Background: The nuclear protein UHRF2 is a member of the RING-type ubiquitin E3 ligase family.
Results: UHRF2 promotes covalent SUMOylation of ZNF131 regardless of its ubiquitin E3 ligase activity.
Conclusion: UHRF2 acts as a novel SUMO E3 ligase for ZNF131.
Significance: UHRF2 is a dual-functional E3 ligase for covalent ubiquitin and SUMO conjugation.
Keywords: Posttranslational Modification, Sumo, Sumoylation, Ubiquitin Ligase, Ubiquitination, SUMO E3 Ligase, UHRF2, ZNF131
Abstract
Small ubiquitin-like modifier (SUMO), a member of the ubiquitin-related protein family, is covalently conjugated to lysine residues of its substrates in a process referred to as SUMOylation. SUMOylation occurs through a series of enzymatic reactions analogous to that of the ubiquitination pathway, resulting in modification of the biochemical and functional properties of substrates. To date, four mammalian SUMO isoforms, a single heterodimeric SUMO-activating E1 enzyme SAE1/SAE2, a single SUMO-conjugating E2 enzyme ubiquitin-conjugating enzyme E2I (UBC9), and a few subgroups of SUMO E3 ligases have been identified. Several SUMO E3 ligases such as topoisomerase I binding, arginine/serine-rich (TOPORS), TNF receptor-associated factor 7 (TRAF7), and tripartite motif containing 27 (TRIM27) have dual functions as ubiquitin E3 ligases. Here, we demonstrate that the ubiquitin E3 ligase UHRF2 also acts as a SUMO E3 ligase. UHRF2 effectively enhances zinc finger protein 131 (ZNF131) SUMOylation but does not enhance ZNF131 ubiquitination. In addition, the SUMO E3 activity of UHRF2 on ZNF131 depends on the presence of SET and RING finger-associated and nuclear localization signal-containing region domains, whereas the critical ubiquitin E3 activity RING domain is dispensable. Our findings suggest that UHRF2 has independent functional domains and regulatory mechanisms for these two distinct enzymatic activities.
Introduction
A number of posttranslational modifications alter the biochemical and functional properties of the substrate. In the same manner as ubiquitin and several ubiquitin-like proteins, small ubiquitin-like modifier (SUMO)2 is covalently conjugated to the lysine residues of its substrates in a process referred to as SUMOylation. SUMOylation regulates many substrates by changing their intrinsic activity, subcellular localization, and/or protein stability (1). SUMOylation occurs through a serial enzymatic cascade, analogous to the ubiquitination pathway (1). Four SUMO paralogs are currently isolated in mammalian system: SUMO1 to SUMO4 (1). Although there are four known mammalian SUMO protein isoforms, only a single heterodimeric SUMO-activating E1 enzyme (SAE1/SAE2) (2–4) and a single SUMO-conjugating E2 enzyme (UBC9) (5–7) have been identified thus far. Because UBC9 directly binds the substrate and facilitates SUMOylation at the consensus SUMOylation motif ΨKX(DE) (8, 9), the absolute requirement for a SUMO E3 ligase in the SUMOylation pathway is controversial (1). Nevertheless, several mammalian SUMO E3 ligase families that efficiently promote the SUMOylation of their substrates have been reported.
SUMO E3 ligases facilitate SUMO protein transfer from the E2-conjugase to a substrate. The first class of mammalian SUMO E3 ligases is the protein inhibitor of activated STAT (PIAS) family, which are homologs of the yeast Siz proteins (10–12). PIAS proteins have an internal Siz/PIAS (SP)-RING domain that is necessary for stimulation of SUMO ligation (13). This SP-RING domain shares sequence similarity to the RING domains of ubiquitin E3 ligases (13). The second SUMO E3 ligase is the RanBP2 protein, a component of the nuclear pore complex (14, 15). RanBP2 also contains a RING domain that is dispensable for SUMO E3 activity, suggesting that RanBP2 acts through a distinct mechanism compared with the PIAS family (14). The third class is polycomb protein 2 (Pc2; also known as CBX4), which is a component of multimeric polycomb repressive complex 1 and is also involved in stable gene repression (16). Human Pc2 (hPc2) contains two SUMO-interacting motifs (17, 18). These motifs contribute to non-covalent SUMO binding and are required for full SUMO E3 activity, as well as for auto-SUMOylation of hPc2 (18). The fourth class of SUMO E3 ligases includes histone deacetylase 4 (HDAC4) (19), TOPORS (20), TRAF7 (21), and TRIM27 (22). This group is considered non-categorized because their molecular mechanisms to promote SUMO modification remain less clear.
Interestingly, various posttranslational modifications that use ubiquitin-like modifiers influence each other (23). In particular, from a biochemical point of view, the SUMOylation pathway closely resembles ubiquitination, and these two pathways are closely related. This notion was further supported by reports that there are currently three dual-functional ubiquitin and SUMO E3 ligases identified, TOPORS (20), TRAF7 (21), and TRIM27 (22).
The nuclear protein UHRF2 (also known as NIRF; Np95/ICBP90-like RING finger protein) consists of an ubiquitin-like domain, a plant homeodomain, a RING finger domain, and a SET and RING finger-associated (SRA) domain (24). Although UHRF2 lacks a canonical nuclear localization signal (NLS) sequence, a recent study revealed that the NLS-containing region (NCR) is located at amino acids 602–693 (25). UHRF2 was initially identified as a cell cycle regulator that induces G1 arrest by interacting with the inactive CDK2-cyclin E complex (26) and associates with multiple cell cycle proteins, including cyclins (A2, B1, D1, and E1), p53, and retinoblastoma protein (pRb) (27). In addition, UHRF2 displays RING-type ubiquitin E3 ligase activity and ubiquitinates cyclins D1 and E1 (27), as well as PEST proteolytic signal-containing nuclear protein (28). Moreover, UHRF2 plays a role in the ubiquitination and degradation of p53 (29) and nuclear aggregates containing polyglutamine repeats (25). Here, we demonstrate that UHRF2 acts as a novel SUMO E3 ligase for zinc finger protein 131 (ZNF131), thus becoming the fourth-identified dual-functional ubiquitin/SUMO E3 ligase.
EXPERIMENTAL PROCEDURES
Materials
The proteasome inhibitor MG-132 was purchased from A. G. Scientific. Rabbit polyclonal actin, HA, FLAG, SUMO1, and mouse monoclonal FLAG antibodies were purchased from Sigma-Aldrich. The mouse monoclonal HA antibody was purchased from Covance. Rabbit polyclonal GFP, Myc, mouse monoclonal GFP, HA, Myc, and ubiquitin antibodies were purchased from Santa Cruz Biotechnology. The rabbit polyclonal V5 antibody was purchased from Abcam. Mouse monoclonal V5 and HRP-conjugated anti-rabbit antibodies were purchased from Invitrogen. Mouse polyclonal anti-ZNF131 antibody was purchased from Abnova. The HRP-conjugated anti-mouse antibody was purchased from Thermo Scientific. DMEM, FBS, and Lipofectamine PLUS were purchased from Invitrogen. Protein A-Sepharose was purchased from Amersham Biosciences. All other chemicals were purchased from Sigma-Aldrich.
DNA Constructions and RNA Interference
Human UHRF2 cDNA (GenBankTM accession no. NM_152896) from HEK293 cells was PCR-amplified and subcloned into modified pRK5-Myc (Stratagene), pRK5-FLAG, or pEGFP-C2 (Clontech) to generate Myc-tagged UHRF2 (Myc-UHRF2), FLAG-tagged UHRF2 (FLAG-UHRF2), or GFP-tagged UHRF2 (GFP-UHRF2), respectively. To construct various UHRF2 deletion and point mutants, site-directed mutagenesis was performed using the QuikChange XL site-directed mutagenesis kit (Stratagene). The GFP plasmid with an artificial NLS (pEGFP-NLS) was generated by inserting the following two annealed oligonucleotides between XhoI and EcoRI sites of the modified pEGFP-C2 vector: 5′-TCGAGAACAGGAAAAAATGAGGCCAAAAAAAGGAAGATTGCAG-3′ and 5′-AATTCTGCAATCTTCCTTTTTTTGGCCTCATTTTTTCCTGTTC-3′. The denoted oligonucleotide encodes a single artificial NLS (TGKNEAKKRKIA). Amplified UHRF2-ΔNCR cDNA was subcloned into the pEGFP-NLS to generate GFP-NLS-tagged UHRF2-ΔNCR (GFP-NLS-UHRF2-ΔNCR).
