Background: HSF1 influences chemoresistance in cancer.
Results: Chemotherapy activates HSF1, leading to direct transcriptional regulation of autophagy related gene, ATG7. In vitro findings are supported by patient sample study.
Conclusion: HSF1 regulates cytoprotective, heat shock-independent autophagy by directly regulating ATG7, which plays an important role in chemoresistance.
Significance: Identification of novel HSF1/ATG7 axis in chemoresistance strongly supports development of robust combination therapies, targeting it in cancer.
Keywords: Autophagy, Cancer, Cell Death, Chemoresistance, Transcription Regulation, ATG7, HSF1, Heat Shock-independent Autophagy
Abstract
Heat shock factor 1 (HSF1), a master regulator of heat shock responses, plays an important role in tumorigenesis. In this study we demonstrated that HSF1 is required for chemotherapeutic agent-induced cytoprotective autophagy through transcriptional up-regulation of autophagy-related gene ATG7. Interestingly, this is independent of the HSF1 heat shock response function. Treatment of cancer cells with the FDA-approved chemotherapeutic agent carboplatin induced autophagy and growth inhibition, which were significantly increased upon knockdown of HSF1. Mechanistic studies revealed that HSF1 regulates autophagy by directly binding to ATG7 promoter and transcriptionally up-regulating its expression. Significantly, breast cancer patient sample study revealed that a higher ATG7 expression level is associated with poor patient survival. This novel finding was further confirmed by analysis of two independent patient databases, demonstrating a prognostic value of ATG7. Furthermore, a strong positive correlation was observed between levels of HSF1 and ATG7 in triple-negative breast cancer patient samples, thus validating our in vitro findings. This is the first study identifying a critical role for HSF1 in controlling cytoprotective autophagy through regulation of ATG7, which is distinct from the HSF1 function in the heat shock response. This is also the first study demonstrating a prognostic value of ATG7 in breast cancer patients. These findings strongly argue that combining chemotherapeutic agents with autophagy inhibition by repressing HSF1/ATG7 axis represents a promising strategy for future cancer treatment.
Introduction
Breast cancer is the most common type of cancer as well as second leading cause of cancer-related deaths in women. The increasing emergence of resistance to commonly used chemotherapeutic agents, leading to low disease-free survival rates for women with advanced and highly metastatic cancer, has warranted the development of novel robust treatment options for these patients. Carboplatin, a second-generation platinum agent, has been approved by FDA for the treatment of non-small cell lung cancers and ovarian cancers. Its reduced side effects compared with cisplatin have led to its increased use in combination with taxanes to treat highly metastatic breast cancers. Moreover, carboplatin has shown promising results as a single agent in previously untreated metastatic breast cancer with response rates of 20–35% (1). As per NIH clinical trial registry, currently there are approximately 52 ongoing clinical trials to examine the expanded use of carboplatin in breast cancer as a combination treatment with taxanes and other drugs. Hence, an important step in improving clinical management of these patients is to identify the mechanism of carboplatin sensitivity in these tumors, to develop more robust combination therapy to overcome drug resistance that may develop.
HSF12 is a transcription factor that functions both as an activator and a repressor of its target genes, depending on spacing of the heat shock elements it binds to and the co-factors it interacts with (2–7). Exposure to a variety of stress leads to oligomerization of HSF1 into active trimers, followed by translocation into the nucleus and binding to the heat shock elements located within the promoter region of the target genes such as Hsp90, Hsp25, and Hsp70 (8–10). Overall, HSF1 mediates cellular recovery under a variety of stress conditions such as thermal injury, ischemia, and age-related neurodegeneration by orchestrating a gamut of cellular processes (11, 12). Although less well understood, HSF1 has also been shown to regulate non-heat shock response genes to support highly malignant human cancers (7, 13, 14). Emerging evidence suggests an important role for HSF1 in cellular transformation and in cancer development (14–17). HSF1 supports malignant transformation by modulating multiple pathways regulating proliferation, survival, protein synthesis, and cellular metabolism (14). We and others have shown that HSF1 regulates glucose metabolism and mediates tumorigenesis and cell growth in cancer cells (14, 15). In addition to association of spontaneous mutations in HSF1 with tumorigenesis, a recent study using a large breast cancer patient sample cohort showed that high levels of HSF1 are associated with poor patient prognosis (18). HSF1 has also been reported to regulate drug resistance. However, the mechanism for HSF1-mediated drug resistance is not well understood.
