro-Gln-Gly775∼Leu776-Ala-Gly |
| MMP-1/8/9/12/13, MT1-MMP, and MT3-MMP | α1(III) | Pro-Leu-Gly775∼Ile776-Ala-Gly |
| Cathepsin K | α1(I) | Gly-Pro-Arg9∼Gly10-Leu-Pro |
| Cathepsin K | α1(I) | Gly-Pro-Gln21∼Gly22-Phe-Gln |
| Cathepsin K | α1(I) | Gly-Leu-Asp96∼Gly97-Ala-Lys |
| Cathepsin K | α1(I) | Gly-Pro-Gln189∼Gly190-Val-Arg |
| Cathepsin K | α1(I) | Gly-Pro-Ser810∼Gly811-Ala-Ser |
| Cathepsin K | α2(I) | Gly-Pro-Arg9∼Gly10-Pro-Pro |
| Cathepsin K | α2(I) | Gly-Pro-Gln21∼Gly22-Phe-Gln |
| Cathepsin K | α2(I) | Gly-Leu-Lys99∼Gly100-Pro-Gln |
| Cathepsin K | α2(I) | Gly-Ala-Arg144∼Gly145-Ser-Asp |
| Cathepsin K | α2(I) | Pro-Pro-Gly814∼Ala815-Arg-Gly |
| Cathepsin K | α1(II) | Lys-Pro-Gly61∼Lys62-Ser-Gly |
| Elastase | α1(III) | Ala-Gly-Ile779∼Thr780-Gly-Arg |
| Crab collagenase 1 | α1(I) | Ala-Gly-Gln779∼Arg780-Gly-Val |
| Crab collagenase 1 | α1(I) | Gly-Gln-Arg780∼Gly781-Val-Val |
| Crab collagenase 1 | α1(I) | Gly-Glu-Arg792∼Gly793-Phe-Hyp |
| Crab collagenase 1 | α1(I) | Arg-Gly-Leu587∼Thr588-Gly-Pro |
| Crab collagenase 1 | α2(I) | Gly-Phe-Leu783∼Gly784-Leu-Pro |
| Cardosin A | α2(I) | Pro-Gly-Phe464∼Asn465-Gly-Leu |
| Kumamolisin-As/ScpA | α1(I) | Gly-Pro-Lys108∼Gly109-Glu-Hyp |
| Kumamolisin-As/ScpA | α1(I) | Gly-Pro-Arg183∼Gly184-Ser-Glu |
| Kumamolisin-As/ScpA | α1(I) | Gly-Ala-Arg396∼Gly397-Gln-Ala |
| Kumamolisin-As/ScpA | α1(I) | Gly-Asp-Ala489∼Gly490-Ala-Hyp |
| Kumamolisin-As/ScpA | α2(I) | Gly-Pro-Arg42∼Gly43-Pro-Ala |
| GP2 | α1(I) | Gly-Pro-Ala285∼Gly286-Glu-Glu |
| GP2 | α1(I) | Gly-Ala-Arg498∼Gly499-Glu-Arg |
| GP2 | α1(I) | Gly-Pro-Ser711∼Gly712-Asn-Ala |
| GP2 | α2(I) | Gly-Pro-Ser285∼Gly286-Glu-Glu |
| GP2 | α2(I) | Gly-Ala-Arg498∼Gly499-Glu-Arg |
| GP2 | α2(I) | Gly-Pro-Ser711∼Gly712-Ile-Ser |
| ColG | α1(II) | Gly-Phe-Gln24∼Gly25-Asn-Pro |
| ColG | α1(III) | Gly-Glu-Arg69∼Gly70-Leu-Hyp |
| ColH | α1(I) | Gly-Ala-Arg396∼Gly397-Gln-Ala |
| ColH | α2(I) | Gly-Ala-Arg396∼Gly397-Glu-Pro |
| ColH | α1(II) | Gly-Phe-Pro405∼Gly406-Pro-Lys |
| ColH | α1(III) | Gly-Pro-Arg399∼Gly400-Gln-Hyp |
a Numbering begins at the N terminus of the triple-helical region of each collagen.
Two membrane-type (MT) MMPs (MT-MMPs), MT1-MMP and MT2-MMP, allow invasion-incompetent cells to penetrate type I collagen matrices (6). MT1-MMP (Fig. 1) processes type I-III collagens at the same site as the classic collagenases (Table 1) (3). MT3-MMP also cleaves type III collagen at the classic site (Table 1) (3). MT6-MMP was initially reported to have little or no collagenolytic activity (7, 8) but was subsequently found to cleave type I and II collagens (albeit at 37 °C) (9) and a triple-helical peptide (THP) model of the classic collagenase cleavage site (10). The catalytic (CAT) domain of MMP-12 processes type I and III collagens, where hydrolysis occurs at the classic cleavage site and numerous other sites (11). The classic collagenase cleavage site seems to be the most sensitive to MMP-12 (Table 1). Xenopus laevis MMP-18 and chicken MMP-22/MMP-27 cleave type I collagen at the same site as the classic collagenases (3).
The interstitial collagen triple helix is cleaved by the Cys protease cathepsin K under acidic conditions (optimum pH 5.0). Five distinct sites of cathepsin K hydrolysis type I collagen have been identified, as well as one in type II collagen (Table 1) (12, 13).
An extracellular Ser protease contributes to collagenolysis by temporomandibular joint fibroblasts (14). Several Ser proteases possess interstitial collagenolytic activity, including human neutrophil elastase, Uca pugilator (fiddler crab) collagenase 1, Hypoderma lineatum (insect) collagenase, Penaeus vanameii (shrimp) chymotrypsin, and Pseudoalteromonas sp. SM9913 deseasin MCP-01 (3, 15–18). However, neutrophil elastase is ineffective toward fibrillar type III collagen (19). For a number of collagenolytic Ser proteases, the site of collagen cleavage is close to the site of MMP action (Table 1). Although initially reported as being collagenolytic, the Ser proteases fibroblast activation protein/separase and trypsin-2 do not cleave interstitial collagens within their triple helices (20).3
Additional interstitial collagenases include several that act under acidic conditions, such as Cynara cardunculus Asp protease cardosin A (22) and Alicyclobacillus sendaiensis Ser-carboxyl protease kumamolisin-As/ScpA (23), as well as the Cys proteases ginger (Zingiber officinale) GP2 and GP3 (24) and Fasciola hepatica FhCL2 and FhCL3 (25). GP2 hydrolyzes type I collagen at three distinct sites (Table 1) (24). FhCL2 cleaves the α1(I) chain at 43 sites and the α2(I) chain at 26 sites, whereas FhCL3 cleaves the α1(I) chain at 24 sites and the α2(I) chain at 24 sites, with only three sites shared by the two proteases (25).
Clostridium histolyticum possesses two zinc proteases with collagenolytic activity: class I (ColG) and class II (ColH) (Fig. 1). ColG cleaves interstitial collagens initially near the N termini, whereas ColH cleaves interstitial collagens near the middle to produce 35- and 62-kDa fragments (Table 1) (26). Clostridium perfringens produces a collagenase that is highly similar to ColG, whereas Vibrio alginolyticus collagenase is a zinc protease that initially processes collagen at a similar site as collagenolytic MMPs (3).
