Background: p21-activated kinase 1 (Pak1) is activated by Cdc42 as well as phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2).
Results: PI(4,5)P2 and Cdc42 both contribute to Pak1 membrane recruitment and synergistically activate Pak1 but not another Cdc42 effector, Ack (activated Cdc42-associated kinase).
Conclusion: Pak1 is a coincidence detector regulated by GTPase and PI(4,5)P2 binding.
Significance: Coincidence detection may allow for Pak1 activation independently from other Cdc42 effectors.
Keywords: Cdc42, Phosphoinositides, Phosphorylation, Rac1, Rho GTPases, Ack, Pak1, Kinase Activation
Abstract
Autoinhibited p21-activated kinase 1 (Pak1) can be activated in vitro by the plasma membrane-bound Rho GTPases Rac1 and Cdc42 as well as by the lipid phosphatidylinositol (4,5)-bisphosphate (PIP2). Activator binding is mediated by a GTPase-binding motif and an adjacent phosphoinositide-binding motif. Whether these two classes of activators play alternative, additive, or synergistic roles in Pak1 activation is unknown, as is their contributions to Pak1 activation in vivo. To address these questions, we developed a system to mimic the membrane anchoring of Rho GTPases by creating liposomes containing both PIP2 and a Ni2+-NTA modified lipid capable of binding hexahistidine-tagged Cdc42. We find that among all biologically relevant phosphoinositides, only PIP2 is able to synergistically activate Pak1 in concert with Cdc42. Membrane binding of the kinase was highly sensitive to the spatial density of PIP2 and Pak1 demonstrated dramatically enhanced affinity for Cdc42 anchored in a PIP2 environment. To validate these findings in vivo, we utilized an inducible recruitment system to drive the ectopic synthesis of PIP2 on Golgi membranes, which normally have active Cdc42 but lack significant concentrations of PIP2. Pak1 was recruited to PIP2-containing membranes in a manner dependent on the ability of Pak1 to bind to both PIP2 and Cdc42. These findings provide a mechanistic explanation for the essential role of both phosphoinositides and GTPases in Pak1 recruitment and activation. In contrast, Ack, another Cdc42 effector kinase that lacks an analogous phosphoinositide-binding motif, fails to show the same enhancement of membrane binding and activation by PIP2, thus indicating that regulation by PIP2 and Cdc42 could provide a combinatorial code for activation of different GTPase effectors in different subcellular locations.
Introduction
It is well established that certain lipid species, particularly phosphoinositides, play active roles in cellular signaling events in addition to structural roles comprising cellular membranes (1–4). Phosphoinositides consist of phosphatidylinositol and seven other variously phosphorylated forms that localize to distinct membrane compartments and fulfill different biological functions (1, 2). For example, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2, or PIP2)3 is predominantly found in the cytosolic leaflet of the plasma membrane where it plays numerous roles regulating endocytosis and the actin cytoskeleton (5–7), whereas phosphatidylinositol 4-phosphate (PI(4)P) is localized to and plays important roles at the Golgi membrane (1, 2). Signaling by phosphoinositides is mediated in part by their binding to a variety of phosphoinositide binding domains, including the pleckstrin homology (PH), phox homology (PX), and espin amino-terminal homology domains (1, 2), however, other proteins can bind these acidic phospholipids through so called “basic regions” (8–11), typically unstructured sequences enriched in positively charged amino acids.
Engagement of phosphoinositide-binding motifs can facilitate membrane translocation of signaling proteins (1). In some cases, phosphoinositide binding has been shown to cooperate with binding to other membrane features, including other lipids or GTP-binding proteins (GTPases) in a process referred to as “coincidence detection” (1, 12–14). For example, FAPP is recruited to the trans-Golgi network due to coordinated binding to both PI(4)P and the GTPase Arf1 (15), whereas the neuronal Wiskott-Aldrich syndrome protein (N-WASP) is targeted to the plasma membrane by coordinated binding to PIP2 and the Rho family GTPase Cdc42 (9, 12).
In addition to regulating membrane recruitment, coincident membrane features can also regulate the biological activity of signaling proteins. N-WASP adopts an autoinhibited conformation that negatively regulates its biochemical function: activation of actin filament assembly by the Arp2/3 complex (16, 17). Binding of N-WASP to PIP2 and membrane-bound Cdc42 cooperatively disrupts this autoinhibited conformation and results in potent stimulation of Arp2/3 activity (9, 12, 18, 19). This mechanism allows for complex spatial and temporal regulation of Arp2/3 activity. It has been theorized that signaling information transmitted by phosphoinositides and GTPase-dependent pathways converge on N-WASP leading to Arp2/3 activation only when both upstream pathways are sufficiently active, thus suppressing inappropriate activation that could be triggered by stochastic activation of either pathway alone (9, 12, 19). Furthermore, dual inputs could provide a combinatorial code for N-WASP activation at specific subcellular locations. Although active, GTP-bound Cdc42 is present at both the plasma membrane and intracellular membranes (1), the primary localization of PIP2 to the plasma membrane may ensure that this is predominantly where N-WASP activation occurs. Unfortunately, whereas biochemical aspects of N-WASP regulation by these activators have been examined exhaustively in vitro, the relative importance of these inputs regulating N-WASP or other Cdc42 effectors in cells remains poorly understood.
The serine/threonine kinase Pak1, which plays a fundamental role in cell morphology and proliferation control (20–23), is an effector of the Rho family GTPases Cdc42 and Rac1 (23, 24). Its dysregulation has recently been clearly linked to human cancers including melanoma (25, 26) and breast cancer (27–29). Like N-WASP, Pak1 exists in an autoinhibited conformation that is relieved by GTPase binding (30–33). A basic region was identified in Pak1 that binds to phosphoinositides, particularly PIP2 (8). Pak1 is activated in vitro by binding of its basic region to liposomes containing PIP2 and a Pak1 mutant lacking the basic region fails to be activated in cells stimulated by growth factors, suggesting that PIP2 may be an important regulator of Pak1 in vivo (8). Here, using experimental modulations of phosphoinositide synthesizing enzymes in cells and the development of a novel in vitro system to recapitulate Cdc42 signaling, we demonstrate for the first time a direct role for PIP2 as a regulator of Pak1 in vivo and reveal its mechanism of action. Thus, like N-WASP, Pak1 serves as an integrator of diverse upstream signaling cascades and is likely activated at the plasma membrane based on the concordance of signals mediated there by phosphoinositides and Rho family GTPases. Importantly, we find that another Cdc42 effector, the nonreceptor tyrosine kinase Ack (activated Cdc42-associated kinase) (34), which lacks an analogous basic region, shows no synergy between Cdc42 and PIP2 for membrane recruitment or activation. Thus, more broadly, our findings, together with previous studies, suggest a specificity mechanism for controlling activation of specific effectors in particular subcellular contexts.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant proteins were prepared as previously described: Pak1 (8), 8T-Pak1 (8), GST-Paktide (8), GST (8), GST-Grp1 (35), GST-dual adaptor of phosphotyrosine and 3-phosphoinositides (35), GST-PHPLCδ (35), and GST-sorting nexin-3 (36). Full-length, human Cdc42 Q61L (constitutively active (37)) was cloned into pET28a to append a C-terminal hexahistidine tag, expressed in Escherichia coli and purified according to the manufacturer's protocols (Qiagen). The peptide sequence GAKVIYDFIEKKKKG was fused in-frame to GST in the pGEX 6P-1 vector to create the GST-Acktide construct. It was expressed and purified according to the manufacturer's protocols (Qiagen).
Ack (residues 117–489) was expressed as a GST-tagged protein in Sf9 cells using the Bac-to-Bac system (Invitrogen). The pFastBac HTB vector (Invitrogen) was first modified to replace the His tag with a GST tag followed by a Pre-Scission protease site by cloning the GST tag and the Pre-Scission site from a pGEX 6P-1 vector (GE Healthcare) into the RsrII and NcoI sites of pFastBac HTB. DNA encoding residues 117 to 489 of human Ack was subsequently cloned into the EcoRI and NotI sites of the modified pFastBac vector. The resultant plasmid was transformed into DH10Bac cells (Invitrogen) and baculovirus was produced using the Bac-to-Bac system. Amplified virus was used to infect Sf9 cells and cells were harvested after 65 h. Cells were thawed and resuspended in 50 mm Tris, pH 7.5, 300 mm NaCl, 10% glycerol, 2 mm DTT, 1% Triton, 1 mm Na3VO4, and Complete-EDTA Free (Roche Applied Science), lysed by sonication, and debris was pelleted at 39,000 × g for 30 min. Soluble lysates were incubated with glutathione-Sepharose 4 Fast Flow (GE Healthcare) for 3 h at 4 °C. Beads were washed extensively with lysis buffer, then resuspended in a buffer containing 50 mm Tris, pH 8, 150 mm NaCl, 10% glycerol, 1 mm DTT, and 1 mm Na3VO4. To remove the GST tag, Pre-Scission protease was added, and beads were incubated overnight at 4 °C. Cleaved Ack was recovered from the supernatant by harvesting the beads at 3,000 × g for 10 min. The purified protein was concentrated, brought up to 50% glycerol, and stored at −80 °C.