Human ubiquitin C cDNA (GenBankTM accession no. NM_021009.5) from HEK293 cells was PCR-amplified and subcloned into modified pRK5-FLAG to generate FLAG-tagged ubiquitin (FLAG-ubiquitin). Plasmids encoding human wild type ZNF131 isoform 2 (GenBankTM accession no. NM_003432) and its K567R point mutant (pRK5-HA-ZNF131-WT and pRK5-HA-ZNF131-K567R), human wild type SUMO1 (GenBankTM accession no. NM_003352) and its conjugation-defective mutant (pRK5-V5-SUMO1-GG and pRK5-V5-SUMO1-AA), and Myc-tagged hPc2 (GenBankTM accession no. NM_003655; pRK5-Myc-hPc2), were constructed as described previously (30). The plasmid encoding 3xMyc-tagged human UBC9 (3×Myc-UBC9) was a kind gift from Dr. Y. K. Jang (Yonsei University, Seoul, Korea). All cDNA sequences were verified by DNA sequencing (COSMO Genetech).
The UHRF2-specific and control siRNAs were designed and synthesized by Bioneer. The UHRF2 siRNA duplex sequences were as follows: 5′-GUACUAUUGUCCCUUCUAA(dTdT)-3′ and 5′-UUAGAAGGGACAAUAGUAC(dTdT)-3′. The control siRNA duplex sequences were 5′-CCUACGCCACCAAUUUCGU(dTdT)-3′ and 5′-ACGAAAUUGGUGGCGUAGG(dTdT)-3′. These siRNA duplexes were transfected into cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions.
Cell Culture and DNA Transfection
HEK293 and COS-7 cells were maintained in DMEM supplemented with 10% FBS and 100 units/ml penicillin-streptomycin (Invitrogen). DNA transfections were performed using either Lipofectamine PLUS reagents (Invitrogen) or polyethylenimine (Sigma-Aldrich), according to the manufacturer's instructions.
In Vivo SUMOylation Assays and Western Blot Analyses
To detect target protein SUMOylation, HEK293 cells were lysed in lysis buffer (10 mm Tris, pH 7.4, 1% Nonidet P40, 150 mm NaCl, 10% glycerol, 20 mm N-ethylmaleimide, 1 mm EDTA, 1 mm EGTA, 1 mm Na3VO4, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 5 mm NaF, and 0.5 mm phenylmethylsulfonyl fluoride) supplemented with 1% SDS. After sonicating to reduce the viscosity, cell lysates were mixed with Benzonase (Sigma-Aldrich) and incubated for 15 min at 37 °C. The samples were clarified by centrifugation at 14,000 × g for 15 min at 14 °C. For immunoprecipitation, ∼100–200 μg of protein lysates were incubated overnight at 4 °C with ∼0.2–0.4 μg of the indicated antibodies. The mixtures were incubated for 2 h at 4 °C with 30 μl of 1:1 Protein A-Sepharose bead suspension. The pellets were washed four times with lysis buffer, resolved by SDS-PAGE, and transferred to nitrocellulose membranes (Whatman). The membranes were blocked with 5% nonfat dry milk (Bio-Rad) in Tris-buffered saline supplemented with 0.1% Tween 20 (TBST) and incubated with the primary antibody in TBST containing 3% nonfat dry milk. After washing three times with TBST, the membranes were incubated with the HRP-conjugated secondary antibody in TBST containing 3% nonfat dry milk. The protein bands were visualized using enhanced chemiluminescence reagent (PerkinElmer Life Sciences), following the manufacturer's instructions.
To detect endogenous ZNF131 protein SUMOylation, HEK293 cells were lysed in lysis buffer supplemented with 0.1% SDS. After incubation for 15 min at 4 °C, cells were sonicated, and 500 μg of the protein lysate was incubated overnight at 4 °C with 1 μg of the ZNF131 antibody with gentle rotation. The samples were incubated with protein A-Sepharose beads, washed with lysis buffer, and subjected to Western blot analyses.
Co-immunoprecipitation Assays
HEK293 cells were washed with ice-cold PBS and lysed in lysis buffer supplemented with 0.1% SDS. After incubation for 15 min at 4 °C, cells were sonicated, and 250 μg of the protein lysate was incubated overnight at 4 °C with 0.5 μg of the indicated antibody with gentle rotation. The samples were incubated with protein A-Sepharose beads, washed with lysis buffer, and subjected to Western blot analyses.
Immunocytochemistry
COS-7 cells were seeded at 60% confluence onto cover glasses in P35 dishes and incubated overnight. Cells were washed with PBS, fixed for 20 min in 4% paraformaldehyde in PBS, and permeabilized for 30 min in 0.2% Triton X-100 in PBS. Cells were blocked for 30 min with 1% BSA in PBS and incubated overnight at 4 °C with mouse monoclonal anti-Myc or rabbit polyclonal anti-HA antibodies. After washing three times with PBS, the cells were incubated for 2 h with Alexa Fluor 488-conjugated anti-mouse and Alexa Fluor 594-conjugated anti-rabbit antibodies (Molecular Probes). To stain the nuclei, cells were incubated for 5 min with 1 μg/ml DAPI in PBS. After washing with PBS three times, cells were analyzed using confocal microscopy (LSM510 META; Carl Zeiss).
Cell Growth Analysis
The number of viable cells was determined using Cell Counting Kit-8 (Dojindo Molecular Technology). Significant differences in cell viability were analyzed using the Student's t test in Sigma Plot (version 11; Systat Software, Inc.).
