Background: Dietary β-carotene is the natural precursor of vitamin A.
Results: The retinoic acid-inducible homeobox transcription factor ISX controls intestinal expression of the vitamin A producing enzyme BCMO1.
Conclusion: Vitamin A production is under negative feedback regulation.
Significance: Large individual variability in intestinal β-carotene conversion is associated with this diet-responsive regulatory network.
Keywords: Carotenoid, Gene Regulation, Intestinal Metabolism, Lipids, Vitamin A, BCMO1, ISX, β-Carotene
Abstract
Low dietary intake of β-carotene is associated with chronic disease and vitamin A deficiency. β-Carotene is converted to vitamin A in the intestine by the enzyme β-carotene-15,15′-monoxygenase (BCMO1) to support vision, reproduction, immune function, and cell differentiation. Considerable variability for this key step in vitamin A metabolism, as reported in the human population, could be related to genetics and individual vitamin A status, but it is unclear how these factors influence β-carotene metabolism and vitamin A homeostasis. Here we show that the intestine-specific transcription factor ISX binds to the Bcmo1 promoter. Moreover, upon induction by the β-carotene derivative retinoic acid, this ISX binding decreased expression of a luciferase reporter gene in human colonic CaCo-2 cells indicating that ISX acts as a transcriptional repressor of BCMO1 expression. Mice deficient for this transcription factor displayed increased intestinal BCMO1 expression and produced significantly higher amounts of vitamin A from supplemental β-carotene. The ISX binding site in the human BCMO1 promoter contains a common single nucleotide polymorphism that is associated with decreased conversion rates and increased fasting blood levels of β-carotene. Thus, our study establishes ISX as a critical regulator of vitamin A production and provides a mechanistic explanation for how both genetics and diet can affect this process.
Introduction
Vitamin A (all-trans-retinol) is required for mammalian vision, reproduction, immune function, and cell differentiation. In the eye, vitamin A is converted to 11-cis-retinal, the visual chromophore, in a complex pathway known as the retinoid cycle (1). Vitamin A can also be oxidized to all-trans-retinoic acid (RA),2 a hormone-like compound that binds to a specialized class of nuclear receptors called retinoic acid receptors (RARs) (2). RARs are ligand-regulated transcription factors, which in conjunction with retinoid X receptors control the activity of genes in diverse cell types (3).
Dietary vitamin A precursors are absorbed in the intestine and delivered systemically in the form of retinyl esters bound to triglyceride-rich lipoproteins. Inadequate dietary supply of vitamin A is a major health problem, causing blindness and increased mortality in children and pregnant women, especially in large regions of Asia and Africa (4). Carotenoids such as β-carotene (BC) from plant food are the natural precursors of vitamin A in the human diet (5). BC must be absorbed in the intestine before it can be metabolically converted to vitamin A. It has long been assumed that such absorption occurs by passive non-ionic diffusion, but emerging evidence indicates that absorption of carotenoids and other lipid soluble vitamins is actively facilitated by class 2 scavenger receptors such as SCARB1 and CD36 (6). The subsequent conversion of BC to retinoids is catalyzed by the enzyme β-carotene-15,15′-monoxygenase (BCMO1) in enterocytes of the small intestine (7). The primary cleavage product of BC, retinaldehyde, is reduced to retinol, which is esterified and incorporated into chylomicrons that are secreted into the lymph for storage in the liver and tissue distribution (8).
Surprisingly, the bioavailability of BC from food is considered to be low and influenced by the food matrix and vitamin A status of the host (9). Additionally, intervention studies have revealed large individual variations in intestinal BC metabolism, leading to identification of a “low converter” phenotype (10). In these individuals, low conversion efficiency occurs after BC supplementation, reflected by the retinyl ester/BC ratio in the chylomicron fraction (10). About 45% of the Western population can be classified as low converters (11, 12). Recent studies provide evidence that genetics may contribute to these differences. A genome-wide study has associated a single nucleotide polymorphism (SNP, rs6564851) 7-kb upstream of the BCMO1 gene with increased fasting BC serum levels (13). A small intervention study provided evidence that this SNP is associated with reduced intestinal BC conversion (14). Additionally, several lines of evidence indicate that conversion of BC is under negative feedback control of vitamin A (15–17). A recent study suggests that this dietary responsiveness is mediated by the intestine-specific homeobox transcription factor ISX (18). ISX harbors an RAR binding motif in its promoter region and its expression can be induced by the BC metabolite RA (19). Upon induction of this transcription factor, intestinal Bcmo1 mRNA expression decreases (18, 19).
The effects of genetics and diet might hold a key to understand individual variability in intestinal BC metabolism in humans, but molecular details of how intestinal BCMO1 expression is controlled and how this regulation might be affected by genetics require elucidation. Additionally, direct evidence from animal models is lacking to evaluate the concept that ISX controls intestinal vitamin A production. A proper description of this regulatory network would help provide appropriate recommendations for BC intake to combat vitamin A deficiency. Thus, we aimed to characterize how ISX controls BCMO1 promoter activity and to study the role of ISX for vitamin A homeostasis in a mouse model.
EXPERIMENTAL PROCEDURES
Materials
Platinum Pfx polymerase, Prolong Gold anti-fade mounting medium, MAX efficiency competent cells DH5α, BL21, BL21(DE3), conjugated secondary antibody Alexa 488, and antibiotics were purchased from Invitrogen. Taq DNA polymerase, X-tremeGENE HD transfection reagents, and protease inhibitor mixture tablets were obtained from Roche Applied Science. Restriction endonucleases were obtained from New England BioLabs (Ipswich, MA). DMEM was obtained from Invitrogen. M-PER (mammalian) and B-PER (bacterial) protein extraction reagents, and the BCA protein assay kit were from Pierce Biotechnology Inc. Primary anti-ISX (C-16) antibodies were from Santa Cruz Biotechnologies (Santa Cruz, CA), loading control anti-α-tubulin was purchased from Cell Signaling Technologies (Danvers, MA), whereas anti-Bcmo1 antibody was obtained as previously described (20). All primers were synthesized and obtained from Integrated DNA Technologies (IDT, Coralville, IA). Secondary antibodies were purchased from Cell Signaling Technologies (Danvers, MA) or Bio-Rad. The pGL3-promoter luciferase vector and relevant reagents for luciferase assays were provided by Promega (Madison, WI). Solvents used for retinoid extraction and chromatography were of HPLC grade (Fisher Scientific).
ISX Protein Extracts for DNA Binding Assays
Total RNA was isolated from mouse intestine with TRIzol reagent (Invitrogen). About 2 μg of total RNA was reverse transcribed to cDNA using the RNA to cDNA kit as outlined by the manufacturer (Applied Biosystems, Carlsbad, CA). Using this cDNA as a template, full-length mouse ISX cDNA was PCR amplified using primer pairs listed in Table 1 and cloned into the pTrcHis2 TOPO TA vector. Murine WT-ISX in the pTrcHis2 TOPO TA vector (pTrcHis2-WT-ISX) was then transformed into Escherichia coli BL21 or XL1 blue cells. We isolated crude protein extracts from three different colonies. Cells were grown in Luria-Bertani (LB) medium with 100 μg/ml of the appropriate antibiotics at 37 °C overnight. After achieving an A600 of 0.8, cells were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside for 6 h at 25 °C for ISX expression. Total protein extracts were then prepared as per published protocols (21). Expression of ISX protein in different supernatant and pellet fractions was determined by immunoblot analysis with an anti-His antibody (Qiagen). Extracts from different colonies yielded similar results in EMSA. As a control protein extract, we isolated crude protein from E. coli transfected with an empty pTrcHis2 vector by the same protocol.