Liposome Preparation
Stocks of phosphatidylcholine, phosphatidylethanolamine, DGS-NTA(Ni) (Ni2+ salt), phosphatidylinositol (4,5)-bisphosphate, and phosphatidylinositol (Avanti Polar Lipids) were stored in chloroform or chloroform:methanol (2:1) at −80 °C. Appropriate mole ratios of each were added in an amber bottle and dried under nitrogen gas. 2.5 mm liposome stock solutions were prepared by addition of the appropriate volume of lipid buffer (20 mm Hepes, pH 7.5, 300 mm NaCl, 200 mm sucrose, adapted from Ref. 38). Then bottles were sonicated at 4 °C for 20 min. Liposomes were stored at 4 °C and used within 10 days.
In Vitro Kinase Assays
Liposomes were incubated with Cdc42-His in kinase buffer (50 mm Hepes, pH 7.5, 12.5 mm NaCl, 650 μm MgCl2, 650 μm MnCl2 (8)) on ice for 30 min to allow binding to DGS-NTA(Ni). 0.4 μg of full-length WT or 8T-Pak1 or equimolar Ack was added along with excess GST-Paktide/Acktide and kinase buffer to a volume of 14 μl. Reactions were started by addition of ATP (1 μCi of [γ-32P]ATP) to a final concentration of 30 μm. After incubation for 15 min at 30 °C, reactions were stopped by addition of loading buffer and heating to 95 °C for 10 min. Substrate and kinase were separated by SDS-PAGE. Gels were Coomassie-stained and dried. The degree of 32P incorporation was determined by exposing the dried gel to a PhosphorImager plate. Bands were visualized and quantitated using ImageGauge (version 4.0, FUJIFILM). The data were analyzed using GraphPad Prism to determine IC50 values. Liposomes used for kinase assays were always PC:PI:DGS-NTA(Ni) (equal molar PC, PI; 7% DGS-NTA(Ni) with or without 6% phosphoinositide). Concentrations of total lipid used in assays varied from 15 to 500 μm.
Liposome Sedimentation Assays
Liposomes were preincubated in lipid buffer (previously defined) containing 20 mm imidazole for 15 min on ice. Cdc42-His was then added. Complexes were allowed to form on ice for 15 min. Finally, 10 μg of Pak1 (or equimolar Ack), 300 μm ATP, and 5 μg of BSA were added to a volume of 40 μl (final buffer: 20 mm Tris, pH 8.5, 300 mm NaCl, 200 mm sucrose, 20 mm imidazole, adapted from Ref. 38). After 15 min on ice, reactions were spun at 100,000 × g with a Beckman TLA 120.2 rotor at 4 °C. Supernatant was removed and pellets were resuspended with loading buffer. Pak1 was resolved by SDS-PAGE and visualized with silver stain or Western blot (α-Pak1, Invitrogen). Bands were quantitated with ImageJ and graphed with GraphPad Prism. Liposomes used for sedimentation assay were always PC:PE:DGS-NTA(Ni) (equal molar PC, PE; 3% DGS-NTA(Ni), with or without 12% phosphoinositide) and held at a constant 400 μm total lipid per reaction.
Pak1/Cdc42-His Pulldown Assays
10 μg of Pak1, 10 μg of 8T-Pak1 was incubated with Cdc42-His in a final buffer of 20 mm Tris, pH 8.5, 300 mm NaCl, 200 mm sucrose, 20 mm imidazole. Complexes were allowed to form during nutation at 4 °C for 1 h. 15 μl of Ni2+-NTA-agarose (Qiagen) was added to each tube and allowed to mix for an additional 15 min. Resin was pelleted at 3,000 × g and washed with excess buffer. Bound proteins were separated by SDS-PAGE and detected by Western blot with α-Pak1 (Invitrogen). Binding was quantified by ImageJ.
iRap-inducible PIP 5-Kinase Plasmids
The recruitable PIP5K constructs were kindly provided by Dr. Tamas Balla and were generated using the mouse PIP5KIα template and PCR amplifying a segment (1–461) using primers containing PvuI and KpnI sites in the 5′ and 3′ ends, respectively. This fragment was subcloned into the mRFPC1 plasmid for determination of localization and catalytic activity and for verifying the sequence. This construct was then used to generate mutants where the Glu-61 residue was replaced with either a Met or Leu residue. These mutations comprise a Rac1 binding site and make the enzyme mostly cytoplasmic (39), although the constructs still bind to the PM at higher expression levels or longer transfections times (>16–20 h). The wild-type and E61L mutant enzymes were then subcloned into the mRFP-FKBP12 backbone (40) with PvuI/KpnI enzymes to generate the recruitable versions of the proteins.
iRap-inducible Recruitment Assays
HEK293 (ATCC) cells grown on coverslips were transfected with Tgn38-FRB-CFP (Golgi targeting plasmid) and FKBP-PIP5K-mRFP (PIP5 kinase) plasmids in addition to GFP-PHPLCδ (40) or full-length HA-tagged WT-Pak1, 8T-Pak1, or LL-Pak1 (8). HA-tagged LL-Pak1 was kindly provided by Dr. Jonathan Chernoff. Transfections were performed with Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). 24 h following transfection, dimerization was induced by the addition of 5 μm rapamycin or 3-methylindole rapamycin (iRap) for the designated time period. iRap was synthesized by the Organic Synthesis Facility at the Fox Chase Cancer Center. Coverslips were removed and cells were then fixed with formaldehyde and then immunostained with α-Pak1 (Invitrogen) and α-rabbit Alexa 488 (Invitrogen). Images were obtained with the Leica TCS SP5 spectral confocal system with a 68× PlanApo objective. Eighteen cells for each set of triply transfected cells were chosen at random. Using the coloc_2 plugin for Fiji/Image,4 the outline of each cell was defined and a Pearson correlation coefficient was determined for both CFP/mRFP and CFP/Alexa 488 colocalization.
RESULTS
Selective Activation of Pak1 by PIP2
Previous in vitro studies showed a weak potentiating effect of PIP2-containing liposomes on catalytic activation of Pak1 by Rho GTPases (8). However, these studies were conducted with soluble, nonprenylated GTPase, whereas activated Rho GTPases in the cell are anchored via a C-terminal geranylgeranyl group to cellular membranes (43–45). In addition, not all of the naturally occurring phosphoinositides were tested for a role in Pak1 binding or activation (8). To exhaustively investigate the relative roles of phosphoinositides and GTPases in regulating Pak1 in a more physiologically relevant setting, we developed a novel experimental system in which synthetic liposomes of defined lipid composition are generated that incorporate lipids bearing a modified Ni2+-NTA head group (DGS-NTA(Ni)). This modified lipid can bind to recombinant GTPases bearing a C-terminal hexahistidine tag, thus tethering these proteins to the liposome surface in a biologically relevant orientation (schematically represented in Fig. 1) (46–51). Importantly, lipid composition and GTPase density on the external leaflet of these liposomes can be easily and precisely controlled. Indeed, sedimentation assays confirmed approximately stoichiometric binding of Cdc42-His to Ni2+-NTA lipid exposed on the outer face of these liposomes (supplemental Fig. S1). We then synthesized Ni2+-NTA-containing liposomes incorporating each of the seven biologically relevant occurring phosphoinositide species and confirmed the proper presentation of incorporated phosphoinositides by documenting the binding of corresponding recombinant phosphoinositide-binding domains of known specificity in liposome sedimentation assays (35, 36) (supplemental Fig. S2).
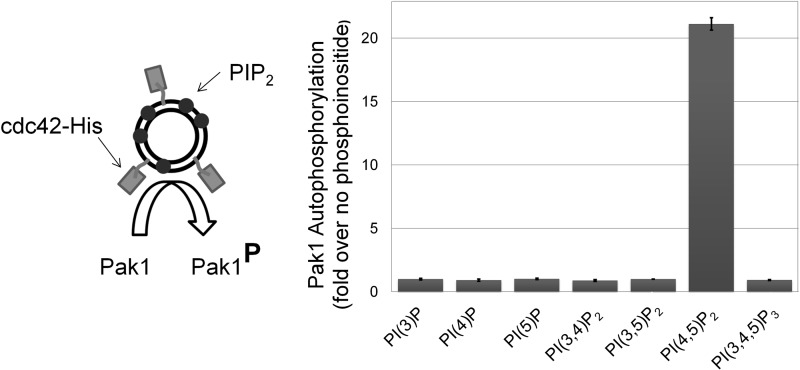
FIGURE 1.