RESULTS
UHRF2 Physically Interacts with ZNF131
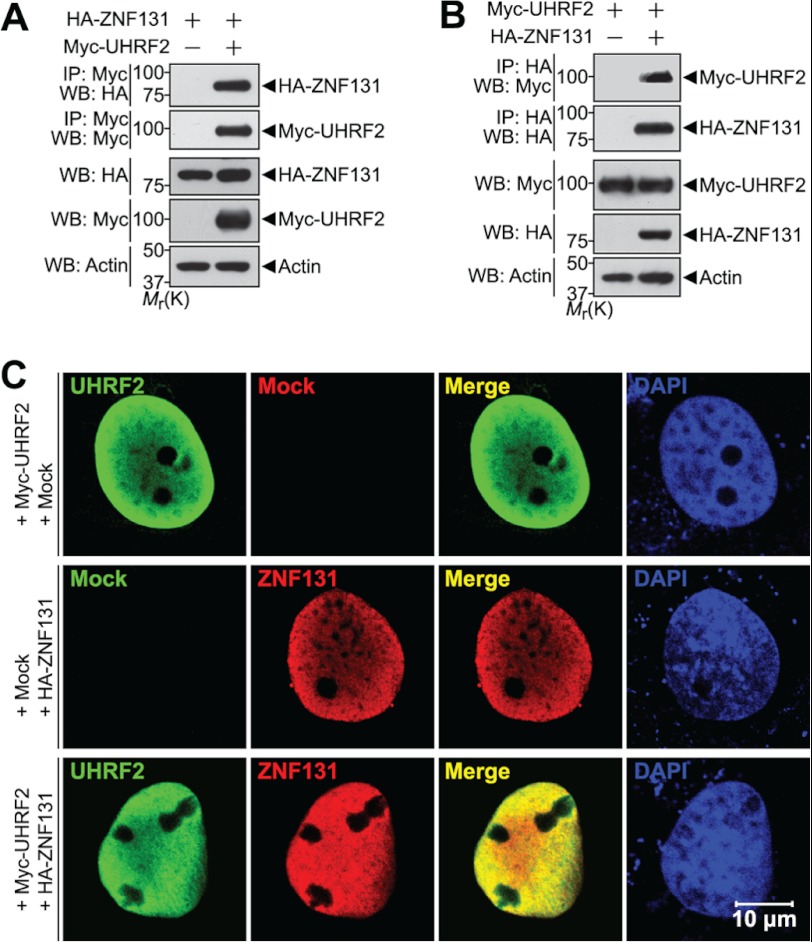
To characterize further functional roles and/or regulatory modes of ZNF131, we performed a yeast two-hybrid assay using full-length ZNF131 as bait. After screening a human fetal brain cDNA library, we isolated several binding partners of ZNF131, including hPc2, a previously reported SUMO E3 ligase of ZNF131 (30), and UHRF2 (data not shown). Because UHRF2 has ubiquitin E3 ligase activity, we examined the relationship between these two proteins. To confirm whether UHRF2 interacts with ZNF131 in mammalian cells, HEK293 cells were transfected with plasmids encoding Myc-tagged UHRF2 and HA-tagged ZNF131. The lysates were co-immunoprecipitated with the Myc (Fig. 1A) or HA (Fig. 1B) antibodies, followed by Western blotting with HA (Fig. 1A) or Myc (Fig. 1B) antibodies, respectively. As shown in Fig. 1, A and B, UHRF2 binds to ZNF131 in HEK293 cells. Immunostaining of COS-7 cells transfected with Myc-UHRF2 or HA-ZNF131 demonstrates that UHRF2 or ZNF131 alone exhibit diffused nuclear localization (Fig. 1C, top and middle panels). This finding is consistent with previous reports that both UHRF2 and ZNF131 are nuclear proteins (24, 31). Moreover, when cells co-express UHRF2 and ZNF131, the proteins are co-localized within the nucleoplasm (Fig. 1C, bottom panel). Overall, these data show that UHRF2 physically interacts with ZNF131 in mammalian cells.
FIGURE 1.
UHRF2 physically interacts with ZNF131. A, HEK293 cells were transfected for 24 h with HA-ZNF131 alone or together with Myc-UHRF2 and immunoprecipitated (IP) with the Myc antibody. Immunoprecipitates were immunoblotted with the indicated antibodies. Actin served as a protein loading control. B, HEK293 cells were transfected for 24 h with Myc-UHRF2 alone or together with HA-ZNF131, and immunoprecipitated with the HA antibody. Immunoprecipitates were immunoblotted with the indicated antibodies as indicated. C, COS-7 cells were transfected for 24 h with Myc-UHRF2 alone or together with HA-ZNF131, and immunocytochemical analyses were performed using the Myc and HA antibodies. Expression of Myc-UHRF2 (green), HA-ZNF131 (red), and DAPI-stained nuclei (blue) were analyzed using confocal microscopy. The merged image (yellow) indicates Myc-UHRF2 and HA-ZNF131 co-localization. WB, Western blot; Mr(K), relative molecular mass.
UHRF2 Does Not Affect Ubiquitination but Induces the SUMOylation of ZNF131
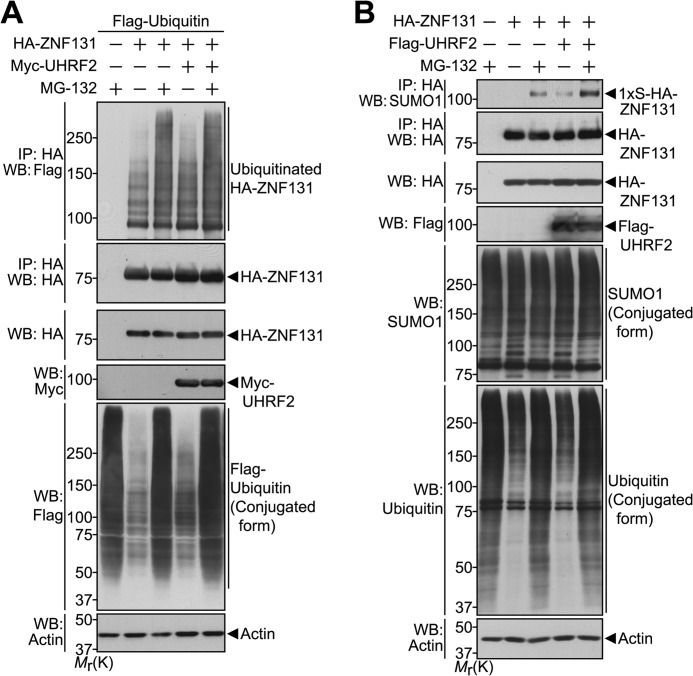
We next assessed whether UHRF2 enhances the ubiquitination of ZNF131. HEK293 cells were transfected with Flag-tagged ubiquitin and HA-tagged ZNF131 alone or together with Myc-tagged UHRF2. The formation of ubiquitinated-ZNF131 was determined by immunoprecipitating cell extracts with the HA antibody followed by Western blotting with the Flag antibody. As shown in Fig. 2A, upon ubiquitin overexpression, ZNF131 was considerably ubiquitinated. In addition, pretreatment with the proteasomal inhibitor MG-132 enhanced ZNF131 ubiquitination (Fig. 2A), supporting data from our previous report (30). However, UHRF2 did not significantly affect the extent of ZNF131 ubiquitination, which was also irrespective of MG-132 treatment (Fig. 2A). Based on our previous findings that impaired proteasomal activity remarkably promotes the formation of SUMOylated ZNF131 (30), we additionally examined whether UHRF2 alters the MG-132-induced SUMOylation pattern of ZNF131. HEK293 cells were transfected with HA-tagged ZNF131 alone or together with FLAG-tagged UHRF2. In a similar manner as the ZNF131 ubiquitination experiments, the formation of SUMOylated ZNF131 was determined by immunoprecipitating cell extracts with the HA antibody followed by Western blotting with the SUMO1 antibody. As expected, the addition of MG-132 specifically induced the mono-SUMOylation of ZNF131 (Fig. 2B). Interestingly, UHRF2 overexpression remarkably increased MG-132-induced mono-SUMOylation of ZNF131 (Fig. 2B). Moreover, we observed that UHRF2 effectively promotes the mono-SUMOylation of ZNF131 in the absence of MG-132 (Fig. 2B). These data demonstrate that UHRF2 does not influence ZNF131 ubiquitination but enhances ZNF131 mono-SUMOylation.
FIGURE 2.
UHRF2 does not affect ubiquitination but induces the SUMOylation of ZNF131. A, HEK293 cells were transfected for 24 h with FLAG-ubiquitin, HA-ZNF131, or Myc-UHRF2 alone or in combination. Cells were left untreated or were treated with MG-132 (20 μm) for 9 h. The cell lysates were immunoprecipitated (IP) with the HA antibody followed by Western blot (WB) with the indicated antibodies. Actin served as a protein loading control. B, HEK293 cells were transfected for 24 h with HA-ZNF131 alone or together with Myc-UHRF2. Cells were left untreated or were treated with MG-132 (20 μm) for 9 h. The cell lysates were immunoprecipitated with the HA antibody followed by Western blot with the indicated antibodies. Mr(K), relative molecular mass.