Pathways of Collagen Catabolism
There are presently four pathways that have been considered for mammalian collagen catabolism: 1) phagocytosis mediated by the α2β1 integrin, where internalized insoluble collagen is transported to lysosomes and degraded by cathepsins (27); 2) cathepsin K collagenolysis in osteoclast-mediated bone resorption (28); 3) extracellular MMP hydrolysis, followed by gelatinolytic MMPs laterally diffusing on collagen extracellularly, finding “tails” from the cleaved sites, denaturing the triple helix, and further proteolyzing the α chains (29, 30); and 4) extracellular MMP hydrolysis, followed by the resulting collagen fragments undergoing endocytosis (mediated by urokinase plasminogen activator receptor-associated protein/Endo180 on mesenchymal cells and mannose receptor on macrophages), lysosomal delivery, and cathepsin-catalyzed degradation (31). Collagen can also be degraded intracellularly by autophagy-mediated lysosomal processes, which may be a form of collagen regulation (32).
In vivo processing of collagen for pathways 1) and 2) above initially involves MMP interaction with fibrils. Hydrolysis of collagen proceeds at the outer edge of the fibril (33, 34). MMP-1 is a diffusion-based “burnt bridge” Brownian ratchet capable of biased diffusion on the surface of collagen fibrils, where the bias is driven by proteolysis (35). Surface-bound MT1-MMP movement is via a similar diffusion mechanism (4). While on collagen fibrils, MMP-1 spends ∼90% of its time in one of two distinct pause classes (36). Class I occurs randomly along the fibril, whereas class II occurs periodically at 1.3 and 1.5 μm along the fibril and exhibits multistep escape kinetics (36). Five percent of the class II pauses result in initiation of processive collagen degradation for ∼15 consecutive cleavage events (36). The temperature dependence of the pauses suggests local unfolding, but the low probability of hydrolysis (∼5%) indicates that local unfolding is not sufficient for hydrolysis (36).
Unique Features of Interstitial Collagen Cleavage Sites
MMPs bind to multiple sites in collagen (37), but hydrolysis ultimately occurs at a single site (Table 1). Collagen primary structure is not the only basis for discriminatory MMP collagenolytic behavior (1). A model of the cleavage sites in interstitial collagens suggested that all of the information necessary for efficient hydrolysis of collagen is contained in a 24-residue stretch (subsites P13–P12′) (2). Cleavage site regions were distinguished by <10% charged residues, being “tightly” triple-helical (high Pro/Hyp content) prior to the cleavage site and being “loosely” triple-helical (low Pro/Hyp content) following the cleavage site (2). Arg residues in the P5′ or P8′ subsite have been proposed to stabilize the triple helix through electrostatic interactions, and these interactions may need to be disrupted for hydrolysis to occur (38).
Soluble collagens are thermally unstable at physiological temperatures, slowly melting unless incorporated into fibrils (39). This instability may lead to local flexibility/microunfolding that is needed for protease processing of collagen. Molecular dynamics simulations indicate microunfolding of interstitial collagens at the MMP cleavage site (40–42). Based on enzyme susceptibility, the type I collagen MMP cleavage site undergoes local reversible relaxation (43), whereas the cleavage site in type III collagen has been proposed to be more flexible than the one in type I collagen (19, 44).
Homotrimeric type I collagen (α1(I)3) is much less susceptible to MMP-1 hydrolysis than heterotrimeric type I collagen, and the effect is not due to binding (45). The homotrimer is more thermally stable than the heterotrimer by ∼2.5 °C and melts 100 times slower (46, 47). The microunfolding patterns of the two collagen subtypes are different (47). Thus, the difference between MMP-1 activity toward homotrimeric versus heterotrimeric type I collagen is due to local triple-helical unwinding at the cleavage site (45). The α2(I) chain also increases hydrophobicity compared with the α1(I) chain, driving out structured water and facilitating hydrolysis (46). Homotrimeric type I collagen is produced by a variety of tumor cells and enhances tumor cell proliferation and migration compared with heterotrimeric type I collagen (48).
The Ile residue in one of the three chains at the site of MMP hydrolysis has a distinct chemical shift, a higher J coupling value, increased dynamics, and decreased local stability (49). This suggests that a single locally dynamic chain, rather than a labile region with three comparably dynamic chains, is a determining factor for collagen to be cleaved by MMPs (49). Also, a Pro residue at the P3 subsite influences the P1′ subsite Ile residue, enhancing its accessibility to collagenolytic MMPs (49).
The collagen cleavage site model would be valid only if collagenases had extended active or substrate-binding sites. Modulation of MMP-1, MMP-8, MMP-13, and MT1-MMP activities was observed in THP substrates spanning subsites P13–P17′ (50–53). Utilizing interstitial collagen sequences inserted into bacterial collagen, the minimum type III collagen sequence necessary for MMP-1 or MMP-13 hydrolysis was found to be 15 residues (subsites P4–P11′), whereas a similar rate of hydrolysis to type III collagen was obtained with a sequence spanning subsites P7–P11′ (54). Thus, both the THP and bacterial collagen studies confirm that the collagenolytic MMPs interact with a significant span of the collagen triple helix.
Molecular Mechanisms of Collagen Catabolism
The 15 Å collagen triple helix does not fit into the 5 Å MMP CAT domain active site cavity (55). Models have generally accounted for this steric clash by (a) requiring active unwinding of the triple helix by an MMP (55–57) and/or (b) considering that the site of hydrolysis within collagen has a distinct conformation or conformational flexibility, rendering it more susceptible to proteolysis than other regions in collagen (2). The “vulnerable” site hypothesis proposes that the distinct cleavage site region within collagen is alone responsible for collagenolysis (58).