PI(4,5)P2 selectively enhances Cdc42-dependent Pak1 activation. Left, the experimental system. Synthetic liposomes (double black circle) with bound Cdc42-His (rectangles) that either contained phosphoinositide (small dark circles) or not, were incubated with Pak1 and [γ-32P]ATP to monitor phosphoinositide-dependent Pak1 autophosphorylation. Right, Pak1 autophosphorylation in kinase assays with Cdc42-His-bound liposomes (comprised of equimolar PC:PI and containing 7% (by mole) DGS-NTA(Ni) with or without 6% of the indicated phosphoinositides) is plotted normalized to control reactions with Cdc42-containing liposomes without the test phosphoinositide. Pak1 autophosphorylation was quantified by PhosphorImager analysis of dried electrophoretic gels of the reaction products. Mean results are shown from two independent replicates ± S.D.
Next, we comprehensively tested biologically relevant phosphoinositides for their ability to modulate Pak1 activation by liposome-bound Cdc42-His. Ni2+-containing liposomes containing 6% (by molar ratio) of phosphoinositide, or control liposomes with no phosphoinositide, were charged with Cdc42-His and used in a [32P]ATP in vitro kinase assay where Pak1 autophosphorylation was monitored by SDS-PAGE and digital autoradiography. The ability of each phosphoinositide to potentiate Pak1 activation by Cdc42-His was assessed by comparison with control liposomes bearing only GTPase. Strikingly, only liposomes with PI(4,5)P2 showed significant enhancement of Pak1 autophosphorylation compared with liposomes without phosphoinositide (Fig. 1). This experiment demonstrates that among biologically relevant phosphoinositides, PIP2 uniquely promotes Pak1 activation by Cdc42. Furthermore, because PIP2 is the most abundant plasma membrane phosphoinositide (11, 52), these results support a potentially predominant role for this phosphoinositide in Pak1 regulation in cells.
Pak1 Is Sensitive to the Spatial Density of Membrane PIP2 in Vitro and in Vivo
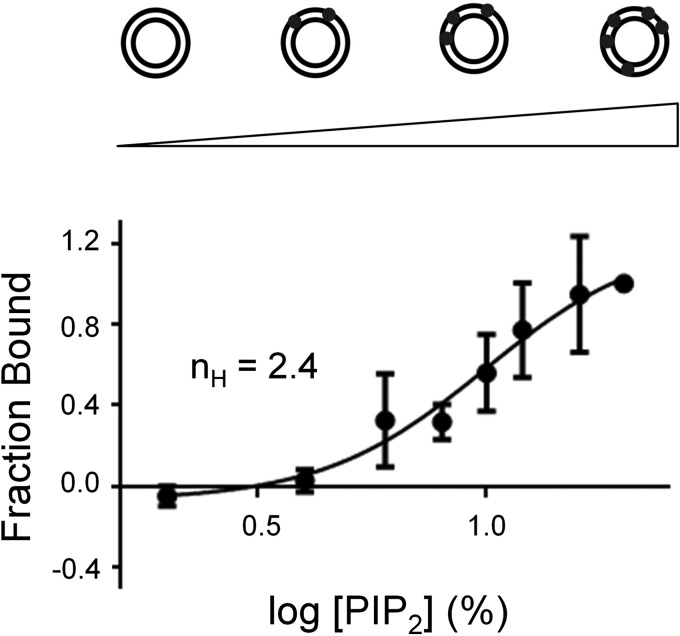
The basic region of Pak1, which is both necessary and sufficient for PIP2 binding (8), is a 25-amino acid region containing nine lysines and three arginines located just N-terminal to the Pak1 Cdc42-binding site (8). Similar positively charged phosphoinositide-binding motifs have been described in other proteins including other Rho GTPases effectors such as N-WASP (9–12). Previous work demonstrated that activation of N-WASP by PIP2 is ultrasensitive with respect to the density of PIP2 in the membrane (9, 12). The N-WASP basic motif contains 15 amino acids of which 10 are positively charged and binds PIP2-containing liposomes of increasing PIP2 density with sigmoidal dose dependence and a Hill coefficient of 3.1 (12). To ask if membrane binding of Pak1 was also sensitive to PIP2 spatial density, we performed liposome sedimentation assays with full-length Pak1 and liposomes in which the mole percent of PIP2 was increased from 0 to 20%. All reactions had a constant total lipid concentration (shown in Fig. 2). We found that Pak1 also bound to PIP2 liposomes with a sigmoidal dependence on PIP2 concentration and with an apparent Hill coefficient of 2.4 (Fig. 2). These data support cooperative, multivalent binding of the basic region of Pak1 to PIP2 as has been observed for the basic region of N-WASP and other phosphoinositide-binding proteins.
FIGURE 2.
Pak1 binding is sensitive to the spatial density of PIP2. Pak1 was incubated with 400 μm PC:PE (equimolar) liposomes containing increasing mole fractions of PIP2 (0–20%). Liposomes were sedimented and bound Pak1 was quantified by Western blotting. Results are shown for five replicate experiments (± S.D.) and the data were fit to a sigmoidal dose-response curve using GraphPad Prism to determine the Hill coefficient (nH).
Prior studies demonstrated an essential role for the basic region in Pak1 membrane recruitment and catalytic activation in cultured cells (8). Together with our data, these findings suggest that PIP2 may regulate Pak1 membrane recruitment in vivo. To test this hypothesis directly, we utilized a rapamycin-based inducible recruitment system to artificially generate PIP2 on membranes of the Golgi apparatus and assessed its impact on membrane recruitment of Pak1. The Golgi apparatus is not thought to harbor a significant fraction of cellular PIP2, although it is enriched in its biological precursor PI(4)P, which is phosphorylated by phosphatidylinositol 4-phosphate 5-kinase (PIP5K) to generate PIP2. In addition, Golgi membranes are well known to contain GTP-bound Cdc42 on their cytoplasmic leaflet (53–59). Interestingly, however, this pool of activated Cdc42 has not been reported to recruit and activate Pak1, which is instead predominantly localized to the cytosol and plasma membrane (60, 61). Thus, we speculated that limiting PIP2 concentrations in the Golgi membrane may prevent the recruitment of Pak1 by Golgi-localized Cdc42. We sought to test this idea by artificially generating PIP2 at Golgi membranes and monitoring Pak1 recruitment and its dependence on PIP2 and Cdc42 binding.
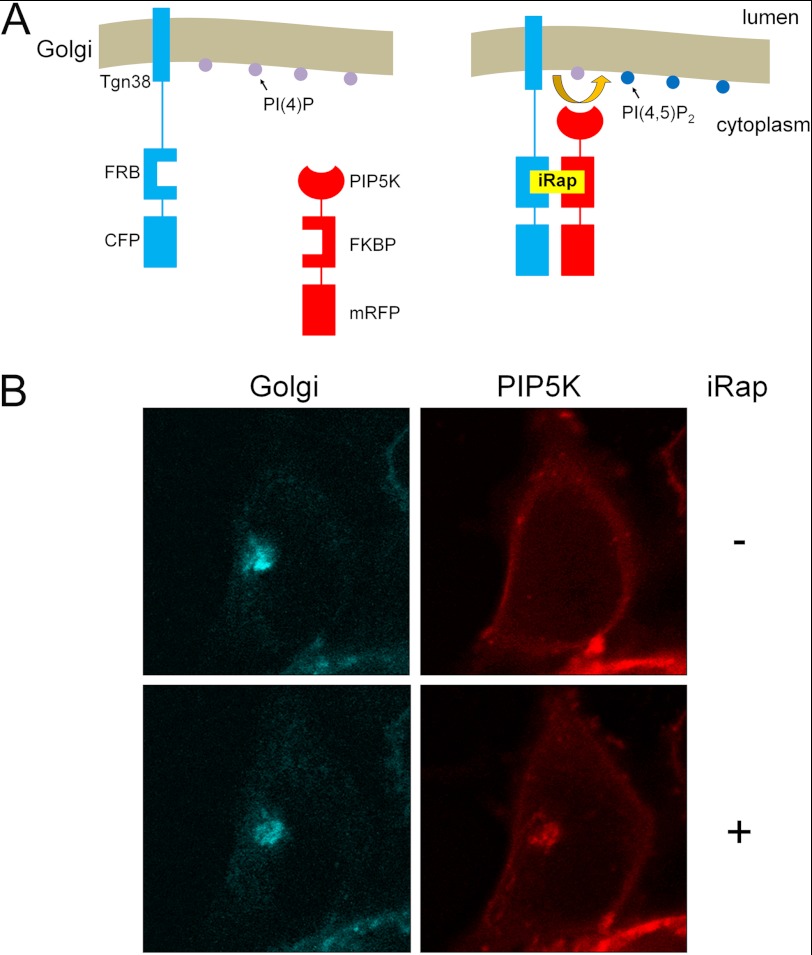
HEK293 cells were co-transfected with plasmids encoding the FRB domain of mammalian target of rapamycin fused to the Golgi targeting sequence of the type 1 transmembrane Golgi-resident protein Tgn38 (42, 62) (cyan fluorescent protein labeled) and the FKBP domain of FKBP12-rapamycin-associated protein fused to PIP5K (red fluorescent protein labeled) (Fig. 3A). Tgn38-FRB exhibited perinuclear Golgi localization and FKBP-PIP5K was diffusely distributed in the cytoplasm before iRap treatment (Fig. 3B). Following rapamycin-induced dimerization for 5 min, however, PIP5K showed strong colocalization with FRB (Fig. 3B, right).
