Background: Whether active DNA demethylase(s) exist in vertebrates is under debate.
Results: Mammalian DNA methyltransferases (DNMTs) can directly convert 5-methylcytosine in DNA to cytosine in vitro.
Conclusion: Vertebrate DNMTs can function as Ca2+- and redox state-dependent DNA demethylases.
Significance: The DNA demethylase activities of vertebrate DNMTs could regulate gene expression, development, neuroplasticity, carcinogenesis, etc., through global and/or local genomic demethylation.
Keywords: Calcium, Calcium Signaling, DNA Methylation, DNA Methyltransferase, Epigenetics, Redox, Redox Regulation, DNA Demethylation
Abstract
Methylation at the 5-position of DNA cytosine on the vertebrate genomes is accomplished by the combined catalytic actions of three DNA methyltransferases (DNMTs), the de novo enzymes DNMT3A and DNMT3B and the maintenance enzyme DNMT1. Although several metabolic routes have been suggested for demethylation of the vertebrate DNA, whether active DNA demethylase(s) exist has remained elusive. Surprisingly, we have found that the mammalian DNMTs, and likely the vertebrates DNMTs in general, can also act as Ca2+ ion- and redox state-dependent active DNA demethylases. This finding suggests new directions for reinvestigation of the structures and functions of these DNMTs, in particular their roles in Ca2+ ion-dependent biological processes, including the genome-wide/local DNA demethylation during early embryogenesis, cell differentiation, neuronal activity-regulated gene expression, and carcinogenesis.
Introduction
In vertebrates, DNA methylation occurs primarily at the 5-position of cytosine (C-5) in CpG dyads, and their genomic methylation patterns are established/maintained by the DNA (C-5)-methyltransferases (DNMTs).2 Of the known vertebrate DNMTs, DNMT1 shows a substrate preference for hemimethylated DNA and maintains the methylation patterns during DNA replication (1). On the other hand, DNMT3A and DNMT3B show equal C-5 methylation activities toward unmethylated and hemimethylated DNA in vitro, and they are essential for de novo genomic DNA methylation as well as development of early embryos (2, 3). The vertebrate DNA methylation system comprising the above three essential DNMTs is indispensable for the establishment of the genomic DNA methylation patterns, globally and locally, and consequently the processes of gene expression (4), neuroplasticity (5), differentiation (6), carcinogenesis (7, 8), imprinting (9), X-inactivation (10), and development (9, 11, 12). A fourth DNMT of vertebrates, DNMT2, can also methylate DNA (13, 14), but no phenotype(s) is associated with DNMT2-deficient mice (15). In cells, the DNA methyltransferase activities of the DNMTs are known to be regulated by a number of factors, including folate intake, redox state of the enzymes, ratio of S-adenosylmethionine (SAM) to S-adenosylhomocysteine, etc. (16).
A concerted action of DNA methylation and demethylation is essential for shaping the vertebrate genomes with specific patterns of 5-methylcytosine (5-mC) distribution. However, in contrast to the DNA C-5 methylation catalyzed by the single family of DNMT proteins mentioned above, multiple pathways have been proposed to carry out demethylation of the vertebrate genomes. The methyl-CpG-binding domain protein MBD2 has been reported to possess demethylase activity (17), but the result is yet to be reproduced by others. On the other hand, in parallel to the active DNA demethylation by the base excision repair (BER) pathways in plants (18), the vertebrate glycosylases appear to initiate DNA demethylation by directly removing the 5-mC base (19). Subsequently, the nucleotide excision repair and conversion of 5-mC to T through deamination reaction in combination with the BER pathway have been suggested to be involved in DNA C-5 demethylation of the vertebrate genomes (20–22). More recently, a family of mammalian TET proteins has been shown to convert 5-mC to 5-hydroxymethylcytosine (5-hmC), which promotes the idea of DNA demethylation through TET action and 5-hmC oxidation (23–25). In an interesting connection, mammalian DNMT3A and DNMT3B are capable of directly removing the hydroxymethyl moiety from 5-hmC in vitro in a redox state-dependent manner (26). Thus, these two DNMTs in combination with TET might actively demethylate 5-mC on DNA.
Despite all of the above, it has remained elusive whether enzyme(s) that can actively and directly convert 5-mC on DNA into C exist in vertebrates. We report here that in in vitro reactions, mouse and/or human DNMT1, DNMT3A, and DNMT3B acted as active DNA demethylases, removing the methyl group from 5-mC on DNA in an Ca2+ ion- and redox state-dependent manner.
EXPERIMENTAL PROCEDURES
Recombinant Plasmids and Recombinant Proteins
Construction of the expression plasmids used in this study was described previously (26). The DNA methylation-inactive mutants of the DNMTs, i.e. DNMT1-PSC, DNMT3A-PS, and DNMT3B-PS, were generated by insertion of a serine residue before Cys-1229 in the catalytic site of DNMT1 or by replacing Cys-706 and Cys-657 in the catalytic domains of DNMT3A and DNMT3B, respectively, with a serine residue. All of the recombinant DNMTs, including human DNMT1 (hDNMT1; purity of ∼78%), hDNMT3A (purity of ∼90%), and mouse DNMT3B (purity of ∼50%) (supplemental Fig. S1), were purchased from BPS Bioscience.
Cell Culture and DNA Transfection
293T cells were cultured under 5% CO2 at 37 °C in DMEM (Invitrogen) supplemented with 10% FBS (Biological Industries) and 1% penicillin/streptomycin (Invitrogen). For DNA transfections, the different expression plasmids were transfected into cells using either Lipofectamine 2000 (Invitrogen) or MAXifect (Omics Bio). The cells were collected 2 days later for further experimentation.