Autophagy, an evolutionary conserved catabolic process, is induced in response to metabolic stress such as nutrient deprivation and hypoxia (19–21). During autophagy, autophagosomes are formed as double-membrane vesicles engulfing cellular organelles and cytoplasm. These autophagosomes fuse with lysosomes, where the sequestered cytoplasmic contents are degraded and recycled for synthesis of protein and ATP (22). Autophagy is genetically regulated by a family of autophagy-related genes (ATGs), which coordinate specific steps in autophagy induction and cytoplasmic sequestration (23). Autophagy has been recognized as a cytoprotective process against environmental stress as well as stress induced by certain chemotherapeutic agents (24–27). Recent studies have demonstrated that inhibition of autophagy sensitizes the cancer cells to DNA-damaging agents, hormonal therapies, radiation therapy, and chemotherapeutics (24, 25, 27–29). Therefore, there has been an increased interest in targeting this process to reduce drug resistance in several different cancers.
This report shows that in cancer cells, HSF1 is required for chemotherapy-induced cytoprotective autophagy, and this is independent of HSF1 function in heat shock response. This is the first study to demonstrate that HSF1, a multifunctional transcription factor, is activated by carboplatin treatment and leads to up-regulation of a novel HSF1 target gene ATG7. This regulation plays a critical role in cancer cell resistance to chemotherapy.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies
MDA-MB-231 and MDA-MB-436 cells, obtained from ATCC, were cultured in DMEM/F-12 (Mediatech) with 10% FBS. Antibodies to HSF1, PARP, cPARP, cleaved caspase 3, p62, and LC3 were purchased from Cell Signaling Technology. Anti-ATG7 was from Millipore, anti-β-actin was from Sigma, and anti-pS326 HSF1 was from Abcam. HRP-conjugated secondary anti-mouse IgG and anti-rabbit IgG were purchased from Bio-Rad. ECL Western blotting substrate was from Pierce.
Flow Cytometry Analysis
Control and carboplatin-treated cells were stained with Annexin V-FITC and 7-AAD according to the manufacturer's protocol (BD Pharmingen). Acridine orange staining was analyzed using flow cytometry. Control, carboplatin, and carboplatin +3-MA-treated stable HSF1-knockdown and scramble MDA-MB-231 cells were stained with 1 μg/ml acridine orange for 15 min at 37 °C. Flow cytometry was done on FACSCanto and analyzed using Diva software.
Stable HSF1-knockdown Cells
Five different seed shRNA sequences specific to HSF1 were obtained from Sigma. Clone NM_005526.1-331s1c1 generated maximum knockdown of HSF1 expression and was used to generate lentiviral particles according to the manufacturer's protocol. MDA-MB-231 and MDA-MB-436 were transduced with shRNA against HSF1 and scramble shRNA. After primary puromycin selection, 3 μg/ml cell pools of MDA-MB-231-shHSF1, MDA-MB-231-shscramble, MDA-MB-436-shHSF1, and MDA-MB-436shscramble were maintained for multiple passages in 1.5 μg/ml puromycin.
siRNA Experiments
Down-regulation of HSF1 and ATG7 was confirmed with two siRNA sequences to avoid off-target effects. HSF1 siRNAs were Sigma 0050 and SASI_Hs02_00339745 and Invitrogen HSF1 stealth siRNA. ATG7 siRNA were 0020 SASI_Hs01_00077648 and 0050 SASI_Hs01_000341471. siRNA 0020 showed significantly more knockdown and was used for all experiments described below. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. 24 h after siRNA transfection, cells were treated with carboplatin (75 mg/ml) for 24 h. Forty-eight hours after transfection, cell lysates were prepared for Western blot analysis.
Cell Counting
MDA-MB-231 control and stable HSF1-knockdown cells (4 × 105 cells/well) were seeded in 6-well plates overnight. Cells were treated with carboplatin for 24 h. Cell viability was measured by trypan blue staining and direct cell counting using a hematocytometer.
Western Blotting
Western blotting was performed as described (30). Immunoblot analysis for HSF1 trimers was performed as described earlier. Cell extracts were treated with cross-linker ethylene glycol bis(succinimidylsuccinate) at a final concentration of 1 mm followed by glycine at a final concentration of 75 mm (9). The reaction mixture was run on an 8% SDS-polyacrylamide gel and immunoblotted with HSF1 antibody.