TABLE 1.
Sequences of primers used for PCR-amplification of mouse Bcmo1 promoter fragments and full length mouse ISX cDNA
| Fragment | Position from ATG | Vector | Forward primer | Reverse primer | Product size |
|---|---|---|---|---|---|
| Mouse | |||||
| Bcmo1, fragment1 | − 1660 to − 2200 | pTrcHis2 | 5′-gagctcttagtgtatcccag-3′ | 5′-ggagggtgagagctgagcag-3′ | 540 bp |
| Bcmo1, fragment 2 | − 1290 to − 1721 | pTrcHis2 | 5′-acagcctgttctagaacgtt-3′ | 5′-cctgctgcctgctgcctgct-3′ | 431 bp |
| Bcmo1, fragment 3 | − 850 to − 1350 | pTrcHis2 | 5′-actgggattgtatacatgca-3′ | 5′-tcgggaacccttcagttcaa-3′ | 500 bp |
| Bcmo1, fragment 4 | − 479 to − 980 | pTrcHis2 | 5′-ctcctggctgagggcgtcac-3′ | 5′-actattgcaagttgtgggct-3′ | 501 bp |
| Bcmo1, fragment 5 | − 180 to − 579 | pTrcHis2 | 5′-ttttaagagtgggaacgaac-3′ | 5′-gctctttgctccttgacatc-3′ | 399 bp |
| Bcmo1, fragment 6 | − 1 to − 340 | pTrcHis2 | 5′-ctcttccgactcagagcgga-3′ | 5′-ggaaccggtgagcggcagca-3′ | 339 bp |
| Full-length Bcmo1, promoter | − 1 to − 2200 | pTrcHis2 and pGL3-promoter | 5′-gagctcttagtgtatcccag-3′ | 5′-ggaaccggtgagcggcagca-3′ | 2200 bp |
| Bcmo1, promoter-F2/F3 fragment | − 1290 to − 1350 | pTrcHis2 and pGL3-promoter | 5′-cctgcctcaacctcctgag-3′ | 5′-gcctgctgcctgctgcctg-3′ | 60 bp |
| Bcmo1, promoter-Hbox | − 1309 to − 1357 | pGL3-promoter | 5′-cctgcctcaacctcctgag-3′ | 5′-tgcctgctgcatgtataca − 3′ | 48 bp |
| ISX | Full-length cDNA | pTRcHis2 | 5′-atggcaggccctacgatcca-3′ | 5′-atggcaggccctacgatccacag-3′ | 2163 bp |
| Human | |||||
| Bcmo1, SNP region P1 | Region − 7943 to − 7893 | ChiP | 5′-tgcctactgtattttaag-3′ | 5′-taaacagtgtaaaactgt-3′ | 50 bp |
| Bcmo1, SNP region P2 | Region − 7943 to − 7893 | ChiP | 5′-tgcctactgtattttaagca-3′ | 5′-taaacagtgtaaaactgt-3′ | 50 bp |
| Mouse | |||||
| Bcmo1 region P1 | Region − 1350 to − 1290 | ChiP | 5′-gctaagctggttggccatcag-3′ | 5′-cctgctgcctgctgcctgct | 119 bp |
| Bcmo1 region P2 | Region − 1350 to − 1290 | ChiP | 5′-gttggccatcagctaaggag-3′ | 5′-ctgtctgctgtctgctgcctgc-3′ | 122 bp |
Cloning of Full-length Bcmo1 Promoter
The mouse Bcmo1 promoter (2.2 kb) was subdivided into six overlapping DNA fragments (339–540 bp in length). Each fragment was PCR amplified with genomic DNA isolated from mouse intestine as a template and the oligonucleotide primer pairs listed in Table 1, essentially as described previously (22). An aliquot of each PCR product was evaluated on 2% agarose gels to determine its size and relative purity. The six individual Bcmo1 promoter fragments were subsequently cloned into the pTrcHis2 TOPO TA vector according to the manufacturer's protocols (Invitrogen). Appropriate construction and cloning of Bcmo1 promoter fragments were confirmed by direct sequencing of both nucleotide strands.
Radioactive Isotope Labeling of Individual Bcmo1 Promoter Fragments
The six individual Bcmo1 promoter fragments cloned into the pTrcHis2 TOPO TA vector were excised with NcoI and EcoRI restriction enzymes. Restricted Bcmo1 promoter fragments were gel-purified (Qiagen, Valencia, CA), and eluted in Tris-EDTA (TE) buffer. Following purification, individual fragments were 5′ end labeled with [α-32P]dATP (Amersham Biosciences) using T4 polynucleotide kinase. TE buffer (100 μl) was then added to the [α-32P]ATP/Bcmo1 promoter fragment reaction mixtures, which then were further purified on Sephadex G-25 spin columns (GE Healthcare). The radioactivity of individually labeled Bcmo1 promoter fragments was determined by measuring the counts per minute (cpm) of 1 μl of radiolabeled double stranded DNA in a scintillation counter (Beckman-Coulter LS6500). About 12,000 cpm of individually labeled 32P-Bcmo1 promoter fragments were used in the EMSA reactions.
Electrophoresis Mobility Shift Assays (EMSAs)
EMSAs were carried out according to published procedures (21). Radiolabeled Bcmo1 promoter fragments (∼12,000 cpm) were incubated for 10 min at room temperature (RT) with crude ISX protein extract from BL21 cells in EMSA buffer containing 10 mm Tris-HCl, pH 7.5, 50 mm NaCl, 3 mm MgC12, 1 mm EDTA, and 10% glycerol. Protein-DNA complexes then were separated from unbound DNA by gel electrophoresis on 6 or 8% nondenaturing polyacrylamide gels. For supershift assays, either 2 μg of anti-ISX antibody or 2 μg of control rabbit IgG antibody were added to the binding reaction and incubated for 10–30 min at RT. In control reactions, radiolabeled Bcmo1 promoter fragments were incubated with an E. coli protein extract without recombinant ISX protein. Gels were dried under vacuum and analyzed by autoradiography. To prevent nonspecific binding of proteins to labeled DNA fragments, the synthetic heteropolymer poly(dI-dC) was added to a concentration of 2 μg/μl in each reaction. To confirm that the observed protein-DNA complexes resulted from specific binding, a 100-fold molar excess of the corresponding unlabeled Bcmo1-promoter fragments also was added to compete with nonspecific protein-DNA binding. All EMSA images in relevant figures are representative of at least two independent experiments.
Plasmid Construction for Luciferase Reporter Assays
The reporter vector used for transfection into mammalian cells was pGL3 (Promega), which provided a basis for the quantitative analysis of factors potentially regulating mammalian gene expression. Taking advantage of this system, we first generated three Bcmo1 promoter fragments by PCR using primer pairs listed in Table 1: the full-length murine Bcmo1 promoter fragment (nucleotides −1 to −2200, of the ATG start site), the 60-bp murine Bcmo1 promoter fragment (nucleotides −1290 to −1350), and the 60-bp murine Bcmo1 promoter fragment containing the SNP in the human Bcmo1 promoter, the last generated by site-directed mutagenesis. We then cloned these three fragments individually into the pGL3-promoter vector and named them pGL3 Bcmo1-full promoter (2200 bp), pGL3 Bcmo1-promoter-F2/F3 fragment (60 bp), and pGL3 Bcmo1-F2/F3 SNP (60 bp fragment with the G/T SNP), respectively. Postcloning, the recombinant plasmids with inserts were sequenced to confirm the integrity of the inserts in each construct.
