Background: A lys141Asn (K141N) missense mutation in heat shock protein (HSP) B8 causes distal hereditary motor neuropathy (HMN).
Results: HSPB8 K141N transgenic mice exhibited mild hypertrophy and apical fibrosis as well as slightly reduced cardiac function.
Conclusion: A single point mutation of HSPB8, such as K141N, can cause cardiac disease.
Significance: The cardiomyopathy phenotype was observed in cardiac-specific HSPB8 K141N transgenic mice.
Keywords: Amyloid, Cardiomyopathy, Heat Shock Protein, Mitochondria, Small Heat Shock Proteins
Abstract
A K141N missense mutation in heat shock protein (HSP) B8, which belongs to the small HSP family, causes distal hereditary motor neuropathy, which is characterized by the formation of inclusion bodies in cells. Although the HSPB8 gene causes hereditary motor neuropathy, obvious expression of HSPB8 is also observed in other tissues, such as the heart. The effects of a single mutation in HSPB8 upon the heart were analyzed using rat neonatal cardiomyocytes. Expression of HSPB8 K141N by adenoviral infection resulted in increased HSPB8-positive aggregates around nuclei, whereas no aggregates were observed in myocytes expressing wild-type HSPB8. HSPB8-positive aggresomes contained amyloid oligomer intermediates that were detected by a specific anti-oligomer antibody (A11). Expression of HSPB8 K141N induced slight cellular toxicity. Recombinant HSPB8 K141N protein showed reactivity against the anti-oligomer antibody, and reactivity of the mutant HSPB8 protein was much higher than that of wild-type HSPB8 protein. To extend our in vitro study, cardiac-specific HSPB8 K141N transgenic (TG) mice were generated. Echocardiography revealed that the HSPB8 K141N TG mice exhibited mild hypertrophy and apical fibrosis as well as slightly reduced cardiac function, although no phenotype was detected in wild-type HSPB8 TG mice. A single point mutation of HSPB8, such as K141N, can cause cardiac disease.
Introduction
Heat shock protein (HSP)2 B8 (also known as HSP22, H11 kinase, or αC-crystallin) is assigned to the small HSP family that also includes HSP20 (HSPB6), HSPB2, HSPB3, HSPB4 (αA-crystallin (CryAA)), HSPB5 (αB-crystallin (CryAB)), and HSP27 (HSPB1) (1, 2). The small HSPs share sequence similarity within the α-crystallin domain but exhibit different patterns of gene expression, transcriptional regulation, and subcellular localization (1, 2). The chaperone-like activity of the small HSPs, which are grouped together based on their ability to prevent protein aggregation and/or restore the biological activity of cell substrates, is widely believed to be a protective mechanism against protein misfolding and denaturation triggered by noxious environmental stimuli such as hyperthermic stress, heavy metals, ischemic injury, and some genetic diseases (1, 3, 4).
Missense mutations in small HSPs, such as CryAA and CryAB, can cause dominant disease, such as cataracts and myofibrillar myopathy (1, 2). These diseases, characterized by the formation of aggregates, are misfolded protein-related diseases that can be recapitulated in transgenic (TG) mice, for example, by the presence of a mutant HSP protein in the lens of the eye or in cardiomyocytes (5, 6). Similar to the effect of missense mutations of CryAA and CryAB, a missense mutation of HSPB8 such as K141N or K141E causes autosomal dominant distal hereditary motor neuropathy (HMN) type II in some European families, which is characterized by the formation of inclusion bodies in cells (7). In another study, HSPB8 K141N was also shown to cause axonal Charcot-Marie-Tooth (CMT) disease type 2L in a large Chinese family (8). The K141N mutation affects a highly conserved residue in the central α-crystallin domain of the protein. Overexpression of the HSPB8 K141N protein in COS cells increased the interaction of HSPB8 and HSPB1, leading to the formation of intracellular aggregates (7). A recent study reported that an HSPB8 gene KO mouse showed normal development and no obvious abnormalities in tissue function, but cardiac dysfunction and remodeling, as well as transition into heart failure, were accelerated by pressure-induced overload to the heart (9). These results suggest that the overexpression of mutant HSPB8 can lead to the induction of some degree of cellular toxicity in cultured cells, although no similar phenotype was detected in the HSPB8 knock-out mouse. Thus, it is unclear whether a missense mutation of HSPB8 alone can cause neurodegenerative diseases such as HMN and CMT disease because of loss of function, particularly in vivo. Furthermore, it is known that the HSPB8 expression pattern is ubiquitous, with the highest expression in muscle tissue (10). This implies that mutant HSPB8 protein can be present in muscle tissue, such as the heart, and can affect cardiac function as well as neuronal tissues.
To study the structure-function relationship of a single missense mutation of HSPB8 in vivo, cardiac-specific TG mice expressing HSPB8 K141N as well as wild-type HSPB8 TG mice as controls were generated using tetracycline-controlled transcriptional activator (tTA) and the attenuated myosin heavy chain system (11, 12). The phenotype of HSPB8 K141N TG mice is an accumulation of aggregates containing HSPB8, causing slightly impaired cardiac function, mild ventricular hypertrophy, and apical cardiac fibrosis with cardiac mitochondrial dysfunction at approximately 6 months of age. Our results indicate that mutant HSPB8 TG mice have a cardiomyopathy phenotype, whereas no abnormality was observed in wild-type HSPB8 TG mice. Thus, our data suggest that the HSPB8 mutation can act in a dominant negative manner and that the cellular toxicity of the mutant protein may play an important role in disease development. Our results imply that the HSPB8 mutation can cause cardiomyopathy as well as neuronal degenerative diseases such as HMN and CMT disease and that a phenotype induced by the mutant HSPB8 may result from the mild toxicity of the mutated HSPB8 protein.
EXPERIMENTAL PROCEDURES
cDNAs
cDNAs of HSPB8 and CryAB were isolated by reverse transcription-PCR and used to generate recombinant protein and adenoviral constructs as described previously (13). The missense mutations HSPB8 K141N and CryAB R120G were introduced using reverse transcription-PCR and subcloned into the pBSKII vector (Agilent Technologies, Palo Alto, CA). To distinguish the transgenic products from endogenous protein, a FLAG epitope was introduced at the N terminus of CryAB R120G. An HA epitope was introduced at the N terminus of HSPB8 as described previously (12, 13).
Recombinant Protein
To produce recombinant protein, His epitope-tagged wild-type HSPB8, HSPB8 K141N, wild-type CryAB, and CryAB R120G were overexpressed in BL21 cells (Invitrogen) using the pET system (Novagen, Madison, WI) and purified with a nickel-nitrilotriacetic acid column (Qiagen) as described previously (13). To measure the amyloid oligomer level (amyloid oligomer is positive immunoreactive material against the anti-oligomer antibody), each recombinant protein was incubated and blotted on a nitrocellulose membrane and quantified as described previously (13). The cellular toxicity of the recombinant HSPB8, HSPB8 K141N, CryAB, and CryAB R120G protein in HEK293T cells as well as cardiomyocytes was determined using a 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay. Recombinant proteins were added to give a final concentration of 0.4 mg/ml in serum-free Dulbecco's modified Eagle's medium and incubated for 12 h. After incubation, MTT assays were performed as described previously (12, 13).
Cardiomyocyte and N1E115 Cell Cultures and Adenovirus Infection
Rat neonatal cardiomyocytes were isolated using the Worthington cardiomyocyte isolation system (Worthington Biochemical Corporation, Lakewood, NJ). After isolation of the rat neonatal cardiomyocytes, the cells were grown on glass slides coated with gelatin as described previously (12, 14). Mouse neuroblastoma N1E-115 cells were cultured as described previously (15). Replication-deficient recombinant adenoviruses were made using an AdEasy system (Agilent Technologies) as described previously (5, 13, 14). Viral titration was determined using an AdEasy viral titer kit (Agilent Technologies), and the manufacturer's protocol was followed to calculate the infectious units/milliliter as well as the multiplicity of infection (MOI), which is the ratio of transfer vector transducing particles to cells. To address the transgene expression level to the endogenous HSPB8 level, cardiomyocytes were infected at a MOI of 1 for both the HSPB8 and HSPB8 K141N virus as described previously (13). The N1E-115 cells were infected at a MOI of 10 for each adenovirus. Cellular viability was measured using an MTT assay (12, 13).
