Background: How cytosolic thioesterases (APT1 and APT2) depalmitoylate their membrane-anchored substrates remains unclear.
Results: APT1 and APT2 require palmitoylation for membrane localization. Although APT1 depalmitoylates itself, H-Ras, and APT2, APT2 depalmitoylates GAP-43.
Conclusion: Dynamic palmitoylation regulates steady-state membrane localization and function of cytosolic thioesterases and their substrates.
Significance: The findings may provide new insight into the regulation and function of cytosolic thioesterases and their substrates.
Keywords: Enzyme Mechanisms, Enzymes, Membrane Trafficking, Oncogene, Ras, Acyl-protein Thioesterases, Cytosol-Membrane Trafficking, Dynamic Palmitoylation, GAP-43, H-Ras
Abstract
Acyl-protein thioesterase-1 (APT1) and APT2 are cytosolic enzymes that catalyze depalmitoylation of membrane-anchored, palmitoylated H-Ras and growth-associated protein-43 (GAP-43), respectively. However, the mechanism(s) of cytosol-membrane shuttling of APT1 and APT2, required for depalmitoylating their substrates H-Ras and GAP-43, respectively, remained largely unknown. Here, we report that both APT1 and APT2 undergo palmitoylation on Cys-2. Moreover, blocking palmitoylation adversely affects membrane localization of both APT1 and APT2 and that of their substrates. We also demonstrate that APT1 not only catalyzes its own depalmitoylation but also that of APT2 promoting dynamic palmitoylation (palmitoylation-depalmitoylation) of both thioesterases. Furthermore, shRNA suppression of APT1 expression or inhibition of its thioesterase activity by palmostatin B markedly increased membrane localization of APT2, and shRNA suppression of APT2 had virtually no effect on membrane localization of APT1. In addition, mutagenesis of the active site Ser residue to Ala (S119A), which renders catalytic inactivation of APT1, also increased its membrane localization. Taken together, our findings provide insight into a novel mechanism by which dynamic palmitoylation links cytosol-membrane trafficking of APT1 and APT2 with that of their substrates, facilitating steady-state membrane localization and function of both.
Introduction
Post-translational modifications such as phosphorylation, glycosylation, ubiquitination, and lipidation play critical roles in regulating the function of many proteins (1). Post-translational lipid modifications of proteins facilitate membrane localization, protein-protein interaction, cell signaling, subcellular trafficking, and vesicular transport (1–8). Some of the common lipid modifications of proteins include N-myristoylation, palmitoylation, and prenylation, which occur in the cytoplasmic face of the cell membrane (1, 5, 6). Among these, palmitoylation (also called S-acylation) is the only reversible lipid modification in which a 16-carbon fatty acid (predominantly palmitate) is attached to cysteine residues of polypeptides via thioester linkage (4, 6). Many soluble proteins require this modification for localization to membranes essential for function especially in the central nervous system (4). Although palmitoylation is required for membrane localization and function of these proteins, depalmitoylation is equally critical for recycling or for degradation by lysosomal hydrolases. Thus, dynamic palmitoylation (palmitoylation-depalmitoylation) has emerged as an important mechanism regulating the function of many important proteins, including the α-subunit of G-proteins and the product of the proto-oncogene H-Ras (2, 3, 5–10).
In mammals, palmitoylation is catalyzed by a family of 23 enzymes called palmitoyl acyltransferases (PATs)3 (7), whereas depalmitoylation is catalyzed by four thioesterases. Two of these thioesterases, acyl-protein thioesterase-1 (APT1) (11) and APT2 (12), are localized predominantly in the cytoplasm, and the other two, palmitoyl-protein thioesterase-1 (PPT1) (13, 14) and PPT2 (15), are lysosomal enzymes (16, 17). The first cytosolic thioesterase to be characterized was APT1, which catalyzed depalmitoylation of the α-subunit of G-proteins and proto-oncogene H-Ras product in vitro (11) as well as synaptosomal-associated protein 23 (SNAP-23) (18). More recently, a second cytosolic thioesterase, APT2, has been reported to depalmitoylate GAP-43 (growth-associated protein 43) (12). Although both H-Ras and GAP-43 undergo palmitoylation (3, 4) for membrane localization, these proteins also require depalmitoylation catalyzed by APT1 and APT2, respectively, to detach from the membrane and to be recycled. However, until now it remained unclear how APT1 and APT2, being cytosolic enzymes, catalyze depalmitoylation of membrane-anchored, palmitoylated proteins such as H-Ras and GAP-43. Resolution of this question is pivotal in understanding the roles of these proteins in health and disease. We hypothesized that both APT1 and APT2 undergo dynamic palmitoylation for steady-state membrane localization to catalyze depalmitoylation of their membrane-anchored, palmitoylated substrates, H-Ras and GAP-43, respectively.
In this study, we demonstrate that both APT1 and APT2 are palmitoylated proteins, and Cys-2 in both proteins is palmitoylated. We also show that blocking palmitoylation adversely affects membrane localization of APT1 and APT2 as well as that of their substrates, H-Ras and GAP-43, respectively. Moreover, APT1 not only catalyzes self-depalmitoylation but also that of APT2. This promotes dynamic palmitoylation of both of these cytosolic thioesterases. Furthermore, we tested shRNA suppression of APT1 expression and inhibition of its thioesterase activity by an inhibitor, Palm B, which showed elevated levels of membrane localization. In addition, as it has been reported that mutation in any amino acid residue in the active-site triad (GXSXG) causes catalytic inactivation of APT1 (19, 20), we performed mutagenesis studies using S119A in APT1, which markedly increased membrane localization of both APT1 and APT2, whereas shRNA suppression of APT2 had virtually no effect on membrane localization of APT1. Taken together, our results provide insight into a novel mechanism by which dynamic palmitoylation of both APT1 and APT2 facilitates their own cytosol-membrane shuttling as well as that of their substrates. This facilitates steady-state membrane localization and function of APT1 and APT2 as well as H-Ras product and GAP-43, respectively.
EXPERIMENTAL PROCEDURES
Neurosphere Culture and Astrocyte Differentiation in Vitro
All animal experiments were conducted under a protocol approved by the NICHD Animal Care and Use Committee. Mouse neurospheres were isolated from the brain tissues derived from 15-day-old fetuses. The cells were cultured in NeuroCult NSC Basal Medium (Stem-Cell Technologies) containing 10% NeuroCult NSC proliferation supplements and human epidermal growth factor (final concentration of 20 ng/ml). To achieve astrocyte differentiation, the proliferating neurospheres were cultured in DMEM supplemented with 10% FBS. The cultures were incubated at 37 °C under an atmosphere of 5% CO2 and 95% air.
Mutagenesis of APT1 and APT2
APT1 and APT2 cDNA constructs tagged with Myc-DDK were bought from OriGene Technologies, Inc. (Rockville, MD). To generate palmitoylation site mutants in both APT1 and APT2 mutants, the cysteine residue in position 2 of the protein sequence (Cys-2) was mutated to serine. The resulting mutant constructs are designated as follows: Myc-DDK-APT1 mutant (C2S) as APT1-M-1 and Myc-DDK-APT2 mutant (C2S) as APT2-M-1. Mutagenesis was performed by PCR using the following primers: APT1-M-1 forward, 5′-GCCGCGATCGCCATGAGCGGCAATAACATGT-3′, and reverse, 5′-ACATGTTATTGCCGCTCATGGCGATCGCGGC-3′; APT2-M-1 forward, 5′-CCGCGATCGCCATGTCTGGTAACACCATGTC-3′, and reverse, 5′-GACATGGTGTTACCAGACATGGCGATCGCGG-3′. For generating APT1 active site mutant (S119A), designated APT1-M-2, we used following primers: forward, 5′-CTTCTAACAGAATTATTTTGGGAGGGTTTGCTCAGGGAGGAGCTTTATCTTTATATACTG-3′, and reverse, 5′-CAGTATATAAAGATAAAGCTCCTCCCTGAGCAAACCCTCCCAAAATAATTCTGTTAGAAG-3′. These mutants were generated by PCR using the method of QuikChange site-directed mutagenesis from Agilent Technologies. The mutations were confirmed by DNA sequencing.
Determination of APT1 and APT2 Palmitoylation by ABE Method
Palmitoylation of APT1 and APT2 was assayed by acyl-biotinyl exchange method (ABE) as described previously (21) with minor modifications. Briefly, HEK293T cells were transfected with APT1-cDNA or APT2-cDNA or APT1-M-1 or APT2-M-1 cDNA constructs. The transfected cells were lysed with RIPA buffer (Pierce), and the lysates were incubated overnight with 10 mm N-ethylmaleimide (Pierce) plus 1× protease inhibitor (PI) mixture (Pierce) at 4 °C with gentle mixing. N-Ethylmaleimide was then removed by three sequential precipitations using the chloroform/methanol (CM) method as described previously (22). Following a third precipitation, the protein precipitate was divided into two equal aliquots. One aliquot was mixed with 1 m hydroxylamine (Sigma), pH 7.4 (freshly prepared), 1 mm N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide-biotin (Pierce), 0.2% Triton X-100 (Sigma), and 1× PI, and the other aliquot was treated with the identical mixture except that it did not contain hydroxylamine. Both aliquots were then incubated for 1 h at room temperature. The proteins were precipitated by CM method and treated with 200 μm N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide-biotin, 0.2% Triton X-100, and 1× PI at room temperature for 1 h. N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide-biotin was then removed by three sequential CM precipitations. Following the third precipitation, proteins were immunoprecipitated with streptavidin-agarose (Pierce) and eluted with SDS-PAGE loading buffer containing 5% β-mercaptoethanol by boiling for 5 min. Samples were then subjected to Western blot analysis with c-Myc antibody (Sigma). Experiments were repeated at least three times.