Fluorescent Microscopy
Transient transfection with EGFP-LC3B (Addgene plasmid 11546) (31) was carried out in scramble and stable HSF1-knockdown breast cancer cells using Lipofectamine 2000. Twenty-four hours after transfection the cells were treated with 75 μg/ml carboplatin or rapamycin for 24 h. Punctate EGFP-LC3 was captured using an Automated Nikon TE2000E at a magnification of ×60. NIS-Elements software used to calculate punctate structures.
Luciferase Reporter Assay
pGL3-promoter-ATG7 luciferase vector (ATG7) was constructed by amplification of HSF1 binding site within the ATG7 promoter region in the human genomic DNA using forward primer ACTGGTACCTGGTTGCCTCCCTTGTTTAC and reverse primer ACTAGATCTCCATCATATCCCCAGTGACC. This amplified region was cloned within BglII and KpnI sites in pGL3 promoter. Mutant luciferase constructs were generated using the site-directed mutagenesis approach. Deletion mutants were generated using pGL3-promoter-ATG7 and forward primer GGTGAGAATTACAAATTAAGGGTGGGGGAGACT and reverse primer AGTCTCCCCCACCCTTAATTTGTAATTCTCACC for mut1 and forward primer GGTGAGAATACAAATTAAGGGTGGGGGAGATT and reverse primer AATCTCCCCCACCCTTAATTTGTAATTCTCACC for mut2. Cells were transfected with 1 μg of luciferase reporters and 8 ng of internal control plasmid pRL-SV40 vector (Promega) using Lipofectamine 2000. Twenty-four hours later cells were treated with 75 μg/ml carboplatin. Forty-eight hours after transfection, cells were harvested and lysed with passive lysis buffer (Promega). Luciferase activity was measured using the Dual Luciferase Reporter Assay System protocol (Promega) and 20/20n luminometer (Turner Biosystems).
Quantitative Real-time PCR
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen). cDNA was generated using a cDNA synthesis kit (ABI) followed by amplification using SYBR Green PCR master mix (Roche Applied Science) and primers specific for ATG7 (26) and GAPDH (forward, ATCATCCCTGCCTCTACTGG and reverse, CTGCTTCACCACCTTCTTGA) All reactions were performed in duplicate. Relative mRNA levels were calculated using three independent experiments. Data were normalized to GAPDH; control groups were set to 1.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described earlier (32). Chromatin was sonicated using the Sonicator Ultrasonic processor with microtip. Antibodies were anti-HSF1, rabbit IgG (control), and anti-H3 acetylation (Millipore).
Immunofluoresence Staining
Staining performed as described previously (32). Nucleus was stained by DAPI (1:10,000) or HSF1 (1:500). Images were captured using Automated Nikon TE2000E at a magnification of ××60. Nuclear staining intensity was quantified using NIS-Elements software.
Immunohistochemical Staining and Analysis
142 randomly selected invasive breast carcinoma patients were surgically resected and archived at M. D. Anderson Cancer Center. Tissue microarray blocks were constructed by taking core samples from representative areas of paraffin-embedded breast tumor tissues and assembling them on a recipient paraffin block. Two 1-mm tissue cores were obtained from each specimen. Paraffin sections of recipient blocks were obtained and stained with hematoxylin and eosin to confirm the presence of neoplastic tissues. Patient information was delinked from the specimens, and the study was approved by the Institutional Review Board of M. D. Anderson Cancer Center. ER/PR/HER2 results were obtained from the original pathology reports for each patient in the tissue microarray. Standard immunohistochemistry was performed on tissue microarray samples for HSF-1 (1:250 dilution, 4356; Cell Signaling Technology) and ATG7 (1:250 dilution; Millipore). Nuclear expression of HSF1 and cytoplasmic expression of ATG7 were assessed semiquantitatively using staining intensity and percentage by two independent pathological investigators. The H score was determined by multiplying staining intensity by percentage of positive tumor cells (33). The Kaplan-Meier method was used to analyze breast cancer patient overall survival. Two-tailed p values ≤0.05 were considered statistically significant. SPSS software and XLSTAT were used for statistical analysis. Tetrachoric correlation analysis was used to analyze the correlation between HSF1 and ATG7 (34).
Survival Analysis using External Breast Cancer Expression Datasets
Microarray gene expression and clinical data (GSE1456 and GSE7390) were downloaded from the NCBI GEO website. Analyses were performed using the R software. Gene expression levels were normalized using the “rma” package from R, and patients were automatically assigned into ATG7-low and -high groups using the kmean function performed on ATG7 expression. Kaplan-Meier analysis was performed using the “survival” package from R.