Determination of Bcmo1 Promoter Activities by the Luciferase Assay
CaCo-2 (human adenocarcinoma) cells (ATCC, Manassas, VA) were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 4.5 mm glutamine, and 100 units/ml of penicillin/streptomycin. CaCo-2 cells were seeded in 6-well plates (density ∼2.0 × 105 cells per well) in DMEM with 10% (v/v) FBS at 37 °C for 24 h or until reaching 50–60% confluence. Each well then was transfected with either 3 μg of empty pGL3-promoter vector (pGL3-promoter vector) or one of the different pGL3-promoter constructs as indicated in the figures and corresponding legends. Specifically to determine Bcmo1 promoter activities, plasmid DNA was mixed with X-tremeGENE HD (Roche Applied Science) transfection reagent at a ratio of 1 μg of DNA to 3 μl of transfection reagent in OptiMEM (Invitrogen) and incubated for 20 min at RT to initiate formation of complexes (19, 23) before transfecting cells with individual plasmids. Forty hours after transfection, cells were harvested with StableGLO cell lysis buffer (Promega). Luciferase activities were measured as luminescent units (relative light units) and data were normalized to protein concentrations determined by the Bradford protein assay (Bio-Rad).
RNA Isolation and Quantitative Real Time (QRT)-PCR Analysis
RNA was isolated from animal tissues or cultured cells as described previously (23). Two μg of total RNA was reverse transcribed with a RNA to cDNA system (Applied Biosystems). QRT-PCR was carried out with TaqMan chemistry and Assays on Demand probes (Applied Biosystems) for mouse Isx (Mm_01243745m1) and mouse Bcmo1 (Mm01251350_m1). The 18S rRNA (4319413E) probe set (Applied Biosystems) was used as the endogenous control. All real time experiments were performed with the ABI Step One Plus QRT-PCR machine in triplicate and repeated twice, using freshly synthesized cDNA.
ChIP Assay and PCR
To evaluate binding of ISX to the Bcmo1 promoter, a ChIP assay was performed with the EZ-ChIP kit (Millipore, Temecula, CA) as previously described (19). Specifically, CaCo-2 cells were cultured in 150-cm2 dishes and transfected with 3 μg of pGL3-promoter plasmid containing the 2.2-kb full-length Bcmo1 promoter (pGL3 Bcmo1-promoter-full) from cells at 60% confluence. Forty hours post-transfection, cells were treated with 1 μm RA for 24 h at 37 °C to induce endogenous ISX expression. Cells then were treated with 1% paraformaldehyde at 37 °C with gentle swirling for 10 min to enable cross-linking of nuclear proteins with genomic DNA, followed by sonication to shear and obtain DNA fragment sizes between 300 and 1000 bp in length. Four μg of human ISX antibody (Santa Cruz Biotechnologies) was used for immunoprecipitation, whereas 4 μg of IgG was used as a control. Immunoprecipitated DNA was PCR amplified with primer pairs listed in Table 1. Mouse Bcmo1 promoter primers used to PCR amplify the 60-bp putative ISX-Bcmo1 binding region (nucleotides −1290 to −1350) and human BCMO1 promoter primers (nucleotides −7943 to −7893) both relevant to the ATG start site, are listed in Table 1. PCR products were analyzed by gel electrophoresis on 2% agarose gels.
Animals, Diets, and Experimental Procedures
Animal experiments were performed in accordance with United States animal protection laws with adherence to guidelines of the institutional animal care and use committee of Case Western Reserve University (Cleveland, OH). These also conformed to recommendations of both the American Veterinary Medical Association Panel on Euthanasia and the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Mice were kept in a room under controlled conditions (24 °C, 12-h light/dark cycle) with free access to food and water. Heterozygous Isx+/− mice (24) were mated with C57BL/6 mice. Heterozygous offspring from this crossing were further bred to obtain Isx−/− (knock-out) and Isx+/+ (wild-type) mice. Five-week-old mice (n = 5 animal per dietary condition and genotype) were fed for 8 weeks with diets supplemented, respectively, with 4 IU of vitamin A/g or 0.5 mg/g of BC provided as beadlets (DSM, Basel, Switzerland). Individual diets were prepared by Research Diets, Inc. (New Brunswick, NJ) with an AIN-93G formulation.
HPLC Separation of Retinoids from Tissues
Retinoids were extracted from tissues of WT and ISX-deficient mice (n = 5 per genotype and diet) under a dim red safety light (600 nm) and quantified by HPLC analysis after direct phase chromatography as previously described (25). For quantification of molar amounts of retinoids, the HPLC analyses were previously scaled with appropriate retinoid standards.
Immunostaining of Mouse Intestinal Tissues
Mouse intestinal tissue was fixed in a 4% paraformaldehyde/PBS solution. After 24 h, samples were placed in a 30% sucrose/PBS solution. When fully saturated with sucrose, tissue samples were mounted in OCT medium (Sakura Fine Chemicals, Japan) and cut into 20-μm thick sections with a cryostat (Leica). Sections were collected in phosphate-based saline (PBS), pH 7.4, and stored at 4 °C before immunostaining for ISX expression. For immunostaining, 20 sections for each experimental condition (n = 3 per genotype and diet) were washed twice in PBS containing 0.01% Triton X-100 (PBS-T) for 10 min at RT. Sections were then placed in blocking buffer (5% bovine serum, 1% BSA in PBS) at RT and shaken for 1–2 h in a thermomixer (Eppendorf, Hauppauge, NY) at 300 rpm, following which they were washed twice with PBS-T for 10 min at RT. Then sections were incubated with shaking in a thermomixer overnight at 4 °C with the appropriate primary antibody diluted (1:250) with the blocking buffer used above, and subsequently washed twice with PBS-T for 10 min at RT. Then tissue sections with primary antibody were shaken overnight at 4 °C in a thermomixer along with a 1:250 dilution of secondary antibody conjugated with Alexa 488 (green fluorescence) in the blocking buffer followed by 2 washes in PBS-T for 10 min each at RT. Finally, these washed samples with secondary antibody were exposed to DAPI (1 μg/ml in PBS-T) for 5 min at RT. Sections were finally washed three times for 10 min each, and mounted in mounting medium (Prolong Gold Antifade Reagent) on frost-free glass slides and allowed to dry. Fluorescent images were obtained by a Zeiss LSM 510 UV Meta confocal microscope equipped with an HCX Plan ×63 numerical aperture 1.4 oil-immersion objective lens. Zeiss confocal software, version 2.0, and a Zeiss AxioCam MR camera were also employed for image acquisition.
Protein Isolation and Western Blot Analyses
Total protein from animal small intestine (n = 5 per genotype and diet) was isolated by using the M-PER mammalian protein extraction reagent with protease inhibitors (Roche Applied Science) according to the manufacturer's instructions (Pierce). One-hundred and fifty μg of total protein from extracts (n = 5 per genotype and diet) were pooled and vortexed. Twenty-five μg of the pooled protein extracts per lane were separated on 5–12% SDS-PAGE gels and then transferred onto PVDF membranes (Bio-Rad). Primary antibodies used anti-α-tubulin (1:10,000 dilutions) as the loading control and anti-ISX or anti-BCMO1, each at 1:1000 dilution. Membranes were incubated with their respective primary antibody overnight at 4 °C, followed by incubation with the appropriate HRP-conjugated secondary antibody for 1 h at RT before being visualized with the enhanced ECL chemiluminescence detection system (Pierce or Pharmacia).