UHRF2 Functions as a SUMO E3 Ligase for ZNF131
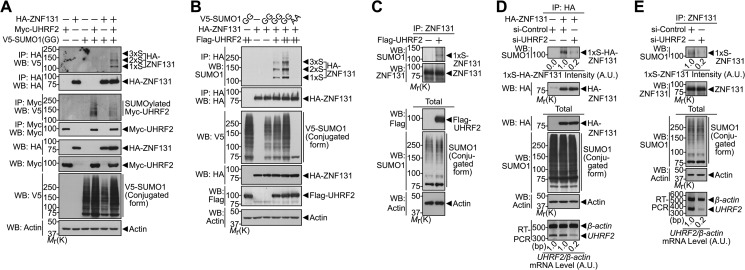
Next, we evaluated how UHRF2 stimulates ZNF131-SUMOylation. After HEK293 cells were transiently transfected with HA-tagged ZNF131 in various combinations with Myc-tagged UHRF2 or/and V5-tagged SUMO1-GG, we assessed the formation of SUMOylated ZNF131 by immunoprecipitation of HA-tagged ZNF131, followed by Western blotting with the V5 antibody. Compared with the ZNF131 SUMOylation pattern in the absence and presence of UHRF2, the level of SUMOylated ZNF131 was relatively undetectable in cells co-expressing ZNF131 and SUMO1-GG. The presence of UHRF2 greatly increased the amount of SUMOylated ZNF131 (Fig. 3A, top panel). In addition, UHRF2 was found to be a target for covalent SUMO modification (Fig. 3A, third panel). Surprisingly, UHRF2 overexpression resulted in increased SUMO-conjugated protein levels (Fig. 3A, seventh panel). Moreover, there was an additive and dose-dependent effect of UHRF2 on ZNF131 SUMOylation, suggesting the possibility that UHRF2 has a novel role as a SUMO E3 ligase (Fig. 3B). As expected, UHRF2 did not stimulate ZNF131 SUMOylation in the presence of a conjugation-defective SUMO1-AA mutant (Fig. 3B). Furthermore, UHRF2 overexpression greatly enhanced SUMO modification of endogenous ZNF131 (Fig. 3C). These results demonstrate that UHRF2 effectively promotes the SUMOylation of ZNF131 and that effect is not an artifact of ZNF131 overexpression.
FIGURE 3.
UHRF2 functions as a SUMO E3 ligase for ZNF131. A, HEK293 cells were transfected for 24 h with HA-ZNF131, Myc-UHRF2, or V5-SUMO1-GG alone or in combination. Cell extracts were immunoprecipitated (IP) with the HA or Myc antibodies and immunoblotted with the indicated antibodies. Actin was used as a protein loading control. B, HEK293 cells were transfected for 24 h with HA-ZNF131, Myc-UHRF2, and either V5-SUMO1-GG or V5-SUMO1-AA. The cell extracts were immunoprecipitated with the HA antibody and immunoblotted with the indicated antibodies. C, HEK293 cells were transfected with either mock vector or FLAG-UHRF2. The cell extracts were immunoprecipitated with the ZNF131 antibody followed by immunoblot with the indicated antibodies. D, HEK293 cells were transfected for 36 h with HA-ZNF131 and either si-control RNA or si-UHRF2 RNA. The cell extracts were immunoprecipitated with the HA antibody, followed by Western blot (WB) with the indicated antibodies. E, HEK293 cells were transfected for 48 h with either si-control RNA or si-UHRF2 RNA. The cell extracts were immunoprecipitated with the ZNF131 antibody, followed by immunoblot with the indicated antibodies. Total cellular RNA was analyzed by RT-PCR using UHRF2 primers, whereas β-actin primers were used as an internal control for RT-PCR efficiency. The intensity of mono-SUMOylated HA-ZNF131 and RT-PCR bands were quantified by densitometry using Multi Gauge software (version 3.1; Fujifilm Corp.). A.U., arbitrary unit; Mr(K), relative molecular mass.
However, we observed multiple UHRF2-mediated ZNF131 SUMOylation bands (Fig. 3, A and B), which corresponded with the sizes of multi-SUMO-conjugated ZNF131 band. As described previously (30), overexpression of the SUMO protein generated di- and tri-SUMOylated forms of ZNF131 in HEK293 cells (2xS, 3xS), which likely occurs because of increased SUMO levels and subsequent excessive SUMOylation. Nevertheless, ZNF131 is mono-SUMOylated at the lysine 567 residue in physiological conditions using either endogenous SUMO1 or SUMO2/3 (30). Similarly, although multiple SUMOylated UHRF2 bands were generated in cells overexpressing SUMO protein (Fig. 3A, third panel), UHRF2 was mono-SUMOylated in physiological conditions using endogenous SUMO1 (supplemental Fig. S1).
To prove that UHRF2 acts as an endogenous SUMO E3 ligase for ZNF131, we examined the effect of UHRF2 knockdown on ZNF131 SUMOylation. To do this, we tested many commercially available UHRF2 antibodies. Unfortunately, none of the tested antibodies successfully detected endogenous UHRF2 protein. Therefore, as an alternative, we assessed the effect of knocking down UHRF2 using UHRF2-specific siRNA on mRNA expression using RT-PCR. Compared with the nonspecific control siRNA, UHRF2 siRNA reduces UHRF2 mRNA expression by ∼80% (Fig. 3, D and E). Furthermore, when UHRF2 was knocked down, the level of mono-SUMOylated ZNF131 protein decreased by ∼80% (Fig. 3, D and E). Overall, these data suggest that UHRF2 is an endogenous SUMO E3 ligase for ZNF131.
UHRF2 Promotes ZNF131 SUMOylation at Lysine 567
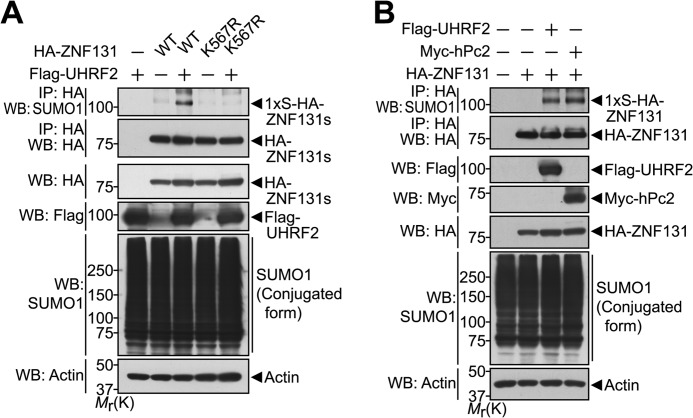
Previously, we demonstrated that the SUMO E3 ligase hPc2 enhances mono-SUMOylation of ZNF131 at the lysine 567 residue (30). To determine the major acceptor site(s) of UHRF2-induced SUMOylation, we first examined whether lysine 567 could also be the main acceptor residue of UHRF2-induced ZNF131-SUMOylation. HEK293 cells were co-transfected with UHRF2 and either wild type ZNF131 or a point mutant that substitutes lysine 567 with conjugation-defective arginine (ZNF131-K567R). The formation of SUMOylated ZNF131 was determined by immunoprecipitating cell extracts with the HA antibody, followed by Western blotting with the SUMO1 antibody. As shown in Fig. 4A, UHRF2 stimulates ZNF131 SUMOylation in cells overexpressing wild type ZNF131. However, SUMOylation was not observed in the presence of ZNF131-K567R, confirming that lysine 567 is a major SUMO acceptor site for UHRF2-induced ZNF131 SUMOylation (Fig. 4A). Moreover, UHRF2 promoted ZNF131 SUMOylation at the same efficiency as hPc2 (Fig. 4B). These data suggest that, similar to hPc2, UHRF2 promotes ZNF131 SUMOylation and modifies the same lysine 567 residue of ZNF131.
FIGURE 4.
UHRF2 promotes ZNF131 SUMOylation at lysine 567. A, HEK293 cells were transfected for 24 h with FLAG-UHRF2, HA-ZNF131-WT, or HA-ZNF131-K567R alone or in combination. The cell extracts were immunoprecipitated (IP) with the HA antibody followed by Western blot (WB) with the indicated antibodies. Actin was used as a protein loading control. B, HEK293 cells were transfected for 24 h with HA-ZNF131 and either FLAG-UHRF2 or Myc-hPc2. Cell extracts were immunoprecipitated with the HA antibody and immunoblotted with the indicated antibodies. Mr(K), relative molecular mass.