Preparation of Nuclear Extracts
Nuclear extracts were prepared from porcine sperm and 293T cells by a modified method (27). Briefly, the porcine semen was washed three times with PBS, and the sperm pellet was isolated using Ficoll (GE Healthcare). The pellet was resuspended in hypotonic buffer (10 mm Tris-HCl (pH 7.4), 10 mm NaCl, 1 mm EDTA, and EDTA-free protease inhibitors (Roche Applied Science)) on ice for 15 min. The resuspended sperm were passed 10 times through a 21-gauge needle and then centrifuged at 13,200 rpm for 10 min at 4 °C. The supernatant was removed, and the pellet of the nuclei was resuspended in resuspension buffer (10 mm Tris-HCl (pH 7.4), 10 mm NaCl, 1.5 mm MgCl2, and EDTA-free protease inhibitors), and an equal volume of 1 m NaCl was added, followed by a 30-min incubation on ice. The solution was centrifuged at 13,200 rpm for 30 min at 4 °C, and the supernatant (nuclear extract) was dialyzed at 4 °C in buffer B (10 mm Tris-HCl (pH 7.4), 50 mm NaCl, 1.5 mm MgCl2, and EDTA-free protease inhibitors) (26) overnight with 2 changes of the dialysis buffer.
Preparation of the nuclear extract from 293T cells followed the procedures described above. The transfected cells were washed three times with PBS and resuspended in hypotonic buffer on ice for 10 min. The solution was centrifuged at 4000 rpm for 10 min, and the supernatant was removed. The nuclear pellet was resuspended in resuspension buffer, and an equal volume of 1 m NaCl was added, followed by a 30-min incubation on ice. The lysate was centrifuged at 13,200 rpm for 30 min at 4 °C, and the supernatant was collected as the nuclear extract, which was then dialyzed overnight at 4 °C in buffer B.
DNA Substrates for in Vitro DNA Demethylation Assay
The 5-mC-containing substrate for DNA demethylation assay of the porcine sperm nuclear extract was prepared from the 2819-bp pMR1-8 plasmid containing 185 CpG dyads and 11 MspI restriction sites. The unmodified pMR1-8 plasmid was amplified in SCS110 bacteria (Stratagene) and then methylated by the bacterial methyltransferase M.SssI (New England Biolabs) in NEB Buffer 2 supplemented with 160 μm SAM. The extent of methylation of the plasmid was checked by HpaII digestion. C-5-methylated double-stranded DNA substrate (26) was used in the demethylation reactions with 293T nuclear extract or recombinant DNMTs (see below) and then analyzed by the hydrolysis-TLC assay.
In Vitro Reactions of Conversion of 5-mC to C on DNA
For DNA demethylation reactions of the porcine sperm nuclear extract, 40 ng of the methylated pMR1-8 plasmid was incubated with 100 μg of the nuclear extract in 50 μl of buffer B with 100 μg/ml BSA at 37 °C for 1–4 h. When needed, one of three divalent cations (Ca2+, Mg2+, or Fe2+; 10 μm to 10 mm), CRT0044876 (10 μm to 1 mm), 3-aminobenzamide (0.5 μm to 50 mm), or tetrahydrouridine (30 μm to 1 mm) was added to the reaction mixtures.
For DNA demethylation reactions of recombinant DNMT proteins or nuclear extracts from 293T cells overexpressing different DNMTs, 40 ng of the 5-mC-containing double-stranded DNA substrate was incubated with 40 nm recombinant DNMTs or 100 μg of the 293T nuclear extracts in 50 μl of buffer B with 100 μg/ml BSA at 37 °C for 0.5–8 h. When required, 10 μm to 10 mm CaCl2, 5 mm DTT, 2 mm tris(2-carboxyethyl)phosphine hydrochloride, or 160 μm SAM was included in the reaction mixtures.
To understand the effect of the oxidation state of the enzymes, 40 nm recombinant mouse DNMT3B was pretreated with 10 μm to 10 mm H2O2 in 49 μl of buffer B at 15 °C for 30 min or with 1–5 mm oxidized glutathione (GSSG) in 49 μl of buffer B at 37 °C for 1 h. After preincubation, 40 ng of the 5-mC-containing double-stranded DNA substrate was added, and the incubation was continued for another 4 h at 37 °C.
All reactions were stopped with 1.3% SDS and then treated with proteinase K for 20 min at 50 °C. The DNA substrates were isolated using the QIAquick nucleotide removal kit (Qiagen) and subjected to the restriction digestion-PCR assay or hydrolysis-TLC assay as described previously (26).
In Vitro Reactions of Conversion of C to 5-mC on DNA
Methylation in vitro of unmodified pMR1-8 plasmid DNA by the DNMTs was carried out and analyzed by the hydrolysis-TLC assay. When needed, 1 mm CaCl2 or 5 mm DTT was also included in the reaction mixture.
Restriction Digestion-PCR Assay of C-5 Methylation on Double-stranded DNA Substrate(s)
The procedures used were those described previously (26). See the legend of supplemental Fig. S2A for more details.
Hydrolysis-Thin Layer Chromatography (TLC) Assay of 5-mC, 5-hmC, and C on DNA
The experimental procedures were similar to those described previously (26). For more details, see the legend of supplemental Fig. S2B.