FIGURE 3.
A system for rapamycin-inducible recruitment of PIP5K to the Golgi membrane for localized PIP2 synthesis. A, schematic diagram: a fusion protein where the transmembrane region of Tgn38 anchors the FRB domain of mammalian target of rapamycin (labeled with cyan fluorescent protein, CFP) to the Golgi where PI(4)P (purple circles) is present. Rapamycin or its analog 3-methylindole rapamycin (iRap) triggers binding and recruitment of a cytosolic fusion protein containing the FKBP binding domain of FKBP12-rapamycin-associated protein (FRAP) fused to PIP5K (and labeled with monomeric red fluorescent protein, mRFP) to Golgi-localized Tgn38-FRB. Golgi recruitment of PIP5K allows for localized phosphorylation of PI(4)P to PIP2 (blue circles). B, HEK293 cells were cotransfected with plasmids encoding Tgn38-FRB-CFP and PIP5K-FKB-mRFP. Transfected cells were stimulated with iRap for 5 min to induce dimerization. CFP and mRFP were visualized by fluorescence microscopy.
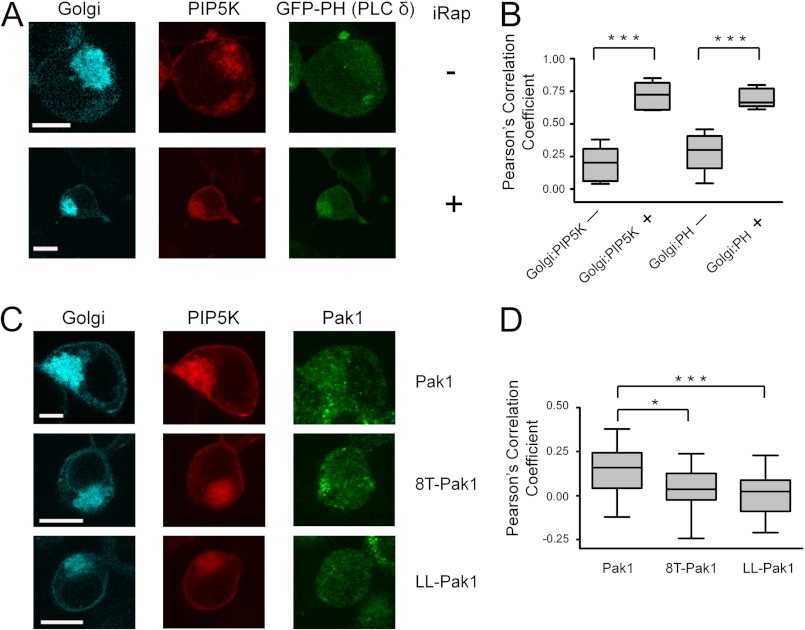
Next, we assessed whether Golgi-targeted PIP5K was capable of phosphorylating Golgi PI(4)P to PIP2. We monitored PIP2 production with a widely used PIP2 biosensor, the pleckstrin homology domain of phospholipase C δ (GFP-PHPLCδ, green fluorescent protein labeled). In addition, we utilized the rapamycin analog 3-methylindole rapamycin (iRap) with improved dimerization activity in cells. Triple transfected cells were stimulated with iRap and transfected proteins were visualized by confocal microscopy of fixed cells. As expected, stimulation with iRap triggered a dramatic relocalization of PIP5K that overlapped with FRB on perinuclear Golgi membranes (Fig. 4A). Importantly, the PIP2 biosensor, GFP-PHPLCδ, was also strongly co-recruited to the Golgi. Quantitative analysis of the Pearson correlation coefficients for these proteins from multiple cells confirmed highly significant iRap-dependent synthesis of PIP2 at the Golgi (Fig. 4B). Thus, this novel system allows for inducible PIP2 generation at an endomembrane system that normally has low concentrations of this phosphoinositide.
FIGURE 4.
Pak1 recruitment to the Golgi membrane is enhanced by Golgi-localized PIP2 and Cdc42. A, HEK293 cells were cotransfected with plasmids encoding Tgn38-FRB-CFP (Golgi targeting protein), PIP5K-FKBP-mRFP (PIP5 kinase), and a GFP-tagged PIP2-specific binding protein (GFP-PH(PLCδ)). Transfected cells were stimulated by iRap for 20 min to induce FRB-FKBP dimerization (bottom panels). The white bars indicate 10 μm. B, Pearson correlation coefficients for the cellular colocalization of FRB and PIP5K were calculated for 5 randomly selected cells before (−) or after (+) 20 min of iRap treatment; colocalization of FRB and GFP-PH before (−) and after (+) iRap treatment was analyzed similarly. The iRap-stimulated colocalization of these proteins is highly significant for both FRB/PIP5K (***, p = 0.0007) and FRB/GFP-PH (***, p = 0.0001). The distribution of Pearson coefficients in each case are shown as a box and whisker plot, where whiskers represent the maximum and minimum values, the box represents the 25th to 75th percentiles, and the central line depicts the median. C, HEK293 cells were transfected with Tgn38-FRB-CFP, PIP5K-FKBP-mRFP, and HA-tagged Pak1, 8T-Pak1 (cannot bind PIP2), or LL-Pak1 (cannot bind Cdc42). Cells were treated for 20 min with iRap and then immunostained for Pak1 using a total Pak1 antibody (Invitrogen). The white bars indicate 10 μm. D, for each condition, 18 cells were randomly chosen and Pearson correlation coefficients were determined for colocalization of Pak1 and FRB. The distribution of Pearson correlation coefficients is shown as box and whisker plots as in B. *, p = 0.0148 and ***, p = 0.0009.
Having validated the experimental system, we next repeated this experiment by replacing the PIP2 reporter plasmid with wild-type and mutant forms of Pak1. Following iRap addition, Pearson correlation analysis revealed modest colocalization of wild-type Pak1 and the Golgi-anchored FRB, consistent with a direct role for PIP2 in the membrane recruitment of Pak1 (Fig. 4, C and D). To confirm that this PIP2-dependent recruitment was mediated by direct binding to the Pak1 basic region, we also tested 8T-Pak1, a mutant deficient in phosphoinositide binding in which 8 lysine residues in the Pak1 basic region are simultaneously mutated to threonine (8). As expected, 8T-Pak1 showed significantly weaker colocalization to the Golgi membranes (p = 0.0148). Finally, we tested LL-Pak1, a mutant that abolishes Pak1 binding to Rho GTPases (21). Strikingly, LL-Pak1 also exhibited significantly weaker Golgi recruitment following iRap treatment (p = 0.0009, Fig. 4, C and D). Taken together, these data support a specific role for PIP2 in Pak1 membrane recruitment in live cells and, importantly, demonstrate that both PIP2 binding and Rho GTPase binding are required to mediate full Pak1 recruitment.
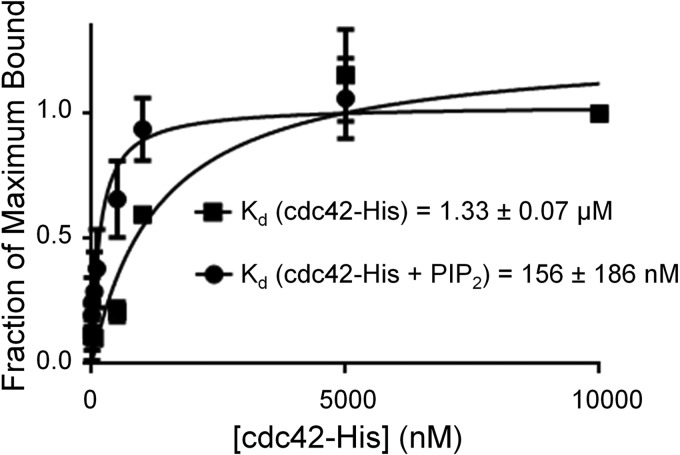
Because the GTPase-binding domain of Pak1 and its basic region lie adjacent to one other (8), we next tested whether the binding of one enhanced the binding to the other. To do so we used liposome sedimentation assays to measure the binding of Pak1 to Cdc42-bound liposomes that either contained PIP2 or did not. Although Pak1 bound Cdc42-bound liposomes without PIP2 with an apparent affinity of 1.33 μm, PIP2 enhanced the apparent affinity of liposome binding by approximately 1 order of magnitude to 0.16 μm (Fig. 5). This suggests that simultaneous binding of the basic region to PIP2 and Cdc42 to the adjacent GTPase binding motif may enhance the affinity of Pak1 for such liposomes. We cannot rule out the alternative possibility, however, that PIP2 in some way orients Cdc42 at the membrane to enhance its affinity for Pak1. Overall our results support a model in which PIP2 plays a biologically important role together with Rho GTPases in membrane binding of Pak1.