Determination of APT1 and APT2 Palmitoylation Using [14C]Palmitate Method
To further confirm that APT1 and APT2 undergo palmitoylation, HEK293T cells in 6-well plates were transfected with APT1-cDNA or APT2-cDNA construct or the vector alone (control). Thirty six hours after transfection, the cells were washed with Opti-MEM once and then starved for 1 h. After starvation, 200 μCi of [14C]palmitic acid was added into the media and incubated for 30 min at 25 °C followed by 5 h of incubation at 37 °C. The labeled cells were gently washed three times before they were harvested. Cells were lysed with RIPA buffer and then split into two equal aliquots in two 1.5-ml centrifuge tubes. One aliquot was treated with hydroxylamine, and the other aliquot was incubated with 1× PBS for 1 h at 4 °C with gentle rotation. After incubation, the lysates were mixed with 1 μg of anti-FLAG antibody (Sigma) in a pull-down assay. The pulled-down proteins were washed six times with ice-cold PBS containing 1× PI. Samples were mixed with equal amounts of 2× loading buffer without β-mecaptomethanol and resolved by SDS-PAGE using 4–12% gradient SDS-polyacrylamide gels (Invitrogen). The gels were dried with the GelDryer system (Bio-Rad). Images of [14C]palmitate-labeled proteins were visualized using the Cyclone PhosphorImager (Packard Instrument Co.). Autoradiographs of the dried gels were also prepared using Blue Lite Autorad Films (GeneMate). Experiments were repeated at least three times.
Transfection of Cultured Astrocytes and NIH 3T3 Cells with cDNA Constructs
Cultured astroglia and NIH 3T3 cells were transfected with APT1, APT2, APT1-M-1 (C2S), APT1-M-2(S119A), or APT2-M-1(C2S) cDNA-constructs using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. The plasmids used for transfection were prepared using a plasmid midi kit (Qiagen). The consistency between plasmid preparations was monitored by determining the concentration of plasmids by both spectrophotometry and agarose gel electrophoresis. The cDNA constructs used in this study were as follows: Myc-DDK-APT1, Myc-DDK-APT1-M-1 (C2S), Myc-DDK-APT1-M-2 (S119A), Myc-DDK-APT2, and Myc-DDK-APT2-M-1(C2S) and GFP-H-Ras (Addgene plasmid 18662).
Cell Fractionation
Cytosolic and membrane fractions from cultured astroglia were prepared using a previously reported protocol (23) with minor modifications. Briefly, cells were harvested and rinsed with PBS before homogenizing at 4 °C followed by a brief centrifugation (500 × g) to pellet the intact cells and nuclei. Supernatants were decanted carefully and centrifuged at 16,000 × g, and the membrane and cytosolic fractions were collected. The supernatant was collected as a cytosolic fraction. The pellet was dissolved in 2% Triton X-100 with PI for an hour on ice and centrifuged at 16,000 × g, and the supernatant containing the membrane fraction was collected.
Western Blot Analyses
Protein samples (20 μg) were resolved by electrophoresis using 4–12% SDS-polyacrylamide gels (Invitrogen) under denaturing and reducing conditions. Proteins were then electrotransferred to nitrocellulose membranes (Invitrogen). The membranes were blocked with 5% nonfat dry milk (Bio-Rad) and then subjected to immunoblot analysis using standard methods. The primary antibodies used for the immunoblots were anti-APT1 (Epitomics), anti-APT2 (Novus), anti-GAP43 (Chemicon), anti-Myc (Sigma), anti-pan-cadherin (Cell Signaling), and anti-β-actin (US Biological). The secondary antibodies were goat anti-rabbit IgG (Santa Cruz Biotechnology) and rabbit anti-mouse IgG (Santa Cruz Biotechnology). Chemiluminescence was detected using SuperSignal west pico luminol/enhancer solution (Pierce) according to the manufacturer's instructions. Experiments were repeated at least three times, and reproducibility was confirmed.
Bromopalmitate and Palm B Treatment of Cultured Astroglia
Astroglia were cultured in DMEM supplemented with 10% FBS at 37 °C under a humidified atmosphere containing 5% CO2. Cells were treated with DMSO and varying concentrations of bromopalmitate (final concentration of 25 or 50 μm) or palmostatin B (final concentration of 0.5, 1, or 2 μm) for 12 h with a change of fresh media containing Palm B every 6 h. Protein samples were prepared from the treated cells and were used for Western blot analysis. To visualize the subcellular localization of APT1, APT2, GAP43, or H-Ras, treated astrocytes were fixed and analyzed immunocytochemically under a confocal microscope.
shRNA-mediated Knockdown of APT1 and APT2 in Astroglia
Cultured astroglia were plated into 75-cm2 flasks and four-chamber slides (Nunc). The following day, the cells in 75-cm2 flasks were transfected with 5 μg each of APT1-shRNA (catalog no. RMM4532-NM_008866) or scrambled shRNA (catalog no. RMM2208) in GIPZ vector (Thermo Fisher Scientific, Huntsville, AL) or APT2-shRNA (catalog no. TF311634) or scrambled shRNA (catalog no. TR30015) in pRFP-C-RS vector (Origene Technologies, Inc., Rockville, MD) using Lipofectamine 2000. However, the cells grown in four-chamber slides were transfected using 200 ng each of the constructs. Protein fractions for Western blot analysis were performed as described above.
Confocal Microscopic Imaging
Cultured astrocytes and NIH 3T3 cells were maintained at 37 °C in an atmosphere of 5% of CO2 and 95% air for 72 h on slide chambers (Nunc). The cells were washed three times with PBS, pH 7.2, and fixed in 4% paraformaldehyde solution for 15 min at room temperature. The primary antibodies used were anti-APT1 (Epitomics), anti-APT2 (Novus), anti-GAP43 (Chemicon), anti-Myc (Sigma), anti-Na+/K+ATPase (Millipore), anti-FLAG (Sigma), and anti-GFP (Abcam). The secondary antibodies used were Alexa Fluor 488-conjugated anti-rabbit, Alexa Fluor 594-conjugated anti-rabbit (Invitrogen), Alexa Fluor 594-conjugated anti-mouse, and Alexa Fluor 488-conjugated anti-mouse (Invitrogen). Nuclei were stained with DAPI (Sigma). Fluorescent images of the cells were captured using a Zeiss LSM 510 inverted meta confocal microscope or Zeiss Axioskop2 plus florescence microscope (Carl Zeiss), and the images were processed using LSM image software (Carl Zeiss). In each experiment, images were acquired using identical settings, and the same standard was applied for all groups. All experiments were repeated at least three times.
Co-localization and Quantitation of Immunofluorescence
Quantification of co-localization was performed with Carl Zeiss AIM 4.2 software (Carl Zeiss). The images of overlapping area of immunofluorescence (FITC) from APT1 or APT1M and APT2 or APT2M with those of the membrane marker, Na+/K+ATPase (rhodamine fluorescence) were selected manually with PROCESS tab of the software, and co-localization of the fluorescence in chosen areas was analyzed with the Image Calculator function, and Pearson correlations were analyzed by intensity-based co-localization function of the software.