RESULTS
In Vitro DNA Demethylation by the Porcine Sperm Extract
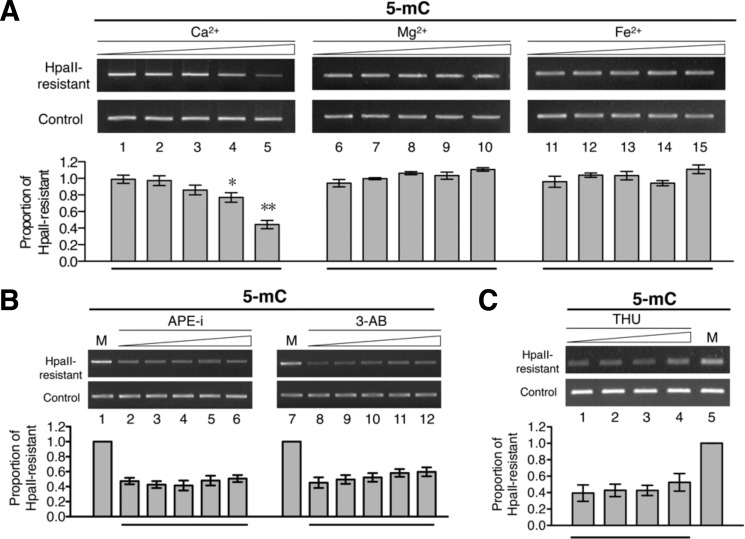
Because the level of 5-mC on the paternal genome in the mammalian pronuclei decreases rapidly after fertilization without the need for DNA synthesis, and sperm-derived factor(s) appear to be involved in this demethylation process (28–30), we carried out in vitro DNA C-5 demethylation reactions using nuclear extract prepared from porcine sperm. We also tested the effect of Ca2+ ion in view of the calcium wave in the oocyte upon fertilization (31). Remarkably, inclusion of 1–10 mm Ca2+, but not Mg2+ (Fig. 1A, compare lanes 7–10 with lane 6) or Fe2+ (compare lanes 12–15 with lane 11), significantly reduced the extent of DNA methylation by 20–50% (compare lanes 4 and 5 with lane 1).
FIGURE 1.
DNA demethylation activity of porcine sperm extracts. A, fully methylated plasmid DNA pMR1-8 was incubated at 37 °C for 2 h in the porcine sperm nuclear extract with increasing concentrations (0, 10, and 100 μm and 1 and 10 mm) of CaCl2 (lanes 1–5), MgCl2 (lanes 6–10), or Fe(NH4SO4)2 (lanes 11–15). After the reactions, the extent of DNA demethylation of the plasmid DNA was analyzed by the restriction digestion-PCR assay outlined in supplemental Fig. S2A. The histograms show the relative proportions of the plasmid DNA resistant to HpaII cleavage. For comparison of bars 2–5 with bar 1, p < 0.05 (*) and p < 0.01 (**) by t test. B, effects of BER inhibitors on the demethylation activities of the porcine sperm nuclear extract. The DNA demethylation reactions were carried out by incubating fully methylated plasmid DNA pMR1-8 and sperm nuclear extract containing 10 mm CaCl2 and increasing concentrations of APE1 inhibitor (APE-i; 0, 1, 10, and 100 μm and 1 mm (lanes 2–6)) or 3-aminobenzamide (3-AB; 0, 5, and 500 μm and 5 and 50 mm (lanes 8–12)). After the reactions, the extent of demethylation of the plasmid DNA was analyzed by the restriction digestion-PCR assay. C, effect of the cytidine deaminase inhibitor tetrahydrouridine (THU) on the demethylation reaction in the porcine sperm nuclear extract. The fully methylated pMR1-8 DNA was subjected to incubation in the extract at 37 °C for 2 h with increasing concentrations of tetrahydrouridine (0, 30, and 100 μm and 1 mm (lanes 1–4)). The extent of demethylation of the plasmid DNA was analyzed by the restriction digestion-PCR assay. The histogram shows the relative proportions of plasmid DNA resistant to HpaII cleavage. Lane M, mock control without incubation. Error bars indicate S.D.
Both DNA cytosine deamination and the BER pathway (in particular, through its two components APE1 and poly(ADP-ribose) polymerases) are involved in active DNA demethylation (22, 32–34). However, inclusion of inhibitors of either the BER pathway (CRT0044876 for APE1 and 3-aminobenzamide for poly(ADP-ribose) polymerases) or the cytidine deaminase (tetrahydrouridine) in the reactions had little effect on the in vitro DNA demethylation activity of the sperm nuclear extract (Fig. 1, B and C). The data of Fig. 1 suggested that Ca2+ ion stimulated a BER- and cytidine deaminase-independent DNA demethylation activity in the nuclear extract of porcine sperm.
DNA 5-mC Demethylase Activities of Wild-type (but Not Mutant) Murine DNMT1, DNMT3A, and DNMT3B with Intact Catalytic Domains
It was not trivial to purify the factor(s)/enzyme(s) in the nuclear extract of the porcine sperm that was responsible for the in vitro conversion of 5-mC to C on DNA. Because the porcine sperm nuclear extract contained DNMT1/DNMT3A/DNMT3B (data not shown), and the murine/human orthologs of the latter two DNMTs act in vitro as DNA 5-hmC dehydroxymethylases under oxidative conditions in the absence of SAM (26), we suspected that under appropriate conditions, these DNMTs might also be capable of converting other modified forms of cytosine, e.g. 5-mC, to C.