FIGURE 5.
The presence of PIP2 in the membrane increases the affinity of Pak1 for Cdc42 bound to liposomes. Cosedimentation of Pak1 with liposomes (equimolar PC:PE, 3% DGS-NTA(Ni)) was assessed in the presence or absence of 12% PIP2 by titrating the reactions with increasing concentrations of Cdc42-His. The data were fit in GraphPad Prism to derive apparent affinities (shown). The average ± S.D. of duplicates is shown.
Pak1 Is Synergistically Activated by Cdc42 and PIP2
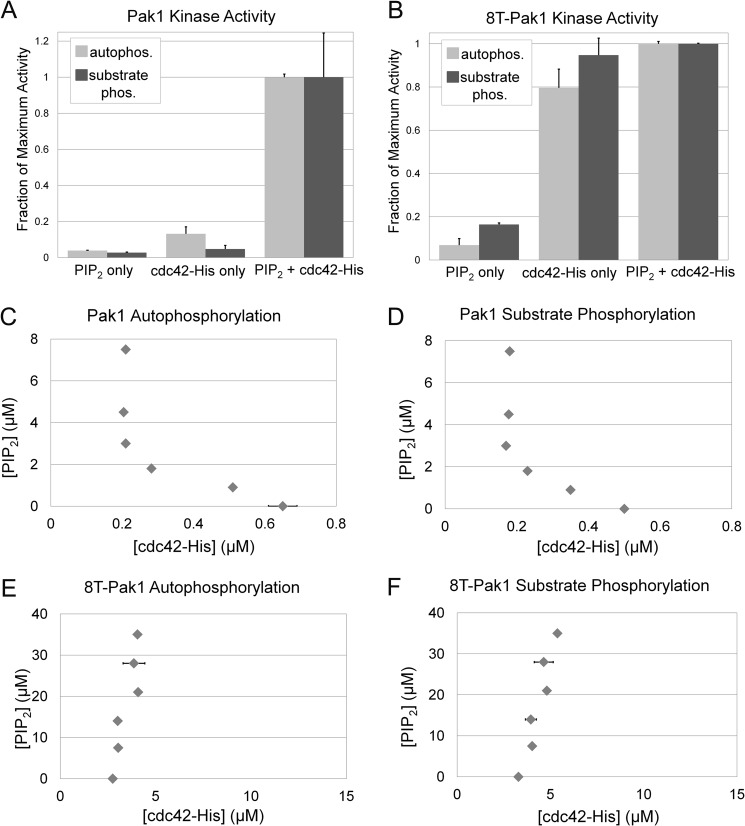
Our previous work established that PIP2 can modestly potentiate catalytic activity in vitro by soluble Rac1 (8), however, the experiments reported here suggested highly synergistic binding interactions between Pak1 and PIP2-containing liposomes bearing Cdc42-His (Figs. 2 and 5). In addition, we observed striking synergistic activation of Pak1 in this system (Fig. 1). Because Rho GTPases are found prenylated at the plasma membrane and our liposome system mimics those conditions, we sought to further explore the synergistic activation of Pak1. We first conducted dose-response experiments in which Cdc42-His was titrated into reactions containing a fixed concentration of PIP2 liposomes (50 μm total lipid, 6% PIP2) containing sufficient DGS-NTA(Ni) lipid to quantitatively bind all added Cdc42-His. These reactions were then incubated with Pak1, [32P]ATP, and peptide substrate of Pak1 (GST-Paktide) to allow Pak1 to undergo autophosphorylation and phosphorylate GST-Paktide. Reaction products were analyzed by SDS-PAGE and digital autoradiography. As expected, the dose-response data revealed a steep dose dependence on Cdc42-His concentration for wild-type Pak1, consistent with a cooperative interaction of Pak1 by Cdc42-His and PIP2 (supplemental Fig. S3A). To obtain further evidence of synergy, we next assessed the individual contributions of Cdc42-His and PIP2 to Pak1 activation. A combination of 3 μm PIP2 (50 μm total lipid, 6% PIP2 by mole percent) and 200 nm Cdc42-His led to maximum activation of Pak1 (supplemental Fig. S3A). We then compared this catalytic activity with the kinase incubated with either activator alone: (a) 3 μm PIP2 in Ni2+-NTA containing liposomes without Cdc42-His, (b) 200 nm Cdc42-His bound to control liposomes lacking PIP2, or (c) 200 nm Cdc42-His bound to liposomes containing PIP2. In each case, [32P]ATP kinase assays were utilized to measure Pak1 autophosphorylation and substrate phosphorylation separately. Strikingly, Pak1 autophosphorylation was enhanced ∼21-fold in the presence of both activators relative to the sum of each activator on its own. Similarly dramatic results were observed for substrate phosphorylation (Fig. 6A).
FIGURE 6.
Pak1 catalytic activity is synergistically enhanced by Cdc42 and PIP2. A, [γ-32P]ATP kinase assays were conducted with Pak1 in the presence of the indicated activators and Pak1 catalytic activity was measured by either Pak1 autophosphorylation or Pak1 phosphorylation of a peptide substrate (GST-Paktide). Activators were either PIP2-containing liposomes (PC:PI:PIP2:DGS-NTA(Ni), 43.5:43.5:6:7 mole ratio) or control liposomes (PC:PI:DGS-NTA(Ni), 46.5:46.5:7 molar ratio) with 200 nm Cdc42-His or their combination. These activator concentrations were selected on the basis of their ability to maximally activate Pak1 when combined (supplemental Fig. S3A). Results are normalized to phosphorylation observed in the reaction containing both activators and are shown for two independent replicates ± S.D. B, activation for 8T-Pak1 was measured as in A. Note that higher concentrations of PIP2 (500 μm total lipid and 15 μm Cdc42-His) were required to achieve full activation of 8T-Pak1 (supplemental Fig. S3B). C, 50% activation contour for Pak1 autophosphorylation. The concentration of Cdc42-His required to achieve 50% activation of Pak1 was determined at each indicated PIP2 concentration. Horizontal error bars (smaller than the data symbol in most cases) represent the S.D. calculated from two replicates. The curve that can be drawn through these points represents all combinations of PIP2 and Cdc42-His that lead to 50% activation of Pak1. Table 1 presents the data used to generate the plots in C–F. D, Pak1 catalytic activity in the reactions in C was also measured by phosphorylation of a Pak1 substrate peptide included in the reaction and the results are presented as a 50% activation contour. The significant departure from linearity of the contour is indicative of synergistic activation. 8T-Pak1 autophosphorylation (E) and 8T-Pak1 substrate phosphorylation (F) is not enhanced by PIP2. The 50% activation contours were determined as in C and D and the data are summarized in Table 1. The vertical nature of the 50% activation contour demonstrates a lack of sensitivity to PIP2.
To confirm that this synergy was due to basic region-mediated binding to PIP2, the same set of experiments was repeated using the 8T-Pak1 mutant. As expected, dose-response curves revealed less cooperativity than wild-type Pak1 (supplemental Fig. S3B). Maximal catalytic activity of 8T-Pak1 was observed in the combined presence of 30 μm PIP2 and 15 μm Cdc42-His (supplemental Fig. S3B) and, unsurprisingly, the degree of catalytic activity observed for both inputs together was only slightly greater than the additive effect of each activator on its own, demonstrating that PIP2 binding to the basic region is required for the synergistic activation of wild-type Pak1 by the combination of phosphoinositide and GTPase (Fig. 6B).
Surprisingly, 8T-Pak1 was significantly less sensitive to Cdc42-bound liposomes as a single agent compared with wild-type Pak1. Prior studies showed that 8T-Pak1 is activated comparably to wild-type Pak1 by soluble, nonprenylated Cdc42-His (8) and we confirmed equivalent binding of wild-type and 8T-Pak1 to soluble Cdc42-His (supplemental Fig. S4). By contrast, we consistently observed significantly weaker activation of 8T-Pak1 by liposome-bound Cdc42 than wild-type Pak1. This unexpected finding, together with the observed sensitivity to PIP2 spatial density (Figs. 4 and 5) suggests that the Pak1 basic region may also promote Pak1-Cdc42 binding independent of its role in PIP2 binding in the physiological context of the membrane.