Statistical Analysis
Statistical evaluation was determined by Student's t test. All data are shown as means ± S.E. A p value <0.05 was considered significant.
RESULTS
HSF1 Expression Protects Cancer Cells from Chemotherapeutic Agent-induced Cell Death
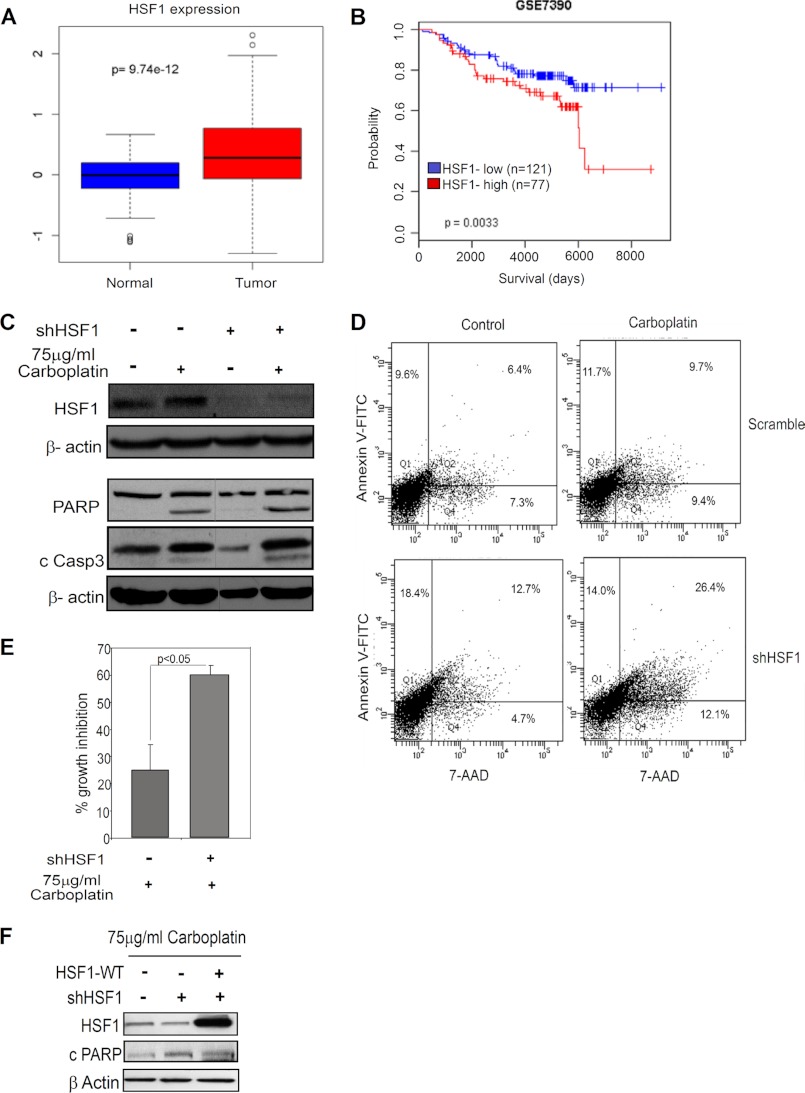
To study the role of HSF1 in cancer cell survival, we first analyzed a published breast cancer patient database. The analysis revealed that HSF1 expression is higher in breast tumors compared with normal tissue (Fig. 1A). Moreover, survival analysis using the patient database indicated an association between high HSF1 expression and poor patient survival (Fig. 1B). This is consistent with the results of a recent study (18). These data indicate that HSF1 expression plays a protective role in cancer cell survival. Furthermore, a published study from our laboratory has shown that HSF1 expression is elevated and contributes to trastuzumab resistance (15, 35), as well as reduces sensitivity to Taxol and doxorubicin treatments (supplemental Fig. S1, A and B). Next, to study the role of HSF1 in sensitivity to carboplatin treatment, we used stable HSF1-knockdown cell lines which were generated using a lentiviral construct targeting HSF1. Treatment with 75 μg/ml carboplatin, a second-generation DNA-alkylating agent, increasingly used in the clinic and in combination with taxanes in several breast cancer clinical trials (1), showed an increase in apoptosis in HSF1-knockdown cells as measured by cleaved PARP and cleaved caspase 3 in MDA-MB-231 and MDA-MB-436 cells (Fig. 1C and supplemental Fig. S1C). An increase in Annexin V-FITC and 7-AAD staining further confirmed that knockdown of HSF1 increased sensitivity to carboplatin (Fig. 1D). Furthermore, reduction of endogenous HSF1 expression also enhanced growth inhibition in MDA-MB-231 cells treated with carboplatin (Fig. 1E).