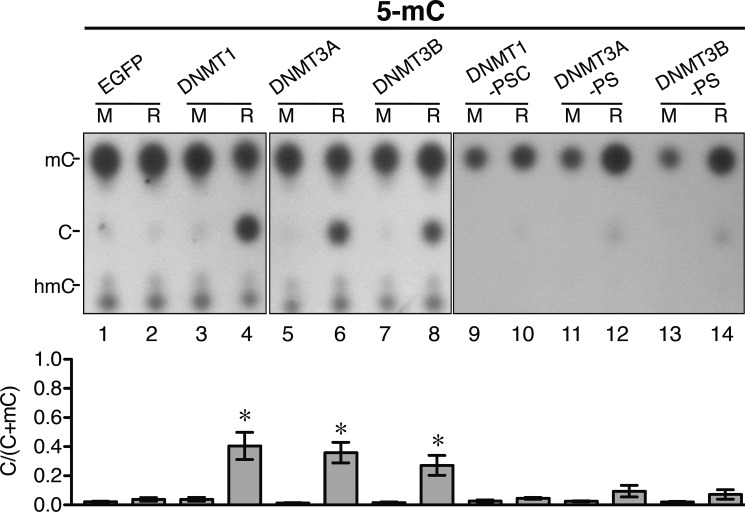
In view of the data of Fig. 1, we carried out in vitro DNA demethylation reactions in the presence of 10 mm Ca2+. The 5-mC-containing double-stranded DNA substrate was incubated with nuclear extracts prepared from 293T cells transfected with plasmids overexpressing enhanced green fluorescent protein (EGFP) and mouse DNMT1, DNMT3A, and DNMT3B, as well as their mutants. After the reactions, the DNA products were hydrolyzed as depicted in supplemental Fig. S2B, and the nucleotides were analyzed by TLC (Fig. 2). As shown in Fig. 2, under the reaction conditions tested, the nuclear extracts containing exogenously expressed DNMT1 (lane 4), DNMT3A (lane 6), and DNMT3B (lanes 8) removed the 5-methyl group from ∼30% of the 5-mC residues at the MspI-cleaved ends of the DNA substrates.
FIGURE 2.
DNA 5-mC demethylation activities of mammalian DNMTs. The 5-mC-containing DNA substrates were subjected to incubation in 293T nuclear extracts in buffer B with 10 mm CaCl2 and exogenously expressed EGFP (lanes 1 and 2) and mouse DNMT1 (lanes 3 and 4), DNMT3A (lanes 5 and 6), and DNMT3B (lanes 7 and 8), as well as their catalytic mutants (site-directed mutants of the DNA C-5 methylation catalytic sites of the three enzymes): DNMT1-PSC, DNMT3A-PS, and DNMT3B-PS (lanes 9–14). The extent of conversion of 5-mC to C was analyzed by the hydrolysis-TLC assay and is shown quantitatively in the histogram. The amounts of the exogenous wild-type enzyme in lanes 4, 6, and 8 were similar to those of the mutant enzymes in lanes 10, 12, and 14, respectively (Western blotting data not shown). M, mock control without incubation; R, with incubation. Error bars indicate S.D. *, p < 0.05 by t test comparing bars 4, 6, and 8 with bar 2.
Remarkably, the DNA demethylase activities of the three mouse DNMTs were greatly diminished (by ∼73–88%) when amino acid substitutions or insertions were introduced into the known catalytic sites of C-5 methylation of these enzymes (Fig. 2, compare lanes 10, 12, and 14 with lanes 4, 6, and 8, respectively). The data of Fig. 2 suggested that the two de novo DNMTs, as well as the maintenance enzyme DNMT1, could act as active DNA 5-mC demethylases under appropriate conditions. In addition, the methylation and demethylation activities of the three DNMTs might share the same catalytic domain(s).
Ca2+- and Redox State-dependent DNA 5-mC Demethylase Activities of Partially Purified Recombinant DNMTs
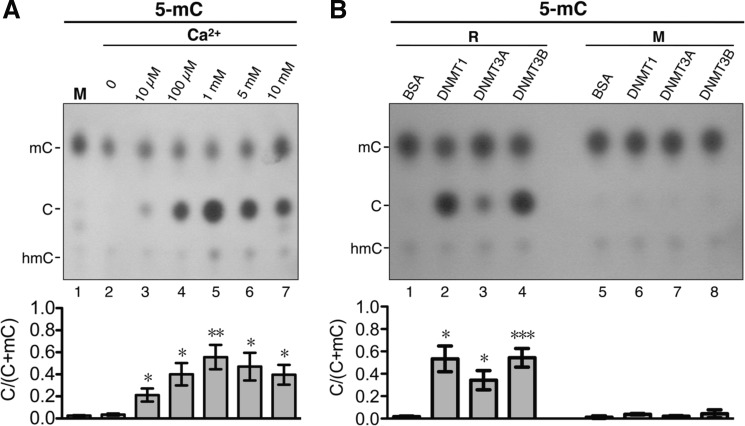
To further confirm the results of Fig. 2, recombinant mouse DNMT3B, hDNMT1, and hDNMT3A partially purified from recombinant baculovirus-infected Sf9 insect cells were examined for their DNA demethylation activities. First, recombinant mouse DNMT3B (∼50% purity) (supplemental Fig. S1) was subjected to incubation with the 5-mC-containing DNA substrate in buffer B containing increasing concentrations (0, 10, and 100 μm and 1, 5, and 10 mm) of CaCl2 (Fig. 3A). As shown in Fig. 3A, recombinant DNMT3B exhibited significant DNA demethylation activity only in the presence of Ca2+ (compare lanes 3–7 with lane 2), with the activity highest in the presence of 1 mm Ca2+ (lane 5). We next tested and compared the DNA demethylation activities of DNMT3B, hDNMT1 (∼70% purity) (supplemental Fig. S1), and hDNMT3A (∼90% purity) (supplemental Fig. S1) in buffer B containing 1 mm CaCl2 (Fig. 3B). Both hDNMT1 and DNMT3B exhibited significant DNA demethylation activity, converting at least 50% of 5-mC at the MspI-cleaved ends on DNA to C (Fig. 3B, lanes 2 and 4), whereas recombinant hDNMT3A showed significantly lower activity (lane 3). The demethylation reaction with recombinant mouse DNMT3B also decreased the resistance to HpaII digestion of the methylated DNA substrate (data not shown). Finally, DNA demethylation by the DNMTs was most likely the result of direct conversion of 5-mC to C because similarly low levels of 5-hmC were present in the mock control samples (Figs. 2 and 3), as well as throughout the time course of the DNA demethylation reactions in vitro (supplemental Fig. S3). These data together demonstrate that the mammalian DNMTs can function as active DNA demethylases in vitro.