To comprehensively investigate the synergy between PIP2 and Cdc42 in Pak1 activation, we conducted dose-response experiments in which Cdc42-His was titrated into Pak1 kinase assays in the presence of PIP2/Ni2+-NTA-containing liposomes (see supplemental Fig. S3). The concentration of PIP2 in those liposomes was systematically varied and the concentration of Cdc42-His required for 50% Pak1 activation at each PIP2 concentration was determined. The experimentally determined IC50 values are listed in Table 1. To visualize the results, we plotted a 50% activation contour on a graph of PIP2 concentration versus Cdc42-His IC50 (12), in which each plotted point denotes a pair of concentrations of Cdc42-His and PIP2 yielding 50% of maximal Pak1 activation. Separate contours were prepared quantifying Pak1 autophosphorylation (Fig. 6C) and substrate phosphorylation (Fig. 6D). The resulting contours reflect the relative importance of each activator alone in regulating Pak1 catalytic activity. Although an additive interaction would be expected to produce an approximately linear contour, the striking concavity of the contour is consistent with the synergy observed in Fig. 6A. Approximately 0.6 μm Cdc42-His is required for 50% Pak1 activation in the absence of PIP2, but 3-fold less Cdc42-His is required in the presence of 3 μm PIP2. Similarly, as the Cdc42-His concentration decreases, increasing PIP2 must compensate to achieve similar levels of Pak1 activity. We were unable to extend this contour to very low concentrations of Cdc42-His because no concentration of PIP2 was able to support 50% Pak1 activation below ∼0.2 μm Cdc42-His. Thus, PIP2 serves as a potent modifier of Cdc42-dependent activation, enhancing Pak1 sensitivity to Cdc42, without a significant ability to activate Pak1 alone. The synergy we observe at moderate levels of each activator likely explains the requirement for both PIP2 and GTPase for Pak1 recruitment to the Golgi in cells (Fig. 4).
TABLE 1.
Quantitative analysis of Pak1 and 8T-Pak1 activation by PIP2 and Cdc42-His
This table represents IC50 values for Pak1 and 8T-Pak1 activation by Cdc42-His in the presence of the indicated concentration of PIP2. PIP2 concentrations were titrated by increasing total liposome concentration rather than the density of PIP2 in individual liposomes. These data are presented graphically in Fig. 6, C-F. The single and double asterisks indicate data sets shown in full for illustrative purposes in supplemental Figs. 3, A and B, respectively.
| Total lipid | PIP2 | IC50 (Cdc42-His) |
|
|---|---|---|---|
| Pak1 autophosphorylation | GST-Paktide phosphorylation | ||
| μm | μm | μm | |
| Pak1 | |||
| 0 | 0 | 0.65 ± 0.16 | 0.50 ± 0.02 |
| 15 | 0.9 | 0.51 ± 0.03 | 0.35 ± 0.01 |
| 30 | 1.8 | 0.282 ± 0.001 | 0.23 ± 0.01 |
| 50* | 3 | 0.21 ± 0.02 | 0.17 ± 0.02 |
| 75 | 4.5 | 0.204 ± 0.004 | 0.1775 ± 0.0004 |
| 125 | 7.5 | 0.21 ± 0.01 | 0.18 ± 0.03 |
| 8T-Pak1 | |||
| 0 | 0 | 2.75 ± 0.38 | 3.29 ± 0.26 |
| 125 | 7.5 | 3.04 ± 0.19 | 4.02 ± 0.27 |
| 200 | 12 | 3.01 ± 0.04 | 3.95 ± 0.59 |
| 300 | 18 | 4.08 ± 0.17 | 4.81 ± 0.12 |
| 400 | 24 | 3.86 ± 1.14 | 4.64 ± 1.01 |
| 500** | 30 | 4.05 ± 0.26 | 5.38 ± 0.04 |
A similar study was conducted with 8T-Pak1 (Fig. 6, E and F). As expected, these contours do not show similar concavity due to the insensitivity of this basic region mutant to PIP2 and supports the loss of synergy observed in Fig. 6B. Instead, Cdc42-His concentrations required for 50% Pak1 activation are similar or even higher as PIP2 concentrations increase. Thus, the synergistic activation observed for wild-type Pak1 is indeed due to PIP2 binding to the Pak1 basic region.
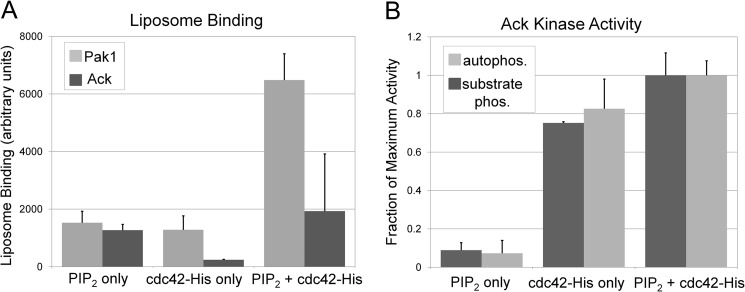
Finally, to address whether synergy between PIP2 and Cdc42 is relevant for other Cdc42 effectors or whether it could be a mechanism to selectively activate some Cdc42 effectors but not others in PIP2-enriched membrane contexts, we assessed the impact of PIP2 on membrane recruitment and kinase activity of a different Cdc42 effector kinase, Ack. Unlike Pak1 and N-WASP, the amino acid sequence of Ack contains no analogous basic region adjacent to the Cdc42-binding motif. We prepared recombinant Ack protein comprising the kinase, SH3, and Cdc42-binding domains. Binding of Ack and Pak1 to liposomes with or without PIP2 and Cdc42 was compared (Fig. 7A). Overall binding of Ack to both PIP2 and Cdc42-His was weaker than Pak1 and Ack displayed additive binding to liposomes containing both PIP2 and Cdc42-His. This was in striking contrast to the synergistic binding of Pak1 (Figs. 5 and 7A). Ack kinase activity was also measured in a radioactive kinase assay for its stimulation by Cdc42 and PIP2 (Fig. 7B). Although Pak1 was synergistically activated by PIP2 and Cdc42 (Fig. 6A), Ack activation was merely additive (Fig. 7B) and similar to the 8T mutant of Pak1 (Fig. 6B).
FIGURE 7.
Ack binding and catalytic activity is not enhanced by PIP2. A, cosedimentation of Pak1 and Ack with liposomes (equimolar PC:PE, 3% DGS-NTA(Ni)) was assessed in the presence or absence of 5 μm Cdc42-His and 12% PIP2. Equimolar amounts of Pak1 and Ack were used. B, [γ-32P]ATP kinase assays were conducted with Ack in the presence of the indicated activators and Ack catalytic activity was measured by either Ack autophosphorylation or Ack phosphorylation of a peptide substrate (GST-Acktide). Activators were either PIP2-containing liposomes (PC:PI:PIP2:DGS-NTA(Ni), 43.5:43.5:6:7 mole ratio), or control liposomes (PC:PI:DGS-NTA(Ni), 46.5:46.5:7 molar ratio) with 300 nm Cdc42-His or their combination. These activator concentrations were selected on the basis of their ability to maximally activate Ack when combined. Results are normalized to phosphorylation observed in the reaction containing both activators and are shown for two independent replicates ± S.D.
DISCUSSION
The Rho GTPases Rac1 and Cdc42 are found anchored to cellular membranes where they are able to activate downstream effector proteins (1). It was previously reported that PIP2 has a weak, synergistic effect on Pak1 activation by Rac1, however, these experiments were performed with constitutively active GTPase and separate PIP2-containing liposomes (8). Intriguingly, cellular studies suggest an absolutely essential role in vivo for PIP2 binding on Pak1 activation (8). To reconcile these findings we developed an in vitro system in which Cdc42 was anchored in its native orientation on the surface of engineered liposomes and used it to measure the impact of phosphoinositides on Cdc42-dependent activation of Pak1 in a more physiologically relevant assay. Our most important discovery is that Pak1 is activated in a highly synergistic manner by PIP2 and Cdc42. This finding provides a mechanistic basis for the in vivo dependence of kinase on PIP2 for activation. Pak1 that is insensitive to PIP2 (8T-Pak1) is unable to be activated in cells despite the presence of active GTPases (8). Furthermore, we validated the specific in vivo role for PIP2 in Pak1 regulation by demonstrating that ectopic synthesis of PIP2 at the Golgi membrane promotes Pak1 recruitment in a manner dependent on the ability of Pak1 to bind both PIP2 and Cdc42. These data demonstrate for the first time an in vivo role for phosphoinositides in Pak1 activation, that PIP2 is the only phosphoinositide able to fulfill this function, and that both PIP2 and Cdc42 are required for Pak1 regulation. Thus, our results identify Pak1 as an integrator of information from phosphoinositide and GTPase-dependent signaling pathways.