Statistical Analyses
Results are presented as mean ± S.D. and the number of experiments is indicated in the figure legends. Statistical significance was assessed by two-way analysis of variance and two-tailed Student's t test using SPSS 14.0 for windows (SPSS, Chicago, IL), with the threshold of significance set at p < 0.05.
RESULTS
Electrophoresis Mobility Shift Assays of the Bcmo1 Promoter Reveal an ISX Binding Region
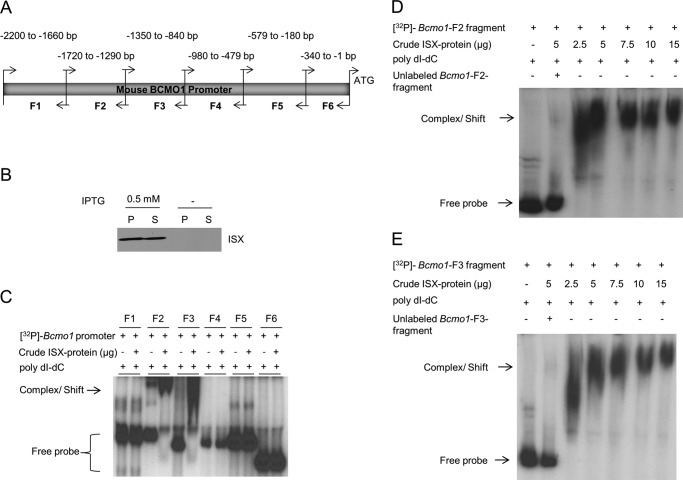
Previous studies indicate that ISX regulates intestinal Bcmo1 expression (18, 19), implying a physical interaction between the transcription factor and the Bcmo1 promoter. To test this hypothesis, we determined the region within the Bcmo1 promoter that might bind ISX. Electrophoretic mobility shift assays (EMSA) with 32P-labeled Bcmo1 promoter fragments and ISX protein extracts (Fig. 1, A and B) revealed that ISX bound two of the six 32P-labeled Bcmo1 promoter fragments (fragments 2 (nucleotides −1720 to −1290) and 3 (nucleotides −1350 to −840)) relative to the ATG initiation codon (Fig. 1C). To confirm that ISX binding to Bcmo1 promoter fragments 2 and 3 was specific, we added 100-fold excess of unlabeled fragments 2 and 3 to the respective reactions as competitors. The excess nonlabeled DNA fragments competed for binding of ISX to Bcmo1 promoter fragments in each case (Fig. 1, D and E, second lanes).
FIGURE 1.
ISX protein binds with BCMO1 promoter fragments. A, genomic DNA was isolated from mouse intestine and, using primer pairs listed in Table 1, the full-length Bcmo1 promoter was PCR amplified into six overlapping fragments (F1–F6) as indicated. Individual fragment sizes and positions are relevant to the Bcmo1 ATG start site. B, Western blot analysis for ISX protein expression in BL21 cells (± isopropyl β-d-thiogalactopyranoside (IPTG) induction), using the anti-His antibody. P, pellet fraction; S, supernatant/soluble fraction. C, in EMSAs, individual 32P-labeled Bcmo1 promoter fragments were incubated with crude ISX protein. Bcmo1 promoter fragments F2 and F3, each capable of interacting with ISX protein are indicated by the complex/shift (arrow) and faster migrating unbound [32P]DNA probes are shown as free probes within a bracket. D and E, ISX protein binds specifically to Bcmo1 promoter fragments 2 and 3. 32P-Labeled Bcmo1-promoter fragment 2 (D) and fragment 3 (E) were incubated with the supernatant fraction of ISX protein. Protein-DNA complexes were resolved on 6% PAGE gels, and results were obtained by autoradiography. Shifts of the ISX protein bound Bcmo1-probe complexes are indicated by arrows. Although the free unbound probe migrated rapidly upon electrophoreses, the retarded, shifted ISX-Bcmo1 bound probe migrated more slowly and above the labeled free DNA probe. The figures show representative autoradiographies of images from at least two independent experiments using different ISX protein crude extract preparations.
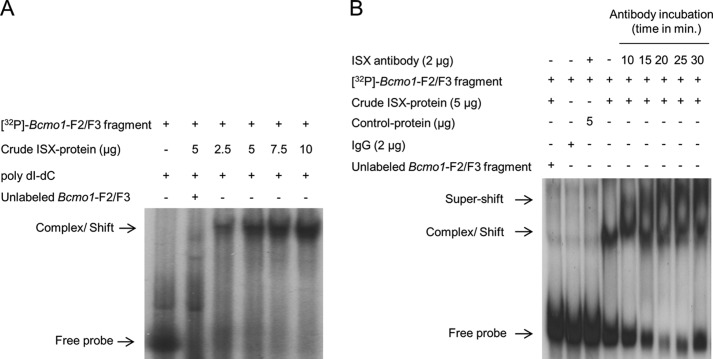
As each of the six Bcmo1 promoter fragments overlapped with neighboring fragments, we reasoned that the putative binding site for ISX might encompass a region shared by fragments 2 and 3 (Fig. 1A). To test this hypothesis, we repeated EMSAs using a 60-bp fragment corresponding to nucleotides −1290 to −1350, which included the overlapping region. Addition of crude extract from ISX-expressing cells shifted this 60-bp Bcmo1 promoter fragment almost entirely to a slower migrating DNA-protein complex (Fig. 2A). The labeled 60-bp Bcmo1 promoter fragment failed to shift when incubated with protein extracts from BL21 cells that did not express ISX, indicating that the shift in migration was due to specific ISX binding (Fig. 2B, third lane). To further demonstrate specificity of ISX binding to the Bcmo1 promoter, we incubated the 32P-labeled 60-bp fragment with crude ISX protein extract in the presence of ISX antibody. This resulted in slower migration indicative of a larger ISX-DNA-antibody supercomplex (Fig. 2B). Thus, nucleotides within the −1290 to −1350 region of the Bcmo1 promoter bind ISX specifically.
FIGURE 2.
Refinement of the potential ISX-Bcmo1 promoter binding region. A, a 60-bp DNA fragment encompassing overlapping regions between Bcmo1 promoter fragments 2 and 3 was 32P-labeled and reacted with the ISX protein. The ISX protein bound Bcmo1-probe complex, indicated by an arrow, is seen migrating higher than the free DNA probe. B, supershift assays were performed with a commercially available anti-human ISX antibody to evaluate the specificity of ISX protein binding to putative sites within the Bcmo1 promoter region. After incubating the 60-bp 32P-labeled Bcmo1 fragment with the crude ISX protein, ISX antibody was added to each reaction as indicated. As shown by arrows, complexes with ISX antibody (ISX-BCMO1-ISX) shows higher migration (super-shift) as compared with those with only ISX-Bcmo1 fragment (complex/shift). The figure shows representative autoradiographies of images from at least two independent experiments using different ISX protein crude extract preparations.