FIGURE 1.
HSF1 confers drug resistance in breast cancer. A, box plot for expression of HSF1 in normal and tumor tissue using TCGA database. B, survival analysis for breast cancer patient using the GSE7390 database. C, immunoblot analysis of HSF1 and total and cleaved PARP (cPARP) and cleaved caspase 3 (c Casp3) in MDA-MB-231 cells after stable HSF1-knockdown and treatment with 75 μg/ml carboplatin for 24 h. D, flow cytometry of Annexin V-FITC and 7-AAD staining in carboplatin-treated MDA-MB-231 cells in the presence or absence of HSF1. E, growth inhibition in the presence or absence of HSF1 in MDA-MB-231 cells treated with 75 μg/ml carboplatin for 24 h. Error bars, S.E. (n = 3). F, immunoblot analysis of HSF1 and cPARP in carboplatin-treated control (−), HSF1-knockdown (+), and HSF1-knockdown transiently overexpressing HSF1. Loading control, β-actin. Vertical gray line in some blots indicates deleted lanes from the blot.
To further confirm that HSF1 increases resistance to carboplatin, we transiently overexpressed HSF1 in stable HSF1-knockdown cells. Restoring HSF1 expression led to decreased apoptosis as measured by cleaved PARP, in presence of carboplatin (Fig. 1F). Collectively, these data indicate that HSF1 plays an important role in protecting cancer cells from carboplatin-induced cell death.
HSF1 Is Required for Induction of Autophagy
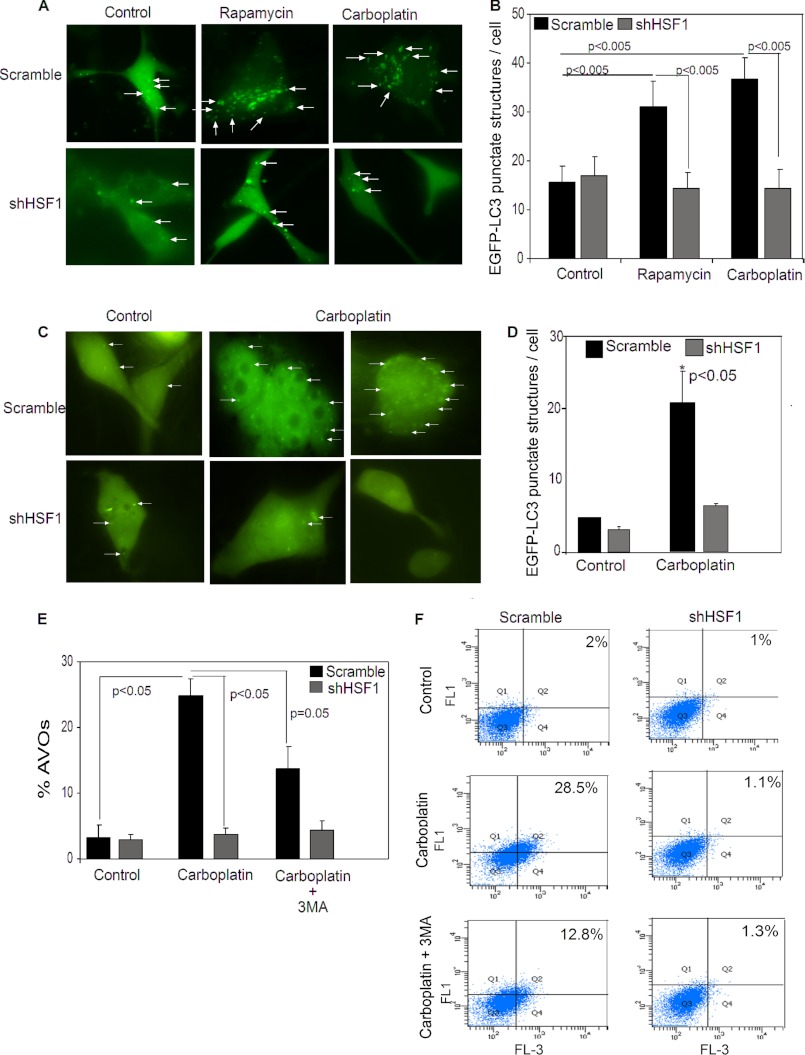
Autophagy is a cellular process that is activated to mitigate cellular damage to allow cells to survive in stressful conditions (36). Cisplatin, a first-generation platinum agent, has been reported to induce autophagy in cancer cells leading to decreased chemosensitivity (36). Similarly, we wanted to test whether carboplatin treatment also induced autophagy in cancer cells. As observed in Fig. 2A, treatment with carboplatin robustly induced autophagy, similar to rapamycin, an mTOR inhibitor and a well known inducer of autophagy (Fig. 2, A and B). Interestingly, knockdown of HSF1 expression in these cells completely blocked the induction of autophagosomes by both rapamycin and carboplatin as indicated by the EGFP-LC3 punctate structures (Fig. 2, A and B). Similarly, treatment with carboplatin in MDA-MB-436 cells also increased formation of EGFP-LC3 punctate structures, which was blocked upon knockdown of HSF1 (Fig. 2, C and D). These data indicate that HSF1 is required for induction of autophagy. Moreover, the requirement