Immunohistochemistry
Immunohistochemical analyses were performed as described previously (12, 14). Alexa 488-conjugated anti-rabbit and Alexa 568-conjugated anti-mouse antibodies and TO-PRO-3 for nuclear staining were purchased from Molecular Probes (Eugene, OR), anti-HSPB8 antibody from Imgenex Corporation (San Diego, CA), anti-HA antibody from MB Laboratories Co., Ltd. (Nagoya, Japan), and anti-cTnI antibody (MAB1691) from Millipore (Billerica, MA). The anti-oligomer antibody (A-11) was generated and used as described previously (12, 14). Image J 1.38x public domain software was used to quantify the immunofluorescent intensity. The results from 30 to 50 cells were averaged for cohort comparison. Areas stained with the oligomer antibody were defined, and the average pixel intensities for the cardiomyocytes were determined for comparison (12, 14).
Isolation of Mitochondrial and Cytosolic Fractions
Isolation of mitochondrial and cytosolic fractions was performed as described previously (14). Hearts were homogenized in buffer containing 250 mm sucrose, 10 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm Na3VO4, and complete protease inhibitor mixture tablets (Roche Applied Science). The homogenates were centrifuged at 1,000 × g for 10 min at 4 °C to remove the nuclei. Supernatant fluids were then centrifuged again at 13,000 × g for 30 min at 4 °C. Pellets were washed extensively in the same buffer and centrifuged at 13,000 × g for 30 min at 4 °C. The mitochondrial fraction of the pellets was then resuspended in lysis buffer containing 150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1 mm Na3VO4, complete protease inhibitor mixture tablets (Roche Applied Science), and 1% Nonidet P-40. Supernatant fluids were further purified at 100,000 × g for 30 min (4 °C) and used as the cytosolic fraction.
Immunoprecipitation Assay
Immunoprecipitation assay was performed as described previously (13, 16). 200 μg of mitochondrial fractions were incubated with 1 μg of anti-voltage dependent anion channel (VDAC) antibody (Ab-5) (Merck) for 2 h at 4 °C. 20 μl of protein A/G-agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) were added, and the specimens were incubated overnight at 4 °C. Pellets were collected by centrifugation at 2,500 × g for 5 min at 4 °C and washed four times with radioimmune precipitation assay buffer. The pellets were resuspended in 40 μl of sample buffer, boiled for 3–5 min, and centrifuged again, and the supernatants were analyzed first by PAGE and then by Western blotting using either anti-HSPB8 (Imgenex Corporation) or anti-VDAC (Ab-5) (Merck). In some experiments, to examine the direct interaction of HSPB8 K141N and VDAC, the recombinant HSPB8 protein, as well as the HSPB8 K141N protein, was treated with the mitochondrial fractions from nontransgenic (NTG) mouse hearts at 25 °C for 1 h and then washed four times with radioimmune precipitation assay buffer. Recombinant proteins were added to give a final concentration of 0.1 mg/ml in the medium.
Preparation of Isolated Mitochondria and Measurement of Mitochondrial Respiratory Function
Preparation of cardiac mitochondria was performed using the method described previously (17). Heart tissue was homogenized in ice-cold buffer containing 180 mm KCl, 10 mm EDTA (pH 7.4), and 0.5% fatty acid-free BSA. The homogenate was then centrifuged at 700 × g for 10 min at 2 °C, and the resulting supernatant fluid was centrifuged at 8,000 × g for 10 min at 2 °C. The crude mitochondria were again suspended in buffer and centrifuged at 8,000 × g for 10 min at 4 °C. The organelles were then resuspended in suspension buffer (20 mm Tris-HCl, pH 6.8, containing 320 mm sucrose and 0.25% BSA) and used to measure mitochondrial activity. The isolated mitochondria were used for the measurement of mitochondrial respiratory function. The mitochondrial state 3 and 4 respiration, respiratory control index, and oxidative phosphorylation rate were determined using the method described previously (17). Isolated mitochondria were incubated in a medium of pH 7.4 that contained 10 mm Tris-HCl, 250 mm sucrose, 10 mm K2HPO4, and 10 mm glutamate and were stirred at 25 °C. The mitochondrial oxygen consumption rate was measured in the chamber using a Clark-type oxygen electrode (Central Kagaku, Tokyo, Japan). The quality of the mitochondrial preparation was evaluated by assessing the respiratory control index, which was determined in the presence of 240 nmol of ADP. In some experiments, to examine the direct effect of HSPB8 K141N on mitochondrial oxygen consumption ability, the recombinant HSPB8 protein, as well as the HSPB8 K141N protein, was added to the medium. Recombinant proteins were added to give a final concentration of 0.1 mg/ml in the medium.
Miscellaneous Methods
Sample preparation for Western blotting, gel preparation, and electrophoretic conditions were carried out as described previously (12, 14). Western blot analyses were performed using anti-GAPDH antibody (Chemicon International, Temecula, CA), anti-HSPB8 antibody (Imgenex Corporation), anti-HA antibody from MB Laboratories Co., Ltd. (Nagoya), anti-VDAC antibody (Ab-5) (Merck), and anti-cytochrome c antibody (BD Bioscience, CA). The band intensity in the immunoblot was semiquantified using Image J. The filter assay for the detection of aggregates was performed as described previously (12). Lysate from both the transfected cells and the transgenic mouse hearts was centrifuged at 12,000 × g for 10 min. The resultant pellet was diluted into 0.2 ml of 2% SDS and boiled for 5 min. After boiling, the sample was filtered through a 0.2-μm nitrocellulose membrane. The aggregate fraction on the membrane was detected with anti-HSPB8 antibody. Echocardiography and trichrome staining were performed as described previously (12).
Transgenic Mice
Female mice with cardiac-specific overexpression of mutant HSPB8 containing the K141N mutation, driven by the modified α-myosin heavy chain promoter, have been described previously (12, 18). The TG mice were identified by PCR analysis of genomic DNA isolated from tail tips. The responder HSPB8 and HSPB8 K141N mice were crossed with tetracycline-controlled tTA TG mice to generate tTA/HSPB8 and tTA/HSPB8 K141N double TG (tTA/HSPB8 TG and tTA/HSPB8 K141N TG) mice (12, 18). The responder HSPB8 and HSPB8 K141N mice used for all experiments had a C57BL/6Cr Slc genetic background (SLC, Shizuoka, Japan). The CryAB R120G and tTA TG mice were backcrossed with C57BL/6Cr Slc mice more than 10 times and maintained on a C57BL/6Cr Slc background as described previously (12). NTG littermates were always used as controls for comparison. The animals were housed in microisolator cages in a pathogen-free barrier facility. All experimentation was performed under approved institutional guidelines.
Statistics
The data are expressed as the means ± standard error. Statistical analysis was performed using the unpaired Student's t test and one-way analysis of variance followed by a post hoc comparison with Scheffe's multiple comparison, using Statview version 5.0 software (Concepts, Inc., Berkeley, CA).
Ethics
This study was approved by the Animal Care Committee of Iwate Medical University (approval identification 21-056). All of the experimental procedures were performed in accordance with the Guidelines of the Iwate Medical University Ethics Committee for Animal Treatment and the Guidelines for Proper Conduct of Animal Experiments by the Science Council of Japan.