RESULTS
Cysteine 2 in APT1 and APT2 Is Palmitoylated
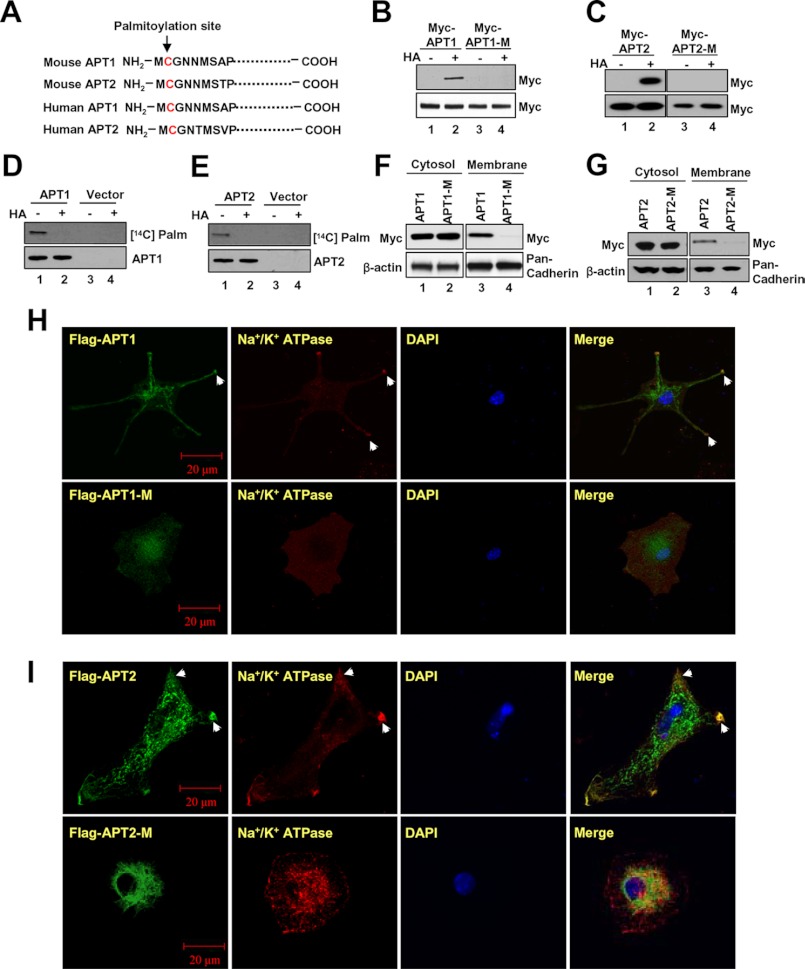
To determine whether APT1 and APT2 undergo palmitoylation, we first analyzed the peptide sequences of both mouse and human APT1 and APT2 by CSS-Palm (24), a computer program that predicts potential palmitoylation site(s) in polypeptides. Our results showed that Cys-2 of both APT1 and APT2 of mice (Fig. 1A, upper rows) and humans (Fig. 1A, lower rows) is a potential palmitoylation site. To delineate whether APT1 and APT2 undergo palmitoylation, we transfected HEK293T cells with Myc-FLAG (DDK)-tagged APT1-cDNA or Myc-FLAG (DDK)-tagged APT2-cDNA construct (herein after called APT1 or APT2 constructs) or Myc-FLAG (DDK)-tagged mutant (C2S) APT1-cDNA or Myc-FLAG (DDK)-tagged mutant (C2S) APT2-cDNA construct (herein after called APT1-M-1 or APT2-M-1 constructs) and analyzed the cell lysates for palmitoylated APT1 and APT2 using the ABE method (21). The results showed that palmitoylated APT1 (Fig. 1B, lane 2) and APT2 (Fig. 1C, lane 2) are readily detectable in total lysates of HEK293T cells transfected with either APT1 or APT2 construct, respectively. However, the palmitoylated APT1 and APT2 protein bands were not detectable in total lysates of cells transfected with APT1-M-1 (Fig. 1B, lane 4) or APT2-M-1 constructs (Fig. 1C, lane 4). To further confirm these results, we also labeled the cells transfected with either APT1 or APT2 construct or vector only with [14C]palmitate. The lysates of the labeled cells were immunoprecipitated with FLAG antibody, and an aliquot of each of the immunoprecipitates was pretreated with hydroxylamine (HA), which cleaves thioester linkage in palmitoylated proteins (5, 6), before they were resolved by SDS-PAGE and Western blot analysis. The results confirmed that [14C]palmitate was incorporated in both APT1 (Fig. 1D, lane 1) and APT2 protein (Fig. 1E, lane 1) via thioester linkage as the radioactive palmitate label was completely removed by HA treatment, and consequently, no protein band was detectable (Fig. 1, D, lane 2, and E, lane 2). As expected, the cells transfected with vector only failed to show radioactive palmitoylated APT1 (Fig. 1D, lane 3) or APT2 (Fig. 1E, lane 3) protein bands. Taken together, these results clearly showed that both APT1 and APT2 undergo S-palmitoylation on Cys-2.
FIGURE 1.
Palmitoylation of APT1 or APT2 promotes their membrane localization. Palmitoylation site (Cys-2) was predicted by CSS-Palm 3.0 analysis at the highest stringency in mouse APT1 and APT2 (A, upper two rows) and human APT1 and APT2 (A, lower two rows). Palmitoylated APT1 (B, lane 2) and APT2 (C, lane 2) were readily detectable by ABE assay, although C2S mutation in both APT1 (B, lane 4) and APT2 (C, lane 4) abrogated palmitoylation. Palmitoylation of APT1 (D, lane 1) and APT2 (E, lane 1) were further confirmed by the presence of [14C]palmitate-labeled APT1 and APT2 rotein bands, which were rendered undetectable by hydroxylamine treatment (D and E, lanes 2). Note that C2S mutation in APT1 (F) and APT2 (G) abrogated membrane ssociation of both APT1 (F, lane 4), and APT2 (G, lane 4). Subcellular localizations of APT1 and APT2 in cells transfected with the APT1-M-1, APT2 or APT2-M-1 construct were analyzed by confocal microscopy. Image data show that although APT1 (H, upper row) and APT2 (I, upper row) were localized to the membrane (arrowheads), the APT1-M-1 (H, lower row) and APT2-M-1 (I, lower row) were predominantly localized to the cytoplasm.
Membrane Localization of APT1 and APT2 Requires Palmitoylation on Cysteine 2
To determine whether APT1 and APT2 are localized to the membrane and whether the membrane association depended on Cys-2 palmitoylation, we fractionated HEK293T cells transfected with either APT1 or APT1-M-1 construct (Fig. 1F) or APT2- or APT2-M-1 construct (Fig. 1G) into cytosolic and membrane fractions. Proteins from these fractions were analyzed by Western blot using Myc antibody. The results clearly showed that APT1 and APT2 protein bands in the cytosolic fractions of both APT1- and APT1-M-1-transfected cells (Fig. 1F, lanes 1 and 2) and APT2- and APT2-M-1-transfected cells (Fig. 1G, lanes 1 and 2) were clearly detectable. Although the membrane fractions from APT1-transfected (Fig. 1F, lane 3) and APT2-transfected (Fig. 1G, lane 3) cells showed the APT1 and APT2 protein bands, those from APT1-M-1-transfected (Fig. 1F, lane 4) or APT2-M-1 transfected cells failed to show those bands (Fig. 1G, lane 4). These results strongly suggested that Cys-2 palmitoylation is required for membrane localization of both APT1 and APT2 proteins.
To confirm the membrane localization of APT1 and APT2, we performed confocal microscopic analysis of cultured mouse astrocytes transfected with either the APT1 or APT2 construct or the APT1-M-1 or APT2-M-1 construct using FLAG and Na+/K+ATPase antibodies, respectively. We used cultured astrocytes because numerous palmitoylated proteins, including the product of H-Ras oncogene and GAP-43, are expressed in the central nervous system (2). The results showed that APT1-transfected (Fig. 1H, upper panels) and APT2-transfected (Fig. 1I, upper panels) astrocytes but not the APT1-M-1-transfected (Fig. 1H, lower panels) and APT2-M-1-transfected (Fig. 1I, lower panels) astrocytes had membrane localization of APT1 or APT2 protein. Membrane localization was ascertained by co-localization (merge) of FLAG immunofluorescence with that of Na+/K+ATPase-specific immunofluorescence. As expected, the tag immunofluorescence in APT1-M-1-transfected (Fig. 1H, lower panels) and APT2-M-1-transfected (Fig. 1I, lower panels) cells failed to co-localize with Na+/K+ATPase-specific immunofluorescence. These results strongly suggested that membrane localization of APT1 and APT2 requires palmitoylation on Cys-2.
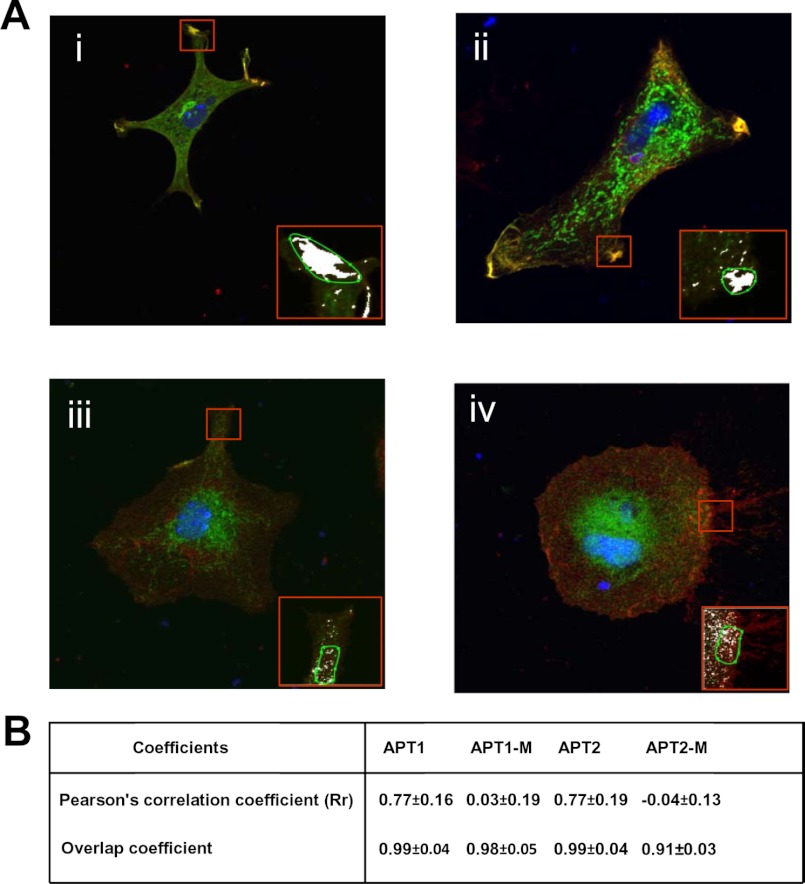
To further confirm that APT1 and APT2 localized on the cell membrane, we performed quantitative analysis of co-localization of APT1 and APT2 immunofluorescence with that of the membrane marker, Na+/K+ATPase, using Carl Zeiss AIM 4.2 software (Carl Zeiss). Although the results confirmed co-localization of APT1 (Fig. 2A, panel i) and APT2 (Fig. 2A, panel ii) with the cell membrane marker Na+/K+ATPase, there was virtually no co-localization of APT1-M-1 (Fig. 2A, panel iii) and APT2-M-1 (Fig. 2A, panel iv) with the cell membrane marker. Pearson's correlation coefficient (Rr) for APT1 and APT2 (Fig. 2B) was greater than 0.75 in the overlapping areas (mean of seven individual measurements). Moreover, there was no significant co-localization of APT1-M-1 and APT2-M-1 immunofluorescence with that of the cell membrane marker Na+/K+ATPase. Taken together, these results strongly suggested co-localization of both the cytosolic thioesterases on the cell membrane and that co-localization is essential for these enzymes to catalyze self-depalmitoylation of APT1 as well as depalmitoylation of APT2 and that of their substrates, H-Ras and GAP-43, respectively.