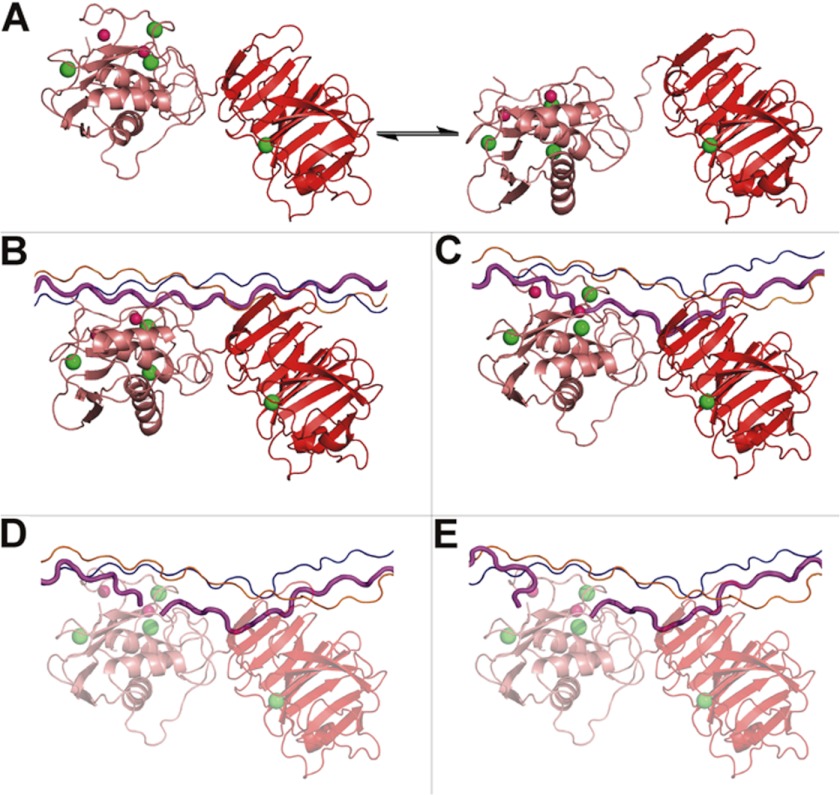
A detailed mechanism of initial collagenolysis was developed from examination of structures of MMP-1 and MMP-1·THP complexes (59). MMP-1 is in equilibrium between open/extended and closed structures (Fig. 2A) (60). An open form of MMP-1 is favored in solution (see below). The hemopexin-like (HPX) domain binds the leading chain (1T) and the middle chain (2T) of the THP, and due to the flexibility of the linker, the CAT domain is guided toward the Gly∼Ile bond of chain 1T (Fig. 2B). This structure would thus correspond to the first event of collagen recognition by MMP-1. The exposure of the MMP cleavage site by removal of the collagen C-terminal telopeptide (34) permits interactions of the MMP-1 HPX and CAT domains with triple helices on the outer edge of the fibril. Visual inspection of the complex at this point suggests that a back-rotation of the CAT and HPX domains would need to occur to achieve the x-ray crystallographic closed MMP-1 conformation. To approximate this action, the residues at the interface between the HPX and CAT domains in the x-ray structure of MMP-1 in the closed form (Protein Data Bank code 1SU3) were imposed as constraints in a docking calculation. In the resulting structure, with the CAT and HPX domains arranged in the x-ray crystallographic closed conformation, the THP is unwound (Fig. 2C). The domain movement drives chain 1T into the active site, allowing the polypeptide to establish a number of H-bonding interactions and the carbonyl oxygen of the cleavage site amide bond to coordinate the metal ion. This result is consistent with the experimentally observed weakening in NOEs for the interaction of chain 1T with chains 2T and 3T (the trailing chain) at the cleavage site. It has been proposed that when released from the triple-helical conformation, the cleavage site sequence in type I collagen has the propensity to form β-bend and β-strand structures (61). Protease active sites appear to accommodate β-strand structures universally (62). Besides liberating the N-terminal region of chain 1T for hydrolysis, the destabilization of the THP also causes a partial detachment of chain 3T near the THP C terminus (Fig. 2C), consistent with the observed lengthening in NOEs of chain 3T. The position that the two peptide fragments assume after cleavage (Fig. 2, D and E) is almost superimposable on the x-ray crystallographic structure of the complex between the MMP-12 CAT domain and the two fragments obtained by enzymatic cleavage of the α1(I) collagen model Pro-Gln-Gly-Ile-Ala-Gly hexapeptide at the Gly∼Ile bond (Protein Data Bank code 2OXZ).
FIGURE 2.
Initial steps of collagenolysis (59). A, closed (left) and open/extended (right) forms of full-length MMP-1 in equilibrium. B, the extended protein binds α1(I)-(772–786) THP chains 1T-2T at Val23–Leu26 with the HPX domain and the residues around the cleavage site with the CAT domain. The THP is still in a compact conformation. C, closed full-length MMP-1 interacting with the released 1T chain (magenta). D, after hydrolysis, both peptide fragments (C- and N-terminal) are initially bound to the active site. E, the C-terminal region of the N-terminal peptide fragment is released. This figure has been reprinted with permission from the Journal of the American Chemical Society.
The MMP-1·THP complex, in which one strand of the triple helix is displaced from the other two (Fig. 2C), is an energetically and mechanistically feasible route between the complexes in Fig. 2 (B and D) (59). MMP-1 may not actively unwind the triple helix (45). Rather, MMP-1 shifts the equilibrium between native helical and locally unwound states, destabilizing the helical state and/or stabilizing the unwound state (45).
The maximum occurrence (MO) of MMP-1 conformations in solution has been recently calculated by paramagnetic NMR and small angle x-ray scattering.4 Many of the MMP-1 conformations with the highest MO value (>35%) were found to have interdomain orientations and positions that could be grouped into a cluster.4 Within this cluster, the collagen-binding residues of the HPX domain are solvent-exposed, and the CAT domain is correctly positioned for its subsequent interaction with the collagen. A ∼50° rotation around a single axis of the CAT domain with respect to the HPX domain positions the CAT domain right in front of the interstitial collagen cleavage site.
Binding sites for the triple helix within the HPX domain have been identified (52, 59, 64, 65). Initially, in MMP-1, Ile290 and Arg291 in the blade I A-B loop were identified as key residues in collagenolysis (52). Subsequently, Phe301, Val319, and Asp338 were implicated in collagen binding (64). Phe320 was found to be an important contributor, along with Ile290 and Arg291, to the S10′ binding pocket (65). The S10′ binding pocket binds the P10′ subsite of collagen, which possesses a conserved Leu residue important for interaction of triple helices with MMP-1 (53, 64, 65). The lack of this Leu residue explains why an earlier study did not observe binding of a THP to the HPX domain (66). Other residues within the HPX domain may also participate in collagen binding (52, 64, 65).
There is some controversy over MMP-1 Phe301, which was identified as a binding site by NMR spectroscopy but was deemed as buried in the CAT domain/HPX domain interface by x-ray crystallography (64, 65). All available x-ray structures of human full-length MMP-1 (Protein Data Bank codes 1SU3, 2CLT, and 4AUO) display relatively closed conformations. The MO values obtained for the x-ray structures for pro-MMP-1 (code 1SU3) and active MMP-1 (2CLT) were 20 and 19%, respectively.4 The x-ray crystallographic structure of an MMP-1·THP complex (code 4AUO) has a more closed structure compared with code 2CLT (65) and has a MO of 18%. Thus, these structures are not the dominant ones sampled by the protein in solution. The radii of gyration (Rg) of the crystallographic structures range from 25.5 to 25.7 Å, whereas the structures with the highest MO (>35%) have Rg of 28.9 ± 1.3 Å. This range of Rg is in better agreement with values from small angle x-ray scattering data, indicating that the x-ray structures are more compact than the average solution conformation. Furthermore, the relative orientations of the HPX and CAT domains in the structures with the highest MO are different from those in the x-ray crystallographic structures. It was reported that the x-ray crystallographic structure of the MMP-1·THP complex is a nonproductive complex (65). Thus, Phe301 probably interacts with the triple helix initially but is then utilized for domain interaction during collagenolysis (64).
Based on chimeric studies using MMP-3 sequences, the active site cleft of MMP-1 is a significant determinant for collagenolytic activity (67, 68). Tyr210 is specifically involved in collagenolytic (but not general peptidase) activity (50, 69).