Human ISX Specifically Interacts with the Murine Bcmo1 Promoter
Human colonic CaCo-2 cells express ISX as an RA-responsive target gene (19). Indeed, treatment of CaCo-2 cells with 1 μm RA for 16 h, followed by immunohistochemistry and confocal microscopy revealed speckled nuclear ISX staining (Fig. 3A). To evaluate the identified Bcmo1-ISX DNA binding site in cells, we cloned the full murine Bcmo1 promoter into the pGL3-promoter/luciferase reporter vector and, after fixing transfected cells with formaldehyde to preserve DNA-protein interactions, performed chromatin immunoprecipitation (ChIP) using antiserum specific for human ISX (Fig. 3B). PCR with primer pairs that flank the candidate ISX binding site in the murine Bcmo1 promoter readily amplified the respective regions in ISX ChIP material from RA-induced CaCo-2 cells (Fig. 3B), but not in IgG control ChIP samples (Fig. 3B). We also cloned the identified ISX binding site (nucleotides −1350 to −1290) into the pGL3 reporter vector. CaCo-2 cells transfected with this plasmid showed increased luciferase activity (Fig. 3C) and treatment of these cells with RA reduced reporter activity (Fig. 3C). Thus, we provide evidence that human ISX binds to the murine Bcmo1 promoter and sequences between nucleotides −1350 and −1290 reduce basal pGL3 luciferase expression in an ISX-dependent manner.
FIGURE 3.
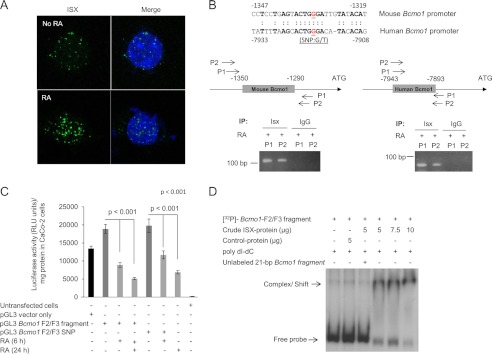
Bcmo1 promoter displays luciferase activity suppressible by ISX. A, induction of endogenous ISX expression in human colonic CaCo-2 cells by RA treatment. CaCo-2 cells were treated or not with 1 μm RA for 24 h. Cells were incubated with anti-human ISX primary antibody and ISX gene expression from the secondary Alexa 488 green fluorescent antibody was visualized by confocal microscopy after staining nuclei with DAPI. Representative images taken at ×63 magnification are shown. Approximately 50 cells from three separate experiments were counted, all of which showed similar staining patterns (B) (upper panel). Sequence comparison between the ISX binding mouse Bcmo1 and human Bcmo1 promoter region are shown. Shown are nucleotide sequences in relationship to the ATG start site from both species. Sequence similarities in Bcmo1 promoter regions between the human and mouse are indicated and the G/T SNP in the human Bcmo1 promoter is underlined. Lower left panel, to show binding of human ISX to the 60-bp putative binding region of the mouse Bcmo1 promoter, CaCo-2 cells were transfected with pGL3 vector containing the full-length Bcmo1-promoter fragment. After 40 h, cells were induced with RA for 24 h. ChIP assays in CaCo-2 cells were performed with the anti-human ISX antibody. IgG was used as a control. Two primer pairs flanking this region were designed for its detection in precipitated DNA fractions, namely P1, primer pair 1, and P2, primer pair 2. Lower right panel, to show binding of human ISX to the human Bcmo1 promoter in CaCo-2 cells, ChIP assays were performed with RA-induced cells. Two primer pairs relevant to the human sequence of Bcmo1 promoter (upper panel) were designed for its detection in precipitated DNA fractions, namely P1, primer pair 1, and P2, primer pair 2. C, human CaCo-2 cells were transfected with either the pGL3 Bcmo1 F2/F3 fragment or the pGL3 Bcmo1 F2/F3 fragment containing the human SNP (pGL3 Bcmo1 F2/F3 SNP). Induction of ISX expression by RA led to a decrease in luciferase activity in transfected cells relative to untreated cells expressing variants of the pGL3 plasmid. Luciferase activity is shown as relative luminescence units (RLU) per mg of protein. Data represent the mean of three independent experiments. p < 0.001 in two-tailed Student's t test. D, a 21-bp oligonucleotide corresponding to nucleotide region −1344 to −1324 (including the consensus homeobox murine sequence and the human G/T SNP) of the Bcmo1 promoter was used in these EMSA competition assays. The 32P-labeled 60-bp DNA fragment used above was incubated in the presence of ISX protein and a 100 m excess of the 21-bp unlabeled DNA.
ISX Binding to the Human BCMO1 Promoter Is Affected by a Single Base Pair Polymorphism
A genome-wide association study revealed that a SNP ((G/T)) in the G allele at rs6564851, near the human BCMO1 gene, is associated with altered levels of circulating carotenoids (13). A comparison of human and mouse genomic sequences revealed that this region is homologous to sequences within the putative ISX binding site (nucleotides −1329 to −1335) in the murine Bcmo1 promoter (Fig. 3B). To evaluate whether ISX can bind to this region of the human BCMO1 promoter, we employed ChIP assays using RA-induced CaCo-2 cells and ISX antiserum. We designed primers to detect the human BCMO1 promoter region containing the SNP and used these primers to amplify the putative BCMO1 promoter in ISX ChIP in RA-induced CaCo-2 cells (Fig. 3B). We detected PCR products when using ISX antibody but not when using IgG for ChIP (Fig. 3B). Thus, ISX can bind specifically in cells to the human BCMO1 promoter within a region encompassing a common SNP associated with plasma carotenoid levels (Fig. 3B).
To determine the functional effect of the rs6564851 polymorphism, we used site-directed mutagenesis to introduce the G/T SNP of the human ISX-Bcmo1 DNA binding site into a homologous sequence of the pGL3-Bcmo1-F2/F3 fragment. We individually transfected the wild-type or mutant pGL3-Bcmo1 F2/F3 plasmids into CaCo-2 cells, treated the cells with RA or vehicle, and measured the luciferase reporter activity. Vehicle-treated cells transfected with either pGL3-Bcmo1-F2/F3 plasmid displayed comparable luciferase activity, and RA treatment reduced this activity in cells treated with either pGL3-Bcmo1-F2/F3 plasmid construct (Fig. 3C). To further evaluate whether the sequence surrounding the SNP is part of the ISX binding site, we synthesized a 21-bp oligonucleotide within the murine Bcmo1 promoter, encompassing nucleotides −1324 to −1344, and incubated the 32P-labeled 60-bp Bcmo1 promoter fragment (Bcmo1-F2/F3) with this unlabeled duplex in the presence of crude ISX protein extract. The unlabeled oligonucleotide prevented binding of ISX to the labeled promoter probe (Fig. 3D). Thus, regions −1324 to −1344 in the Bcmo1 promoter serve as a preferential binding site for ISX (Fig. 3D).