of HSF1 for induction of autophagy was further confirmed by flow cytometry using acridine orange to stain for acidic vesicular organelles (AVOs) in the cells. Carboplatin treatment increased formation of AVOs in cells expressing HSF1, but failed to increase accumulation of AVOs in HSF1-knockdown cells (Fig. 2, E and F). Because AVOs are a measure of all acidic vesicular organelles including but not limited to autophagolysosome, we treated cells with 3-MA, an autophagy inhibitor, to inhibit formation of AVOs caused by autophagy. Inhibition of autophagy reduced AVOs in the scramble cell population (Fig. 2, E and F), indicating that the increase in AVOs in carboplatin-treated cells is mainly due to the induction of autophagy. Taken together, these results demonstrate that HSF1 expression is essential for induction of autophagy in cancer cells.
FIGURE 2.
HSF1 influences induction of autophagy in breast cancer cells. A, microscopy of punctate EGFP-LC3 in scramble and HSF1-knockdown MDA-MB-231 cells in untreated (control) or treated with 10 nm rapamycin or 75 μg/ml carboplatin for 24 h. Magnification, ×60. White arrows, punctate EGFP-LC3 structures. B, bar graph represents average EGFP-LC3 punctate structures per cell in MDA-MB-231 cells expressing HSF1 (black bars) or HSF1-knockdowns (gray bars), upon treatment with rapamycin or carboplatin. Error bars, S.E. Data were collected for two independent experiments, and at least 10 images per slide were analyzed for each condition. C, microscopy of punctate EGFP-LC3 in presence or absence of HSF1 in MDA-MB-436 cells untreated (control) or treated with 75 μg/ml carboplatin for 24 h. Magnification, ×60. D, average EGFP-LC3 punctate structures per cell in MDA-MB-436 cells expressing HSF1 (black bars) or HSF1-knockdown (gray bars), upon treatment with carboplatin. Error bars, S.E. Data for three independent experiments and at least 30 cells for rapamycin treatment and carboplatin treatment were analyzed for each condition. *, p < 0.05). Flow cytometry of acridine orange staining of AVO in scramble and stable HSF1-knockdown MDA-MB-231 cells after treatment with 75 μg/ml carboplatin and carboplatin and 3-MA for 24 h is shown. E, mean AVO-positive cells (n = 3). Error bars, S.E. F, representative flow cytometric profile of AVO-positive cells. FL1 detects green intensity, and FL3 detects red intensity. Cells in right quadrants were considered AVO-positive.