MMP-3 binds to type I collagen but does not cleave the native triple helix (70). However, the MMP-3 CAT domain can cleave collagen when the triple helix is unwound by catalytically inactive MMP-1 (55). Thus, MMP-3 is entirely competent to cleave type I collagen but does not. Based on the MMP collagenolysis mechanism, the linker needs to be able to properly orient the CAT and HPX domains (59). Gly272 is critical for the collagenolytic activity of MMP-1, with its role proposed to be the linker-bending motion that allows the HPX domain to present collagen to the CAT domain (71). The MMP-1 and MMP-8 linkers are considerably shorter than the MMP-3 linker, whereas the MT1-MMP linker is very long (33 residues), with significant and heterogeneous O-glycosylation (72). An MMP-8 chimera with the linker region (16 residues) replaced with the corresponding MMP-3 sequence (25 residues) loses activity toward collagen (73). In a similar fashion, MMP-1/MMP-3 chimeras possessing the MMP-3 linker are not active toward collagen (67, 70). The linker appears to be critical for proper alignment of the CAT and HPX domains during collagenolysis. Ultimately, there may be negative regulation of collagenolytic activity due to (mis)alignment of the CAT and HPX domains in the case of MMP-3 and other non-collagenolytic MMPs.
The experimentally determined mechanism is not consistent with the vulnerable site hypothesis, as fluctuations in the triple helix were not observed until after MMP-1 binding (59). As indicated earlier in the studies of MMP movement on fibrils, local unfolding was not sufficient for hydrolysis (36). Support for the vulnerable site hypothesis comes from the action of the CAT domains of MMP-1 and MMP-8 against type I collagen (58). However, high concentrations of CAT domains were utilized to obtain hydrolysis, and a prior study demonstrated that the MMP-8 CAT domain had a different pattern of type I collagen hydrolysis compared with full-length MMP-8 (74). A highly temperature-dependent collagenolytic activity was observed for the MMP-13 CAT domain but not for full-length MMP-13, indicating that the activity of the CAT domain was based on partial denaturation of the substrate (54). Although the MMP-12 CAT domain hydrolyzes interstitial collagens (11), it appears to possess unique properties that allow it to destabilize the triple helix (75, 76).
It has been noted that although MMP-1 and MMP-8 have similar collagenolytic mechanisms, MMP-2 and MT1-MMP have mechanisms distinct from MMP-1 and MMP-8 (50, 56, 77, 78). In the case of MMP-2 (and MMP-9 as well), interaction with collagen is primarily via the fibronectin type II-like modules within the CAT domain, not the HPX domain (3). All three fibronectin type II-like modules contribute to collagen binding, with the greatest effects observed for modules 2 and 3 (56, 79). Individual residues involved in collagen binding are primarily Arg (positions 252, 296, and 368) and aromatics (Phe297, Tyr302, Tyr323, Tyr329, Trp374, and Tyr381) (79). It has been proposed that MMP-2 preferentially binds the α1(I) chain and grossly distorts the triple helix, followed by initial hydrolysis of the α2(I) chain (77).
A mechanism has been proposed for C. histolyticum collagenolysis based on x-ray crystallographic analysis of the collagenase module (activator + CAT domains), the polycystic kidney disease-like domain, and one or both of the collagen-binding domains (CBDs) and mutagenesis analysis of substrate binding and/or hydrolysis of C. histolyticum class I collagenase (ColG) (80–82). The CBDs of ColG promote interaction with fibrils, not individual triple helices, along the fibril axis. Mutagenesis analysis of ColG CBD binding to THP Gly-(Pro-Hyp-Gly)8 revealed Thr957, Tyr970, Leu992, Tyr994, and Tyr996 as participating in binding, with all of these residues centrally located on one face of the CBD (80). The ColG CBD binds unidirectionally to the undertwisted C terminus of the triple helix but does not facilitate unwinding (83). The polycystic kidney disease-like domain swells the collagen but does not unwind it (18, 82). The ColG collagenase module forms a saddle-shaped two-domain architecture that “squeezes” the fibril, facilitating enzyme (CAT domain) accessibility to monomeric triple helices (81). Initial collagen contact is made with the CAT domain, followed by a closing of the saddle and contact by the activator domain. Only the open state was observed in x-ray crystallographic analysis, whereby the opening between the CAT and activator domains matched that of collagen microfibrils (40 Å) (81). Removal of the activator domain or the Gly-rich hinge region between the activator and CAT domains greatly decreased collagenolytic activity. The mechanical energy for substrate unwinding comes from the release of stored ordered water upon hydrolysis (81). This is consistent with force measurement studies that concluded that C. histolyticum collagenase processes collagen independent of an unwinding transition (84). Supporting this notion is the much lower activation energy for C. histolyticum collagenase hydrolysis of fibrillar type I collagen compared with MMP-1 (33).
The entrance to the cathepsin K active site is 5 Å wide (85), and thus, manipulation of the triple helix in a similar fashion as in MMPs is anticipated. Collagenolytic activity of cathepsin K is lost by the T67L/L205A double mutation, as this mutation renders the S2 subsite unable to accommodate Pro (86). Efficient collagenolytic activity requires a complex between cathepsin K and chondroitin sulfate (87).
Facilitation of Collagen Catabolism
Cell surface collagenolysis may be facilitated by collagen-binding integrins providing strain on the collagen, protease-binding partners, and/or protease dimerization. Binding of a THP by the α2β1 integrin results in disruption of interactions between Arg and Glu side chains in the ligand and significant changes in main chain conformation, reflected in the bending of the triple helix (88). Strain could be induced by integrin-collagen interactions and/or cellular traction forces (57).
The reported effects of strain on collagenolysis have been contradictory. Molecular dynamics simulations indicated that force stabilizes the MMP cleavage site in heterotrimeric type I collagen, slowing proteolysis (89). However, homotrimeric type I collagen possesses a more stable cleavage site, so force enhances proteolysis by destabilizing the cleavage site (89). MMP-1 hydrolysis of a homotrimeric model of type I collagen was increased by 81-fold by a mechanical load (57), whereas similar force enhanced MMP-1 catalysis of heterotrimeric type I collagen by 8-fold (84). These data suggested that heterotrimeric type I collagen was more unwound than homotrimeric type I collagen, and hence, the effect of strain on further unwinding the triple helix was less pronounced in the former case (84). Conversely, strain of reconstituted type I collagen fibrils increased degradation time by MMP-8 (90). The discrepancy between the single-molecule study (84) and the fibrillar collagen study (90) may be due to effects on diffusive transport of the MMP in fibrils (84).
In one study, applied force had little effect on C. histolyticum collagenase processing of heterotrimeric type I collagen (84), whereas in another, the application of force significantly reduced C. histolyticum collagenase activity (91). The different results could be due to the mixtures of collagenases used. C. histolyticum collagenase activity was decreased by increasing strain in corneal tissue (92). As the enzyme processed the tissue, the same applied load strained the remaining tissue to a greater degree, limiting diffusion and slowing collagenolysis (92).
Related to strain, fibronectin binds to type I collagen at Gly788–Gly799, near the classic collagenase cleavage site (93). Fibronectin binding destabilizes the collagen triple helix, potentially facilitating MMP collagenolysis (93).
Many “soluble” collagenolytic MMPs have cell surface binding partners, including the α2β1 integrin (MMP-1) and CD44 (MMP-9) (94). MT1-MMP has numerous cell surface binding partners, including tetraspanins, the α2β1 and αvβ3 integrins, and CD44 (94–96). The HPX domain of MT1-MMP binds to tetraspanins CD63 and CD151 (96). MT1-MMP association with CD151 modulates collagenolysis in that knockdown of CD151 decreases collagenolysis (96).