FIGURE 2.
Co-localization of APT1 and APT2 immunofluorescence with that of Na+/K+ATPase. Quantitation of APT1 or APT2 immunofluorescence with Na+/K+ATPase was performed using AIM 4.2 software (Carl Zeiss) to identify co-localization in the overlapping areas of the image. Insets show co-localization of APT1 (A, panel i) and APT2 (A, panel ii) with the cell membrane marker Na+/K+ATPase. There were no co-localizations of APT1-M-1 (A, panel iii) and APT2-M-1 (A, panel iv) with the cell membrane marker. Pearson's correlation coefficient (Rr) of APT1 or APT2 is greater than 0.75 in the overlapping areas (mean from seven individual measurements) (B). Moreover, there were no significant correlations of APT1-M-1 and APT2-M-1 with cell membrane marker, Na+/K+ATPase.
Endogenous Apt1 and Apt2 Undergoes Palmitoylation for Membrane Localization
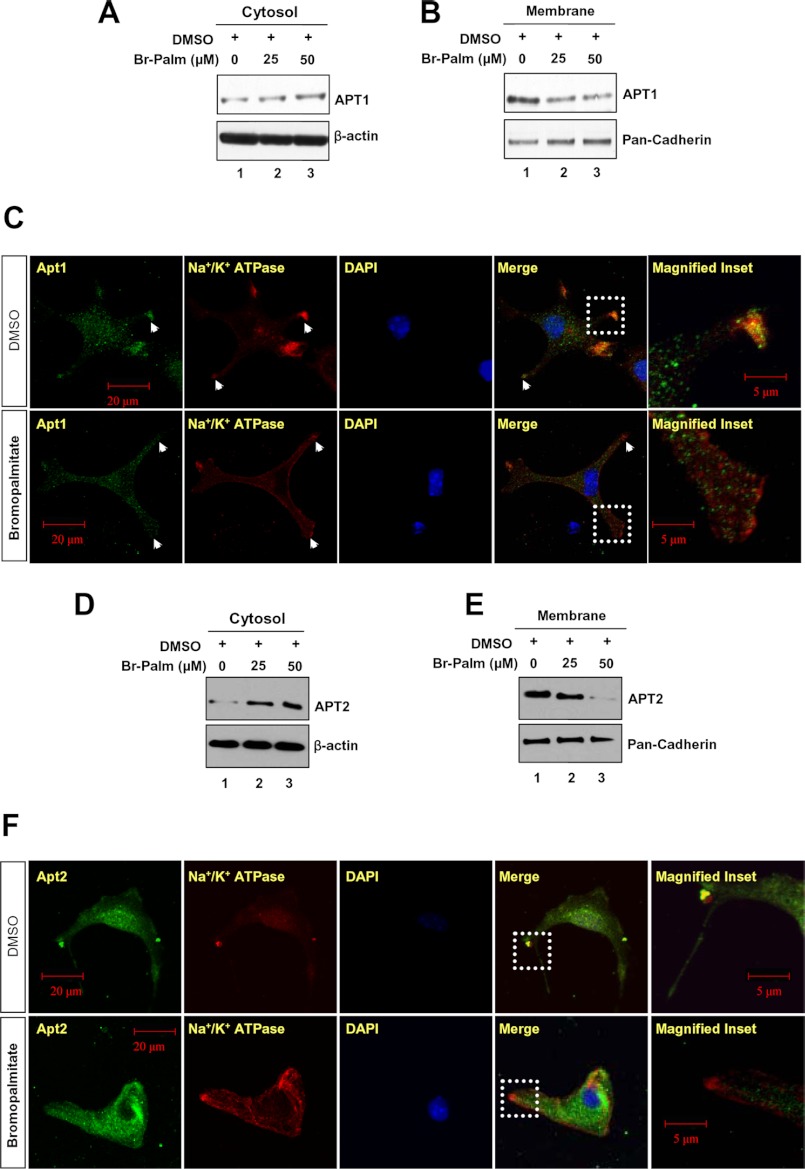
Thus far, we have used overexpression of APT1 and APT2 in HEK293T cells to experimentally demonstrate that both proteins undergo palmitoylation and that palmitoylation of Cys-2 is required for membrane localization. However, it was not clear whether endogenous APT1 and APT2 (hereafter designated as Apt1 and Apt2) require palmitoylation for membrane association. To determine this, we first treated cultured astrocytes with bromopalmitate, a potent inhibitor of palmitoylation (6), and we then resolved the proteins from cytosolic and membrane fractions by SDS-PAGE and Western blot analysis using either Apt1 or Apt2 antibody. The results showed that compared with the levels of Apt1 and Apt2 in cytosolic fractions (Fig. 3, A and D) those in the membrane fractions gradually declined correlating with increased doses of bromopalmitate (Fig. 3, B and E). These results indicated that inhibition of palmitoylation adversely affected membrane localization of both Apt1 and Apt2 suggesting an essential role of palmitoylation in facilitating translocation of these cytosolic thioesterases to the membrane.
FIGURE 3.
Suppression of Apt1 and Apt2 palmitoylation by bromopalmitate. Suppression of Apt1 palmitoylation by bromopalmitate dose-dependently increased the level of cytosolic Apt1 (A) and reduced its membrane localization (B). Immunocytochemical analyses were performed using Apt1 and Na+/K+ATPase antibodies, which showed that Apt1 is localized predominantly in the cytoplasm of the cells treated with bromopalmitate (C, lower row), although accumulation of Apt1 is clearly detected on the membrane of the cells treated with DMSO (C, upper row). The cytosolic and membrane fractions from bromopalmitate-treated cells were probed with APT2 antibody. Suppression of Apt2 palmitoylation by bromopalmitate markedly increased the cytosolic Apt2 (D) and decreased its membrane localization (E). Immunocytochemical analyses were performed using Apt2 or Na+/K+ATPase- antibodies. Note that the Apt2 signal on the cell membrane is virtually abolished in bromopalmitate-treated cells (F, lower row) compared with DMSO-treated control cells (F, upper row).
To further confirm that Apt1 and Apt2 require palmitoylation for membrane localization, we first pretreated the astrocytes with bromopalmitate and then performed confocal microscopic analyses of immunofluorescence using antibodies against Apt1, Apt2, and Na+/K+ATPase. Remarkably, although in DMSO-treated cells (control) co-localization of Apt1 and Apt2 fluorescence with that of Na+/K+ATPase was clearly detectable (Fig. 3, C, upper panels, and F, upper panels), such co-localization was not appreciable in cells treated with bromopalmitate (Fig. 3, C, lower panels, and F, lower panels). Taken together, these results confirmed that palmitoylation of Apt1 and Apt2 is at least one of the requirements for their membrane localization.
Apt1 and Apt2 Are Dynamically Palmitoylated for Steady-state Membrane Localization
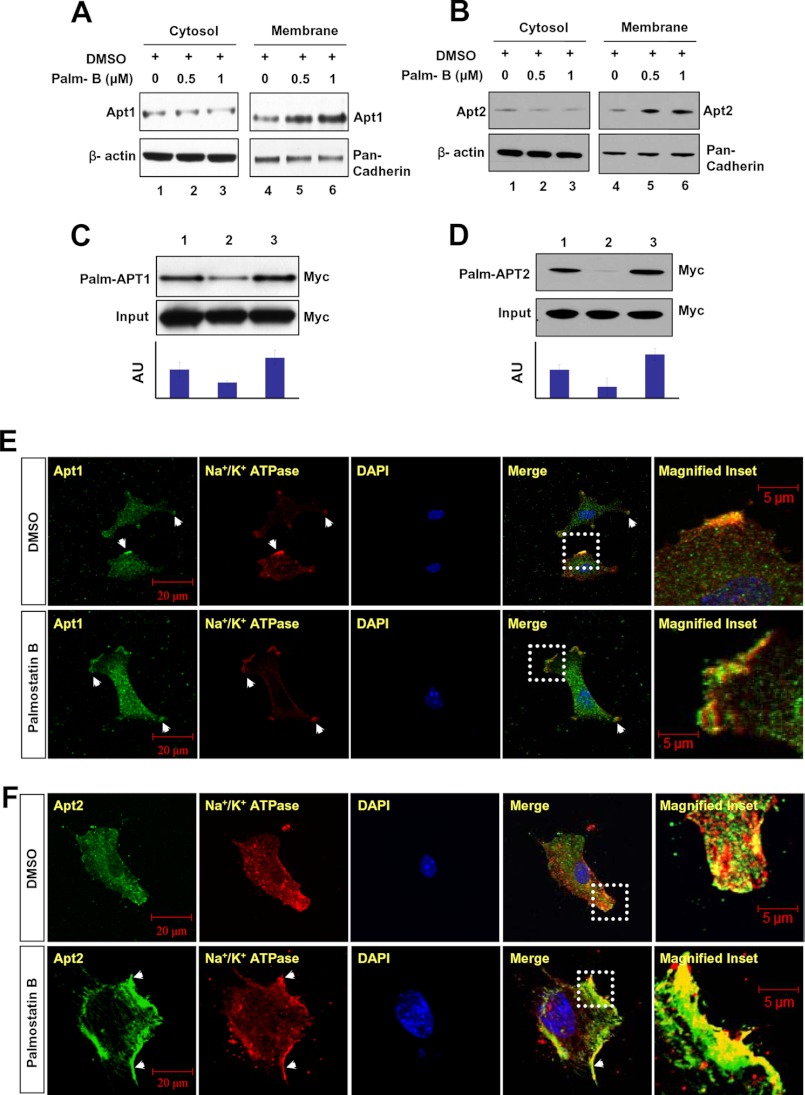
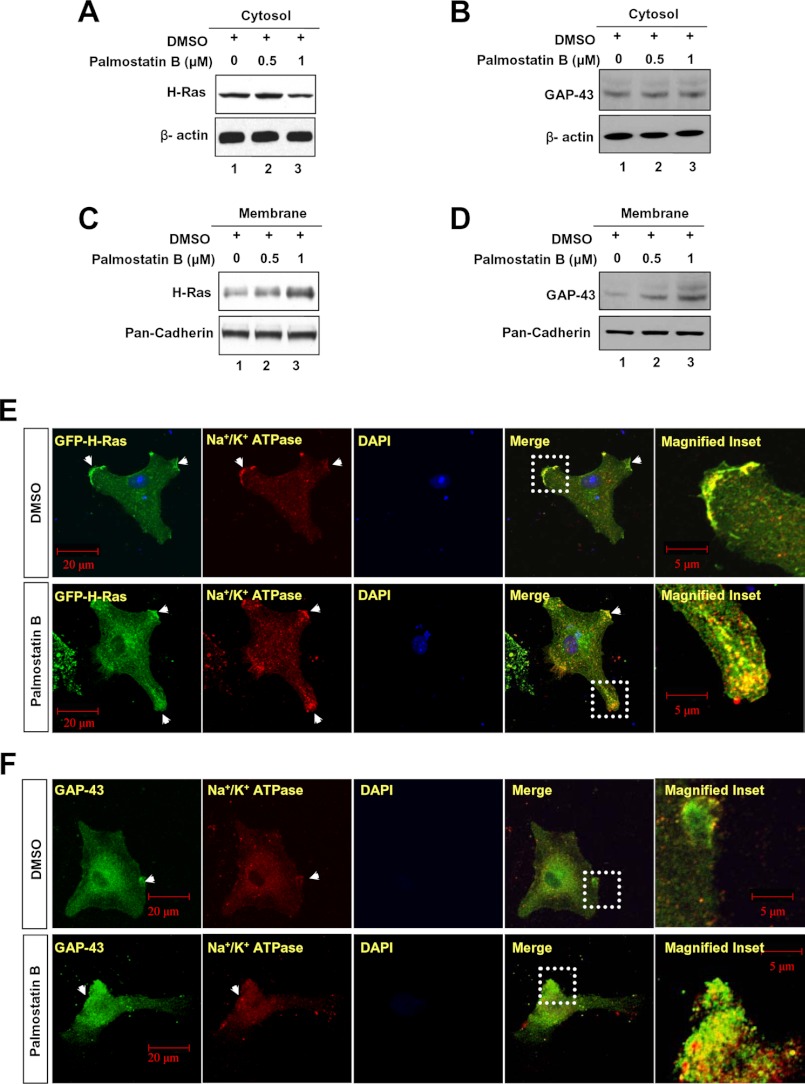
One of the suggested functions of dynamic palmitoylation is to regulate protein sorting (9, 10) and to achieve steady-state membrane localization (2–8, 25, 26). However, recent reports indicate that palmitoylation also regulates protein trafficking to many distinct intracellular compartments due to its sorting role (7, 9). For example, it has been reported that specific PATs and thioesterases regulate surface expression of important proteins such as calcium-activated potassium channels (27). Moreover, palmitoylation increases the affinity for membrane localization, and depalmitoylation plays a vital role in recycling and/or degradation of proteins that undergo S-palmitoylation (9, 10). For example, dynamic