FIGURE 3.
Calcium dependence of DNA 5-mC demethylation activities of recombinant DNMTs. A, results from the hydrolysis-TLC assay of conversion of 5-mC to C by recombinant DNMT3B. The DNA demethylation activity of recombinant mouse DNMT3B was assayed by incubation of 40 ng of 5-mC-containing DNA substrate with 40 nm enzyme in buffer B containing 100 μg/ml BSA and increasing concentrations (0, 10, and 100 μm and 1, 5, and 10 mm) of CaCl2. The incubations were all performed at 37 °C for 4 h. The quantitative results are presented in the histogram. M, mock control without incubation. Error bars indicate S.D. *, p < 0.05; **, p < 0.01 by t test comparing bars 3–7 with bar 2. B, comparison of the DNA demethylation activities of recombinant hDNMT1, hDNMT3A, and DNMT3B by the hydrolysis-TLC assay. The 5-mC-containing substrate was incubated at 37 °C for 4 h with 40 nm each recombinant hDNMT1 (lane 2), hDNMT3A (lane 3), and DNMT3B (lane 4) in buffer B containing 100 μg/ml BSA and 1 mm CaCl2 and then analyzed by the hydrolysis-TLC assay. The quantitative analysis is presented in the histogram. M, mock control without incubation; R, with incubation. Error bars indicate S.D. *, p < 0.05; ***, p < 0.005 by t test comparing bars 2–4 with bar 1.
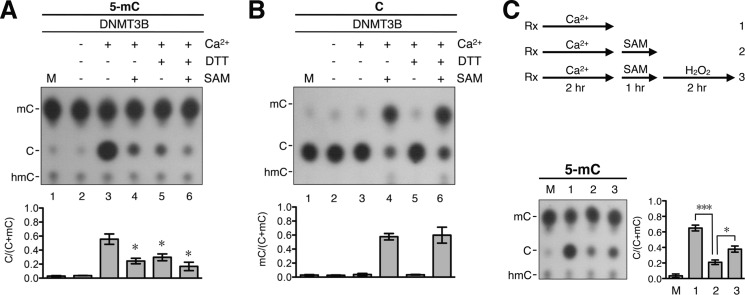
We showed previously that the redox state of de novo DNMT3A and DNMT3B can influence their DNA dehydroxymethylase activities (26). Interestingly, the DNA demethylation activities of the DNMTs also appeared to be affected by the redox state of the enzymes. As exemplified by DNMT3B, preincubation of the enzyme with the reducing DTT (Fig. 4A, compare lane 5 with lane 3) or tris(2-carboxyethyl)phosphine hydrochloride (supplemental Fig. S4A, compare lanes 2 and 3) greatly decreased the extent of conversion of 5-mC to C. On the other hand, preincubation with 10 mm H2O2 or 5 mm GSSG did not affect the DNA demethylation activity of the recombinant enzyme (data not shown). In interesting contrast to 5-mC demethylation, the C-5 methylation reaction of DNMTs did not require Ca2+ (data not shown) (3, 35, 36), nor was it affected by DTT (Fig. 4B, compare lane 6 with lane 4) (26).
FIGURE 4.
Inhibition effects of reducing reagents and SAM on DNA demethylation activity of DNMT3B. A, the 5-mC-containing DNA substrate was subjected to the demethylation reactions with 40 nm recombinant DNMT3B in buffer B containing 100 μg/ml BSA with or without the inclusion of 1 mm CaCl2, 5 mm DTT, or 160 μm SAM. After incubation at 37 °C for 4 h, the DNA products were analyzed by the hydrolysis-TLC assay. The results are presented quantitatively in the histogram. Error bars indicate S.D. *, p < 0.05 by t test comparing bars 4–6 with bar 3. B, unmethylated pMR1-8 plasmid DNA was incubated with 40 nm recombinant DNMT3B in buffer B containing 100 μg/ml BSA with or without 1 mm CaCl2, 5 mm DTT, or 160 μm SAM. After incubation at 37 °C for 4 h, the extent of C methylation of the DNAs from the different reactions was determined by the hydrolysis-TLC assay and is compared quantitatively in the histogram. Error bars indicate S.D. C, strategy of a series of reactions testing the reversibility of DNA demethylation and methylation (see “Results” for more details). Briefly, the 5-mC-containing DNA substrate was incubated at 37 °C for 2 h with 40 nm recombinant DNMT3B in buffer B containing 100 μg/ml BSA and 1 mm CaCl2 (reaction 1). Then, 160 μm SAM was added, and the incubation was continued for another 1 h (reaction 2). Finally, 1 mm H2O2 was added to the reaction mixture, and the incubation was continued for another 2 h (reaction 3). Rx, reactions. The DNA products from the three reactions outlined in the upper panel were purified and analyzed by the hydrolysis-TLC assay. The data are quantitatively compared in the histogram. M, mock control without incubation. Error bars indicate S.D. *, p < 0.05; ***, p < 0.005.
Reversibility of the DNA 5-mC Demethylation and 5-C Methylation Reactions Catalyzed by DNMTs
As exemplified for DNMT3B in Fig. 4A, the inclusion of SAM, the methyl donor need for DNA 5-C methylation by the DNMTs, in the reaction mixture greatly reduced the extent of conversion of 5-mC to C (compare lanes 4 and 6 with lane 3). This could be due to inhibition of the demethylation activity of the DNMTs by SAM. Alternatively, the presence of SAM in the demethylation reaction might favor the methylation function of DNMTs, thus pushing the demethylation backwards. The latter scenario was confirmed with inclusion of radioactive [3H]SAM in the reaction mixtures and quantitation of 3H-labeled CH3 on DNA after the reactions (supplemental Fig. S4B, lanes 4 and 6). The result suggested that the DNA 5-mC demethylation reaction was reversible, with the presence SAM pushing DNMT3B to remethylate the demethylated cytosines on the DNA substrate.