RESULTS
HSPB8 K141N in Cardiomyocytes and N1E115 Cells
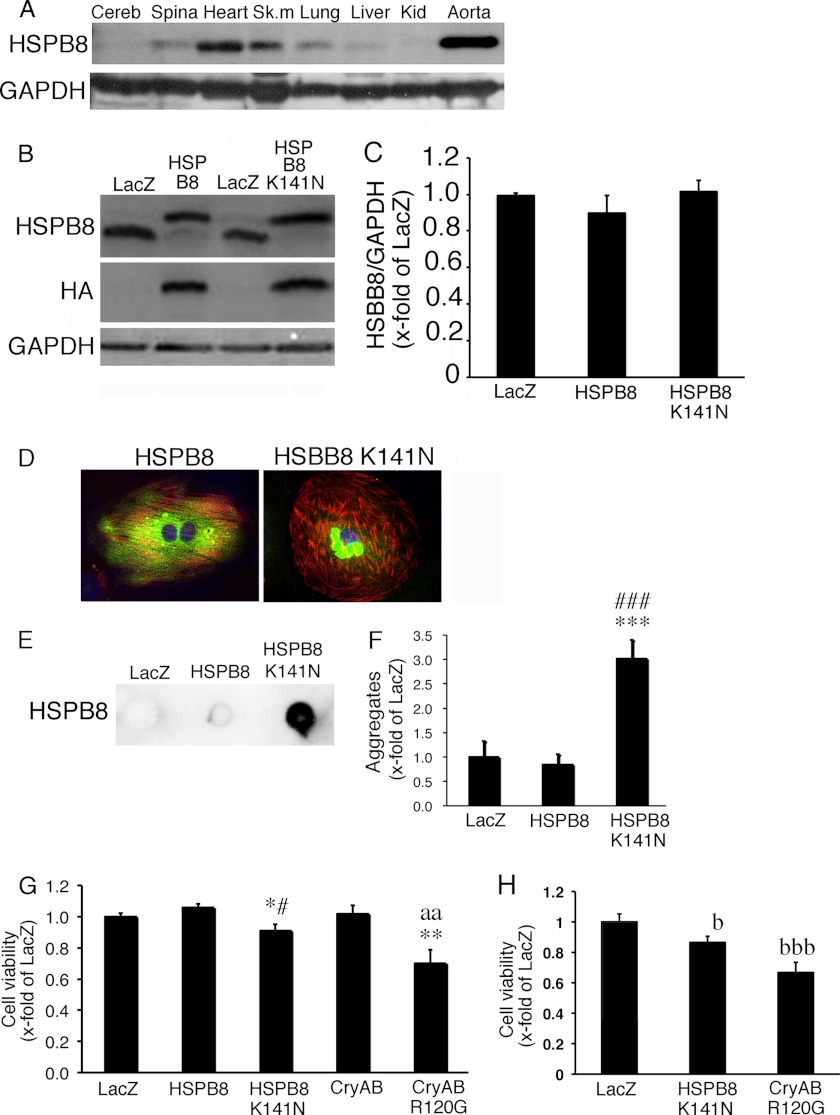
Although a missense mutation in HSPB8, a member of the small HSP family, causes HMN and CMT disease, which is characterized by the formation of inclusion bodies in cells, the role of the mutant HSPB8 protein in other tissues remains uncertain. To study this, we analyzed the distribution of the HSPB8 protein in mouse tissue (Fig. 1A). HSPB8 protein was detectable in most tissues, such as the cerebrum, spinal cord, heart, and aorta (Fig. 1A), although the levels were not uniform among these tissues. The levels of HSPB8 in the heart and aorta were much higher than those in the cerebrum and spinal cord. These results imply that the HSPB8 missense mutation that causes HMN and CMT disease may affect the tissue function of heart and vascular tissues.
FIGURE 1.
Characterization of HSPB8 K141N. A, tissue distribution of endogenous HSPB8 in the mouse showing analysis of several tissues, with the heart and aorta displayed prominently. B and C, typical images of HSPB8 and GAPDH Western blot in cardiomyocytes infected using an adenoviral vector containing HSPB8 and HSPB8 K141N at a multiplicity of infection of 1 (B) and the quantitative analysis (C). D, a typical image of immunohistochemistry in cardiomyocytes expressing HSPB8 (green) and HSPB8 K141N (green). HSPB8-positive aggregates were observed in cardiomyocytes expressing HSPB8 K141N. To distinguish cardiomyocytes, cardiac troponin I was stained (red), and TO-PRO-3 was used for nuclear staining (blue). E and F, quantitative analysis of aggregates. Typical images of the filtration assay (E) and the quantitative analysis of aggregates (F) are shown. G, cell viability in cardiomyocytes. Mild cellular toxicity was observed with HSPB8 K141N overexpression, whereas a significant decrease in cell viability was observed with CryAB R120G overexpression. H, cell viability in N1E115 neuroblastoma cells. The values are the fold increases relative to cardiomyocytes (G) and N1E115 cells (H) expressing LacZ (LacZ), whose value was arbitrarily set to 1. Cereb, cerebrum; Spina, spinal cord; Sk. m, skeletal muscle; Kid, kidney. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus cardiomyocytes expressing LacZ. #, p < 0.05; ###, p < 0.001 versus cardiomyocytes expressing HSPB8. aa, p < 0.01 versus cardiomyocytes expressing CryAB; b, p < 0.05; bbb, p < 0.001 versus N1E115 cells expressing LacZ.
To address the effect of the HSPB8 mutant protein in cardiomyocytes, we expressed HSPB8 and HSPB8 K141N using an adenoviral vector. In this experiment, we used adenoviral vector at a MOI of 1 to express HSPB8 as well as HSPB8 K141N (Fig. 1B). Under this experimental condition, the protein level of the expressed HSPB8 protein, as well as that of the HSPB8 K141N protein, was similar to the endogenous HSPB8 protein level in cardiomyocytes expressing LacZ, because viral expression of HSPB8 resulted in the down-regulation of the endogenous HSPB8 protein (Fig. 1, B and C). Similar HSPB8 protein levels were observed among wild-type HSPB8, HSPB8 K141N, and LacZ cardiomyocytes (Fig. 1, B and C), and cardiomyocytes expressing the wild-type HSPB8 and LacZ showed no detectable aggregates. Despite this, perinuclear aggregates that were immunoreactive against an HSPB8 antibody were observed in cardiomyocytes expressing HSPB8 K141N (Fig. 1, D–F). These results suggest that, in a manner similar to that of the CryAB missense mutation (5), the HSPB8 missense mutation can induce aggregate formation in cardiomyocytes. A slight reduction in cellular viability was observed in cardiomyocytes expressing HSPB8 K141N (Fig. 1G), whereas the CryAB missense mutation led to a significant reduction in cellular viability, as described previously (5). Similar to the results for cardiomyocytes, mild cellular toxicity of HSPB8 K141N and severe cellular toxicity of CryAB R120G were observed in N1E115 mouse neuroblastoma cells (Fig. 1H). This finding may imply that mutant HSPB8 is less toxic than mutant CryAB in cultured cells such as cardiomyocytes and neuroblastoma cells.
Amyloid Oligomer Formation of HSPB8 K141N
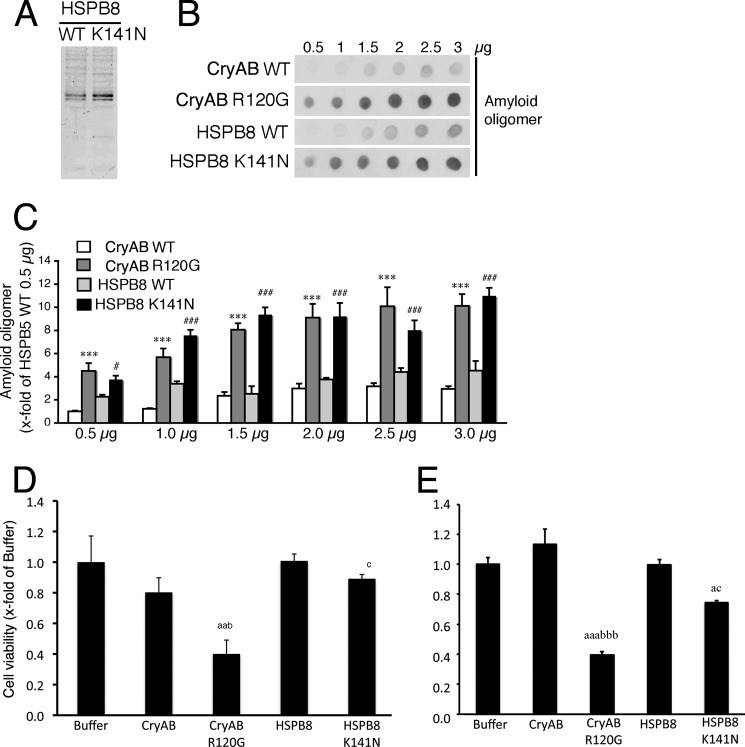
To compare the cellular toxicity and amyloid oligomer formation of the HSPB8 K141N and CryAB R120G proteins, we generated recombinant proteins (Fig. 2A). Recombinant HSPB8 K141N protein showed immunoreactivity against an anti-amyloid oligomer antibody, and its reactivity was similar to that of the recombinant CryAB R120G protein. The immunoreactivities of recombinant wild-type HSPB8 protein and recombinant wild-type CryAB protein were weaker than those of the recombinant mutant HSPB8 and the mutant CryAB proteins (Fig. 2, B and C). When the recombinant HSPB8 K141N protein was added to the culture medium, mild cellular toxicity in 293T cells as well as in cardiomyocytes was observed, but this cytotoxicity was less than that of the recombinant CryAB R120G protein (Fig. 2, D and E). These results suggest that although both the HSPB8 K141N protein and the CryAB R120G protein can form an amyloid oligomer at a level detectable by the anti-oligomer antibody, the cytotoxicity of HSPB8 K141N is milder than that of the CryAB R120G protein.