UHRF2 Interacts with SUMO1 and the SUMO E2 Conjugase UBC9
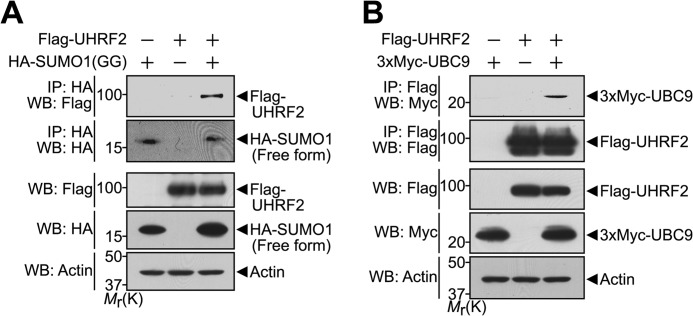
Because UHRF2 enhances ZNF131 SUMOylation in the same manner as hPc2, we speculated that UHRF2 is a novel SUMO E3 ligase. Based on the finding that all SUMO E3 ligases directly bind both SUMO and the SUMO E2 conjugase UBC9 (32), we examined whether UHRF2 binds to SUMO and UBC9. HEK293 cells were co-transfected with plasmids encoding FLAG-tagged UHRF2 and either HA-tagged SUMO1-GG (Fig. 5A) or 3×Myc-tagged UBC9 (Fig. 5B). The lysates were co-immunoprecipitated with the HA (Fig. 5A) or FLAG (Fig. 5B) antibodies, followed by Western blotting with FLAG (Fig. 5A) or Myc (Fig. 5B) antibodies, respectively. The results reveal that UHRF2 interacts with both SUMO1 and UBC9 proteins, suggesting that UHRF2 is a novel SUMO E3 ligase.
FIGURE 5.
UHRF2 interacts with the SUMO1 protein and SUMO E2 conjugase UBC9. A, HEK293 cells were transfected for 24 h with FLAG-UHRF2 alone or together with HA-SUMO1-GG, and immunoprecipitated (IP) with the HA antibody. Immunoprecipitates were immunoblotted with the indicated antibodies. Actin served as a protein loading control. B, HEK293 cells were transfected for 24 h with 3×Myc-UBC9 alone or together with FLAG-UHRF2 and immunoprecipitated with the FLAG antibody. Immunoprecipitates were immunoblotted with the indicated antibodies. WB, Western blot; Mr(K), relative molecular mass.
The SRA and NCR Domains of UHRF2 Are Required for SUMO E3 Ligase Activity on ZNF131
To determine which region(s) of UHRF2 is required for ZNF131 SUMOylation, several UHRF2 deletion and point mutation mutants were constructed, and their catalytic activities were measured (Fig. 6A). Compared with full-length UHRF2, ZNF131 SUMOylation was greatly reduced in the presence of UHRF2 mutants deleting the SRA (UHRF2-ΔSRA) or nuclear localization signal-containing region (UHRF2-ΔNCR) (Fig. 6B). Interestingly, UHRF2 mutants with the RING domain deleted (UHRF2-ΔRING) or with the cysteine 735 residue substituted with serine (UHRF2-C735S), which were previously shown to lack ubiquitin E3 ligase activity (25), did not significantly affect ZNF131 SUMOylation (Fig. 6B). These data suggest that the SRA and NCR domains of UHRF2 are necessary for the SUMO E3 ligase activity on ZNF131, which is independent of the RING domain that is critical for its ubiquitin E3 ligase activity. In the same manner, we also determined that the mono-SUMOylation of UHRF2 is dependent on its SRA and NCR domains (supplemental Fig. S1).
FIGURE 6.
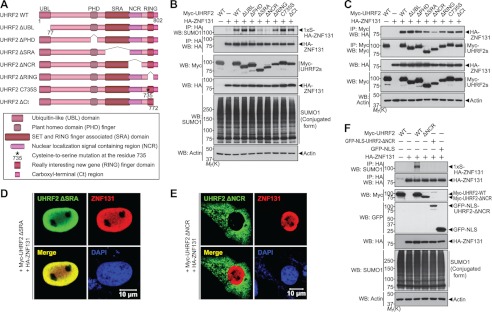
The SRA and NCR domains of UHRF2 are required for SUMO E3 ligase activity on ZNF131. A, schematic diagram of full-length UHRF2 and several deletion and point mutants. B, HEK293 cells were transfected for 24 h with HA-ZNF131 alone or together with either Myc-tagged wild type or mutant UHRF2, as indicated. The cell extracts were immunoprecipitated (IP) with the HA antibody followed by Western blot (WB) with the indicated antibodies. Actin was used as a protein loading control. C, HEK293 cells were transfected for 24 h with HA-ZNF131 alone or together with wild type or mutant Myc-UHRF2, as indicated. The cell extracts were immunoprecipitated with the Myc antibody, followed by Western blot with the indicated antibodies. D and E, COS-7 cells were transfected for 24 h with HA-ZNF131 and either Myc-UHRF2-ΔSRA (D) or Myc-UHRF2-ΔNCR (E), and immunocytochemically analyzed using the Myc and HA antibodies. Expression of Myc-UHRF2 (green), HA-ZNF131 (red), and DAPI-stained nuclei (blue) was analyzed using confocal microscopy. The merged image (yellow) indicates the co-localization of Myc-UHRF2 and HA-ZNF131. F, HEK293 cells were transfected for 24 h with HA-ZNF131 and Myc-UHRF2-WT, Myc-UHRF2-ΔNCR, GFP-NLS-UHRF2-ΔNCR, or GFP-NLS, as indicated. Cell extracts were immunoprecipitated with the HA antibody and immunoblotted with the indicated antibodies. Mr(K), relative molecular mass.
To assess how deletion of SRA or NCR domains diminishes UHRF2 enzymatic activity, we examined whether these mutants have decreased binding affinity to the substrate. Co-immunoprecipitation experiments revealed that the binding affinity of UHRF2-ΔSRA to ZNF131 was slightly decreased, whereas UHRF2-ΔNCR showed a remarkable decrease in binding affinity to ZNF131 (Fig. 6C). Moreover, immunostaining of COS-7 cells co-transfected with HA-ZNF131 and either Myc-UHRF2-ΔSRA or Myc-UHRF2-ΔNCR demonstrated that SRA deletion did not alter the co-localization of UHRF2 and ZNF131 (Fig. 6D). In contrast, expression of Myc-UHRF2-ΔNCR caused a distinct and spatial separation between the two proteins (Fig. 6E). These results demonstrate that the reduced SUMO E3 ligase activity of UHRF2-ΔNCR is predominantly caused by decreased substrate binding, whereas deletion of the SRA region may result in the loss of essential UHRF2 enzymatic function.
Because deletion of the NCR domain is assumed to primarily block the nuclear localization of UHRF2, we tested whether the nuclear localization signal peptide is required for the SUMO E3 ligase activity of UHRF2. We used an artificial NLS-fused UHRF2-ΔNCR mutant generated by inserting an NLS peptide into the N-terminal site of UHRF2-ΔNCR. As expected, this mutant properly led the relocation of UHRF2 protein into the nucleus similar to wild type UHRF2 protein (supplemental Fig. S2). However, the nuclear re-entry of UHRF2 did not rescue its SUMO E3 ligase function on ZNF131 (Fig. 6F). These data provide evidence that the diminished SUMO E3 ligase activity of the UHRF2-ΔNCR mutant is not simply caused by the lack of nuclear localization signal, but rather, the presence of an intact NCR domain is required for proper substrate recognition. Taken together, these data show that the SRA and NCR domains of UHRF2 are necessary for its SUMO E3 ligase activity toward ZNF131.