The reversibility of the DNA methylation-demethylation reactions as catalyzed by the mammalian DNMTs was further studied by an analysis of the dynamic changes of the DNA methylation in vitro. As outlined in Fig. 4C, double-stranded DNA substrate containing 5-mC was first incubated with recombinant DNMT3B in the demethylation buffer for 2 h. SAM was then added, and the incubation was continued for 1 h. Finally, H2O2, which is known to inhibit the methylation reaction (26), was added, and the reaction was continued for another 2 h. As exemplified in the TLC plate and statistically presented in the histogram of Fig. 4C, ∼60% of 5-mC on the MspI-cleaved ends of DNA substrate was demethylated by DNMT3B at the end of the first reaction (compare lane 1/bar 1 with lane M/bar M). The continued 1-h incubation in the presence of SAM converted >60% of C back to 5-mC (Fig. 4C, compare lane 2/bar 2 with lane 1/bar 1). Finally, the addition of H2O2 led to an apparent switch in the enzyme activity of DNMT3B from methylation to demethylation again (Fig. 4C, compare lane 3/bar 3 with lane 2/bar 2), presumably due to loss of the DNA methylation function of the oxidized enzyme. The data of Figs. 3 and 4 and supplemental Fig. S4 suggest that the switch in the catalytic functions of the DNMTs between DNA methylation and demethylation is flexible, subjected to regulation by a range of factors, including the local concentration of Ca2+, the presence of SAM, and the redox state of the DNMTs.
DISCUSSION
This study has revealed an unexpected and novel characteristic of the mammalian DNMTs and likely those of vertebrates in general, i.e. the vertebrate DNMTs, in addition to converting C to 5-mC on DNA, can also demethylate 5-mC on DNA under specific conditions (Figs. 2 and 3), in particular in the presence of Ca2+ ion and in the absence of reducing reagents (Figs. 3 and 4A and supplemental Fig. S4A). In other words, the covalent addition of the methyl group to the 5-position of cytosine on DNA, as catalyzed by DNMTs, is reversible (Fig. 4C). The loss of the DNA demethylation activities of the mutant forms in comparison with the wild-type enzymes (Fig. 2) also suggests that each of the three DNMTs utilizes the same domain or overlapping domains to catalytically methylate and demethylate DNA.
The Ca2+ ion and redox state dependence of the DNA demethylation activities of the DNMTs is especially intriguing. With respect to this point, it is interesting to note that a change in the intracellular concentration of Ca2+ ion presents a key cellular signal, and the Ca2+ flux/influx wave in the cytosol could eventually communicate with the nucleus through a number of ways (37). Significantly, dynamic changes in the DNA methylation patterns, globally or locally, occur in close association with calcium signaling in fertilization/early embryo development (31), synaptic transmission (38, 39), and tumorigenesis (40). Among these processes, the Ca2+ wave during fertilization triggers activation of the oocyte and its cell cycle resumption (41). In addition, this increased concentration of Ca2+ in the zygote is maintained for at least 6 h (42), during which time, genome-wide demethylation of both the paternal and maternal genomes occurs (43, 44). Notably, DNMT1, DNMT3A, and DNMT3B are all expressed in the zygote (45). On the basis of our data presented above, in particular those in Fig. 1, we suggest that, in addition to other pathways, e.g. the conversion of 5-mC to 5-hmC by TET (23) and 5-hmC to C by DNMT3A and DNMT3B (26), the direct conversion of 5-mC to C by the active DNA demethylation activities of the three DNMTs and their isoforms may also play a role in the genome-wide demethylation during early embryonic development of vertebrates.
In addition, the elevation of calcium at synapses can serve as a spark for signal transmission among the neuronal network (46). Concurrently, transcription of specific neuronal genes could be activated, and this is associated with promoter DNA demethylation (39). In view of the above, we suggest that, besides the other previously known pathways of DNA demethylation, the DNA demethylation events in activated neurons could also be accomplished by the Ca2+ ion-stimulated DNA demethylase activities of DNMT3A, DNMT3B, and/or DNMT1.
Finally, calcium signaling in cancer cells, as the result of increased influx of the ion by the Ca2+ channel/pumps and release from the endoplasmic reticulum (40), is involved in aberrant transcription, the disregulated cell cycle, genotoxicity, and tumor invasion/metastasis (47, 48). In parallel, imbalance of CpG methylation in cancer cells leads to a genome-wide hypomethylation (8) accompanied by regional hypermethylation on the promoters of specific tumor suppressor genes (49). Curiously, the elevation, instead of reduction, of the levels of DNMT1, DNMT3A, and DNMT3B in cancer cells (50–52) could not be easily correlated with the global hypomethylation of the cancer genomes (53). The current finding of the Ca2+-dependent oxidation state-facilitated DNA demethylase activities of the three enzymes, in contrast to their DNA methyltransferase activities, provides a plausible basis for the above seemingly paradoxical correlation. Future studies should also be carried out to look into the likelihood of demethylation, in addition to methylation (8), of the promoters of specific tumor suppressor genes/oncogenes by the DNMTs during carcinogenesis.
In relation to the dependence of the DNA demethylation activities of the three DNMTs on Ca2+ and on the redox condition, it is interesting to note that elevation of the Ca2+ ion concentration and the redox state of the cells are interdependent in vivo. For instance, the calcium wave induced during fertilization stimulates the ATP supply and the generation of the reactive oxygen species from the mitochondria during oocyte activation and maintenance (54). The high concentration of intracellular Ca2+ in cancer cells is also associated with the oxidation stress (55). Furthermore, the reciprocal relation between the excitatory event and reactive oxygen species generation exists in neuronal cells (56, 57).