FIGURE 2.
Formation of amyloid oligomer using the recombinant HSPB8 K141N protein. A, typical SDS-PAGE images of the recombinant wild-type HSPB8 (HSPB8) and HSPB8 K141N. B, dot blotting shows the presence of the amyloid oligomer in the recombinant HSPB8 K141N and CryAB R120G proteins, but a lower amyloid oligomer level is detected in wild-type HSPB8 (HSPB8 WT) and CryAB (CryAB WT). C, quantitative amyloid oligomer analysis. The values are the fold increases relative to 0.5 μg of CryAB WT, whose value was arbitrarily set to 1. D and E, cellular toxicity of recombinant proteins. The cellular HSPB8 K141N protein toxicity in HEK293 cells (D) and cardiomyocytes (E) was determined using the MTT method. The values are the fold increase relative to cells with the buffer, added without recombinant proteins (Buffer), whose value was arbitrarily set to 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus CryAB WT; #, p < 0.05; ####, p < 0.001 versus HSPB8 WT. a, p < 0.05; aa, p < 0.01; aaa, p < 0.001 versus buffer. b, p < 0.05; bbb, p < 0.001 versus CryAB. c, p < 0.05 versus HSPB8.
tTA/HSPB8K141N Double TG Mouse
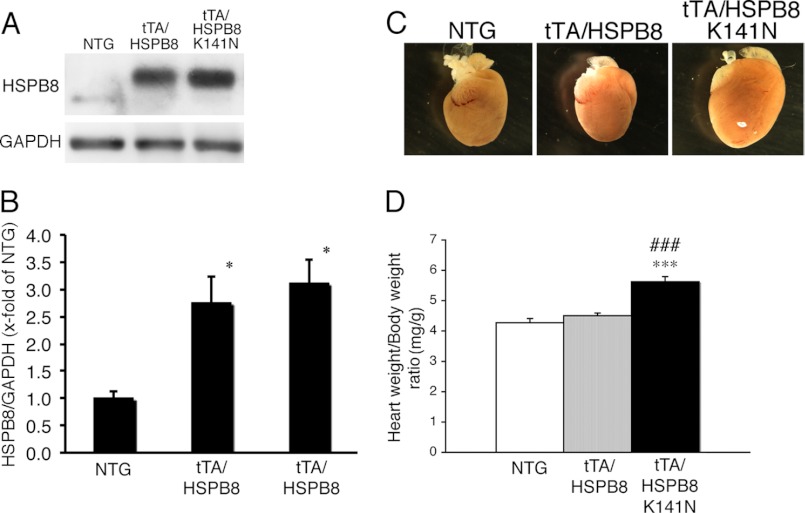
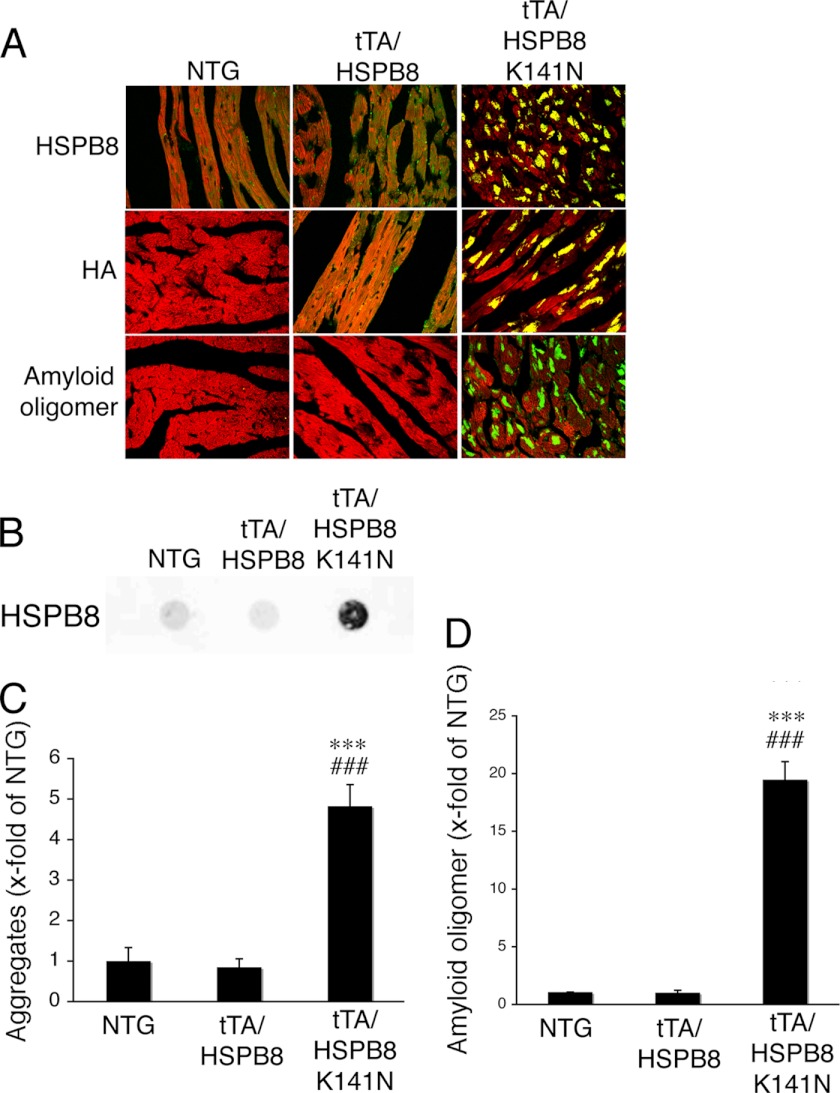
Our in vitro study showed that the missense mutation of HSPB8 led to the formation of a toxic amyloid oligomer and that HSPB8 K141N showed mild cellular toxicity in vitro. To further study the effect of the HSPB8 missense mutation on cardiomyocytes in vivo, we generated a cardiac-specific TG mouse that overexpressed the HSPB8 K141N mutation or wild-type HSPB8 using an inducible cardiac-specific α-myosin heavy chain promoter as described previously (11, 12). The level of HSPB8 proteins observed in the HSPB8 and HSPB8 K141N TG mice was 3-fold higher when they were cross-bred with tTA TG mice than that of NTG mice (12); HSPB8 K141N proteins were also observed (Fig. 3, A and B). No differences were observed in the HSPB8 protein level between tTA/HSPB8 double TG mice and tTA/HSPB8 K141N double TG mice (Fig. 3, A and B). Furthermore, although the protein levels were similar, the tTA/HSPB8 K141N double TG mice showed increased heart weight/body weight ratios compared with those of tTA/HSPB8 double TG mice and NTG mice at 6 months of age (Fig. 3, C and D). At 6 months, intracellular aggregates that were immunoreactive against anti-HSPB8, as well as an anti-HA epitope that was introduced into the transgene products to distinguish them from endogenous protein, were detected in the hearts of the tTA/HSPB8 K141N double TG mice, whereas no aggregates were observed in the hearts of tTA/HSPB8 and NTG mice (Fig. 4, A and B). These aggregates contained the amyloid oligomer in the tTA/HSPB8 K141N double TG mice (Fig. 4, A and D). Thus, concomitant with cardiac hypertrophy, the mutant HSPB8 protein can result in the formation of aggresomes in the heart that are immunoreactive against the anti-oligomer antibody.
FIGURE 3.
Characterization of the tTA/HSPB8 K141N double TG mouse. A, typical images of HSPB8 Western blot analysis. An increase in the transgene product, which had an HA epitope tag inserted at the N terminus, was observed. Anti-GAPDH antibody was used as a loading control. B, quantitative analysis of HSPB8. The values are the fold increases relative to values in NTG mouse hearts, whose values were arbitrarily set to 1. C, representative images of hearts from 6-month-old TG mice. D, ratios of heart weight to body weight. A significant increase in heart weight was observed in the tTA/HSPB8 K141N double TG mouse. *, p < 0.05; ***, p < 0.001 versus the NTG. ###, p < 0.001 versus tTA/HSPB8 double TG mice.