palmitoylation regulates recycling of proteins such as the α-subunit of G-proteins (11), the products of H- and N-Ras proto-oncogenes (13), as well as endothelial nitric-oxide synthase (28). Recently, it has been reported that dynamic palmitoylation regulates T cell activation and anergy (29). Although the specific PATs that catalyze palmitoylation of APT1 and APT2 have yet to be identified, we speculated that APT1 and APT2 may catalyze their own depalmitoylation or they catalyze depalmitoylation of each other to promote their dynamic palmitoylation, promoting steady-state membrane localization and function. Accordingly, we performed Western blot analysis of cytosolic and membrane fractions of cultured astrocytes that were treated with either DMSO (control) or with varying doses of Palm B, first reported to be a catalytic inhibitor of APT1 (30) but subsequently found to inhibit both APT1 and APT2 (31). The results suggested that although Apt1 (Fig. 4A, lane 1) and Apt2 (Fig. 4B, lane 1) levels in the cytosolic fraction of the cells treated with DMSO alone (control) were virtually identical to those in the cytosolic fractions of Palm B-treated cells (Fig. 4, A and B, lanes 2 and 3), the membrane fractions of these cells showed a dose-dependent elevation of Apt1 (Fig. 4A, lanes 5 and 6) and Apt2 (Fig. 4B, lanes 5 and 6) protein levels. These results suggested that inhibition of thioesterase activity markedly increased membrane localization of both Apt1 and Apt2 raising the possibilities that these two thioesterases may catalyze their own depalmitoylation or that of each other or both.
FIGURE 4.
Membrane localization of APT1 and APT2 in Palm B-treated cells. Palm B treatment in a dose-dependent manner elevated the levels of both Apt1 (A) and Apt2 (B) in membrane fractions of the cells. Palmitoylation status of APT1 (C) and APT2 (D) was checked in cultured astroglial cells expressing APT1 or APT2 in which either palmitoylation or depalmitoylation was inhibited by bromopalmitate and Palm B treatment, respectively. Lysates from 3 × 106 cells for each treatment were used for Western blot analysis. The densitometric quantitation of the protein bands in Western blots from three independent experiments were performed, and the results are presented graphically as the mean ± S.D. Note that Palm B treatment markedly elevated the levels of palmitoylated APT1 (C, lane 3, and bar graph) as well as APT2 (D, lane 3, and bar graph) as compared with their respective controls (C and D, lane 1), although bromopalmitate treatment markedly reduced the level of palmitoylated APT1 (C, lane 2) and APT2 (D, lane 2). Palm B treatment prevents dissociation of both Apt1 (E) and Apt2 (F) from the membrane. Compared with DMSO-treated cells (E and F, upper panels, arrowheads), the Palm B-treated cells had markedly elevated levels of membrane-localized Apt1 (E, lower panel, arrowheads) and Apt2 fluorescence (F, lower panel, arrowheads).
Depalmitoylation of Palmitoylated APT1 and APT2
To confirm these results, we transfected HEK293T cells with the APT1 or APT2 construct and treated these cells with DMSO (control), bromopalmitate, or Palm B and determined the levels of palmitoylated APT1 (Palm-APT1) and Palm-APT2 in these cells by the ABE method (21). The results showed that compared with the membrane fractions of DMSO-treated cells (Fig. 4, C, lane 1, and D, lane 1), bromopalmitate-treated cells had lower levels of both Palm-APT1 (Fig. 4C, lane 2) and Palm-APT2 (Fig. 4D, lane 2). However, in cells treated with the thioesterase inhibitor, Palm B, the levels of both Palm-APT1 (Fig. 4C, lane 3) and Palm-APT2 (Fig. 4D, lane 3) were much higher. These results suggested that inhibition of palmitoylation suppresses membrane localization of both APT1 and APT2 and that inhibition of thioesterase activity leads to higher levels of membrane-associated APT1 and APT2. Next, we immunohistochemically analyzed the Palm B-treated cells for cellular distribution of endogenous Apt1 and Apt2. The results showed that compared with the DMSO-treated cells (Fig. 4, E and F, upper panels), Palm B treatment markedly increased both Apt1 (Fig. 4E, lower panels) and Apt2 immunofluorescence (Fig. 4F, lower panels), which merged with that of the membrane marker Na+/K+ATPase. To confirm these results, we also performed the same experiments using pan-cadherin as an alternative membrane marker and the results confirmed the findings obtained with Na+/K+ATPase (data not shown). These results suggested that Apt1 and Apt2 undergo palmitoylation and that either these enzymes self-catalyze depalmitoylation or depalmitoylate each other or both.