In summary, we have discovered that mammalian DNMT1, DNMT3B, and, to a much less extent, DNMT3A, contrary to the conventional thought of their being mainly DNA methyltransferases, can also act in vitro as DNA demethylases in a Ca2+ ion- and redox state-dependent manner, albeit at relatively low efficiencies under the current conditions. How the DNA methylation patterns, globally and locally, of the vertebrate genomes in different cell types under various physiological conditions are generated and maintained may need to be re-evaluated in relation to the interplay between the two totally opposite DNA modification activities of the enzymes. It should be noted that the Ca2+ concentration (≥10 μm) needed for the in vitro DNA demethylation reaction (Fig. 3A) is relatively high in comparison with the intracellular levels of Ca2+, which range from nm to μm even during activation of different Ca2+-mediated biological processes, e.g. fertilization (58). Thus, an efficient in vivo demethylation reaction may require other essential cofactors and/or specific signal transduction pathways. How the Ca2+ ion, the redox condition of the environment, and other factors modulate the structures/functions, in vitro and in vivo, of the vertebrate DNMTs as DNA methyltransferases and DNA demethylases will need to be investigated.
Acknowledgments
We thank Jim Wang (Harvard University) and Alan Lin (National Yang-Ming University) for continued interests and encouragements regarding this project. We also appreciate the discussions with and help from Dr. Tao-Shih Hsieh (Institute of Cellular and Organism Biology, Academia Sinica), Dr. Sheng-Chung Lee (National Taiwan University), Dr. Hung-Ta Chen (Institute of Molecular Biology, Academia Sinica), and Shun-Hsiao Lee (Institute of Cellular and Organism Biology, Academia Sinica).
This work was supported in part by the Frontier of Science Award from the National Science Council and Academia Sinica (Taipei, Taiwan).

This article contains supplemental Figs. S1–S4.
- DNMT
- DNA methyltransferase
- SAM
- S-adenosylmethionine
- 5-mC
- 5-methylcytosine
- BER
- base excision repair
- 5-hmC
- 5-hydroxymethylcytosine
- hDNMT
- human DNMT.
REFERENCES
- 1. Bestor T. H. (2000) The DNA methyltransferases of mammals. Hum. Mol. Genet. 9, 2395–2402 [DOI] [PubMed] [Google Scholar]
- 2. Okano M., Bell D. W., Haber D. A., Li E. (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 3. Hsieh C. L. (1999) In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19, 8211–8218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaenisch R., Bird A. (2003) Epigenetic regulation of gene expression:how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 5. Day J. J., Sweatt J. D. (2011) Epigenetic mechanisms in cognition. Neuron 70, 813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hirabayashi Y., Gotoh Y. (2010) Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 11, 377–388 [DOI] [PubMed] [Google Scholar]
- 7. Robertson K. D. (2005) DNA methylation and human disease. Nat. Rev. Genet. 6, 597–610 [DOI] [PubMed] [Google Scholar]
- 8. You J. S., Jones P. A. (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki M. M., Bird A. (2008) DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 10. Gartler S. M., Riggs A. D. (1983) Mammalian X-chromosome inactivation. Annu. Rev. Genet. 17, 155–190 [DOI] [PubMed] [Google Scholar]
- 11. Feng S., Jacobsen S. E., Reik W. (2010) Epigenetic reprogramming in plant and animal development. Science 330, 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li E. (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 [DOI] [PubMed] [Google Scholar]
- 13. Hermann A., Schmitt S., Jeltsch A. (2003) The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J. Biol. Chem. 278, 31717–31721 [DOI] [PubMed] [Google Scholar]
- 14. Tang L. Y., Reddy M. N., Rasheva V., Lee T. L., Lin M. J., Hung M. S., Shen C. K. (2003) The eukaryotic DNMT2 genes encode a new class of cytosine-5 DNA methyltransferases. J. Biol. Chem. 278, 33613–33616 [DOI] [PubMed] [Google Scholar]
- 15. Okano M., Xie S., Li E. (1998) Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 26, 2536–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ulrey C. L., Liu L., Andrews L. G., Tollefsbol T. O. (2005) The impact of metabolism on DNA methylation. Hum. Mol. Genet. 14, R139–R147 [DOI] [PubMed] [Google Scholar]
- 17. Bhattacharya S. K., Ramchandani S., Cervoni N., Szyf M. (1999) A mammalian protein with specific demethylase activity for mCpG DNA. Nature 397, 579–583 [DOI] [PubMed] [Google Scholar]
- 18. Law J. A., Jacobsen S. E. (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhu B., Zheng Y., Hess D., Angliker H., Schwarz S., Siegmann M., Thiry S., Jost J. P. (2000) 5-Methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl. Acad. Sci. U.S.A. 97, 5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ooi S. K., Bestor T. H. (2008) The colorful history of active DNA demethylation. Cell 133, 1145–1148 [DOI] [PubMed] [Google Scholar]
- 21. Wu S. C., Zhang Y. (2010) Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 11, 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bhutani N., Burns D. M., Blau H. M. (2011) DNA demethylation dynamics. Cell 146, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen C. C., Wang K. Y., Shen C. K. J. (2012) The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J. Biol. Chem. 287, 33116–33121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Har-Vardi I., Mali R., Breietman M., Sonin Y., Albotiano S., Levitas E., Potashnik G., Priel E. (2007) DNA topoisomerases I and II in human mature sperm cells: characterization and unique properties. Hum. Reprod. 22, 2183–2189 [DOI] [PubMed] [Google Scholar]