UHRF2 Mediates Auto-SUMOylation within the SRA and NCR Domains
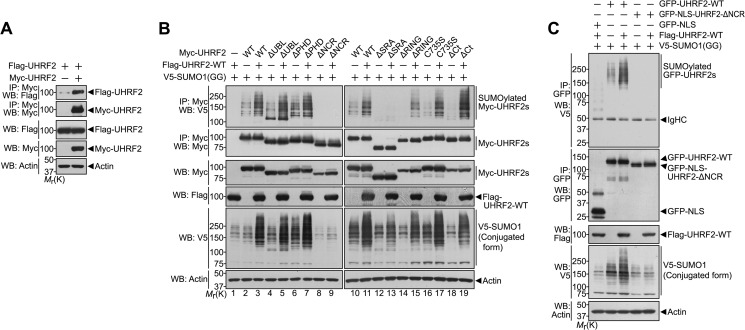
UHRF2 is a substrate for covalent SUMO modification (Fig. 3A, third panel; supplemental Fig. S1) and promotes ZNF131 SUMOylation (Fig. 3). Therefore, we determined whether UHRF2 directly enhances auto-SUMOylation. To examine whether UHRF2 interacts with itself, HEK293 cells were co-transfected with plasmids encoding Myc- or FLAG-tagged UHRF2, respectively. The lysates were immunoprecipitated with the Myc antibody, followed by Western blotting with the FLAG antibody. As shown in Fig. 7A, UHRF2 binds itself in HEK293 cells (Fig. 7A). This result suggests that UHRF2 could be a substrate for itself. Next, we examined whether UHRF2 directly promotes auto-SUMOylation. HEK293 cells were transfected with Myc-tagged wild type UHRF2 and V5-tagged SUMO1-GG in the absence or presence of FLAG-tagged wild type UHRF2. The formation of SUMOylated UHRF2 was determined by immunoprecipitating cell extracts with the Myc antibody, followed by Western blotting with the V5 antibody. As shown in Fig. 7B, wild type UHRF2 effectively enhances intermolecular auto-SUMOylation of full-length UHRF2 (compare lanes 2 and 3 or lanes 10 and 11 in Fig. 7B, top panel).
FIGURE 7.
UHRF2 mediates auto-SUMOylation within the SRA and NCR domains. A, HEK293 cells were transfected for 24 h with FLAG-UHRF2 alone or together with Myc-UHRF2 and immunoprecipitated (IP) with the Myc antibody. Immunoprecipitates were immunoblotted with the indicated antibodies. Actin served as a protein loading control. B, HEK293 cells were transfected for 24 h with V5-SUMO1-GG, various Myc-tagged UHRF2 mutants, or/and FLAG-UHRF2-WT, as indicated. The cell extracts were immunoprecipitated with the Myc antibody, followed by Western blot (WB) with the indicated antibodies. C, HEK293 cells were transfected for 24 h with V5-SUMO1-GG, GFP-UHRF2-WT, GFP-NLS-UHRF2-ΔNCR, GFP-NLS, or/and FLAG-UHRF2-WT, as indicated. Cell extracts were immunoprecipitated with the GFP antibody and immunoblotted with the indicated antibodies. IgHC, immunoglobulin heavy chain; Mr(K), relative molecular mass.
Next, we examined which site(s) or region(s) of UHRF2 is a target or important for auto-SUMOylation. The human UHRF2 protein contains a single consensus SUMOylation motif ΨKX(DE) at amino acids 306–309 as well as a single inverted SUMOylation motif (DE)XK(FILPV) at amino acids 546–549, both of which are well conserved across species (supplemental Fig. S3, A and B). However, mutation of either of these sites had no significant effect of UHRF2 auto-SUMOylation, suggesting that neither of the well conserved SUMOylation motifs are the major SUMO acceptor site(s) (supplemental Fig. S3, C and D). To further clarify the self-SUMOylation region(s) of UHRF2, we generated several constructs encoding Myc-tagged deletion or point mutants of UHRF2. After each of these mutants was co-transfected into HEK293 cells with V5-tagged SUMO1-GG in the absence or presence of FLAG-tagged wild type UHRF2, the formation of SUMOylated UHRF2 was determined by immunoprecipitating the cell extracts with the Myc antibody and immunoblotting with the V5 antibody. As shown in Fig. 7B, the addition of wild type UHRF2 failed to enhance SUMOylation of UHRF2-ΔSRA and -ΔNCR mutants, whereas other UHRF2 mutants did not show any considerable change in auto-SUMOylation pattern (Fig. 7B). Furthermore, co-expressing wild type UHRF2 with either UHRF2-ΔNCR or -ΔSRA did not increase the levels of SUMO-conjugated proteins, compared with cells transfected with wild type UHRF2 alone (compare the intensities of input V5-SUMO bands in the lanes 1 and 9 or lanes 10 and 13 in Fig. 7B, fifth panel). In addition, SUMOylation of the NLS-fused UHRF2-ΔNCR mutant, which leads to re-entry of UHRF2 into the nucleus, was not enhanced by wild type UHRF2 (Fig. 7C). These results suggest that both the SRA and NCR domains of UHRF2 are necessary for auto-SUMOylation. These results are coincident with the finding that UHRF2 mono-SUMOylation is dependent on the presence of the SRA or NCR domain (supplemental Fig. S1). Accordingly, we speculate that mono-SUMOylation of UHRF2 in physiological conditions may be caused by UHRF2 itself.
Taken together, our data suggest that UHRF2 directly enhances auto-SUMOylation at the SRA and NCR domains. Moreover, the auto-SUMOylation of UHRF2 appears to enhance its SUMO E3 ligase activity toward ZNF131.
DISCUSSION
In the present study, we demonstrate that UHRF2 functions as a SUMO E3 ligase for ZNF131. The identification of this new enzyme activity is based on the following observations: (i) UHRF2 binds to the SUMO-conjugating E2 enzyme UBC9 and SUMO protein; (ii) UHRF2 overexpression increases SUMO-conjugated protein levels; (iii) UHRF2 binds ZNF131, the substrate of SUMOylation (30); (iv) UHRF2 overexpression stimulates SUMO modification of its binding partners, including ZNF131; (v) knockdown of UHRF2 dramatically reduces ZNF131 SUMOylation; (vi) similar to other SUMO E3 ligases, UHRF2 becomes auto-SUMOylated, which largely contributes to the activation of its SUMO E3 ligase activity; (vii) the SRA and NCR domains of UHRF2 are specifically required for the SUMO E3 activity on ZNF131, whereas the RING domain of UHRF2, which is critical for ubiquitin E3 ligase activity, is dispensable.
TOPORS was the first reported dual-functional ubiquitin/SUMO E3 ligase for p53 (20). TOPORS belongs to a class of RING-dependent ubiquitin E3 ligases (33), but its SUMO E3 ligase activity on p53 is independent of the RING finger domain (20), which is identical to UHRF2 activity toward ZNF131 SUMOylation. The second reported dual-functional E3 ligase is TRAF7, which promotes SUMOylation of c-Myb (21) as well as the auto-ubiquitination of TRAF7 (34). Interestingly, although TRAF7 possesses the RING domain, it did not enhance c-Myb ubiquitination (21). The third identified dual-functional E3 ligase is TRIM27 (22). TRIM27 is also classified as a RING-type ubiquitin E3 ligase, which mediates p53 ubiqutination as well as Mdm2 SUMOylation (22). However, in contrast to TOPORS (20), the SUMO E3 activity of TRIM27 relies on the presence of the RING domain (22). The present data indicate that the RING-type ubiquitin E3 ligase UHRF2 also acts as a novel SUMO E3 ligase for ZNF131. We also demonstrate that the SUMO E3 ligase activity of UHRF2 is independent from its ubiquitin E3 activity because a UHRF2 mutant lacking the RING domain still enhances ZNF131 SUMOylation. In addition, we found that both the SRA and NCR domains are critical regions of UHRF2 for SUMO E3 ligase activity on ZNF131, although the NCR domain predominantly allows UHRF2 to bind ZNF131 properly. These data strongly indicate that the essential SUMO E3 ligase function of UHRF2 depends primarily on the SRA domain.