Effects of ISX Deficiency on Intestinal Retinoid Metabolism and Vitamin A Homeostasis
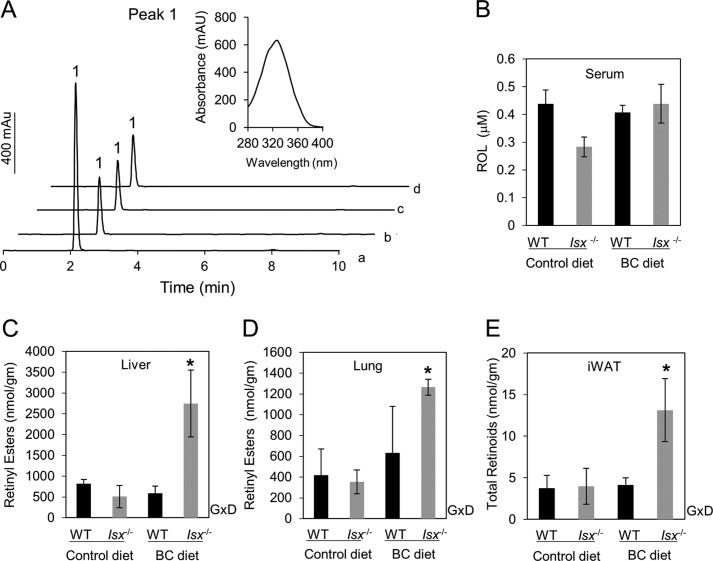
Taken together, the above findings suggest that ISX represses intestinal Bcmo1 expression. To provide evidence for this function in vivo, we propagated Isx−/− and wild-type (WT) mice on a similar genetic background and supplemented the diets of 5-week-old mice (n = 5 per genotype and dietary condition) with either retinyl esters (control diet) or BC (BC diet) as the source of vitamin A. During the feeding period, mice displayed no differences in weight gain attributable to genotype and/or diet. We euthanized mice after 8 weeks and collected blood, intestine, and the major organs used to store vitamin A: the liver, white adipose tissue, and lungs. Immunofluorescence, qRT-PCR, and immunoblotting verified the absence of ISX protein and mRNA in Isx−/− mice (Fig. 4). Duodenal tissue from WT mice showed a speckled pattern of staining for ISX in enterocyte nuclei, similar to observations in human colonic CaCo-2 cells (Figs. 3A and 4A). QRT-PCR analysis further revealed significantly higher Bcmo1 mRNA levels in Isx−/− compared with WT mice. In Isx−/− mice fed the control diet, Bcmo1 mRNA levels were about 9 and 61 times higher in the duodenum and jejunum, respectively (Fig. 4, C and D). In Isx−/− mice fed the BC diet, Bcmo1 mRNA levels were 16.3 times higher in the duodenum and 83.4 times higher in the jejunum (Fig. 4, B and C). This increased expression was also evident in immunoblot analyses of the mutant duodenum and jejenum (Fig. 4, E and F), with more pronounced differences in the latter.
FIGURE 4.
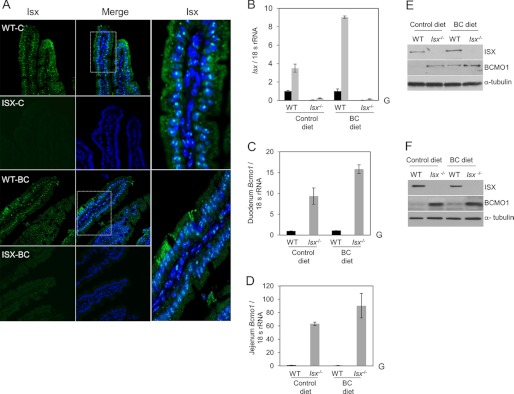
Consequences of ISX deficiency for BC metabolism. A, mouse duodenum from wild-type (WT) and Isx-deficient (Isx−/−) mice (n = 3 per genotype and diet) were immunostained with anti-ISX primary antibody, and ISX was visualized by using a secondary conjugated Alexa 488 antibody (green speckled fluorescence) and confocal microscopy. Nuclei were stained with DAPI. Representative images taken at ×40 magnification are shown. Speckled pattern of staining for ISX was observed in enterocyte nuclei. Approximately 20 sections per genotype and diet were scored for ISX localization. B–D, total RNA was isolated from intestinal tissue and expression of relevant genes was determined with gene-specific probe sets (ABI) and qRT-PCR. Values represent mean ± S.D. from 2 independent experiments carried out in triplicate. G, genotype effect in two way analysis of variance analysis (p < 0.05). B, solid black bars indicate the duodenal and gray bars indicate the jejunal mRNA expression levels. E and F, protein extracts from mice (n = 5 per genotype and diet) were obtained from intestinal tissue (duodenum and jejunum, respectively), pooled together, and 25 μg of protein per lane was subjected to immunoblot analysis. Immunoblots were probed with the relevant antibodies as indicated. α-Tubulin was used as the protein loading control. Representative immunoblots from three separate analysis are shown. WT, wild-type mice; Isx−/−, Isx deficient mice; BC diet, β-carotene diet.
To determine the consequences of ISX deficiency on vitamin A homeostasis, we measured retinoid levels in blood and tissues of WT and Isx−/− mice (Fig. 5, A–E). Serum levels of all-trans-retinol were comparable between diets and genotypes (Fig. 5B). Isx−/− and WT mice supplemented with preformed vitamin A also showed similar retinoid levels in the liver, lung, or white adipose tissues (Fig. 5, C–E), indicating that ISX deficiency does not interfere with the metabolism of preformed dietary vitamin A. This finding, however, contrasted sharply with results in BC-supplemented animals. Whereas WT mice on the BC diet displayed similar tissue retinoid stores as animals supplemented with preformed vitamin A (Fig. 5, C–E), Isx−/− mice on this diet showed nearly 3-fold higher retinoid stores in all tissues compared with their vitamin A-fed counterparts (Fig. 5, C–E).
FIGURE 5.
HPLC analysis for retinoids in various tissues of WT- and Isx-deficient mice. A, representative HPLC traces at 325 nm from liver extracts. The inset shows the spectral characteristics of the retinyl ester peak (peak 1). Trace a, Isx−/− mice (BC diet); trace b, Isx−/− mice (control diet); trace c, WT (control diet); trace d, WT (BC diet). B, all-trans-retinol (ROL) levels in serum. C, retinyl ester levels in liver, and D, lung are shown as well as, E, total retinoids (retinyl esters and all-trans-retinol) in inguinal white tissue (iWAT). For retinoid quantification, the HPLC was previously scaled with the appropriate standards. Isx−/− mice on the BC diet showed a significant increase of non-polar retinoids in the tissues examined. GxD, interaction between genotype and diet in two-way analysis of variance analysis (*, p < 0.05). WT, wild-type mice; Isx−/−, Isx-deficient mice; BC diet, β-carotene diet.
DISCUSSION
Here we show that the homeobox transcription factor ISX is an integral regulator of vitamin A production. We identify a binding site for ISX in the Bcmo1 promoter and provide evidence that ISX is a transcriptional repressor of Bcmo1 expression. Moreover, Isx knock-out mice displayed highly elevated intestinal mRNA and protein levels of Bcmo1 and produced significantly higher amounts of vitamin A from supplemented BC compared with wild-type mice. We also showed the ISX binding site in the human BCMO1 promoter contains a common sequence polymorphism.
A Homeobox Transcription Factor Controls Intestinal Vitamin A Production
ISX is a 242-amino acid homeodomain transcription factor expressed highly selectively in the intestine, where it is restricted to epithelial cells and appears during development just prior to the transition of gut endoderm to a columnar epithelium (24). Isx−/− mice are grossly normal and they are healthy when raised on standard vitamin A-rich mouse chow (24). Analysis of their intestinal transcriptome suggested that ISX regulates intestinal lipid metabolism, because expression of the scavenger receptor, class B type 1 (Scarb1) is significantly elevated (24). Studies in cell culture and animal models indicate that SCARB1 facilitates absorption not only of dietary BC but also other isoprenoid lipids such as tocopherols (vitamin E) and carotenoids, including lutein (26–29). Similar to Scarb1, intestinal expression of Bcmo1 is significantly elevated in Isx−/− mice, suggesting that ISX is involved in regulating vitamin A production. This conclusion is supported by intestinal expression of Isx as a function of dietary vitamin A levels (18). An explanation for this dietary responsiveness was found in an RAR binding element identified in the Isx promoter (19). Thus, RA produced from dietary precursors can induce intestinal Isx expression accompanied by a decrease of Scarb1 and Bcmo1 expression (19). In Bcmo1−/− mice, BC absorption is suppressed by preformed vitamin A that eventually is converted to RA (19).