Mechanism of Dynamic Palmitoylation of APT1 and APT2
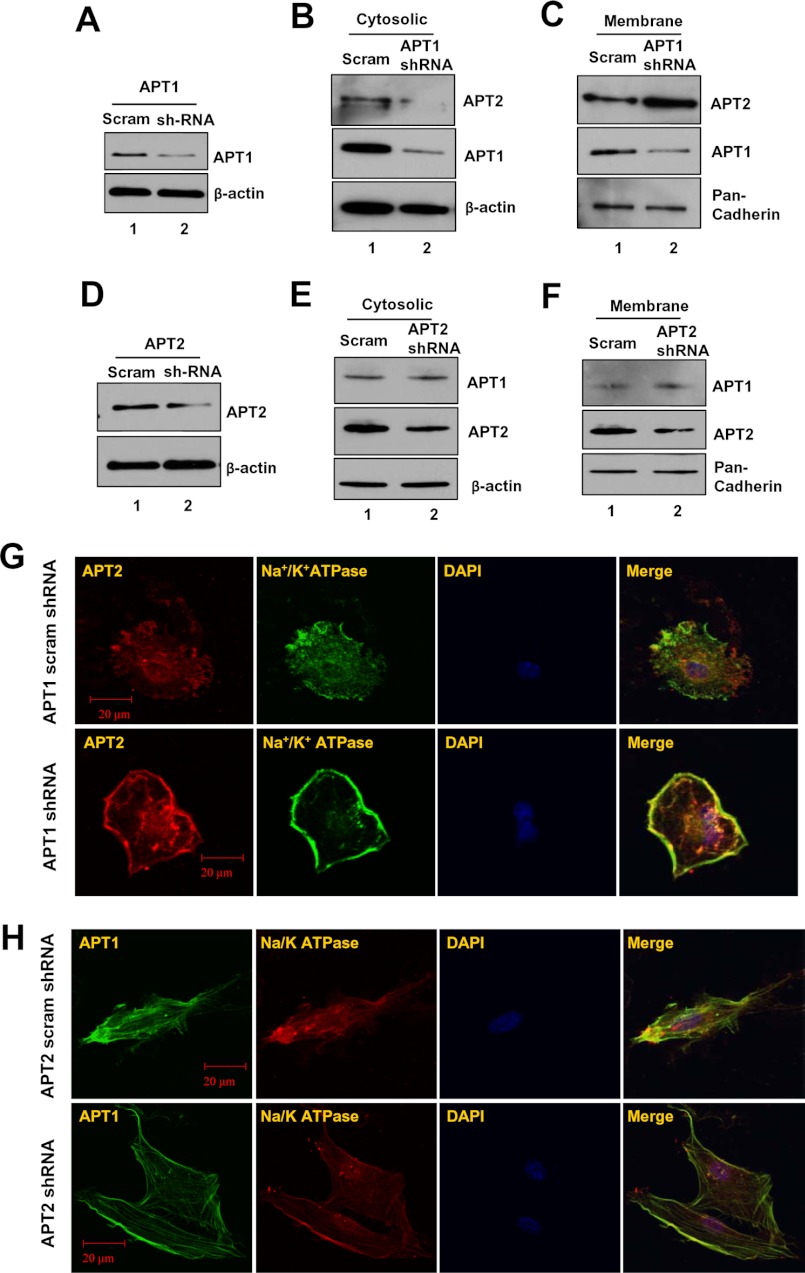
We next sought to determine which of the above possible mechanisms may actually regulate dynamic palmitoylation and steady-state membrane localization of Apt1 and Apt2. However, because Palm B inhibits both Apt1 and Apt2 (30, 31), generalized inhibition of these cytosolic thioesterases may not clearly answer our question. Therefore, we first performed experiments in which cultured astrocytes were transfected with either scrambled shRNA (control) or APT1 shRNA and determined the levels of Apt1 expression. The results show that compared with the control (Fig. 5A, lane 1), the APT1-shRNA-transfected cells had a marked inhibition of Apt1 expression (Fig. 5A, lane 2). We then fractionated the control and APT1-shRNA-transfected cells and determined the levels of Apt2-protein levels in the cytosolic and membrane fractions by Western blot analysis. The results showed that compared with the cytosolic fractions of the control cells (Fig. 5B, lane 1), those of the APT1-shRNA-treated cells had a lower level of Apt2 protein (Fig. 5B, lane 2). Interestingly, compared with the level of Apt2 in the membrane fraction of scrambled shRNA-transfected cells (Fig. 5C, lane 1), the level of APT1-shRNA-transfected cells showed a markedly higher level of Apt2 (Fig. 5C, lane 2). These results indicated that Apt1 catalyzes depalmitoylation of Apt2 promoting its dynamic palmitoylation critical for steady-state membrane localization.
FIGURE 5.
Effects of shRNA suppression of Apt1 or Apt2 on membrane localization of Apt1 and Apt2. Compared with scrambled shRNA-transfected cells (A, lane 1), those transfected with APT1-shRNA (A, lane 2) had appreciably decreased levels of Apt1 protein. Apt1 knockdown reduced the level of Apt2 in the cytosolic fractions (B) but increased that of Apt2 in the membrane fractions (C). APT2-shRNA transfection markedly decreased Apt2 expression (D, lane 2) compared with that of scrambled shRNA-transfected cells (D, lane 1). Western blot analysis of cytosolic (E) and membrane fractions (F) from APT2-shRNA-transfected cells showed virtually no alteration in Apt1 protein level. Immunocytochemical analysis was performed with either Apt1 or Apt2 and Na+/K+ATPase antibody. Compared with scrambled shRNA-transfected cells (G, upper row), those transfected with APT1-shRNA had a markedly higher Apt1 signal co-localized with that of Na+/K+ATPase (G, lower row). However, compared with its scrambled shRNA-transfected cells (H, upper panel), those transfected with APT2-shRNA had very similar Apt1 signal co-localization with that of the membrane marker Na+/K+ATPase (H, lower row).
We then performed similar experiments in which the cells were transfected with either scrambled shRNA (control) or APT2-shRNA, and we determined the level of Apt1 protein in the cytosolic and membrane fractions by Western blot analysis. First, we checked the effectiveness of the APT2-shRNA in suppressing Apt2 expression, and the results showed that, compared with the scrambled shRNA-transfected cells (control) (Fig. 5D, lane 1), those transfected with APT2-shRNA had a marked suppression of Apt2 expression (Fig. 5D, lane 2). We then checked the Apt1 protein levels in the cytosolic and membrane fractions of control and APT2-shRNA-transfected cells by Western blot analysis. The results showed that the levels of Apt1 protein in the cytosolic fractions of control (Fig. 5E, lane 1) and APT2-shRNA-transfected cells (Fig. 5E, lane 2) were virtually identical. Moreover, compared with the membrane fractions of the control cells (Fig. 5F, lane 1), those of the APT2-shRNA-transfected cells (Fig. 5F, lane 2) appeared to contain virtually identical levels of membrane-associated Apt1. To confirm these results, we performed confocal microscopic analysis of the cells transfected with scrambled shRNA (control) or with either APT2- or APT1-shRNA for membrane localization of Apt1 or Apt2 immunofluorescence, respectively. Co-localization of Apt1 and Apt2 immunofluorescence with that of Na+/K+ATPase was considered evidence of membrane localization. The results showed that compared with the level of membrane-localized Apt2 immunofluorescence in control cells (Fig. 5G, upper panels), the levels of APT1-shRNA-transfected cells (Fig. 5G, lower panels) were markedly higher. However, compared with the control cells (Fig. 5H, upper panels), the APT2-shRNA-transfected cells (Fig. 5H, lower panels) showed virtually identical intensity of membrane-associated Apt1 immunofluorescence. Taken together, these results demonstrated that although Apt1 depalmitoylates Apt2 regulating its steady-state membrane localization, Apt2 does not regulate that of Apt1.
Dynamic Palmitoylation of APT1 and APT2 Promotes That of H-Ras and GAP-43, Respectively
To determine whether inhibition of thioesterase activity of Apt1 and Apt2 affected membrane localization of H-Ras and GAP-43, respectively, we treated cultured astrocytes with varying doses of Palm B and performed Western blot analysis of cytosolic and membrane fractions of the cells using antibodies to either H-Ras or GAP-43. The results showed that whereas treatment of the cells with DMSO alone or with 0.5 μm Palm B did not appreciably alter the levels of cytosolic H-Ras (Fig. 6A, lanes 1 and 2) and GAP-43 (Fig. 6B, lanes 1 and 2) proteins, treatment with 1 μm Palm B resulted in a slightly lower level of H-Ras (Fig. 6A, lane 3), although the level of GAP-43 remained virtually unaltered (Fig. 6B, lane 3). In contrast, compared with the levels of membrane-associated H-Ras (Fig. 6C, lane 1) and GAP-43 (Fig. 6D, lane 1) in DMSO-treated cells, the membrane fractions of the cells treated with 1 μm Palm B contained appreciably higher levels of H-Ras (Fig. 6C, lane 3), as well as GAP-43 (Fig. 6D, lane 3). These results provided strong evidence that inhibition of either Apt1 or Apt2 enzymatic activity by Palm B impaired depalmitoylation of palmitoylated H-Ras and GAP-43, respectively, and as a result, these palmitoylated proteins remained anchored to the membrane.
FIGURE 6.
Dynamic palmitoylation of Apt1 and Apt2 regulates steady-state membrane localization of H-Ras and GAP-43, respectively. The cytosolic fractions of cultured astroglia treated with DMSO or Palm B were probed with H-Ras or GAP-43 antibodies as indicated. Note a slight reduction in H-Ras signal in the cytosolic fraction of 1 μm Palm B-treated cells (A). There is, however, no apparent difference in the levels of GAP-43 (B) in the cytosolic fractions of Palm B-treated and control cells. The membrane fractions from astroglial cells treated with DMSO (control) or varying concentrations of Palm B were probed with either H-Ras or GAP-43 antibodies. Note that both H-Ras (C) and GAP-43 (D) levels in the membrane fractions of the cells treated with Palm B were elevated in a dose-dependent manner. Immunocytochemical analysis of cultured astroglia were performed using antibodies to either H-Ras (E) or GAP-43 (F) and Na+/K+ATPase. Compared with DMSO-treated cells (E and F, upper row, arrowheads), those treated with Palm B showed markedly increased membrane localization of H-Ras (E, lower row, arrowheads) as well as that of GAP-43 (F, lower row, arrowheads).
To further confirm the membrane localization of H-Ras and GAP-43, we performed experiments identical to those described above and determined co-localization of H-Ras and GAP-43 immunofluorescence with that of the membrane marker Na+/K+ATPase using confocal microscopy. The results showed that although in DMSO-treated (control) cells a very low level of GFP-H-Ras (Fig. 6E, upper panels) and GAP-43 (Fig. 6F, upper panels) immunofluorescence co-localized with that of Na+/K+ATPase, in Palm B-treated cells the levels of H-Ras (Fig. 6E, lower panels) and GAP-43 (Fig. 6F, lower panels) co-localizing with Na+/K+ATPase were markedly higher. Cumulatively, these results suggested that inhibition of either Apt1 or Apt2 enzymatic activity disrupts the steady-state membrane localization of their respective endogenous substrates H-Ras and GAP-43.