FIGURE 4.
Representative immunohistochemistry images. A, hearts of tTA/HSPB8 K141N double TG mice at 6 months of age. Aggregates containing HSPB8 K141N (upper panel, green, HSPB8; middle panel, green, HA epitope tag) and amyloid oligomer (lower panel, green) were observed in the HSPB8 K141N double TG mice. Cardiac troponin I stain was used to distinguish the cardiomyocytes (red). B, typical image of the filter assay for the detection of aggregates. C and D, quantitative analysis of aggregates containing mutant HSPB8 K141N protein (C) and amyloid oligomer (D). (n = 4). ***, p < 0.001 versus NTG mice. ###, p < 0.001 versus tTA/HSPB8 double TG mice.
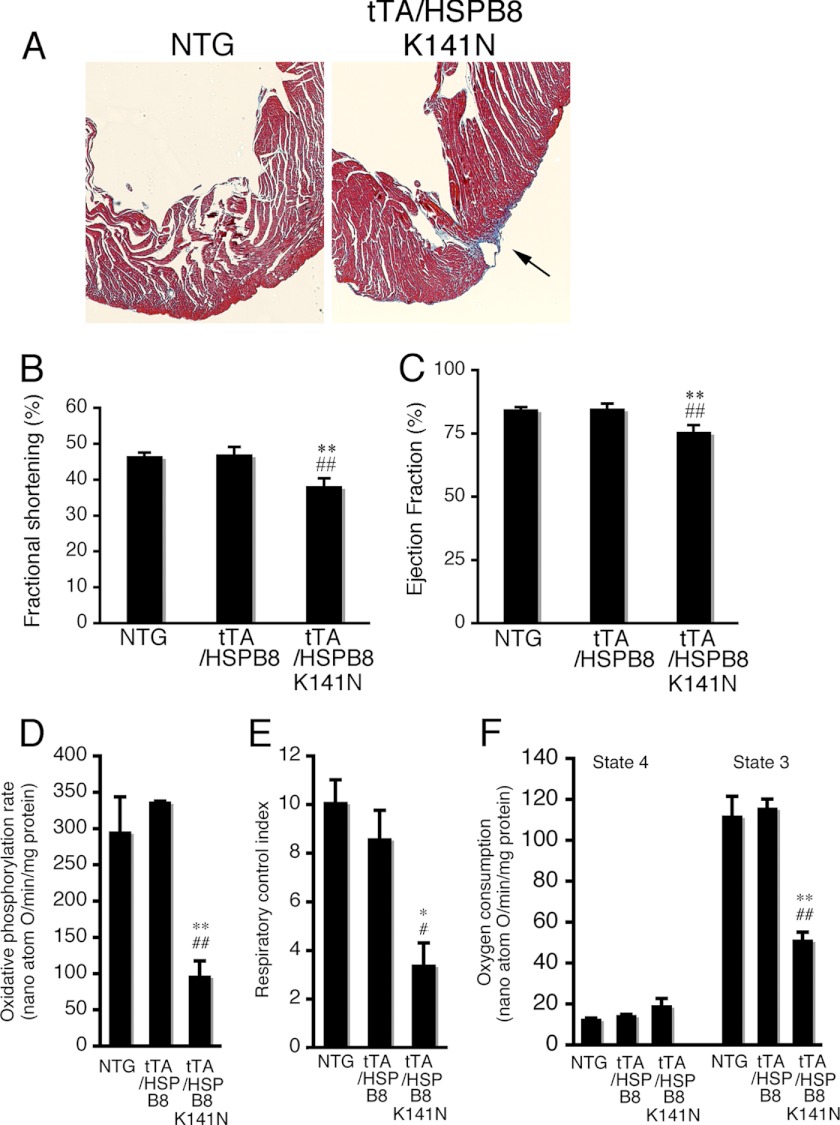
The hearts of the HSPB8 K141N double TG mice exhibited mild apical cardiac fibrosis at 6 months of age (Fig. 5A). Cardiac performance, such as fractional shortening and ejection fraction, was slightly reduced in tTA/HSPB8 double TG mice compared with that in tTA/HSPB8 TG and NTG mice at 6 months of age (Table 1 and Fig. 5, B and C). At 6 months, the oxidative phosphorylation ratio, respiratory control index, and oxygen consumption in state 3 were markedly decreased in the mitochondrial fraction from tTA/HSPB8 K141N TG mice compared with tTA/HSPB8 and NTG mice (Fig. 5, D–F). No distinguishable difference in terms of premature death was detected up to 2 years of age, and no distinguishable difference in terminal TUNEL-positive apoptosis of cardiomyocytes was observed at 6 months of age among tTA/HSPB8 K141N double TG mice, tTA/HSPB8 double TG mice, or NTG mice (data not shown). These results suggest that cardiac disease, such as cardiac fibrosis and reduced cardiac function, occurs in response to the presence of the mutant HSPB8 protein and that this cardiac disease may be associated with mitochondrial dysfunction in the heart.
FIGURE 5.
Cardiac disease in 6-month-old tTA/HSPB8 K141N TG mice. A, Masson's trichrome staining. TG mice exhibited mild apical cardiac fibrosis at 6 months of age (arrow). B and C, cardiac function. The fractional shortening (B) and ejection fraction (C) assessed by echocardiography are shown. Cardiac functional measurements were made (n = 8). D–F, the mitochondrial oxidative phosphorylation parameters in tTA/HSPB8 K141N TG mouse hearts. The oxidative phosphorylation rate (D), respiratory control index (E), and oxygen consumption (F) are shown. Significant reductions in the mitochondrial oxidative phosphorylation parameters were observed in tTA/HSPB8 double TG mice. *, p < 0.05; **, p < 0.01 versus the NTG. #, p < 0.05; ##, p < 0.01 versus the tTA/HSPB8 TG mice (n = 6–8).
TABLE 1.
Echocardiographic parameters at 6 months of age
The values are the means ± S.E. (n = 6–10 in each group). tTA/HSPB8 TG, tTA/HSPB8 double transgenic mouse; tTA/HSPB8 K141N TG, tTA/HSPB8 K141N double transgenic mouse; LVIDd, left ventricular internal end diastolic dimension; LVIDs, left ventricular internal end systolic dimension; PW, posterior wall thickness; Septum, septal wall thickness; LV, left ventricular.
| Body weight | LVIDd | LVIDs | PW | Septum | LV mass/body weight | |
|---|---|---|---|---|---|---|
| g | mm | mm | mm | mm | mg/g | |
| NTG | 33 ± 2 | 3.4 ± 0.2 | 1.8 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.1 | 3.4 ± 0.1 |
| tTA/HSPB8 TG | 34 ± 2 | 3.5 ± 0.2 | 1.9 ± 0.1 | 1.0 ± 0.1 | 1.1 ± 0.1 | 3.5 ± 0.2 |
| tTA/HSPB8 K141N TG | 35 ± 2 | 3.7 ± 0.1a | 2.3 ± 0.1b,c | 1.1 ± 0.1 | 1.2 ± 0.1 | 4.7 ± 0.2d,e***### |
a p < 0.05 versus NTG.
b p < 0.01 versus NTG.
c p < 0.01 versus tTA/HSP B8 TG.
d p < 0.001 versus NTG.
e p < 0.001 versus tTA/HSP B8 TG.