Our data show that ISX is indeed a critical regulator of intestinal vitamin A production. Isx−/− mice produce significantly higher amounts of vitamin A from dietary BC than WT mice and amounts of stored vitamin A also far exceed the levels seen in animals fed a diet supplemented with preformed vitamin A. The absolute increase of retinyl esters was most pronounced in the liver, where ∼80% of postprandial retinyl esters are stored (8), although peripheral tissues such as the lung and white adipose tissue also showed a significant increase in retinyl ester accumulation. In contrast, serum vitamin A levels were similar between different mouse genotypes and diets, as expected, because serum levels are controlled by release from hepatic stores of vitamin A bound to the retinol-binding protein (8). Importantly, because we provided a low-fat diet, the amount of BC available was within the physiological range, corresponding to the amount in carotenoid-rich vegetables such as spinach.
For ISX to control production of vitamin A from dietary BC, it must interact with the promoter of its target genes. Using gel retardation of the murine Bcmo1 promoter with recombinant ISX protein, we narrowed down the ISX binding region to a 21-bp stretch about 1.3 kb upstream of the start ATG of the Bcmo1 gene. ChIP experiments suggested that human ISX also binds this site in the murine Bcmo1 promoter. Studies in CaCo-2 cells further revealed that the identified promoter element decreased expression of a reporter gene in an ISX-dependent fashion, corroborating ISX activity in transcriptional repression. This role for ISX was supported by the observation that Bcmo1 expression was highly increased in the intestine of Isx−/− mice. Such increased expression of Bcmo1 is likely driven by other transcription factors with specific binding sites in the Bcmo1 promoter (16, 30).
Implications for the Regulation of Vitamin A Homeostasis
Our study indicates that mammalian genomes devote a transcription factor to control vitamin A production in a specialized epithelium. Vitamin A plays a variety of important roles throughout life, and both deficiency and excess are associated with adverse health effects (2, 31). Dietary regulation of vitamin A production helps to prevent both scenarios and to adapt to fluctuating amounts of BC in the diet in natural environments. In vitamin A deficiency, increased expression of Sr-b1/Scrab1 and Bcmo1 in distal parts of the intestine allows efficient utilization of dietary BC. On BC-rich diets, induction of Isx expression by the BC derivative RA has an allosteric effect to ensure that vitamin A is not overproduced.
Our findings also shed light on inter-individual variability of intestinal BC metabolism in humans. A large genome-wide association study identified a SNP about 7 kb upstream of the BCMO1 gene that strongly correlates (p = 1.6 × 10−24) with increased fasting BC levels in serum (13). A small intervention study with female volunteers revealed that carriers of this SNP evidence a reduced BC conversion rate by about 50% (14). These observations could be explained by altered intestinal BCMO1 activity, an assumption supported by the finding that a rare missense mutation in human BCMO1 as well as genetic disruption of BCMO1 in mice lead to highly elevated BC blood levels and hypovitaminosis A (32, 33). We found that the sequence surrounding a human SNP showed perfect identity to an 8-bp sequence within the 21 nucleotides to which we localized ISX binding in the murine Bcmo1 promoter. ChIP experiments in CaCo-2 cells showed that, upon RA induction, human ISX binds to this DNA region. Notably, homeobox transcription factors use core DNA recognition sites of about 8 bp (34), and binding of human and murine ISX to DNA fragments containing the identical sequence support the proposal that it is part of the ISX DNA binding site. We introduced the relevant SNP into the pGL3 Bcmo1 F2/F3 reporter plasmid. Transactivation experiments in CaCo-2 cells showed that upon RA induction, ISX reduced activity of the basal promoter to the level of the control plasmid lacking the SNP, indicating that it does not significantly affect luciferase reporter gene expression. Nevertheless, subtle changes in ISX affinity for this binding site may result in reduced intestinal BCMO1 activity, as indicated by significant but small changes in BC serum levels of the SNP carriers (13). Thus, the binding of ISX to this specific sequence and the putative consequences of the SNP for Bcmo1 promoter activity need further experimental evaluation.
In conclusion, we show that ISX is a key component in control of vitamin A homeostasis in vivo. Studies with human CaCo-2 cells and the presence of a phenotypic SNP in the ISX binding site of the human BCMO1 gene suggest that a vitamin A/carotenoid diet-responsive regulatory network is conserved in mammals. Finally, this study provides evidence that the intestinal metabolism of vitamin A can be regulated in mammals and adapted to actual bodily requirements. Such regulation of intestinal vitamin A production, as well as genetic polymorphisms that affect this process, should be considered when making future recommendations for BC intake in health and disease.
Acknowledgment
We are grateful to Dr. Leslie Webster for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants EY019641 and EY020551.
- RA
- all-trans-retinoic acid
- BC
- β,β-carotene
- BCMO1
- β,β-carotene-15,15′-monooxygenase 1
- ISX
- intestinal specific homeobox transcription factor
- RAR
- retinoic acid receptor
- Scarb1/SR-B1
- scavenger receptor class B type I
- SNP
- single nucleotide polymorphism
- qRT-PCR
- quantitative-real time PCR.