We report here the development of two novel experimental systems with broad utility. In the first, we utilized DGS-NTA(Ni) functionalized lipids to immobilize Cdc42 in a signaling-competent manner on the surface of synthetic liposomes. Given the importance of phosphoinositides in the recruitment and regulation of many Ras superfamily GTPases (5), this highly controlled system could find broad use in the elucidation of membrane-associated GTPase signaling. Second, we employed a rapamycin-inducible recruitment system to drive the localization of PIP5K to the Golgi membrane for localized synthesis of PIP2 where it is not normally enriched. To our knowledge, this is the first example of an experimental system to probe PIP2 function in the cellular context using such a gain of function strategy at the Golgi membrane. Here, we applied this tool to demonstrate a specific and functional in vivo role for PIP2 in Pak1 localization.
The cooperative regulation of Pak1 kinase activity by phosphoinositides and GTPases is an evolutionarily ancient regulatory mechanism. Cla4, a Saccharomyces cerevisiae Pak1 orthologue, is coordinately regulated by Cdc42 and phosphoinositides, however, this interaction is mediated by a PH domain, rather than a basic-rich region and is specific for PI(4)P (13) rather than PIP2. Similarly, Schizosaccharomyces pombe Pak2b contains tandem PH-p21 binding domain domains and both are essential for Pak2p function in vivo (41). Like Pak1, STE20 kinase from Saccharomyces cerevisiae contains a basic-rich region preceding its p21-protein binding domain and requires both domains for biological function, although there does not appear to be a specific phosphoinositide requirement because a variety of lipid binding domains can functionally substitute for the basic region (38). Here, we extend these prior studies in yeast to human Pak1 and elucidate the molecular details of these interactions. Importantly, human Pak2 and Pak3 also share similar domain organization with Pak1, suggesting that the regulatory mechanisms defined here may also apply to other Group I Paks (8).
Pak1 activation is mediated by trans-autophosphorylation of one dimer subunit by another (31). Such an autocatalytic, feed-forward type mechanism is potentially prone to dramatic amplification. The requirement for two distinct activators for kinase activation may provide a mechanism to prevent inappropriate triggering of a Pak1 activation cascade by transient, stochastic increases in PIP2 or active GTPase levels. Indeed, noise dampening is an inherent feature of coincidence detectors in biological systems (19). Spatially coordinating phosphoinositide and GTPase signals to converge on Pak1 activation also ensures that the kinase is activated only in certain subcellular localizations at appropriate times.
Signaling pathways have evolved a variety of mechanisms to avoid inappropriate cross-talk between pathways that share common components. GTPases like Cdc42 regulate many diverse downstream effector proteins and an important gap in our understanding is how specific effector pathways are activated when needed, whereas other effector pathways are suppressed. Regulation of Pak1 by PIP2 and Cdc42 suggests one possible resolution for this problem through combinatorial control by multiple activators. According to this model, Pak1 recruitment and activation would be restricted to Rho GTPases active in a PIP2-rich environment, such as the plasma membrane, whereas GTPases localized to the Golgi membrane, normally lacking in large concentrations of PIP2, would selectively activate other downstream effectors. Such a specificity mechanism would allow for selective activation of discrete GTPase effectors in distinct spatial or temporal contexts. Indeed, this concept is strongly supported by our finding that recruitment and activation of Ack, another Cdc42 kinase effector, does not show PIP2-mediated synergy. The fact that effectors of other GTPases, such as FAPP, oxysterol binding protein, and the adaptor complex AP-1, also require both GTPases as well as certain phosphoinositides (1), suggests that this mechanism is a common regulatory theme in GTPase signaling.
Acknowledgments
We gratefully acknowledge T. Balla for providing plasmid constructs for inducible recruitment of PIP5K to the Golgi prior to their publication. We thank J. Chernoff and A. Andrews for comments on the manuscript and A. O'Reilly for use of the confocal microscope.
This work was supported, in whole or in part, by National Institutes of Health Grants GM083025 (to J. R. P.), T32 CA009035 (to K. Malecka), and P30 CA006927 to the Fox Chase Cancer Center. Additional support was provided by the FCCC Keystone Program in Head & Neck Cancer.

This article contains supplemental Figs. S1–S4.
D. J. White, T. Kazimiers, and J. Schindelin, ImageJ software.
- PIP2
- phosphatidylinositol (4,5)-bisphosphate
- Pak1
- p21-activated kinase
- PI(4)P
- phosphatidylinositol 4-phosphate
- PH
- pleckstrin homology domain
- PX
- phox homology domain
- N-WASP
- neuronal Wiskott-Aldrich syndrome protein
- PIP5K
- phosphatidylinositol 5-phosphate kinase
- iRap
- 3-methylindole rapamycin
- Grp1
- general receptor for 3-phosphoinositides
- FAPP
- four-phosphate-adaptor protein
- FRB
- FKBP-rapamycin binding domain
- FKBP
- FK506-binding protein
- Tgn38
- trans-Golgi network protein 38
- DGS-NTA(Ni)
- 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl]-nitrilotriacetic acid (nickel salt)
- Ack
- activated Cdc42-associated kinase
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol.
REFERENCES
- 1. Carlton J. G., Cullen P. J. (2005) Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 15, 540–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 3. Groves J. T., Kuriyan J. (2010) Molecular mechanisms in signal transduction at the membrane. Nat. Struct. Mol. Biol. 17, 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Balla T., Szentpetery Z., Kim Y. J. (2009) Phosphoinositide signaling. New tools and insights. Physiology 24, 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Heo W. D, Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. (2006) PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 314, 1458–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van den Bout I., Divecha N. (2009) PIP5K-driven PtdIns(4,5)P2 synthesis. Regulation and cellular functions. J. Cell Sci. 122, 3837–3850 [DOI] [PubMed] [Google Scholar]
- 7. McLaughlin S., Murray D. (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 8. Strochlic T. I., Viaud J., Rennefahrt U. E., Anastassiadis T., Peterson J. R. (2010) Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol. Cell 40, 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Papayannopoulos V., Co C., Prehoda K. E., Snapper S., Taunton J., Lim W. A. (2005) A polybasic motif allows N-WASP to act as a sensor of PIP2 density. Mol. Cell. 17, 181–191 [DOI] [PubMed] [Google Scholar]
- 10. Wang J., Arbuzova A., Hangyás-Mihályné G., McLaughlin S. (2001) The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 276, 5012–5019 [DOI] [PubMed] [Google Scholar]
- 11. McLaughlin S., Wang J., Gambhir A., Murray D. (2002) PIP(2) and proteins. Interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31, 151–175 [DOI] [PubMed] [Google Scholar]
- 12. Prehoda K. E., Scott J. A., Mullins R. D., Lim W. A. (2000) Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science 290, 801–806 [DOI] [PubMed] [Google Scholar]
- 13. Wild A. C., Yu J. W., Lemmon M. A., Blumer K. J. (2004) The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 279, 17101–17110 [DOI] [PubMed] [Google Scholar]
- 14. Koronakis V., Hume P. J., Humphreys D., Liu T., Hørning O., Jensen O. N., McGhie E. J. (2011) WAVE rgulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U. S. A. 108, 14449–14454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Godi A., Di Campli A., Konstantakopoulos A., Di Tullio G., Alessi D. R., Kular G. S., Daniele T., Marra P., Lucocq J. M., De Matteis M. A. (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol. 6, 393–404 [DOI] [PubMed] [Google Scholar]
- 16. Hemsath L., Dvorsky R., Fiegen D., Carlier M. F., Ahmadian M. R. (2005) An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome protein. Mol. Cell 20, 313–324 [DOI] [PubMed] [Google Scholar]
- 17. Cory G. O., Cramer R., Blanchoin L., Ridley A. J. (2003) Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol. Cell 11, 1229–1239 [DOI] [PubMed] [Google Scholar]
- 18. Rohatgi R., Ho H. Y., Kirschner M. W. (2000) Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol. 150, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prehoda K. E., Lim W. A. (2002) How signaling proteins integrate multiple inputs. A comparison of N-WASP and Cdk2. Curr. Opin. Cell Biol. 14, 149–154 [DOI] [PubMed] [Google Scholar]
- 20. Arias-Romero L. E., Chernoff J. (2008) A tale of two Paks. Biol. Cell 100, 97–108 [DOI] [PubMed] [Google Scholar]