Highly efficient collagenolysis requires homodimerization of MT1-MMP, where association includes interactions of the HPX domain (3). Homodimerization is symmetrical, involving Asp385, Lys386, Thr412, and Tyr436 in blades II and III of the HPX domain (97). MT6-MMP also forms homodimers through a disulfide bond in the stem region (98). MMP-1 and MMP-9 can form an active heterodimeric complex capable of fibrillar type I collagen catabolism (63).
Cell surface-bound MT1-MMP has only a partially decreased collagenolytic activity upon deletion of the HPX domain (21). This suggests that other factors contribute to enzyme activity on the cell surface, possibly by straining the collagen.
Conclusion
Interstitial collagenolytic activity is a convergent evolutionary process. The initial steps of MMP-1 collagenolysis have been experimentally derived, and individual residues involved in this process have been identified. In addition, the roles of specific collagen residues in MMP substrate specificity have been quantified. Binding partners that modulate the activity of collagenolytic enzymes have begun to be identified. Collagenolytic MMPs utilize subtly different mechanisms for processing triple helices, and these differences may be exploited to develop selective inhibitors. As further information on interstitial collagenolytic processes is obtained, inhibition can be fined-tuned to be disease- or pathogen-specific.
Acknowledgment
I thank Dr. Anna Knapinska for constructing Fig. 1.
This work was supported, in whole or in part, by National Institutes of Health Grants CA98799 and MH78948 and Contract 268201000036C from NHLBI. This work was also supported by the Multiple Sclerosis National Research Institute.
L. S. Mirigian, E. Makareeva, H. Koistinen, O. Itkonen, T. Sorsa, U.-H. Stenman, T. Salo, and S. Leikin, manuscript submitted for publication.
I. Bertini, L. Cerofolini, G. B. Fields, M. Fragai, C. F. G. C. Geraldes, C. Luchinat, G. Parigi, E. Ravera, D. I. Svergun, and J. M. C. Teixeira, manuscript submitted for publication.
- MMP
- matrix metalloproteinase
- MT
- membrane-type
- THP
- triple-helical peptide
- CAT
- catalytic
- HPX
- hemopexin-like
- MO
- maximum occurrence
- CBD
- collagen-binding domain.
REFERENCES
- 1. Lauer-Fields J. L., Juska D., Fields G. B. (2002) Matrix metalloproteinases and collagen catabolism. Biopolymers 66, 19–32 [DOI] [PubMed] [Google Scholar]
- 2. Fields G. B. (1991) A model for interstitial collagen catabolism by mammalian collagenases. J. Theor. Biol. 153, 585–602 [DOI] [PubMed] [Google Scholar]
- 3. Barrett A. J., Rawlings N. D., Woessner J. F. (2004) Handbook of Proteolytic Enzymes, 2nd Ed., Elsevier Academic Press, Amsterdam [Google Scholar]
- 4. Collier I. E., Legant W., Marmer B., Lubman O., Saffarian S., Wakatsuki T., Elson E., Goldberg G. I. (2011) Diffusion of MMPs on the surface of collagen fibrils: the mobile cell surface-collagen substratum interface. PLoS ONE 6, e24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bigg H. F., Rowan A. D., Barker M. D., Cawston T. E. (2007) Activity of matrix metalloproteinase-9 against native collagen types I and III. FEBS J. 274, 1246–1255 [DOI] [PubMed] [Google Scholar]
- 6. Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang T., Yi J., Guo A., Wang X., Overall C. M., Jiang W., Elde R., Borregaard N., Pei D. (2001) Subcellular distribution and cytokine- and chemokine-regulated secretion of leukolysin/MT6-MMP/MMP-25 in neutrophils. J. Biol. Chem. 276, 21960–21968 [DOI] [PubMed] [Google Scholar]
- 8. Radichev I. A., Remacle A. G., Shiryaev S. A., Purves A. N., Johnson S. L., Pellecchia M., Strongin A. Y. (2010) Biochemical characterization of the cellular glycosylphosphatidylinositol-linked membrane type-6 matrix metalloproteinase. J. Biol. Chem. 285, 16076–16086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Starr A. E., Bellac C. L., Dufour A., Goebeler V., Overall C. M. (2012) Biochemical characterization and N-terminomics analysis of leukolysin, the membrane-type 6 matrix metalloprotease (MMP25). Chemokine and vimentin cleavages enhance cell migration and macrophage phagocytic activities. J. Biol. Chem. 287, 13382–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Amar S., Fields G. B. (2012) Production and characterization of matrix metalloproteinases implicated in multiple sclerosis. in Peptides 2012: Proceedings of the Thirty-Second European Peptide Symposium (Kokotos G., Constantinou-Kokotou V., Matsoukas J. eds) pp. 102–103, European Peptide Society, Athens, Greece [Google Scholar]
- 11. Taddese S., Jung M. C., Ihling C., Heinz A., Neubert R. H. H., Schmelzer C. E. H. (2010) MMP-12 catalytic domain recognizes and cleaves at multiple sites in human skin collagen type I and type III. Biochim. Biophys. Acta 1804, 731–739 [DOI] [PubMed] [Google Scholar]
- 12. Garnero P., Borel O., Byrjalsen I., Ferreras M., Drake F. H., McQueney M. S., Foged N. T., Delmas P. D., Delaissé J.-M. (1998) The collagenolytic activity of cathepsin K is unique among mammalian proteinases. J. Biol. Chem. 273, 32347–32352 [DOI] [PubMed] [Google Scholar]
- 13. Kafienah W., Brömme D., Buttle D. J., Croucher L. J., Hollander A. P. (1998) Human cathepsin K cleaves native type I and II collagens at the N-terminal end of the triple helix. Biochem. J. 331, 727–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song F., Bergdoll A. S., Windsor L. J. (2006) Temporomandibular joint synovial fibroblasts mediate serine proteinase-dependent type I collagen degradation. Biochim. Biophys. Acta 1760, 1521–1528 [DOI] [PubMed] [Google Scholar]
- 15. Mainardi C. L., Hasty D. L., Seyer J. M., Kang A. H. (1980) Specific cleavage of human type III collagen human polymorphonuclear leukocyte elastase. J. Biol. Chem. 255, 12006–12010 [PubMed] [Google Scholar]
- 16. Lecroisey A., Gilles A.-M., De Wolf A., Keil B. (1987) Complete amino acid sequence of the collagenase from the insect Hypoderma lineatum. J. Biol. Chem. 262, 7546–7551 [PubMed] [Google Scholar]
- 17. Tsu C. A., Perona J. J., Schellenberger V., Turck C. W., Craik C. S. (1994) The substrate specificity of Uca pugilator collagenolytic serine protease 1 correlates with the bovine type I collagen cleavage sites. J. Biol. Chem. 269, 19565–19572 [PubMed] [Google Scholar]
- 18. Wang Y.-K., Zhao G.-Y., Li Y., Chen X.-L., Xie B.-B., Su H.-N., Lv Y.-H., He H.-L., Liu H., Hu J., Zhou B.-C., Zhang Y.-Z. (2010) Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913. J. Biol. Chem. 285, 14285–14291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birkedal-Hansen H., Taylor R. E., Bhown A. S., Katz J., Lin H.-Y., Wells B. R. (1985) Cleavage of bovine skin type III collagen by proteolytic enzymes. J. Biol. Chem. 260, 16411–16417 [PubMed] [Google Scholar]