REFERENCES
- 1. von Lintig J. (2012) Metabolism of carotenoids and retinoids related to vision. J. Biol. Chem. 287, 1627–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rhinn M., Dollé P. (2012) Retinoic acid signalling during development. Development 139, 843–858 [DOI] [PubMed] [Google Scholar]
- 3. Mark M., Ghyselinck N. B., Chambon P. (2006) Function of retinoid nuclear receptors. Lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 46, 451–480 [DOI] [PubMed] [Google Scholar]
- 4. Sommer A. (2008) Vitamin A deficiency and clinical disease. An historical overview. J. Nutr. 138, 1835–1839 [DOI] [PubMed] [Google Scholar]
- 5. Grune T., Lietz G., Palou A., Ross A. C., Stahl W., Tang G., Thurnham D., Yin S. A., Biesalski H. K. (2010) β-Carotene is an important vitamin A source for humans. J. Nutr. 140, 2268S–2285S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reboul E., Borel P. (2011) Proteins involved in uptake, intracellular transport and basolateral secretion of fat-soluble vitamins and carotenoids by mammalian enterocytes. Prog. Lipid Res. 50, 388–402 [DOI] [PubMed] [Google Scholar]
- 7. Wyss A., Wirtz G. M., Woggon W. D., Brugger R., Wyss M., Friedlein A., Riss G., Bachmann H., Hunziker W. (2001) Expression pattern and localization of β,β-carotene 15,15′-dioxygenase in different tissues. Biochem. J. 354, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. D'Ambrosio D. N., Clugston R. D., Blaner W. S. (2011) Vitamin A metabolism. An update. Nutrients 3, 63–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Castenmiller J. J., West C. E. (1998) Bioavailability and bioconversion of carotenoids. Annu. Rev. Nutr. 18, 19–38 [DOI] [PubMed] [Google Scholar]
- 10. Borel P., Grolier P., Mekki N., Boirie Y., Rochette Y., Le Roy B., Alexandre-Gouabau M. C., Lairon D., Azais-Braesco V. (1998) Low and high responders to pharmacological doses of β-carotene. Proportion in the population, mechanisms involved and consequences on β-carotene metabolism. J. Lipid Res. 39, 2250–2260 [PubMed] [Google Scholar]
- 11. Hickenbottom S. J., Follett J. R., Lin Y., Dueker S. R., Burri B. J., Neidlinger T. R., Clifford A. J. (2002) Variability in conversion of β-carotene to vitamin A in men as measured by using a double-tracer study design. Am. J. Clin. Nutr. 75, 900–907 [DOI] [PubMed] [Google Scholar]
- 12. Lin Y., Dueker S. R., Burri B. J., Neidlinger T. R., Clifford A. J. (2000) Variability of the conversion of β-carotene to vitamin A in women measured by using a double-tracer study design. Am. J. Clin. Nutr. 71, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 13. Ferrucci L., Perry J. R., Matteini A., Perola M., Tanaka T., Silander K., Rice N., Melzer D., Murray A., Cluett C., Fried L. P., Albanes D., Corsi A. M., Cherubini A., Guralnik J., Bandinelli S., Singleton A., Virtamo J., Walston J., Semba R. D., Frayling T. M. (2009) Common variation in the β-carotene 15,15′-monooxygenase 1 gene affects circulating levels of carotenoids. A genome-wide association study. Am. J. Hum. Genet. 84, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lietz G., Oxley A., Leung W., Hesketh J. (2012) Single nucleotide polymorphisms upstream from the β-carotene 15,15′-monoxygenase gene influence provitamin A conversion efficiency in female volunteers. J. Nutr. 142, 161S-165S [DOI] [PubMed] [Google Scholar]
- 15. von Lintig J. (2010) Colors with functions. Elucidating the biochemical and molecular basis of carotenoid metabolism. Annu. Rev. Nutr. 30, 35–56 [DOI] [PubMed] [Google Scholar]
- 16. Lietz G., Lange J., Rimbach G. (2010) Molecular and dietary regulation of β,β-carotene 15,15′-monooxygenase 1 (BCMO1). Arch. Biochem. Biophys. 502, 8–16 [DOI] [PubMed] [Google Scholar]
- 17. Bachmann H., Desbarats A., Pattison P., Sedgewick M., Riss G., Wyss A., Cardinault N., Duszka C., Goralczyk R., Grolier P. (2002) Feedback regulation of β,β-carotene 15,15′-monooxygenase by retinoic acid in rats and chickens. J. Nutr. 132, 3616–3622 [DOI] [PubMed] [Google Scholar]
- 18. Seino Y., Miki T., Kiyonari H., Abe T., Fujimoto W., Kimura K., Takeuchi A., Takahashi Y., Oiso Y., Iwanaga T., Seino S. (2008) Isx participates in the maintenance of vitamin A metabolism by regulation of β-carotene 15,15′-monooxygenase (Bcmo1) expression. J. Biol. Chem. 283, 4905–4911 [DOI] [PubMed] [Google Scholar]
- 19. Lobo G. P., Hessel S., Eichinger A., Noy N., Moise A. R., Wyss A., Palczewski K., von Lintig J. (2010) ISX is a retinoic acid-sensitive gatekeeper that controls intestinal β,β-carotene absorption and vitamin A production. FASEB J. 24, 1656–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lampert J. M., Holzschuh J., Hessel S., Driever W., Vogt K., von Lintig J. (2003) Provitamin A conversion to retinal via the β,β-carotene-15,15′-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development 130, 2173–2186 [DOI] [PubMed] [Google Scholar]
- 21. von Lintig J., Kreusch D., Schröder J. (1994) Opine-regulated promoters and LysR-type regulators in the nopaline (noc) and octopine (occ) catabolic regions of Ti plasmids of Agrobacterium tumefaciens. J. Bacteriol. 176, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nassif N. T., Lobo G. P., Wu X., Henderson C. J., Morrison C. D., Eng C., Jalaludin B., Segelov E. (2004) PTEN mutations are common in sporadic microsatellite stable colorectal cancer. Oncogene 23, 617–628 [DOI] [PubMed] [Google Scholar]
- 23. Lobo G. P., Waite K. A., Planchon S. M., Romigh T., Houghton J. A., Eng C. (2008) ATP modulates PTEN subcellular localization in multiple cancer cell lines. Hum. Mol. Genet. 17, 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Choi M. Y., Romer A. I., Hu M., Lepourcelet M., Mechoor A., Yesilaltay A., Krieger M., Gray P. A., Shivdasani R. A. (2006) A dynamic expression survey identifies transcription factors relevant in mouse digestive tract development. Development 133, 4119–4129 [DOI] [PubMed] [Google Scholar]
- 25. Amengual J., Golczak M., Palczewski K., von Lintig J. (2012) Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 287, 24216–24227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kiefer C., Sumser E., Wernet M. F., Von Lintig J. (2002) A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. 99, 10581–10586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Bennekum A., Werder M., Thuahnai S. T., Han C. H., Duong P., Williams D. L., Wettstein P., Schulthess G., Phillips M. C., Hauser H. (2005) Class B scavenger receptor-mediated intestinal absorption of dietary β-carotene and cholesterol. Biochemistry 44, 4517–4525 [DOI] [PubMed] [Google Scholar]
- 28. During A., Dawson H. D., Harrison E. H. (2005) Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is down-regulated in Caco-2 cells treated with ezetimibe. J. Nutr. 135, 2305–2312 [DOI] [PubMed] [Google Scholar]
- 29. Reboul E., Klein A., Bietrix F., Gleize B., Malezet-Desmoulins C., Schneider M., Margotat A., Lagrost L., Collet X., Borel P. (2006) Scavenger receptor class B type I (SR-BI) is involved in vitamin E transport across the enterocyte. J. Biol. Chem. 281, 4739–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boulanger A., McLemore P., Copeland N. G., Gilbert D. J., Jenkins N. A., Yu S. S., Gentleman S., Redmond T. M. (2003) Identification of β-carotene-15,15′-monooxygenase as a peroxisome proliferator-activated receptor target gene. FASEB J. 17, 1304–1306 [DOI] [PubMed] [Google Scholar]
- 31. Hall J. A., Grainger J. R., Spencer S. P., Belkaid Y. (2011) The role of retinoic acid in tolerance and immunity. Immunity 35, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hessel S., Eichinger A., Isken A., Amengual J., Hunzelmann S., Hoeller U., Elste V., Hunziker W., Goralczyk R., Oberhauser V., von Lintig J., Wyss A. (2007) CMO1 deficiency abolishes vitamin A production from β-carotene and alters lipid metabolism in mice. J. Biol. Chem. 282, 33553–33561 [DOI] [PubMed] [Google Scholar]
- 33. Lindqvist A., Sharvill J., Sharvill D. E., Andersson S. (2007) Loss-of-function mutation in carotenoid 15,15′-monooxygenase identified in a patient with hypercarotenemia and hypovitaminosis A. J. Nutr. 137, 2346–2350 [DOI] [PubMed] [Google Scholar]
- 34. Berger M. F., Badis G., Gehrke A. R., Talukder S., Philippakis A. A., Peña-Castillo L., Alleyne T. M., Mnaimneh S., Botvinnik O. B., Chan E. T., Khalid F., Zhang W., Newburger D., Jaeger S. A., Morris Q. D., Bulyk M. L., Hughes T. R. (2008) Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133, 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]