APT1 Catalyzes Its Own De-palmitoylation as Well as That of APT2
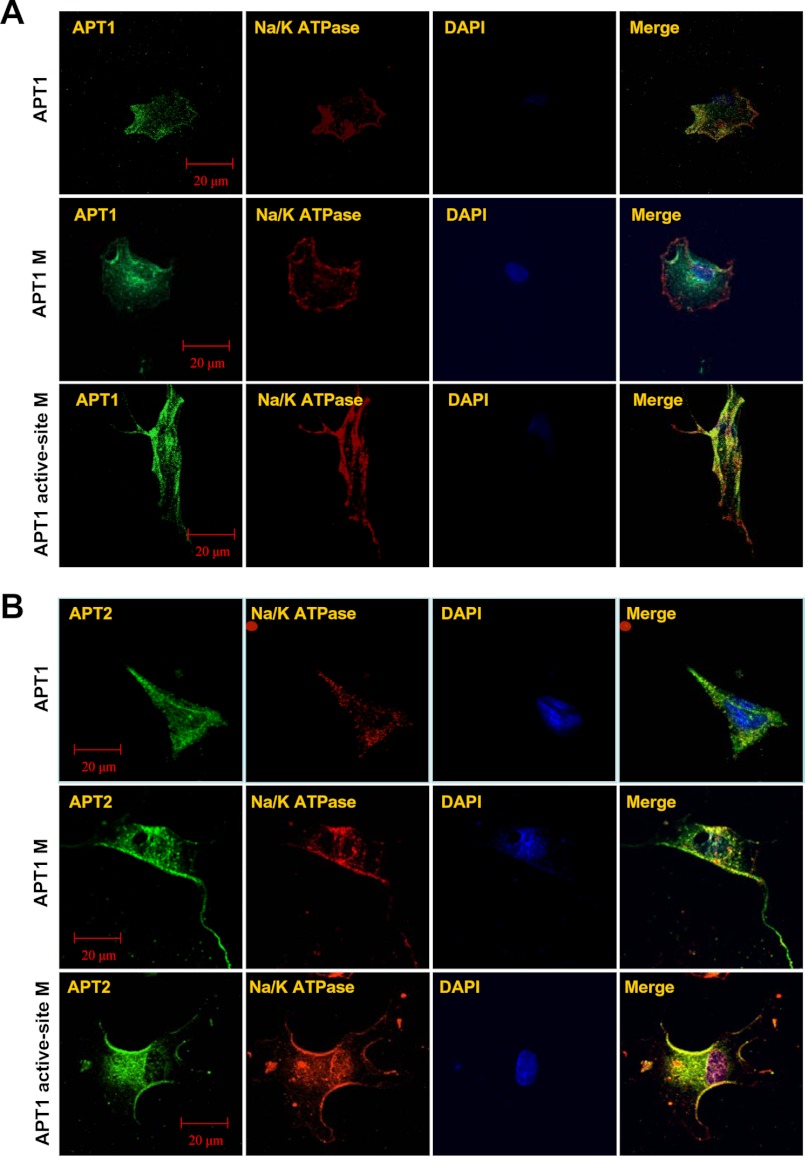
Although Palm B treatment catalytically inactivates APT1, it may also inactivate APT2 (31). To circumvent this problem, we performed mutagenesis studies in which Ser-119 in APT1 is mutated to Ala-119 as it has been previously reported that mutation of any one amino acid in the active site triad of APT1, GXSXG, leads to inactivation of this enzyme (19, 20). To further confirm that APT1 catalyzes its own depalmitoylation promoting its dynamic palmitoylation, which is required for the depalmitoylation of APT2, we performed three separate experiments. In one experiment, cultured astroglia were transfected with a construct containing APT1-cDNA; in the second experiment, cells were transfected with the APT1-M-1 (C2S), and in the third experiment, the cells were transfected with the constructs containing the APT1 active-site mutation APT1-M-2 (S119A). The transfected cells were then analyzed by confocal microscopy to determine co-localization of APT1 fluorescence with that of the membrane marker Na+/K+ATPase. The results showed that in cells transfected with the APT1 construct, APT1 immunofluorescence co-localized with that of the membrane marker (Fig. 7A, upper row), although some fluorescence was also present in the cytoplasm. However, in the cells transfected with the APT1-M-1 construct, APT1 immunofluorescence predominantly localized in the cytoplasm (Fig. 7A, middle row), and in those cells transfected with the APT1 active-site (S119A) mutant, APT1-M-2, high intensity APT1 immunofluorescence co-localized predominantly with the membrane marker Na+/K+ATPase (Fig. 7A, lower row). These results strongly suggested that catalytically inactive APT1 preferentially localized to the membrane suggesting that depalmitoylation of APT1 is self-catalyzed and that self-depalmitoylation promotes dynamic palmitoylation of APT1, which maintains steady-state membrane localization of this thioesterase.
FIGURE 7.
Active site mutation of APT1 increases membrane localization of both APT1 and APT2. Cultured astroglia were transfected with the APT1, APT1-M-1, or APT1-M-2 construct. Note that in cells transfected with the APT1 construct, APT1 fluorescence co-localized with that of Na+/K+ATPase (A, upper row). The cells transfected with APT1-M-2 construct showed increased co-localization with the cell membrane marker (A, lower row). However, in APT1-M-1-transfected cells, the APT1 fluorescence predominantly localized in the cytosol and perinuclear areas (A, middle row). Compared with NIH3T3 cells transfected with wild type APT1 (B, upper row), those transfected with the APT1-M-1 construct showed increased co-localization of APT-2 fluorescence with that of the membrane marker Na+/K+-ATPase (B, middle row). The cells transfected with APT1-M-2 construct also showed increased co-localization of APT2 immunofluorescence with that of Na+/K+-ATPase (B, lower row).
To further confirm that self-catalyzed depalmitoylation of APT1 is required for steady-state membrane localization of APT2, we transfected NIH3T3 cells with each of the three constructs described above and analyzed the cells for membrane localization of APT2 immunofluorescence. Our results showed that when the cells were transfected with the APT1 construct, the APT2 immunofluorescence was co-localized with the membrane marker, although some immunofluorescence was still detectable in the cytoplasm (Fig. 7B, upper row). However, in APT1-M-1-transfected cells, APT2 immunofluorescence was localized mostly on the membrane, but some also appeared to be localized in the cytoplasm (Fig. 7B, middle row). Remarkably, the cells transfected with Apt1 active-site mutation (S119A) showed that the intense APT2 immunofluorescence co-localized predominantly with that of the membrane marker Na+/K+ATPase (Fig. 7B, lower row). These results clearly showed that APT1 catalyzed depalmitoylation of APT2 promoting dynamic palmitoylation of APT2.
DISCUSSION
Several years ago, it was demonstrated that APT1, the first cytosolic thioesterase to be characterized, catalyzed depalmitoylation of the α-subunits of G-proteins in vitro (11). Moreover, APT1 has also been reported to catalyze depalmitoylation of the proto-oncogene H-Ras product in cellulo (30). Recently, it has been reported that whereas endogenous and overexpressed human APT1 were predominantly localized to the cytosol, APT1 signals were also detectable on the plasma membrane, the nuclear membrane, and in the endoplasmic reticulum in HEK293 cells (32). However, it remained unclear how cytosolic thioesterases, Apt1 and Apt2, could catalyze depalmitoylation of their palmitoylated substrates like H-Ras and GAP-43 that are anchored to the cell membrane. Interestingly, a recent study of a proteome scale characterization of human palmitoylated proteins in lipid raft-enriched membranes suggested that APT1 is a palmitoylated protein, and thus, it is likely to be targeted to the membranes (33). Our results are consistent with this notion and for the first time provide experimental evidence that both APT1 and APT2 undergo palmitoylation on N-terminal Cys-2. Moreover, our results show that palmitoylation on Cys-2 facilitates membrane localization of these cytosolic thioesterases. Furthermore, we demonstrated for the first time that Apt1 not only catalyzed its own depalmitoylation but also that of Apt2. Thus, dynamic palmitoylation of Apt1 also facilitated that of Apt2. More importantly, dynamic palmitoylation of Apt1 and Apt2 promoted that of their substrates, H-Ras and GAP-43, respectively. Interestingly, it has been reported that surface expression of calcium-activated potassium channels are regulated by two of the 23 mammalian PATs (DHHC22 and DHHC23), which catalyze palmitoylation, although depalmitoylation is catalyzed only by APT1 and not by APT2 (27). We propose that both of these cytosolic thioesterases require dynamic palmitoylation for their own steady-state membrane localization, which is essential for their function in promoting dynamic palmitoylation and function of their substrates, H-Ras and GAP-43, respectively.