Although several SUMO E3 ligases were reported to be auto-SUMOylated, the functional relevance between their SUMO E3 ligase activities and auto-SUMOylation status has not yet been clarified. Nonetheless, a few reports indicated that similar to many ubiquitin E3 ligases, the auto-SUMOylation of some SUMO E3 ligases seems to directly affect their enzymatic activities. For example, Ihara et al. (35) demonstrated that the auto-SUMOylation of PIASy is necessary for SUMOylation of its substrate, Tcf-4. In addition, Ivanov et al. (36) showed that the interaction of TRIM28 (also known as KAP1) with its effector proteins is dependent on the extent of TRIM28 auto-SUMOylation. Here, we present evidence that UHRF2 becomes auto-SUMOylated. In addition, although we failed to identify the exact lysine site(s) for auto-SUMOylation of UHRF2, the SUMO E3 ligase activity of UHRF2 is largely dependent upon auto-SUMOylation. This hypothesis is based on the finding that the UHRF2-ΔNCR or UHRF2-ΔSRA mutants, both of which are defective in auto-SUMOylation (Fig. 7B and supplemental Fig. S1), did not enhance the SUMOylation of substrate(s), such as ZNF131, UHRF2, and uncharacterized proteins (Figs. 6B and 7B).
UHRF family members, including UHRF1 and UHRF2, are multidomain ubiquitin E3 ligases uniquely possessing the SRA domain that participates in methylation-dependent transcriptional regulation (37). UHRF2 is highly similar to UHRF1 in both sequence and structure; therefore, they are considered to have similar cellular function. A recent study revealed that, similar to UHRF1, UHRF2 binds preferentially to methylated histone H3 lysine 9 (H3K9) through its conserved Tudor domain and hemimethylated DNA through the SRA domain (38). However, UHRF2 and UHRF1 are not functionally redundant in DNA methylation maintenance (38). In addition, UHRF2 acts an intermodular hub that occupies a central position in the multicellular network and facilitates their coordination among the cell cycle machinery, the ubiquitin-proteasome system, and the epigenetic system (39). Many proteins involved in epigenetic regulation are SUMOylated by various epigenetic cues and/or have SUMO E3 ligase functions themselves. For example, histone deacetylase HDAC4 (19) and HDAC7 (40), as well as polycomb protein hPc2 (16) are reported SUMO E3 ligases. Here, we demonstrate that the ubiquitin E3 ligase involved in DNA methylation maintenance, UHRF2, functions as a SUMO E3 ligase. Due to the structural similarity of UHRF1 and UHRF2, it will be interesting to test whether UHRF1 also has a SUMO E3 ligase function. Furthermore, it would be interesting to elucidate whether the SUMOylation pathway is closely associated with DNA methylation maintenance and/or epigenetic regulation by UHRF family proteins.
ZNF131 is a member of a zinc finger protein superfamily with BTB/POZ domains and was initially identified based on its association with developmental disorders (41). Previous reports, including ours, demonstrated that ZNF13 acts as a repressor of estrogen receptor α-mediated trans-activation (30, 42, 43). In addition, we previously reported that the SUMOylation of ZNF131 negatively regulates estrogen receptor α-mediated signaling and estrogen-induced cell proliferation in breast cancer cells (30). Based on the recent report that UHRF2, as a putative oncogene, inactivates tumor suppressor gene activity in breast cancer progression (44), it does not seem likely that ZNF131 SUMOylation by UHRF2 is directly involved with breast cancer occurrence and/or development. Rather, it is possible that UHRF2 regulates the function of the master factors, such as p53 and pRb, in breast cancer progression in a direct or indirect way. The regulatory mode could be either ubiquitination or SUMOylation. Similar to the finding by Wu et al. (44), we also observed that knockdown of UHRF2 reduces breast cancer cell proliferation (supplemental Fig. S4A). Interestingly, overexpression of wild type UHRF2 promotes breast cancer cell proliferation, whereas the deletion of SRA domain significantly attenuates UHRF2-induced cell growth (supplemental Fig. S4B). Because SRA domain is required for full SUMO E3 activity, as well as for SUMOylation of UHRF2 itself, we speculate that SUMO E3 ligase activity of UHRF2 somehow plays a role to regulate cell proliferation. Nevertheless, UHRF2-mediated SUMOylation might regulate other ZNF131 functions, including its putative transcriptional activator function (45). Although additional functions of ZNF131 remain unclear, it will be interesting to test whether SUMOylation may affect those activities of ZNF131.
UHRF2, as an ubiquitin E3 ligase, mediates the ubiquitination of various substrates, including cyclin D1 and E1 (27), p53 (29), PEST proteolytic signal-containing nuclear protein, as well as UHRF2 itself (28). In addition, various UHRF2 binding partners are reported, such as inactive CDK2-cyclin E complex (26), cyclin A2 and B1, and pRb (27). Here, we identify an additional function of UHRF2 as a novel SUMO E3 ligase. Furthermore, we elucidate that ZNF131 and UHRF2 itself are substrates for UHRF2-mediated SUMOylation. Because many proteins are substrates for both SUMOylation and ubiquitination, it would be interesting to test whether UHRF2 enhances SUMOylation of reported binding partners or the substrates for ubiquitination. Moreover, because the SUMOylation of many undefined proteins is remarkably promoted by UHRF2 (Fig. 3A, seventh panel; Fig. 3B, third panel; Fig. 7B, fifth panel), some may be additional SUMOylation substrates for UHRF2. Therefore, further experiments should focus on identifying and testing whether UHRF2 enhances SUMOylation of those putative substrates.
In summary, the current study shows that the ubiquitin E3 ligase UHRF2 acts as a dual-functional SUMO E3 ligase. In addition, UHRF2 effectively enhances the SUMOylation of ZNF131, whereas it does not enhance ZNF131 ubiquitination. Moreover, the SUMO E3 ligase activity of UHRF2 on ZNF131 is dependent on the SRA and NCR domains, whereas the RING domain of UHRF2 is not necessary. Taken together, our data reveal that UHRF2 may have substrate-specific ubiquitin or SUMO E3 ligase function, and these two enzymatic activities work through each independent functional domains and/or regulatory mechanisms. To our knowledge, UHRF2 constitutes the fourth example of a dual-functional ubiquitin/SUMO E3 ligase.
Acknowledgment
We thank Y. K. Jang for providing plasmids.
This work was supported by National Research Foundation Grant 2012R1A1A2021749 (to K. C. C.) funded by the Ministry of Education, Science, and Technology (MEST) and by the Basic Science Research Program through NRF (Korea) 2012-0000810 (to K. C. C.). This work was also supported by the Brain Research Center of the 21st Century Frontier Research Program Technology (2009K-001251 (to K. C. C.) funded by MEST. This work was also supported in part by Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Korea Grants A092004 and A111653 (to K. C. C.).

This article contains supplemental Figs. S1–S4.
- SUMO
- small ubiquitin-like modifier
- ZNF131
- zinc finger protein 131
- Pc2
- polycomb protein 2
- hPc2
- human polycomb protein 2
- SRA
- SET and RING finger-associated
- NLS
- nuclear localization signal
- NCR
- nuclear localization signal-containing region
- UBC9
- ubiquitin-conjugating enzyme E2I
- TOPORS
- topoisomerase I binding, arginine/serine-rich
- TRAF7
- TNF receptor-associated factor 7
- PIAS
- protein inhibitor of activated STAT
- TRIM27
- tripartite motif containing 27.