- 28. Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. (2000) Demethylation of the zygotic paternal genome. Nature 403, 501–502 [DOI] [PubMed] [Google Scholar]
- 29. Reik W., Dean W., Walter J. (2001) Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 30. Beaujean N., Taylor J. E., McGarry M., Gardner J. O., Wilmut I., Loi P., Ptak G., Galli C., Lazzari G., Bird A., Young L. E., Meehan R. R. (2004) The effect of interspecific oocytes on demethylation of sperm DNA. Proc. Natl. Acad. Sci. U.S.A. 101, 7636–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Santella L., Lim D., Moccia F. (2004) Calcium and fertilization: the beginning of life. Trends Biochem. Sci. 29, 400–408 [DOI] [PubMed] [Google Scholar]
- 32. Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., Surani M. A. (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo J. U., Su Y., Zhong C., Ming G. L., Song H. (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L. K., Bartolomei M. S., Rambow F., Bassi M. R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A. J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bestor T., Laudano A., Mattaliano R., Ingram V. (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J. Mol. Biol. 203, 971–983 [DOI] [PubMed] [Google Scholar]
- 36. Okano M., Xie S., Li E. (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19, 219–220 [DOI] [PubMed] [Google Scholar]
- 37. Clapham D. E. (2007) Calcium signaling. Cell 131, 1047–1058 [DOI] [PubMed] [Google Scholar]
- 38. Berridge M. J. (1998) Neuronal calcium signaling. Neuron 21, 13–26 [DOI] [PubMed] [Google Scholar]
- 39. Greer P. L., Greenberg M. E. (2008) From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860 [DOI] [PubMed] [Google Scholar]
- 40. Monteith G. R., McAndrew D., Faddy H. M., Roberts-Thomson S. J. (2007) Calcium and cancer: targeting Ca2+ transport. Nat. Rev. Cancer 7, 519–530 [DOI] [PubMed] [Google Scholar]
- 41. Homa S. T., Carroll J., Swann K. (1993) The role of calcium in mammalian oocyte maturation and egg activation. Hum. Reprod. 8, 1274–1281 [DOI] [PubMed] [Google Scholar]
- 42. Sardet C., Roegiers F., Dumollard R., Rouviere C., McDougall A. (1998) Calcium waves and oscillations in eggs. Biophys. Chem. 72, 131–140 [DOI] [PubMed] [Google Scholar]
- 43. Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. (2000) Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 [DOI] [PubMed] [Google Scholar]
- 44. Wossidlo M., Nakamura T., Lepikhov K., Marques C. J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. (2011) 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. [DOI] [PubMed] [Google Scholar]
- 45. Ko Y. G., Nishino K., Hattori N., Arai Y., Tanaka S., Shiota K. (2005) Stage-by-stage change in DNA methylation status of Dnmt1 locus during mouse early development. J. Biol. Chem. 280, 9627–9634 [DOI] [PubMed] [Google Scholar]
- 46. Hardingham G. E., Chawla S., Johnson C. M., Bading H. (1997) Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature 385, 260–265 [DOI] [PubMed] [Google Scholar]
- 47. Prevarskaya N., Skryma R., Shuba Y. (2011) Calcium in tumour metastasis: new roles for known actors. Nat. Rev. Cancer 11, 609–618 [DOI] [PubMed] [Google Scholar]
- 48. Roderick H. L., Cook S. J. (2008) Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat. Rev. Cancer 8, 361–375 [DOI] [PubMed] [Google Scholar]
- 49. Issa J. P. (2004) CpG island methylator phenotype in cancer. Nat. Rev. Cancer 4, 988–993 [DOI] [PubMed] [Google Scholar]
- 50. Mizuno S., Chijiwa T., Okamura T., Akashi K., Fukumaki Y., Niho Y., Sasaki H. (2001) Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97, 1172–1179 [DOI] [PubMed] [Google Scholar]
- 51. Girault I., Tozlu S., Lidereau R., Bièche I. (2003) Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin. Cancer Res. 9, 4415–4422 [PubMed] [Google Scholar]
- 52. Lin R. K., Wu C. Y., Chang J. W., Juan L. J., Hsu H. S., Chen C. Y., Lu Y. Y., Tang Y. A., Yang Y. C., Yang P. C., Wang Y. C. (2010) Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 70, 5807–5817 [DOI] [PubMed] [Google Scholar]
- 53. Feinberg A. P., Tycko B. (2004) The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153 [DOI] [PubMed] [Google Scholar]
- 54. Dumollard R., Marangos P., Fitzharris G., Swann K., Duchen M., Carroll J. (2004) Sperm-triggered [Ca2+] oscillations and Ca2+ homeostasis in the mouse egg have an absolute requirement for mitochondrial ATP production. Development 131, 3057–3067 [DOI] [PubMed] [Google Scholar]
- 55. Yan Y., Wei C. L., Zhang W. R., Cheng H. P., Liu J. (2006) Cross-talk between calcium and reactive oxygen species signaling. Acta Pharmacol. Sin. 27, 821–826 [DOI] [PubMed] [Google Scholar]
- 56. Bondy S. C., LeBel C. P. (1993) The relationship between excitotoxicity and oxidative stress in the central nervous system. Free Radic. Biol. Med. 14, 633–642 [DOI] [PubMed] [Google Scholar]
- 57. Hernández-Fonseca K., Cárdenas-Rodríguez N., Pedraza-Chaverri J., Massieu L. (2008) Calcium-dependent production of reactive oxygen species is involved in neuronal damage induced during glycolysis inhibition in cultured hippocampal neurons. J. Neurosci. Res. 86, 1768–1780 [DOI] [PubMed] [Google Scholar]
- 58. Taylor C. T., Lawrence Y. M., Kingsland C. R., Biljan M. M., Cuthbertson K. S. (1993) Oscillations in intracellular free calcium induced by spermatozoa in human oocytes at fertilization. Hum. Reprod. 8, 2174–2179 [DOI] [PubMed] [Google Scholar]