- 20. Christiansen V. J., Jackson K. W., Lee K. N., McKee P. A. (2007) Effect of fibroblast activation protein and α2-antiplasmin cleaving enzyme on collagen types I, III, and IV. Arch. Biochem. Biophys. 457, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li X.-Y., Ota I., Yana I., Sabeh F., Weiss S. J. (2008) Molecular dissection of the structural machinery underlying the tissue-invasive activity of membrane type-1 matrix metalloproteinase. Mol. Biol. Cell 19, 3221–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duarte A. S., Pereira A. O., Cabrita A. M. S., Moir A. J. G., Pires E. M. V., Barros M. M. T. (2005) The characterisation of collagenolytic activity of cardosin A demonstrates its potential application for extracellular matrix degradative processes. Curr. Drug Discov. Technol. 2, 37–44 [DOI] [PubMed] [Google Scholar]
- 23. Tsuruoka N., Nakayama T., Ashida M., Hemmi H., Nakao M., Minakata H., Oyama H., Oda K., Nishino T. (2003) Collagenolytic serine-carboxyl proteinase from Alicyclobacillus sendaiensis strain NTAP-1: purification, characterization, gene cloning, and heterologous expression. Appl. Environ. Microbiol. 69, 162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim M., Hamilton S. E., Guddat L. W., Overall C. M. (2007) Plant collagenase: unique collagenolytic activity of cysteine proteases from ginger. Biochim. Biophys. Acta 1770, 1627–1635 [DOI] [PubMed] [Google Scholar]
- 25. Robinson M. W., Corvo I., Jones P. M., George A. M., Padula M. P., To J., Cancela M., Rinaldi G., Tort J. F., Roche L., Dalton J. P. (2011) Collagenolytic activities of the major secreted cathepsin L peptidases involved in the virulence of the helminth pathogen, Fasciola hepatica. PLoS Negl. Trop. Dis. 5, e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. French M. F., Bhown A., Van Wart H. E. (1992) Identification of Clostridium histolyticum collagenase hyper-reactive sites in type I, II, and III collagens: lack of correlation with local triple helical stability. J. Protein Chem. 11, 83–97 [DOI] [PubMed] [Google Scholar]
- 27. Arora P. D., Manolson M. F., Downey G. P., Sodek J., McCulloch C. A. (2000) A novel model system for characterization of phagosomal maturation, acidification, and intracellular collagen degradation in fibroblasts. J. Biol. Chem. 275, 35432–35441 [DOI] [PubMed] [Google Scholar]
- 28. Costa A. G., Cusano N. E., Silva B. C., Cremers S., Bilezikian J. P. (2011) Cathepsin K: its skeletal actions and role as a therapeutic target in osteoporosis. Nat. Rev. Rheumatol. 7, 447–456 [DOI] [PubMed] [Google Scholar]
- 29. Atkinson S. J., Patterson M. L., Butler M. J., Murphy G. (2001) Membrane type 1 matrix metalloproteinase and gelatinase A synergistically degrade type I collagen in a cell model. FEBS Lett. 491, 222–226 [DOI] [PubMed] [Google Scholar]
- 30. Rosenblum G., Van den Steen P. E., Cohen S. R., Bitler A., Brand D. D., Opdenakker G., Sagi I. (2010) Direct visualization of protease action on collagen triple helical structure. PLoS ONE 5, e11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Madsen D. H., Ingvarsen S., Jürgensen H. J., Melander M. C., Kjøller L., Moyer A., Honoré C., Madsen C. A., Garred P., Burgdorf S., Bugge T. H., Behrendt N., Engelholm L. H. (2011) The non-phagocytic route of collagen uptake: a distinct degradation pathway. J. Biol. Chem. 286, 26996–27010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim S. I., Na H.-J., Ding Y., Wang Z., Lee S. J., Choi M. E. (2012) Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-β1. J. Biol. Chem. 287, 11677–11688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welgus H. G., Jeffrey J. J., Eisen A. Z. (1981) Human skin fibroblast collagenase. Assessment of activation energy and deuterium isotope effect with collagenous substrates. J. Biol. Chem. 256, 9516–9521 [PubMed] [Google Scholar]
- 34. Perumal S., Antipova O., Orgel J. P. R. O. (2008) Collagen fibril architecture, domain organization, and triple-helical conformation govern its proteolysis. Proc. Natl. Acad. Sci. U.S.A. 105, 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saffarian S., Collier I. E., Marmer B. L., Elson E. L., Goldberg G. (2004) Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science 306, 108–111 [DOI] [PubMed] [Google Scholar]
- 36. Sarkar S. K., Marmer B., Goldberg G., Neuman K. C. (2012) Single-molecule tracking of collagenase on native type I collagen fibrils reveals degradation mechanism. Curr. Biol. 22, 1047–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun H. B., Smith G. N., Jr., Hasty K. A., Yokota H. (2000) Atomic force microscopy-based detection of binding and cleavage site of matrix metalloproteinase on individual type II collagen helices. Anal. Biochem. 283, 153–158 [DOI] [PubMed] [Google Scholar]
- 38. Salsas-Escat R., Stultz C. M. (2010) Conformational selection and collagenolysis in type III collagen. Proteins 78, 325–335 [DOI] [PubMed] [Google Scholar]
- 39. Leikina E., Mertts M. V., Kuznetsova N., Leikin S. (2002) Type I collagen is thermally unstable at body temperature. Proc. Natl. Acad. Sci. U.S.A. 99, 1314–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stultz C. M. (2002) Localized unfolding of collagen explains collagenase cleavage near imino-poor sites. J. Mol. Biol. 319, 997–1003 [DOI] [PubMed] [Google Scholar]
- 41. Ravikumar K. M., Humphrey J. D., Hwang W. (2007) Spontaneous unwinding of a labile domain in a collagen triple helix. J. Mech. Mater. Struct. 2, 999–1010 [Google Scholar]
- 42. Suárez E., Díaz N., Suárez D. (2008) Entropic control of the relative stability of triple-helical collagen peptide models. J. Phys. Chem. B 112, 15248–15255 [DOI] [PubMed] [Google Scholar]
- 43. Ryhänen L., Zaragoza E. J., Uitto J. (1983) Conformational stability of type I collagen triple helix: evidence for temporary and local relaxation of the protein conformation using a proteolytic probe. Arch. Biochem. Biophys. 223, 562–571 [DOI] [PubMed] [Google Scholar]
- 44. Welgus H. G., Burgeson R. E., Wootton J. A. M., Minor R. R., Fliszar C., Jeffrey J. J. (1985) Degradation of monomeric and fibrillar type III collagens by human skin collagenase. J. Biol. Chem. 260, 1052–1059 [PubMed] [Google Scholar]
- 45. Han S., Makareeva E., Kuznetsova N. V., DeRidder A. M., Sutter M. B., Losert W., Phillips C. L., Visse R., Nagase H., Leikin S. (2010) Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. J. Biol. Chem. 285, 22276–22281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miles C. A., Sims T. J., Camacho N. P., Bailey A. J. (2002) The role of the α2 chain in the stabilization of the collagen type I heterotrimer: a study of the type I homotrimer in oim mouse tissues. J. Mol. Biol. 321, 797–805 [DOI] [PubMed] [Google Scholar]
- 47. Kuznetsova N. V., McBride D. J., Jr., Leikin S. (2003) Changes in thermal stability and microunfolding pattern of collagen helix resulting from the loss of α2(I) chain in osteogenesis imperfecta murine. J. Mol. Biol. 331, 191–200 [DOI] [PubMed] [Google Scholar]