REFERENCES
- 1. Wilkinson K. A., Henley J. M. (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 428, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Johnson E. S., Schwienhorst I., Dohmen R. J., Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Desterro J. M., Rodriguez M. S., Kemp G. D., Hay R. T. (1999) Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 274, 10618–10624 [DOI] [PubMed] [Google Scholar]
- 4. Okuma T., Honda R., Ichikawa G., Tsumagari N., Yasuda H. (1999) In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254, 693–698 [DOI] [PubMed] [Google Scholar]
- 5. Johnson E. S., Blobel G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802 [DOI] [PubMed] [Google Scholar]
- 6. Desterro J. M., Thomson J., Hay R. T. (1997) Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417, 297–300 [DOI] [PubMed] [Google Scholar]
- 7. Gong L., Kamitani T., Fujise K., Caskey L. S., Yeh E. T. (1997) Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272, 28198–28201 [DOI] [PubMed] [Google Scholar]
- 8. Rodriguez M. S., Dargemont C., Hay R. T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 9. Sampson D. A., Wang M., Matunis M. J. (2001) The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276, 21664–21669 [DOI] [PubMed] [Google Scholar]
- 10. Sachdev S., Bruhn L., Sieber H., Pichler A., Melchior F., Grosschedl R. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15, 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kahyo T., Nishida T., Yasuda H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 [DOI] [PubMed] [Google Scholar]
- 12. Schmidt D., Müller S. (2002) Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. U.S.A. 99, 2872–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hochstrasser M. (2001) SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107, 5–8 [DOI] [PubMed] [Google Scholar]
- 14. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 15. Kirsh O., Seeler J. S., Pichler A., Gast A., Müller S., Miska E., Mathieu M., Harel-Bellan A., Kouzarides T., Melchior F., Dejean A. (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21, 2682–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kagey M. H., Melhuish T. A., Wotton D. (2003) The polycomb protein Pc2 is a SUMO E3. Cell 113, 127–137 [DOI] [PubMed] [Google Scholar]
- 17. Wotton D., Merrill J. C. (2007) Pc2 and SUMOylation. Biochem. Soc. Trans. 35, 1401–1404 [DOI] [PubMed] [Google Scholar]
- 18. Merrill J. C., Melhuish T. A., Kagey M. H., Yang S. H., Sharrocks A. D., Wotton D. (2010) A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE 5, e8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., Yao T. P. (2005) Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell Biol. 25, 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weger S., Hammer E., Heilbronn R. (2005) Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 579, 5007–5012 [DOI] [PubMed] [Google Scholar]
- 21. Morita Y., Kanei-Ishii C., Nomura T., Ishii S. (2005) TRAF7 sequesters c-Myb to the cytoplasm by stimulating its sumoylation. Mol. Biol. Cell 16, 5433–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chu Y., Yang X. (2011) SUMO E3 ligase activity of TRIM proteins. Oncogene 30, 1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ulrich H. D. (2005) Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol. 15, 525–532 [DOI] [PubMed] [Google Scholar]
- 24. Mori T., Li Y., Hata H., Ono K., Kochi H. (2002) NIRF, a novel RING finger protein, is involved in cell-cycle regulation. Biochem. Biophys. Res. Commun. 296, 530–536 [DOI] [PubMed] [Google Scholar]
- 25. Iwata A., Nagashima Y., Matsumoto L., Suzuki T., Yamanaka T., Date H., Deoka K., Nukina N., Tsuji S. (2009) Intranuclear degradation of polyglutamine aggregates by the ubiquitin-proteasome system. J. Biol. Chem. 284, 9796–9803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Y., Mori T., Hata H., Homma Y., Kochi H. (2004) NIRF induces G1 arrest and associates with Cdk2. Biochem. Biophys. Res. Commun. 319, 464–468 [DOI] [PubMed] [Google Scholar]
- 27. Mori T., Ikeda D. D., Fukushima T., Takenoshita S., Kochi H. (2011) NIRF constitutes a nodal point in the cell cycle network and is a candidate tumor suppressor. Cell Cycle 10, 3284–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mori T., Li Y., Hata H., Kochi H. (2004) NIRF is a ubiquitin ligase that is capable of ubiquitinating PCNP, a PEST-containing nuclear protein. FEBS Lett. 557, 209–214 [DOI] [PubMed] [Google Scholar]
- 29. Bai L., Wang X., Jin F., Yang Y., Qian G., Duan C. (2012) UHRF2, another E3 ubiquitin ligase for p53. Biochem. Biophys. Res. Commun. 425, 908–911 [DOI] [PubMed] [Google Scholar]
- 30. Oh Y., Chung K. C. (2012) Small ubiquitin-like modifier (SUMO) modification of zinc finger protein 131 potentiates its negative effect on estrogen signaling. J. Biol. Chem. 287, 17517–17529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Donaldson N. S., Daniel Y., Kelly K. F., Graham M., Daniel J. M. (2007) Nuclear trafficking of the POZ-ZF protein Znf131. Biochim. Biophys. Acta 1773, 546–555 [DOI] [PubMed] [Google Scholar]
- 32. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rajendra R., Malegaonkar D., Pungaliya P., Marshall H., Rasheed Z., Brownell J., Liu L. F., Lutzker S., Saleem A., Rubin E. H. (2004) Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J. Biol. Chem. 279, 36440–36444 [DOI] [PubMed] [Google Scholar]
- 34. Bouwmeester T., Bauch A., Ruffner H., Angrand P. O., Bergamini G., Croughton K., Cruciat C., Eberhard D., Gagneur J., Ghidelli S., Hopf C., Huhse B., Mangano R., Michon A. M., Schirle M., Schlegl J., Schwab M., Stein M. A., Bauer A., Casari G., Drewes G., Gavin A. C., Jackson D. B., Joberty G., Neubauer G., Rick J., Kuster B., Superti-Furga G. (2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 [DOI] [PubMed] [Google Scholar]
- 35. Ihara M., Yamamoto H., Kikuchi A. (2005) SUMO-1 modification of PIASy, an E3 ligase, is necessary for PIASy-dependent activation of Tcf-4. Mol. Cell Biol. 25, 3506–3518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd. (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bronner C., Achour M., Arima Y., Chataigneau T., Saya H., Schini-Kerth V. B. (2007) The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol. Ther. 115, 419–434 [DOI] [PubMed] [Google Scholar]
- 38. Zhang J., Gao Q., Li P., Liu X., Jia Y., Wu W., Li J., Dong S., Koseki H., Wong J. (2011) S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 21, 1723–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mori T., Ikeda D. D., Yamaguchi Y., Unoki M. (2012) NIRF/UHRF2 occupies a central position in the cell cycle network and allows coupling with the epigenetic landscape. FEBS Lett. 586, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 40. Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. (2008) Histone deacetylase 7 promotes PML sumoylation and is essential for PML nuclear body formation. Mol. Cell Biol. 28, 5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tommerup N., Vissing H. (1995) Isolation and fine mapping of 16 novel human zinc finger-encoding cDNAs identify putative candidate genes for developmental and malignant disorders. Genomics 27, 259–264 [DOI] [PubMed] [Google Scholar]
- 42. Han X., Guo J., Deng W., Zhang C., Du P., Shi T., Ma D. (2008) High-throughput cell-based screening reveals a role for ZNF131 as a repressor of ERα signaling. BMC Genomics 9, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oh Y., Chung K. C. (2013) Zinc finger protein 131 inhibits estrogen signaling by suppressing estrogen receptor α homo-dimerization. Biochem. Biophys. Res. Commun. 430, 400–405 [DOI] [PubMed] [Google Scholar]
- 44. Wu J., Liu S., Liu G., Dombkowski A., Abrams J., Martin-Trevino R., Wicha M. S., Ethier S. P., Yang Z. Q. (2012) Identification and functional analysis of 9p24 amplified genes in human breast cancer. Oncogene 31, 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donaldson N. S., Nordgaard C. L., Pierre C. C., Kelly K. F., Robinson S. C., Swystun L., Henriquez R., Graham M., Daniel J. M. (2010) Kaiso regulates Znf131-mediated transcriptional activation. Exp. Cell Res. 316, 1692–1705 [DOI] [PubMed] [Google Scholar]