- 21. Sells M., Knaus U. G., Bagrodia S., Ambrose D., Bokoch G. M., Chernoff J. (1997) Human p21-activated kinase (Pak1) regulates actin organization in mammlian cells. Curr. Biol. 7, 202–210 [DOI] [PubMed] [Google Scholar]
- 22. Manser E., Huang H. Y., Loo T. H., Chen X. Q., Dong J. M., Leung T., Lim L. (1997) Expression of constitutively active α-PAK reveals effects of the kinase on actin and focal complexes. Mol. Cell. Biol. 17, 1129–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bokoch G. M. (2003) Biology of the p21-activated kinases. Annu. Rev. Biochem. 72, 743–781 [DOI] [PubMed] [Google Scholar]
- 24. Manser E., Leung T., Salihuddin H., Zhao Z. S., Lim L. (1994) A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367, 40–46 [DOI] [PubMed] [Google Scholar]
- 25. Jung I. D., Lee J., Lee K. B., Park C. G., Kim Y. K., Seo D. W., Park D., Lee H. W., Han J. W., Lee H. Y. (2004) Activation of p21-activated kinase 1 is required for lysophosphatidic acid-induced focal adhesion kinase phosphorylation and cell motility in human melanoma A2058 cells. Eur. J. Biochem. 271, 1557–1565 [DOI] [PubMed] [Google Scholar]
- 26. Pavey S., Zuidervaart W., van Nieuwpoort F., Packer L., Jager M., Gruis N., Hayward N. (2006) Increased p21-activated kinase-1 expression is associated with invasive potential in uveal melanoma. Melanoma Res. 16, 285–296 [DOI] [PubMed] [Google Scholar]
- 27. Yi C., Maksimoska J., Marmorstein R., Kissil J. L. (2010) Development of small-molecule inhibitors of the group I p21-activated kinases, emerging therapeutic targets in cancer. Biochem. Pharmacol. 80, 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arias-Romero L. E., Chernoff J. (2010) p21-activated kinases in ErbB2-positive breast cancer. Small GTPases 1, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Arias-Romero L. E., Villamar-Cruz O., Pacheco A., Kosoff R., Huang M., Muthuswamy S. K., Chernoff J. (2010) A Rac-Pak signaling pathway is essential for ErbB2-mediated transformation of human breast epithelial cancer cells. Oncogene 29, 5839–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Knaus U. G., Wang Y., Reilly A. M., Warnock D., Jackson J. H. (1998) Structural requirements for PAK activation by Rac GTPases. J. Biol. Chem. 273, 21512–21518 [DOI] [PubMed] [Google Scholar]
- 31. Pirruccello M., Sondermann H., Pelton J. G., Pellicena P., Hoelz A., Chernoff J., Wemmer D. E., Kuriyan J. (2006) A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases. J. Mol. Biol. 361, 312–326 [DOI] [PubMed] [Google Scholar]
- 32. Parrini M. C., Lei M., Harrison S. C., Mayer B. J. (2002) Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9, 73–83 [DOI] [PubMed] [Google Scholar]
- 33. Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. (2000) Structure of PAK1 in an autoinhibited conformation reveals a multistage activation Switch. Cell 102, 387–397 [DOI] [PubMed] [Google Scholar]
- 34. Yokoyama N., Miller W. T. (2003) Biochemical properties of the Cdc42-associated tyrosine kinase ACK1. Substrate specficity, autophosphorylation, and interaction with Hck. J. Biol. Chem. 278, 47713–47723 [DOI] [PubMed] [Google Scholar]
- 35. Ferguson K. M., Kavran J. M., Sankaran V. G., Fournier E., Isakoff S. J., Skolnik E. Y., Lemmon M. A. (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell 6, 373–384 [DOI] [PubMed] [Google Scholar]
- 36. Braun V., Wong A., Landekic M., Hong W. J., Grinstein S., Brumell J. H. (2010) Sorting nexin 3 (SNX3) is a component of a tublar endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell. Microbiol. 12, 1352–1367 [DOI] [PubMed] [Google Scholar]
- 37. Ziman M., O'Brien J. M., Ouellette L. A., Church W. R., Johnson D. I. (1991) Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol. Cell. Biol. 11, 3537–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takahashi S., Pryciak P. (2007) Identification of novel membrane-binding domains in multiple yeast Cdc42 effectors. Mol. Biol. Cell 18, 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Halstead J. R., Savaskan N. E., van den Bout I., Van Horck F., Hajdo-Milasinovic A., Snell M., Keune W. J., Ten Klooster J. P., Hordijk P. L., Divecha N. (2010) Rac controls PIP5K localisation and PtdIns(4,5)P synthesis, which modulates vinculin localisation and neurite dynamics. J. Cell Sci. 123, 3535–3546 [DOI] [PubMed] [Google Scholar]
- 40. Varnai P., Thyagarajan B., Rohacs T., Balla T. (2006) Rapidly inducible changes in phosphatidylinositol 4,5-bisphosphate levels influence multiple regulatory functions of the lipid in intact living cells. J. Cell Biol. 175, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sells M. A., Barratt J. T., Caviston J., Ottilie S., Leberer E., Chernoff J. (1998) Characterization of Pak2p, a pleckstrin homology domain-containing, p21-activated protein kinase from fission yeast. J. Biol. Chem. 273, 18490–18498 [DOI] [PubMed] [Google Scholar]
- 42. Banting G., Ponnambalam S. (1997) TGN38 and its orthologues. Roles in post-TGN vesicle formation and maintenance of TGN morphology. Biochim. Biophys. Acta 1355, 209–217 [DOI] [PubMed] [Google Scholar]
- 43. Adamson P., Marshall C. J., Hall A., Tilbrook P. A. (1992) Post-translational modifications of p21rho proteins. J. Biol. Chem. 267, 20033–20038 [PubMed] [Google Scholar]
- 44. Armstrong S. A., Hannah V. C., Goldstein J. L., Brown M. S. (1995) CAAX geranylgeranyl transferase tranfers farnesyl as efficiently as geranylgeranyl to RhoB. J. Biol. Chem. 270, 7864–7868 [DOI] [PubMed] [Google Scholar]
- 45. Cox A. D., Der C. J. (1992) Protein prenylation. More than just glue? Curr. Opin. Cell Biol. 4, 1008–1016 [DOI] [PubMed] [Google Scholar]
- 46. Zhang H., Shen W., Rempel D., Monsey J., Vidavsky I., Gross M., Bose R. (2011) Carboxyl-group footprinting maps the dimerization interface and phosphorylation-induced conformational changes of a membrane-associated tyrosine kinase. Mol. Cell. Proteomics 10, doi: 10.1074/mcp.M110.005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Merrill S. A., Hanson P. I. (2010) Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme inhibition. J. Biol. Chem. 285, 35428–35438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Monsey J., Shen W., Schlesinger P., Bose R. (2010) Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system. J. Biol. Chem. 285, 7035–7044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montefusco D. J., Asinas A. E., Weis R. M. (2007) Liposome-mediated assembly of receptor signaling complexes. Methods Enzymol. 423, 267–298 [DOI] [PubMed] [Google Scholar]
- 50. Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 51. Shrout A. L., Montefusco D. J., Weis R. M. (2003) Template-directed assembly of receptor signaling complexes. Biochemistry 42, 13379–13385 [DOI] [PubMed] [Google Scholar]
- 52. Lemmon M. A. (2003) Phosphoinositide recognition domains. Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 53. Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K. M. (2004) Activation of endogenous Cdc42 visualized in living cells. Science 305, 1615–1619 [DOI] [PubMed] [Google Scholar]
- 54. Kodani A., Kristensen I., Huang L., Sütterlin C. (2009) GM130-dependent control of Cdc42 activity at the Golgi regulates centrosome organization. Mol. Biol. Cell 20, 1192–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dubois T., Paléotti O., Mironov A. A., Fraisier V., Stradal T. E., De Matteis M. A., Franco M., Chavrier P. (2005) Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 56. Müsch A., Cohen D., Kreitzer G., Rodriguez-Boulan E. (2001) cdc42 regulates the exit of apical and basolateral proteins from the trans-Golgi network. EMBO J. 20, 2171–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Osmani N., Peglion F., Chavrier P., Etienne-Manneville S. (2010) Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 191, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Matas O. B., Martínez-Menárguez J. A., Egea G. (2004) Association of Cdc42/N-WASP/Arp2/3 signaling pathway with Golgi membranes. Traffic 5, 838–846 [DOI] [PubMed] [Google Scholar]
- 59. Estrada L., Caron E., Gorski J. (2001) Fgd1, the Cdc42 guanine nucleotide exchange factor responsible for faciogenital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum. Mol. Genet. 10, 485–495 [DOI] [PubMed] [Google Scholar]
- 60. Sells M. A., Pfaff A., Chernoff J. (2000) Temporal and spatial distribution of activated Pak1 in fibroblasts. J. Cell Biol. 151, 1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dharmawardhane S., Sanders L. C., Martin S. S., Daniels R. H., Bokoch G. M. (1997) Localization of p21-activated kinase 1 (PAK1) to pinocytic vesicles and cortical actin structures in stimulated cells. J. Cell Biol. 138, 1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szentpetery Z., Várnai P., Balla T. (2010) Acute manipulations of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 8225–8230 [DOI] [PMC free article] [PubMed] [Google Scholar]