Interaction of HSPB8 K141N Protein with Mitochondrial Protein
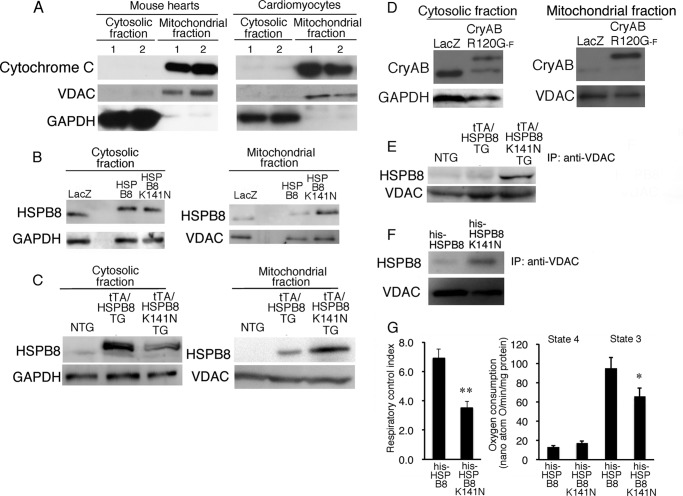
A missense mutation of CryAB, such as R120G, results in the accumulation of a mutant protein around nuclei, called an aggresome; this mutant protein can bind directly to VDAC protein (16). Because marked mitochondrial dysfunction of the heart in CryAB R120G TG mice was clearly observed, this protein interaction between the mutant CryAB and VDAC may play an important role in the cellular toxicity of the mutant CryAB (16). HSPB8, a member of the small HSP family along with CryAB, may be directly associated with mitochondria. To examine this hypothesis, we analyzed the distribution of HSPB8 in cardiomyocytes, as well as in double TG mouse hearts (Fig. 6, B and C). Typical mitochondrial markers, such as cytochrome c and VDAC, were enriched in the mitochondrial fraction from mouse hearts and cardiomyocytes, whereas the cytosolic marker GAPDH was enriched in the cytosolic fraction (Fig. 6A). Similar amounts of wild-type HSPB8 or HSPB8 K141N proteins were detected in the cytosolic fraction after expression of HSPB8 or HSPB8 K141N in the cardiomyocytes infected at a MOI of 1 for each adenovirus, whereas a higher level of the HSPB8 K141N protein was observed in the mitochondrial fraction compared with that expressing the wild-type HSPB8 (Fig. 6B). This difference in the HSPB8 K141N protein distribution pattern was also observed in the hearts of double TG mice (Fig. 6C). A distribution pattern, namely, a trend toward high amounts of the mutant protein in the mitochondrial fraction, was also detected in the CryAB R120G protein (Fig. 6D). These results suggest that a missense mutation of small HSPs, such as HSPB8 K141N and CryAB R120G proteins, can change their localization from cytoplasm to mitochondria, probably because of alteration of their affinity for partner proteins.
FIGURE 6.
Subcellular HSPB8 K141N distribution in hearts of 6-month-old tTA/HSPB8 K141N TG mice and in cardiomyocytes expressing HSPB8 K141N. A, a representative image of mitochondrial markers, such as cytochrome c and VDAC, with GAPDH used as a cytosolic marker. B and C, mutant HSPB8 subcellular distribution in cardiomyocytes (B) and tTA/HSPB8 K141N in double TG mice (C). D, mutant CryAB R120G-FLAG protein (CryAB R120G-F) subcellular distribution in cardiomyocytes. E, immunoprecipitation (IP) analysis using anti-VDAC antibody. F, interaction of VDAC with recombinant HSPB8 (his-HSPB8) and HSPB8 K141N (his-HSPB8 K141N) proteins. G, direct effect of recombinant HSPB8 (his-HSPB8) as well as HSPB8 K141N (his-HSPB8 K141N) proteins on oxidative phosphorylation ability in isolated mitochondria isolated from NTG mice (n = 6). *, p < 0.05; **, p < 0.01 versus mitochondria treated with his-HSPB8.
In a previous study, we showed that CryAB R120G can bind directly to VDAC protein with high affinity compared with wild-type CryAB (16). This result may imply that the HSPB8 missense mutation may be able to bind to VDAC protein. To examine this hypothesis, we performed immunoprecipitation assays using an anti-VDAC antibody (Fig. 6E), which showed a detectable interaction with HSPB8, but almost no interaction with HSPB8 derived from hearts in which NTG or wild-type HSPB8 was overexpressed (Fig. 6E). This result suggests that HSPB8 K141N can interact with VDAC protein in mice. To confirm this result, recombinant HSPB8 and HSPB8 K141N proteins were added to the mitochondrial sample from NTG mice. Similar to the TG mouse study, a detectable interaction with the recombinant HSPB8 K141N but almost no interaction with the recombinant HSPB8 was observed (Fig. 6F). Mitochondrial oxidative phosphorylation was reduced by treatment with recombinant HSPB8 K141N protein compared with wild-type HSPB8-treated mitochondria (Fig. 6G). These results suggest that HSPB8 K141N protein can interact with mitochondria and that this interaction may be associated with a reduced mitochondrial oxygen consumption rate.
DISCUSSION
In the present study, we showed that cardiac-specific overexpression of HSPB8 K141N can cause cardiomyopathy, whereas no obvious phenotype was observed in the overexpression of wild-type HSPB8. Similar results were observed in other TG mice in which disease is caused by missense mutations of small HSPs, such as CryAB R120G (5), CryAA R116C (6), HSPB1 S135F, and P182L (19). The phenotype of the cardiac-specific CryAB R120G TG mouse includes the accumulation of CryAB aggregates as well as severe cardiac disease at approximately 6 months of age (5). Studies have shown that expression of CryAA R116C in lens tissue results in posterior cortical cataracts and structural abnormalities (6) and that the neuronal-specific overexpression of HSPB1 S135F and P182L leads to CMT disease or distal HMN (19). In contrast to TG mice expressing the mutant HSPs, TG mice expressing wild-type small HSPs, such as CryAB, CryAA, and HSPB1, were indistinguishable from age-matched NTG control mice (5, 6, 19). Furthermore, phenotypes were seldom observed in CryAB and HSPB2 KO mice (20) or HSPB8 KO mice (9). These small HSP KO mice showed normal development and no obvious abnormalities in tissue function, but the stress response to pressure-induced overload to the heart was impaired (9, 21). These data indicate that the HSPB8 K141N mutation can cause cardiomyopathy on its own, is dominant negative, and results in cardiac hypertrophy. The mutant protein may play an important role in neurodegenerative disease formation, such as HMN and CMT disease.
There has been no published research regarding the cardiac phenotype induced by the missense HSPB8 mutation in humans because HSPB8 missense mutations were shown to cause HMN and CMT disease (8, 22, 23). The expression pattern of HSPB8 is ubiquitous, and the highest expression occurs in muscle tissue (10). This implies that mutant HSPB8 protein can be present in muscle tissues, such as the heart. The HSPB8 K141N protein showed mild cellular toxicity in both rat neonatal cardiomyocytes and cultured mouse neuroblastoma cells, N1E115 cells. Cardiac toxicity of the mutant HSPB8 was also detected in TG mice in our study. Because the cardiac HSPB8 expression level is higher than in most other human tissues (8, 22, 23) and because this expression pattern is similar to the results obtained in mice in the present study, these results imply that the HSPB8 missense mutations, such as K141N and K141E, are present in cardiomyocytes and peripheral neuronal cells. HSPB8 K141N protein overexpression using an α-myosin heavy chain promoter can cause aggresomal accumulation and amyloid formation in cardiomyocytes without any modification of noncardiomyocytes. Furthermore, no obvious phenotype was observed with wild-type HSPB8 overexpression. Thus, the possibility that the present experimental approach, which included overexpression of the disease causing gene, led to exaggerated aggresomal formation by the mutant HSPB8 cannot be excluded; HSPB8 K141N cardiac expression can affect cellular function by aggresomal formation in the cardiomyocytes and may cause cardiomyopathy in addition to HMN and CMT disease.
Because it is known that Arg-116 in CryAA and Arg-120 in CryAB correspond to Lys-141 in HSPB8 in the α-crystallin domain of small HSPs and because disease-causing missense mutations, such as R120G, R116C, and K141N, were found in these amino acids, similar underlying mechanisms of cellular toxicity may be present among these mutant small HSPs (7). To address this, we examined amyloid oligomer formation for the recombinant HSPB8 K141N and CryAB R120G proteins. Recombinant HSPB8 K141N protein showed immunoreactivity against an anti-amyloid oligomer antibody that detected the structural characteristics of amyloid protein (24), and its reactivity was similar to that of the recombinant CryAB R120G protein, whereas the immunoreactivities of recombinant wild-type HSPB8 protein and recombinant wild-type CryAB protein were weaker than those of the mutant recombinant proteins. Because it is hypothesized that amyloid oligomers can permeabilize cellular membranes and lipid bilayers, which may represent the primary toxic mechanism of amyloid pathogenesis (14, 25), cellular toxicity induced by the amyloid oligomers is associated with mitochondrial function as well as induction of apoptotic cell death by cytochrome c release from mitochondria (12, 16). Marked mitochondrial dysfunction was observed in the hearts of HSPB8 K141N TG mice in this study as well as in CryAB R120G TG mice (12, 16). As we have shown, mutant HSPB8 and mutant CryAB proteins (16) can be associated with mitochondrial protein, including VDAC, a regulatory protein of mitochondria transition pores. The recombinant HSPB8 K141N protein can also be associated with VDAC protein; this association may play a role in the reduction in mitochondrial oxidative phosphorylation.