HSF1 Transcriptionally Up-regulates ATG7
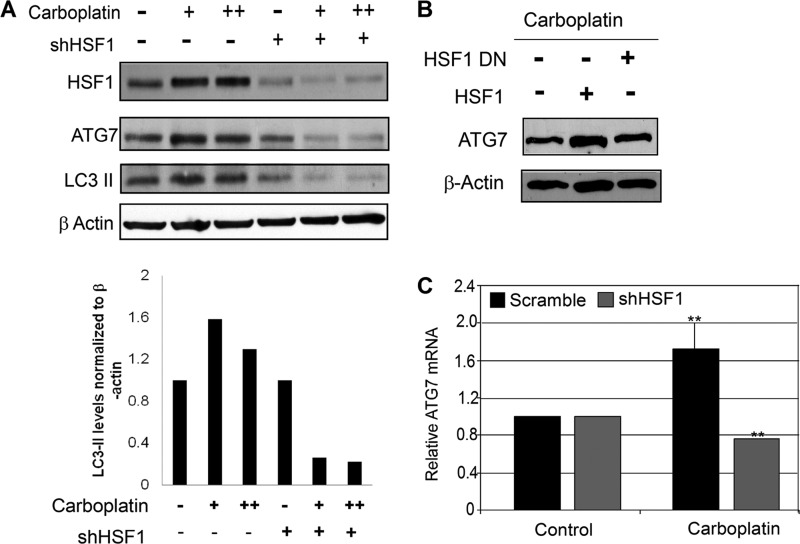
HSF1 is a transcription factor that regulates expression of its target genes under conditions of cellular stress, specifically heat shock. In addition to its ability to regulate heat shock proteins, HSF1 has been shown to regulate the expression of a distinct set of genes in cancer cells (7). A genome-wide analysis of human HSF1 using ChIP microarray studies revealed several gene promoters that can be bound by HSF1 regulating cellular adaptation and survival (13). Of these possible HSF1 targets only two autophagy-related genes, ATG7 and ATG4b, were identified (13). Upon further promoter sequence analysis using the TFsearch database, we identified two potential HSF1 binding sites with adjacent inverted repeats of 5′-nGAAn-3′ at positions −1570 and −1602 within the ATG7 promoter. Next, to determine whether HSF1 regulates autophagy by targeting the expression of ATG7, we assessed the expression of ATG7 in cancer cells that express different levels of HSF1. Carboplatin treatment increased expression of ATG7 protein (Fig. 3A), but knockdown of HSF1 reduced ATG7 protein expression in the presence of carboplatin (Fig. 3A). To further confirm this, we overexpressed wild type HSF1 and dominant negative (DN) HSF1 in stable HSF1-knockdown cells. Reexpression of wild type HSF1 restored the expression of ATG7 in these cells, whereas DN-HSF1 did not (Fig. 3B). The regulation of ATG7 at the protein level upon carboplatin treatment was a direct effect of regulation of ATG7 mRNA expression (Fig. 3C). These results further confirmed that HSF1 regulates the expression of ATG7 and may subsequently regulate autophagy.
FIGURE 3.
HSF1 influences expression of autophagy-related gene ATG7. A, top panel, immunoblot analysis of HSF1, ATG7, and LC3-II in MDA-MB-231 scramble (−) and stable HSF1-knockdown (+) cells in the presence of 75 and 100 μg/ml carboplatin for 24 h. Bottom panel, quantification of LC3-II expression using ImageJ software. LC3-II levels were normalized to β-actin. B, immunoblot analysis for ATG7 upon carboplatin treatment in stable HSF1-knockdowns (+) transiently overexpressing wild type HSF1 (HSF1) or dominant negative HSF1 (HSF1 DN). Loading control was β-actin. C, mRNA levels of ATG7 in Scramble (black bars) and stable HSF1-knockdown (gray bars) MDA-MB-231 cells treated with 75 μg/ml carboplatin for 24 h. Error bars, S.E. (n = 3). **, p < 0.02.
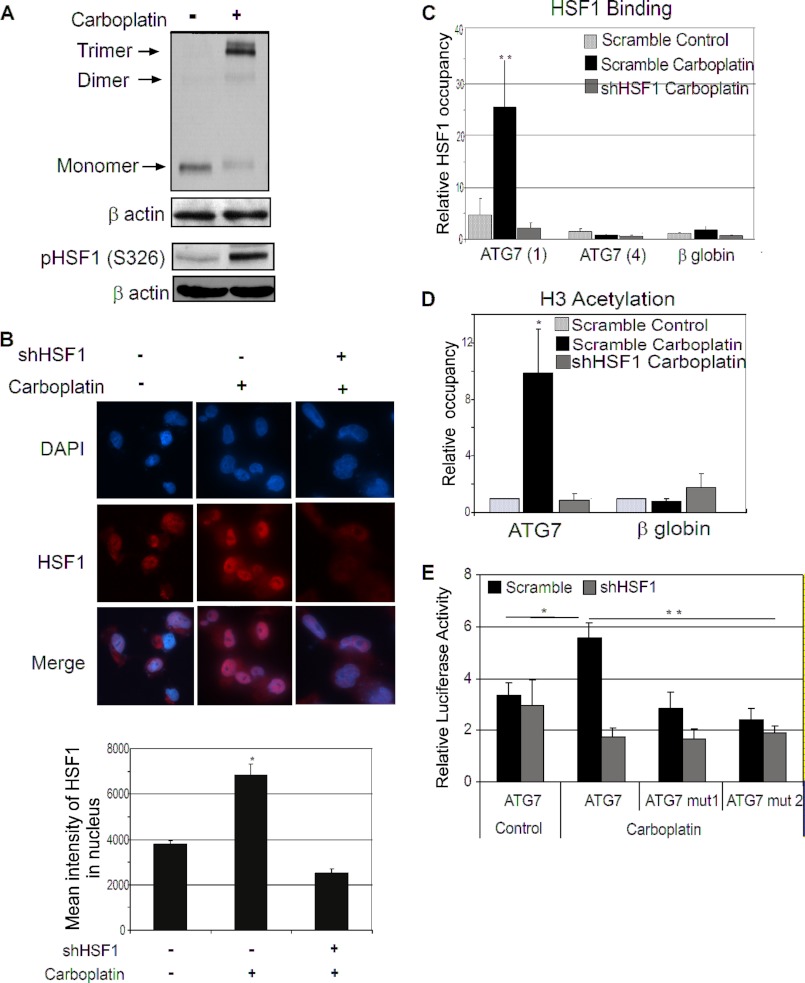
The regulation of ATG7 expression by HSF1 was only observed after treatment with carboplatin. To function as an active transcription factor cytoplasmic HSF1 translocates into the nucleus, phosphorylates and trimerizes, allowing it to bind to its target genes and subsequently regulating their expression (8). To test whether carboplatin can activate HSF1, which could explain why carboplatin induces up-regulation of ATG7 in cancer cells (Fig. 3A), we examined HSF1 trimer formation in HSF1-expressing cells upon carboplatin