- 48. Makareeva E., Han S., Vera J. C., Sackett D. L., Holmbeck K., Phillips C. L., Visse R., Nagase H., Leikin S. (2010) Carcinomas contain a matrix metalloproteinase-resistant isoform of type I collagen exerting selective support to invasion. Cancer Res. 70, 4366–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao J., Addabbo R. M., Lauer J. L., Fields G. B., Baum J. (2010) Local conformation and dynamics of isoleucine in the collagenase cleavage site provide a recognition signal for matrix metalloproteinases. J. Biol. Chem. 285, 34181–34190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Visse R., Nagase H., Fields G. B. (2006) The roles of substrate thermal stability and P2 and P1′ subsite identity in matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 281, 38302–38313 [DOI] [PubMed] [Google Scholar]
- 51. Minond D., Lauer-Fields J. L., Cudic M., Overall C. M., Pei D., Brew K., Moss M. L., Fields G. B. (2007) Differentiation of secreted and membrane-type matrix metalloproteinase activities based on substitutions and interruptions of triple-helical sequences. Biochemistry 46, 3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lauer-Fields J. L., Chalmers M. J., Busby S. A., Minond D., Griffin P. R., Fields G. B. (2009) Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J. Biol. Chem. 284, 24017–24024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Robichaud T. K., Steffensen B., Fields G. B. (2011) Exosite interactions impact matrix metalloproteinase collagen specificities. J. Biol. Chem. 286, 37535–37542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu Z., Visse R., Inouye M., Nagase H., Brodsky B. (2012) Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 287, 22988–22997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chung L., Dinakarpandian D., Yoshida N., Lauer-Fields J. L., Fields G. B., Visse R., Nagase H. (2004) Collagenase unwinds triple helical collagen prior to peptide bond hydrolysis. EMBO J. 23, 3020–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tam E. M., Moore T. R., Butler G. S., Overall C. M. (2004) Characterization of the distinct collagen binding, helicase and cleavage mechanisms of matrix metalloproteinases 2 and 14 (gelatinase A and MT1-MMP). The differential roles of the MMP hemopexin C domains and the MMP-2 fibronectin type II modules in collagen triple helicase activities. J. Biol. Chem. 279, 43336–43344 [DOI] [PubMed] [Google Scholar]
- 57. Adhikari A. S., Chai J., Dunn A. R. (2011) Mechanical load induces a 100-fold increase in the rate of collagen proteolysis by MMP-1. J. Am. Chem. Soc. 133, 1686–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Salsas-Escat R., Nerenberg P. S., Stultz C. M. (2010) Cleavage site specificity and conformational selection in type I collagen degradation. Biochemistry 49, 4147–4158 [DOI] [PubMed] [Google Scholar]
- 59. Bertini I., Fragai M., Luchinat C., Melikian M., Toccafondi M., Lauer J. L., Fields G. B. (2012) Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis. J. Am. Chem. Soc. 134, 2100–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bertini I., Fragai M., Luchinat C., Melikian M., Mylonas E., Sarti N., Svergun D. I. (2009) Interdomain flexibility in full-length matrix metalloproteinase-1 (MMP-1). J. Biol. Chem. 284, 12821–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bhatnagar R. S., Qian J. J., Gough C. A. (1997) The role in cell binding of a β-bend within the triple helical region in collagen α1(I) chain: structural and biological evidence for conformational tautomerism on fiber surface. J. Biomol. Struct. Dyn. 14, 547–560 [DOI] [PubMed] [Google Scholar]
- 62. Tyndall J. D. A., Nall T., Fairlie D. P. (2005) Proteases universally recognize β-strands in their active sites. Chem. Rev. 105, 973–999 [DOI] [PubMed] [Google Scholar]
- 63. Goldberg G. I., Strongin A., Collier I. E., Genrich L. T., Marmer B. L. (1992) Interaction of 92-kDa type IV collagenase with the tissue inhibitor of metalloproteinases prevents dimerization, complex formation with interstitial collagenase, and activation of the proenzyme with stromelysin. J. Biol. Chem. 267, 4583–4591 [PubMed] [Google Scholar]
- 64. Arnold L. H., Butt L. E., Prior S. H., Read C. M., Fields G. B., Pickford A. R. (2011) The interface between catalytic and hemopexin domains in matrix metalloproteinase 1 conceals a collagen binding exosite. J. Biol. Chem. 286, 45073–45082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Manka S. W., Carafoli F., Visse R., Bihan D., Raynal N., Farndale R. W., Murphy G., Enghild J. J., Hohenester E., Nagase H. (2012) Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc. Natl. Acad. Sci. U.S.A. 109, 12461–12466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ottl J., Gabriel D., Murphy G., Knäuper V., Tominaga Y., Nagase H., Kröger M., Tschesche H., Bode W., Moroder L. (2000) Recognition and catabolism of synthetic heterotrimeric collagen peptides by matrix metalloproteinases. Chem. Biol. 7, 119–132 [DOI] [PubMed] [Google Scholar]
- 67. Chung L., Shimokawa K., Dinakarpandian D., Grams F., Fields G. B., Nagase H. (2000) Identification of the RWTNNFREY(183–191) region as a critical segment of matrix metalloproteinase 1 for the expression of collagenolytic activity. J. Biol. Chem. 275, 29610–29617 [DOI] [PubMed] [Google Scholar]
- 68. Knäuper V., Patterson M. L., Gomis-Rüth F. X., Smith B., Lyons A., Docherty A. J. P., Murphy G. (2001) The role of exon 5 in fibroblast collagenase (MMP-1) substrate specificity and inhibitor selectivity. Eur. J. Biochem. 268, 1888–1896 [PubMed] [Google Scholar]
- 69. Pelman G. R., Morrison C. J., Overall C. M. (2005) Pivotal molecular determinants of peptidic and collagen triple helicase activities reside in the S3′ subsite of matrix metalloproteinase 8 (MMP-8). J. Biol. Chem. 280, 2370–2377 [DOI] [PubMed] [Google Scholar]
- 70. Murphy G., Allan J. A., Willenbrock F., Cockett M. I., O'Connell J. P., Docherty A. J. P. (1992) The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem. 267, 9612–9618 [PubMed] [Google Scholar]
- 71. Fasciglione G. F., Gioia M., Tsukada H., Liang J., Iundusi R., Tarantino U., Coletta M., Pourmotabbed T., Marini S. (2012) The collagenolytic action of MMP-1 is regulated by the interaction between the catalytic domain and the hinge region. J. Biol. Inorg. Chem. 17, 663–672 [DOI] [PubMed] [Google Scholar]