Although the physiological significance of the protein interaction between mutant small HSPs and VDAC protein remains unclear, and the molecular mechanisms of mitochondrial dysfunction induced by mutant small HSPs are currently unknown, the association of mutant small HSPs with mitochondria may play an important role in cellular toxicity as well as the formation of diseases such as cardiomyopathy, cataracts, HMN, and CMT disease. In our previous study, we showed that nicorandil, a mitochondrial KATP-sensitive channel opener, could partially inhibit disease progression in CryAB R120G TG mice without any reduction in mitochondrial CryAB translocation (14). This suggests that the translocation of mutant HSP is not associated with disease progression in CryAB R120G TG mice hearts. Similar to mitochondrial CryAB R120G translocation, the physiological significance of mutant HSPB8 translocation in cardiac disease in HSPB8 K141N TG mice remains unclear. Recently, an interaction between STAT3 (a transcription factor) and HSPB8 (which is related to STAT3 translocation to the mitochondria) has been shown in HSPB8 knock-out mice, where both mitochondrial STAT3 translocation and respiration were significantly decreased (9). Thus, it is possible that interaction between HSPB8 and STAT3 or VDAC plays an important role in the regulation of transcription and in mitochondrial oxidative phosphorylation ability and that these protein interactions can be altered by missense mutation such as K141N. Further research is needed to clarify the cause-and-effect relationship in HSPB8 K141N cellular toxicity.
Although the mutant HSPB8 showed immunoreactivity against the anti-oligomer antibody similar to that of the mutant CryAB, the cellular toxicity of the mutant HSPB8 was milder than that of the mutant CryAB. Similar to the degrees of cellular toxicity observed in vitro, the cardiac disease observed in HSPB8 K141N TG mice was more benign than that in CryAB R120G TG mice, although amyloid oligomer-positive aggresomes were detected in cardiomyocytes from the TG mouse hearts (5, 12, 14). These in vivo and in vitro results indicate that the immunoreactivity against the anti-oligomer antibody is somewhat dissociated from the cellular toxicity. Another study also suggested that immunoreactivity against an anti-oligomer antibody is observed in native wild-type HSPs, such as human HSP27, HSP40, HSP70, HSP90, yeast HSP104, and bovine Hsc70 (23). Thus, anti-oligomer antibody immunoreactivity can be present in native proteins, particularly HSPs. Missense mutations from arginine or lysine (a positive-charged amino acid), to glycine or cysteine can markedly alter protein structure, because it is known that a positive charge must be preserved at this position for the structural and functional integrity of small HSPs (26, 27). Thus, higher immunoreactivity against an anti-oligomer antibody in mutant small HSPs may result from altered protein structure because of the loss of charge and changes in overall surface hydrophobicity of the protein (26).
The reason for the higher cellular toxicity of the mutant CryAB compared with that of the mutant HSPB8 remains uncertain in this study. One possible explanation is that the cellular toxicity of the mutant CryAB R120G protein may be induced by multiple factors. Previous studies showed that CryAB can bind to many contractile proteins, such as desmin, actin (28), and titin (29). Thus, the CryAB R120G protein may retain the ability to interact with contractile proteins, and this interaction may be enhanced relative to the normal affinity of the protein, as is the case with FBX4, a member of the F-box family of proteins (30). In addition, CryAB negatively regulates apoptosis by inhibiting caspase-3 activation (31). CryAB R120G, which is defective in chaperone activity, binds tightly to nascent contractile proteins, preventing them from folding correctly and integrating into productive sarcomeres (5). The presence of contractile protein fragments within the aggregates suggests that mutant CryAB binding may directly disturb contractile protein function, rendering muscle tissue particularly sensitive to the action of CryAB R120G (16). Thus, the pathogenesis of mutant CryAB probably reflects a synergistic combination of these mechanisms. Further study will be needed to address these structure-function relationships in mutated small HSPs.
A previous study showed that overexpression of wild-type HSPB8/H11 kinase resulted in cardiac hypertrophy in mice (23). In contrast, our previous study (12), as well as the present study, showed that no obvious phenotype is observed by overexpression of HSPB8 in the heart. The reasons for the different findings between the previous study by another group and our results are uncertain. One possibility involves gene constructs as the previous study used human HSPB8/H11 kinase followed by a C-terminal HA tag to generate TG mice, whereas we used mouse HSPB8/HA followed by an N-terminal HA tag. The genetic background of the mice is another difference between the two studies (FVb/n strain in the previous study and C57BL/6 background in our study). All of the results suggest that overexpression of mouse mutant missense HSPB8, such as K141N ∼3-fold higher than that of NTG, can result in mild cardiac hypertrophy, whereas no obvious phenotype is observed by the same level of overexpression of wild-type mouse HSPB8 in the C57Bl/6 strain.
The hearts of HSPB8 K141N double TG mice exhibited mild apical cardiac fibrosis at 6 months of age. The aggresomal and amyloidal formation in cardiomyocytes can induce mechanical deficits in passive cytoskeletal stiffness in the heart and can observed cardiac wall stress (32). An increase in wall stress, which can cause relatively hypoxic conditions particularly at the apex and subendocardial region of the heart, may be associated with cardiac fibrosis. Further study on the observed in wall stress and cardiac fibrosis is required.
HSPB8 expression induced by an adenoviral vector led to a reduction in endogenous HSPB8 in cardiomyocytes (Fig. 1B). This result is also observed in mouse hearts that overexpress HSPB8 using an α-myosin heavy chain promoter (12), whereas no alteration in endogenous gene expression was observed by the overexpression of other small HSPs, such as CryAB, using the same system (13). Thus, HSPB8 may modify its own gene expression. Because HSPB8 can translocate to the nuclei and can modulate the function of transcription factors such as STAT3, HSPB8 may play a self-regulating role in its gene expression (9).
Summary
Overexpressing HSPB8 K141N resulted in increased perinuclear HSPB8-positive aggregates containing amyloid oligomer and mild cellular toxicity, whereas no aggregates or cellular toxicity were observed in myocytes overexpressing wild-type HSPB8 in vitro and in vivo. Recombinant HSPB5 K141N protein showed reactivity against an anti-oligomer antibody. The reactivity of the mutant HSPB8 protein was much higher than that of the wild-type HSPB8 protein. Thus, a missense mutation of HSPB8 such as K141N can affect cellular function in cardiomyocytes and may cause cardiomyopathy as well as HMN and CMT disease.
This work was supported in part by research grants from the Scientific Fund of the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Ministry of Health, Labour and Welfare, the Japan Health Sciences Foundation, the Astellas Foundation for Research on Metabolic Disorders, the Mitsubishi Pharma Research Foundation, the Naito Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Suzuken Memorial Foundation.
- HSP
- heat shock protein
- CryAA
- αA-crystallin
- CryAB
- αB-crystallin
- TG
- transgenic
- NTG
- nontransgenic
- HMN
- distal hereditary motor neuropathy
- CMT disease
- Charcot-Marie-Tooth disease
- tTA
- tetracycline-controlled transcriptional activator
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- MOI
- multiplicity of infection
- VDAC
- voltage dependent anion channel.