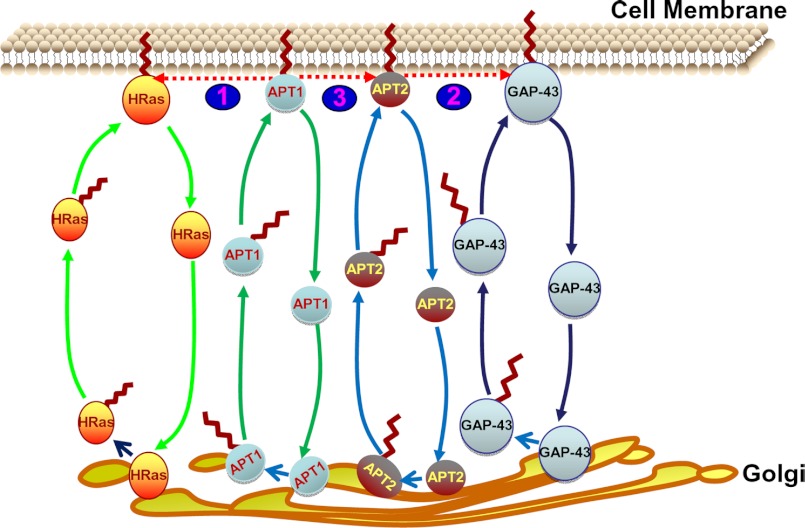
On the basis of these results, we propose a model (Fig. 8) for the role of dynamic palmitoylation of APT1 and APT2 in their steady-state membrane localization and function in regulating those of their substrates, H-Ras and GAP-43, respectively. In this model, cytosolic thioesterases, APT1 and APT2, and their respective substrates undergo palmitoylation in the Golgi by as yet unknown PATs, which facilitates their membrane localization. Membrane-associated APT1 and APT2 then depalmitoylate H-Ras and GAP-43, respectively, detaching them from the cell membrane, which promotes translocation to the Golgi where they are re-palmitoylated and translocated to the membrane, which is essential for their function. APT1 also depalmitoylates APT2 detaching it from the membrane for another round of palmitoylation. Following depalmitoylation of APT2, APT1 then catalyzes its own depalmitoylation for translocation to the Golgi to undergo re-palmitoylation, which is required for membrane localization. However, although APT1 catalyzes depalmitoylation of APT2, APT2 does not depalmitoylate APT1. Taken together, based upon our findings this model provides insight into a novel mechanism in which dynamic palmitoylation links the cytosol-membrane translocation of the two cytosolic thioesterases with that of their substrates H-Ras and GAP-43.
FIGURE 8.
Schematic model explaining how dynamic palmitoylation might regulate steady-state membrane localization and function of APT1 and APT2 and those of H-Ras and GAP-43. In this model, APT1 and APT2 undergo palmitoylation in the Golgi by as yet unknown PATs, which facilitates their membrane localization. Membrane-associated APT1 and APT2 then depalmitoylate H-Ras (1) and GAP-43 (2), respectively, detaching them from the cell membrane and promoting translocation to the Golgi where they are re-palmitoylated and translocated to the membrane to manifest their function. Importantly, APT1 also depalmitoylates APT2 (3) by detaching it from the membrane for another cycle of palmitoylation in the Golgi. Following depalmitoylation of APT2, APT1 then catalyzes its own depalmitoylation for translocation to the Golgi to undergo re-palmitoylation as required for membrane localization. However, although in this model APT1 catalyzes depalmitoylation of APT2, APT2 does not depalmitoylate APT1.
Acknowledgments
We thank Dr. H. Waldmann for the generous gift of Palm B and for reviewing a preliminary draft of the manuscript. We also thank Drs. S. W. Levin, I. Owens, and J. Y. Chou for critical review of the manuscript and helpful suggestions. We are grateful to Dr. R. Yasuda (Duke University Medical Center) and Addgene for providing the GFP-H-Ras plasmid.
This work was supported in whole by National Institutes of Health Intramural Program of the Eunice Kennedy-Shriver NICHD.
- PAT
- palmitoyl acyltransferase
- APT1
- acyl-protein thioesterase-1
- APT2
- acyl-protein thioesterase-2
- GAP-43
- growth-associated protein-43
- PPT1
- palmitoyl-protein thioesterase-1
- PPT2
- palmitoyl-protein thioesterase-2
- Palm B
- palmostatin B
- Br-Palm
- Bromopalmitate
- ABE
- acyl-biotinyl exchange
- PI
- protease inhibitor.
REFERENCES
- 1. Schmidt M. F. (1989) Fatty acylation of proteins. Biochim. Biophys. Acta 988, 411–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bijlmakers M. J., Marsh M. (2003) The on-off story of protein palmitoylation. Trends Cell Biol. 13, 32–42 [DOI] [PubMed] [Google Scholar]
- 3. Smotrys J. E., Linder M. E. (2004) Palmitoylation of intracellular signaling proteins' regulation and function. Annu. Rev. Biochem. 73, 559–587 [DOI] [PubMed] [Google Scholar]
- 4. el-Husseini Ael-D., Bredt D. S. (2002) Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 5. Resh M. D. (2006). Palmitoylation of ligands receptors and intracellular signaling molecules. Sci. STKE. 2006, re14. [DOI] [PubMed] [Google Scholar]
- 6. Linder M. E., Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 7. Fukata Y., Fukata M. (2010) Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 [DOI] [PubMed] [Google Scholar]
- 8. Resh M. D. (2012) Targeting protein lipidation in disease. Trends Mol. Med. 18, 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greaves J., Chamberlain L. H. (2007) Palmitoylation-dependent protein sorting. J. Cell Biol. 176, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salaun C., Greaves J., Chamberlain L. H. (2010) Intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duncan J. A., Gilman A. G. (1998) A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21 (RAS). J. Biol. Chem. 273, 15830–15837 [DOI] [PubMed] [Google Scholar]
- 12. Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L. (2010). Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PloS One 5, e15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Camp L. A., Hofmann S. L. (1993) Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 268, 22566–22574 [PubMed] [Google Scholar]
- 14. Camp L. A., Verkruyse L. A., Afendis S. J., Slaughter C. A., Hofmann S. L. (1994) Molecular cloning and expression of palmitoyl-protein thioesterase. J. Biol. Chem. 269, 23212–23219 [PubMed] [Google Scholar]
- 15. Soyombo A. A., Hofmann S. L. (1997) Molecular cloning and expression of palmitoyl-protein thioesterase 2 (PPT2), a homolog of lysosomal palmitoyl-protein thioesterase with distinct substrate specificity. J. Biol. Chem. 272, 27456–27463 [DOI] [PubMed] [Google Scholar]
- 16. Verkruyse L. A., Hofmann S. L. (1996) Lysosomal targeting of palmitoyl-protein thioesterase. J. Biol. Chem. 271, 15831–15836 [DOI] [PubMed] [Google Scholar]
- 17. Hellsten E., Vesa J., Olkkonen V. M., Jalanko A., Peltonen L. (1996) Human palmitoyl protein thioesterase: evidence for lysosomal targeting of the enzyme and disturbed cellular routing in infantile neuronal ceroid lipofuscinosis. EMBO J. 15, 5240–5245 [PMC free article] [PubMed] [Google Scholar]
- 18. Zeidman R., Jackson C. S., Magee A. I. (2009) Protein acyl thioesterases. Mol. Membr. Biol. 26, 32–41 [DOI] [PubMed] [Google Scholar]
- 19. Wang A., Deems R. A., Dennis E. A. (1997) Cloning, expression and catalytic mechanism of murine lysophospholipase 1. J. Biol. Chem. 272, 12723–12729 [DOI] [PubMed] [Google Scholar]
- 20. Wang A., Loo R., Chen Z., Dennis E. A. (1997) Regiospecificity and catalytic triad of lysophospholipase 1. J. Biol. Chem. 272, 22030–22036 [DOI] [PubMed] [Google Scholar]
- 21. Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007) Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 22. Wessel D., Flügge U. I. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 23. Adam R. M., Yang W., Di Vizio D., Mukhopadhyay N. K., Steen H. (2008) Rapid preparation of nuclei-depleted detergent-resistant membrane fractions suitable for proteomics analysis. BMC Cell Biol. 9, 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou F., Xue Y., Yao X., Xu Y. (2006) CSS-Palm: palmitoylation site prediction with a clustering and scoring strategy (CSS). Bioinformatics 22, 894–896 [DOI] [PubMed] [Google Scholar]
- 25. Rocks O., Peyker A., Kahms M., Verveer P. J., Koerner C., Lumbierres M., Kuhlmann J., Waldmann H., Wittinghofer A., Bastiaens P. I. (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307, 1746–1752 [DOI] [PubMed] [Google Scholar]
- 26. Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- 27. Tian L., McClafferty H., Knaus H. G., Ruth P., Shipston M. J. (2012) Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium (BK) channels. J. Biol. Chem. 287, 14718–14725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yeh D. C., Duncan J. A., Yamashita S., Michel T. (1999) Depalmitoylation of endothelial nitric-oxide synthase by acyl-protein thioesterase 1 is potentiated by Ca2+-calmodulin. J. Biol. Chem. 274, 33148–33154 [DOI] [PubMed] [Google Scholar]
- 29. Ladygina N., Martin B. R., Altman A. (2011) Dynamic palmitoylation and the role of DHHC protein in T cell activation and anergy. Adv. Immunol. 109, 1–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dekker F. J., Rocks O., Vartak N., Menninger S., Hedberg C., Balamurugan R., Wetzel S., Renner S., Gerauer M., Schölermann B., Rusch M., Kramer J. W., Rauh D., Coates G. W., Brunsveld L., Bastiaens P. I., Waldmann H. (2010) Small molecule inhibition of APT1 affects Ras localization and signaling. Nat. Chem. Biol. 6, 449–456 [DOI] [PubMed] [Google Scholar]
- 31. Rusch M., Zimmermann T. J., Bürger M., Dekker F. J., Görmer K., Triola G., Brockmeyer A., Janning P., Böttcher T., Sieber S. A., Vetter I. R., Hedberg C., Waldmann H. (2011) Identification of acyl-protein thioesterase 1 and 2 as the cellular targets of the Ras-signaling modulators palmostatin B and M. Angew. Chem. Int. Ed. Engl. 50, 9838–9842 [DOI] [PubMed] [Google Scholar]
- 32. Hirano T., Kishi M., Sugimoto H., Taguchi R., Obinata H., Ohshima N., Tatei K., Izumi T. (2009) Thioesterase activity and subcellular localization of acyl-protein thioesterase 1/lysophospholipase 1. Biochim. Biophys. Acta 1791, 797–805 [DOI] [PubMed] [Google Scholar]
- 33. Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Proteome scale characterization of human palmitoylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]