- 72. Shuo T., Koshikawa N., Hoshino D., Minegishi T., Ao-Kondo H., Oyama M., Sekiya S., Iwamoto S., Tanaka K., Seiki M. (2012) Detection of the heterogeneous O-glycosylation profile of MT1-MMP expressed in cancer cells by a simple MALDI-MS method. PLoS ONE 7, e43751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hirose T., Patterson C., Pourmotabbed T., Mainardi C. L., Hasty K. A. (1993) Structure-function relationship of human neutrophil collagenase: identification of regions responsible for substrate specificity and general proteinase activity. Proc. Natl. Acad. Sci. U.S.A. 90, 2569–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gioia M., Fasciglione G. F., Marini S., D'Alessio S., De Sanctis G., Diekmann O., Pieper M., Politi V., Tschesche H., Coletta M. (2002) Modulation of the catalytic activity of neutrophil collagenase MMP-8 on bovine collagen I. J. Biol. Chem. 277, 23123–23130 [DOI] [PubMed] [Google Scholar]
- 75. Bhaskaran R., Palmier M. O., Lauer-Fields J. L., Fields G. B., Van Doren S. R. (2008) MMP-12 catalytic domain recognizes triple-helical peptide models of collagen V with exosites and high activity. J. Biol. Chem. 283, 21779–21788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Palmier M. O., Fulcher Y. G., Bhaskaran R., Duong V. Q., Fields G. B., Van Doren S. (2010) NMR and bioinformatics discovery of exosites that tune metalloelastase specificity for solubilized elastin and collagen triple helices. J. Biol. Chem. 285, 30918–30930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gioia M., Monaco S., Fasciglione G. F., Coletti A., Modesti A., Marini S., Coletta M. (2007) Characterization of the mechanisms by which gelatinase A, neutrophil collagenase, and membrane-type metalloproteinase MMP-14 recognize collagen I and enzymatically process two α-chains. J. Mol. Biol. 368, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 78. Lauer-Fields J. L., Whitehead J. K., Li S., Hammer R. P., Brew K., Fields G. B. (2008) Selective modulation of matrix metalloproteinase 9 (MMP-9) functions via exosite inhibition. J. Biol. Chem. 283, 20087–20095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xu X., Mikhailova M., Ilangovan U., Chen Z., Yu A., Pal S., Hinck A. P., Steffensen B. (2009) Nuclear magnetic resonance mapping and functional confirmation of the collagen binding sites of matrix metalloproteinase-2. Biochemistry 48, 5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wilson J. J., Matsushita O., Okabe A., Sakon J. (2003) A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J. 22, 1743–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Eckhard U., Schönauer E., Nüss D., Brandstetter H. (2011) Structure of collagenase G reveals a chew-and-digest mechanism of bacterial collagenolysis. Nat. Struct. Mol. Biol. 18, 1109–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Eckhard U., Brandstetter H. (2011) Polycystic kidney disease-like domains of clostridial collagenases and their role in collagen recruitment. Biol. Chem. 392, 1039–1045 [DOI] [PubMed] [Google Scholar]
- 83. Philominathan S. T. L., Koide T., Matsushita O., Sakon J. (2012) Bacterial collagen-binding domain targets undertwisted regions of collagen. Protein Sci. 21, 1554–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Adhikari A. S., Glassey E., Dunn A. R. (2012) Conformational dynamics accompanying the proteolytic degradation of trimeric collagen I by collagenases. J. Am. Chem. Soc. 134, 13259–13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. McGrath M. E., Klaus J. L., Barnes M. G., Brömme D. (1997) Crystal structure of human cathepsin K complexed with a potent inhibitor. Nat. Struct. Biol. 4, 105–109 [DOI] [PubMed] [Google Scholar]
- 86. Lecaille F., Choe Y., Brandt W., Li Z., Craik C. S., Brömme D. (2002) Selective inhibition of the collagenolytic activity of human cathepsin K by altering its S2 subsite specificity. Biochemistry 41, 8447–8454 [DOI] [PubMed] [Google Scholar]
- 87. Cherney M. M., Lecaille F., Kienitz M., Nallaseth F. S., Li Z., James M. N., Brömme D. (2011) Structure-activity analysis of cathepsin K/chondroitin 4-sulfate interactions. J. Biol. Chem. 286, 8988–8998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Emsley J., Knight C. G., Farndale R. W., Barnes M. J. (2004) Structure of the integrin α2β1-binding collagen peptide. J. Mol. Biol. 335, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 89. Chang S.-W., Flynn B. P., Ruberti J. W., Buehler M. J. (2012) Molecular mechanism of force-induced stabilization of collagen against enzymatic breakdown. Biomaterials 33, 3852–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Flynn B. P., Bhole A. P., Saeidi N., Liles M., DiMarzio C. A., Ruberti J. W. (2010) Mechanical strain stabilizes reconstituted collagen fibrils against enzymatic degradation by mammalian collagenase matrix metalloproteinase 8 (MMP-8). PLoS ONE 5, e12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Camp R. J., Liles M., Beale J., Saeidi N., Flynn B. P., Moore E., Murthy S. K., Ruberti J. W. (2011) Molecular mechanochemistry: low force switch slows enzymatic cleavage of human type I collagen monomer. J. Am. Chem. Soc. 133, 4073–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zareian R., Church K. P., Saeidi N., Flynn B. P., Beale J. W., Ruberti J. W. (2010) Probing collagen/enzyme mechanochemistry in native tissue with dynamic, enzyme-induced creep. Langmuir 26, 9917–9926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Erat M. C., Slatter D. A., Lowe E. D., Millard C. J., Farndale R. W., Campbell I. D., Vakonakis I. (2009) Identification and structural analysis of type I collagen sites in complex with fibronectin fragments. Proc. Natl. Acad. Sci. U.S.A. 106, 4195–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Murphy G., Nagase H. (2011) Localizing matrix metalloproteinase activities in the pericellular environment. FEBS J. 278, 2–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gálvez B. G., Matías-Román S., Yáñez-Mó M., Sánchez-Madrid F., Arroyo A. G. (2002) ECM regulates MT1-MMP localization with β1 or αvβ3 integrins at distinct cell compartments modulating its internalization and activity on human endothelial cells. J. Cell Biol. 159, 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yañez-Mó M., Barreiro O., Gonzalo P., Batista A., Megías D., Genís L., Sachs N., Sala-Valdés M., Alonso M. A., Montoya M. C., Sonnenberg A., Arroyo A. G., Sánchez-Madrid F. (2008) MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood 112, 3217–3226 [DOI] [PubMed] [Google Scholar]
- 97. Tochowicz A., Goettig P., Evans R., Visse R., Shitomi Y., Palmisano R., Ito N., Richter K., Maskos K., Franke D., Svergun D., Nagase H., Bode W., Itoh Y. (2011) The dimer interface of the membrane type 1 matrix metalloproteinase hemopexin domain. Crystal structure and biological functions. J. Biol. Chem. 286, 7587–7600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhao H., Sohail A., Sun Q., Shi Q., Kim S., Mobashery S., Fridman R. (2008) Identification and role of homodimerization interface of the glycosylphosphatidylinositol-anchored membrane type 6 matrix metalloproteinase (MMP25). J. Biol. Chem. 283, 35023–35032 [DOI] [PMC free article] [PubMed] [Google Scholar]