REFERENCES
- 1. Taylor R. P., Benjamin I. J. (2005) Small heat shock proteins. A new classification scheme in mammals. J. Mol. Cell. Cardiol. 38, 433–444 [DOI] [PubMed] [Google Scholar]
- 2. Sun Y., MacRae T. H. (2005) Small heat shock proteins. Molecular structure and chaperone function. Cell Mol. Life Sci. 62, 2460–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee J. S., Samejima T., Liao J. H., Wu S. H., Chiou S. H. (1998) Physiological role of the association complexes of α-crystallin and its substrates on the chaperone activity. Biochem. Biophys. Res. Commun. 244, 379–383 [DOI] [PubMed] [Google Scholar]
- 4. Muchowski P. J., Wacker J. L. (2005) Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6, 11–22 [DOI] [PubMed] [Google Scholar]
- 5. Sanbe A., Osinska H., Saffitz J. E., Glabe C. G., Kayed R., Maloyan A., Robbins J. (2004) Desmin-related cardiomyopathy in transgenic mice. A cardiac amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10132–10136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hsu C. D., Kymes S., Petrash J. M. (2006) A transgenic mouse model for human autosomal dominant cataract. Invest. Ophthalmol. Vis. Sci. 47, 2036–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Irobi J., Van Impe K., Seeman P., Jordanova A., Dierick I., Verpoorten N., Michalik A., De Vriendt E., Jacobs A., Van Gerwen V., Vennekens K., Mazanec R., Tournev I., Hilton-Jones D., Talbot K., Kremensky I., Van Den Bosch L., Robberecht W., Van Vandekerckhove J., Van Broeckhoven C., Gettemans J., De Jonghe P., Timmerman V. (2004) Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 36, 597–601 [DOI] [PubMed] [Google Scholar]
- 8. Tang B. S., Zhao G. H., Luo W., Xia K., Cai F., Pan Q., Zhang R. X., Zhang F. F., Liu X. M., Chen B., Zhang C., Shen L., Jiang H., Long Z. G., Dai H. P. (2005) Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum. Genet. 116, 222–224 [DOI] [PubMed] [Google Scholar]
- 9. Qiu H., Lizano P., Laure L., Sui X., Rashed E., Park J. Y., Hong C., Gao S., Holle E., Morin D., Dhar S. K., Wagner T., Berdeaux A., Tian B., Vatner S. F., Depre C. (2011) H11 kinase/heat shock protein 22 deletion impairs both nuclear and mitochondrial functions of STAT3 and accelerates the transition into heart failure on cardiac overload. Circulation 124, 406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kappé G., Verschuure P., Philipsen R. L., Staalduinen A. A., Van de Boogaart P., Boelens W. C., De Jong W. W. (2001) Characterization of two novel human small heat shock proteins. Protein kinase-related HspB8 and testis-specific HspB9. Biochim. Biophys. Acta 1520, 1–6 [DOI] [PubMed] [Google Scholar]
- 11. Sanbe A., Gulick J., Hanks M. C., Liang Q., Osinska H., Robbins J. (2003) Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ. Res. 92, 609–616 [DOI] [PubMed] [Google Scholar]
- 12. Sanbe A., Daicho T., Mizutani R., Endo T., Miyauchi N., Yamauchi J., Tanonaka K., Glabe C., Tanoue A. (2009) Protective effect of geranylgeranylacetone via enhancement of HSPB8 induction in desmin-related cardiomyopathy. PLoS One 4, e5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanbe A., Yamauchi J., Miyamoto Y., Fujiwara Y., Murabe M., Tanoue A. (2007) Interruption of CryAB-amyloid oligomer formation by HSP22. J. Biol. Chem. 282, 555–563 [DOI] [PubMed] [Google Scholar]
- 14. Sanbe A., Marunouchi T., Yamauchi J., Tanonaka K., Nishigori H., Tanoue A. (2011) Cardioprotective effect of nicorandil, a mitochondrial ATP-sensitive potassium channel opener, prolongs survival in HSPB5 R120G transgenic mice. PLoS One 6, e18922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamauchi J., Miyamoto Y., Sanbe A., Tanoue A. (2006) JNK phosphorylation of paxillin, acting through the Rac1 and Cdc42 signaling cascade, mediates neurite extension in N1E-115 cells. Exp. Cell Res. 312, 2954–2961 [DOI] [PubMed] [Google Scholar]
- 16. Maloyan A., Sanbe A., Osinska H., Westfall M., Robinson D., Imahashi K., Murphy E., Robbins J. (2005) Mitochondrial dysfunction and apoptosis underlie the pathogenic process in αB-crystallin desmin-related cardiomyopathy. Circulation 112, 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Motegi K., Tanonaka K., Takenaga Y., Takagi N., Takeo S. (2007) Preservation of mitochondrial function may contribute to cardioprotective effects of Na+/Ca2+ exchanger inhibitors in ischaemic/reperfused rat hearts. Br. J. Pharmacol. 151, 963–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanbe A., Osinska H., Villa C., Gulick J., Klevitsky R., Glabe C. G., Kayed R., Robbins J. (2005) Reversal of amyloid-induced heart disease in desmin-related cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 102, 13592–13597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. d'Ydewalle C., Krishnan J., Chiheb D. M., Van Damme P., Irobi J., Kozikowski A. P., Vanden Berghe P., Timmerman V., Robberecht W., Van Den Bosch L. (2011) HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat. Med. 17, 968–974 [DOI] [PubMed] [Google Scholar]
- 20. Brady J. P., Garland D. L., Green D. E., Tamm E. R., Giblin F. J., Wawrousek E. F. (2001) αB-Crystallin in lens development and muscle integrity. A gene knockout approach. Invest. Ophthalmol. Vis. Sci. 42, 2924–2934 [PubMed] [Google Scholar]
- 21. Pinz I., Robbins J., Rajasekaran N. S., Benjamin I. J., Ingwall J. S. (2008) Unmasking different mechanical and energetic roles for the small heat shock proteins CryAB and HSPB2 using genetically modified mouse hearts. FASEB J. 22, 84–92 [DOI] [PubMed] [Google Scholar]
- 22. Evgrafov O. V., Mersiyanova I., Irobi J., Van Den Bosch L., Dierick I., Leung C. L., Schagina O., Verpoorten N., Van Impe K., Fedotov V., Dadali E., Auer-Grumbach M., Windpassinger C., Wagner K., Mitrovic Z., Hilton-Jones D., Talbot K., Martin J. J., Vasserman N., Tverskaya S., Polyakov A., Liem R. K., Gettemans J., Robberecht W., De Jonghe P., Timmerman V. (2004) Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat. Genet. 36, 602–606 [DOI] [PubMed] [Google Scholar]
- 23. Depre C., Hase M., Gaussin V., Zajac A., Wang L., Hittinger L., Ghaleh B., Yu X., Kudej R. K., Wagner T., Sadoshima J., Vatner S. F. (2002) H11 kinase is a novel mediator of myocardial hypertrophy in vivo. Circ. Res. 91, 1007–1014 [DOI] [PubMed] [Google Scholar]
- 24. Kayed R., Head E., Thompson J. L., McIntire T. M., Milton S. C., Cotman C. W., Glabe C. G. (2003) Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300, 486–489 [DOI] [PubMed] [Google Scholar]
- 25. Glabe C. G. (2006) Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol. Aging 27, 570–575 [DOI] [PubMed] [Google Scholar]
- 26. Bera S., Thampi P., Cho W. J., Abraham E. C. (2002) A positive charge preservation at position 116 of αA-crystallin is critical for its structural and functional integrity. Biochemistry 41, 12421–12426 [DOI] [PubMed] [Google Scholar]
- 27. Braun N., Zacharias M., Peschek J., Kastenmüller A., Zou J., Hanzlik M., Haslbeck M., Rappsilber J., Buchner J., Weinkauf S. (2011) Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc. Natl. Acad. Sci. U.S.A. 108, 20491–20496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bennardini F., Wrzosek A., Chiesi M. (1992) αB-crystallin in cardiac tissue. Association with actin and desmin filaments. Circ. Res. 71, 288–294 [DOI] [PubMed] [Google Scholar]
- 29. Golenhofen N., Arbeiter A., Koob R., Drenckhahn D. (2002) Ischemia-induced association of the stress protein αB-crystallin with I-band portion of cardiac titin. J. Mol. Cell. Cardiol. 34, 309–319 [DOI] [PubMed] [Google Scholar]
- 30. den Engelsman J., Keijsers V., de Jong W. W., Boelens W. C. (2003) The small heat-shock protein αB-crystallin promotes FBX4-dependent ubiquitination. J. Biol. Chem. 278, 4699–4704 [DOI] [PubMed] [Google Scholar]
- 31. Kamradt M. C., Chen F., Cryns V. L. (2001) The small heat shock protein αB-crystallin negatively regulates cytochrome c- and caspase-8-dependent activation of caspase-3 by inhibiting its autoproteolytic maturation. J. Biol. Chem. 276, 16059–16063 [DOI] [PubMed] [Google Scholar]
- 32. Maloyan A., Osinska H., Lammerding J., Lee R. T., Cingolani O. H., Kass D. A., Lorenz J. N., Robbins J. (2009) Biochemical and mechanical dysfunction in a mouse model of desmin-related myopathy. Circ. Res. 104, 1021–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]