treatment. As seen in Fig. 4, treatment with carboplatin increased HSF1 trimer formation along with phosphorylation at residue Ser-326 (Fig. 4A) and enhanced its nuclear translocation (Fig. 4B), indicating activation of HSF1. These data indicate that carboplatin treatment increased the transcription activity of HSF1. Next, ChIP analysis was used to confirm that activated HSF1 regulates ATG7 expression at the level of transcription by directly binding to the ATG7 promoter region. ChIP assay followed by quantitative PCR analysis using primers flanking the two potential HSF1 binding sites (ATG7 (1)) shows a significant increase in HSF1 binding upon carboplatin treatment which was lost upon knockdown of HSF1 (Fig. 4C). Furthermore, no HSF1 binding was detected using ATG7 primer set flanking region ∼780 bp upstream of the HSF1 binding sites (ATG7 (4)) (Fig. 4C). This indicates that HSF1 binding was specific to the ATG7 promoter region containing potential HSF1 binding sites. The increased binding of HSF1 at the ATG7 promoter was accompanied by an increase in the histone H3 acetylation mark (Fig. 4D), which is an indication of active gene transcription. This change in acetylation was specific to recruitment of HSF1 at the ATG7 promoter, because no change in histone H3 acetylation was observed in HSF1-knockdown cells. The β-globin gene is a negative control that shows no binding of HSF1 as well as no H3 acetylation (Fig. 4, C and D). To further validate that the two potential HSF1 binding sites are required for regulation of ATG7, we cloned the HSF1 binding region within the human ATG7 promoter region upstream of the luciferase gene and assessed the luciferase activity. Two ATG7 promoter deletion constructs were also generated. The HSF1 binding site at −1602 was deleted in ATG7 mutant 1 (ATG7 mut1), whereas both of the HSF1 binding sites were deleted in ATG7 mutant 2 (ATG7 mut2). Carboplatin treatment significantly increased the ATG7 promoter luciferase activity only in the presence of HSF1 (Fig. 4E). Deletion of HSF1 binding sites reduced the promoter activity by ∼50% in presence of carboplatin (Fig. 4E). A similar degree of inhibition of luciferase activity for the two deletion constructs suggests that HSF1 binding site at −1602 is more important for the ATG7 promoter activity. Furthermore, down-regulation of HSF1 by shRNA dramatically decreased the luciferase activity of the wild type promoter by 70% and to a lesser extent in the HSF1 binding site mutants (Fig. 4E). Because HSF1 is a key regulator of heat shock response, we next wanted to identify whether ATG7 was also regulated under heat shock conditions. As shown in supplemental Fig. S2A heat shock at 42 °C induced expression of Hsp70, a classical heat shock response gene, but failed to increase the expression of ATG7. Furthermore, chemotherapeutic treatment activated HSF1 (supplemental Fig. S2B) and increased ATG7 expression (supplemental Fig. S2A) but did not induce a heat shock response as shown by no change in Hsp70 expression (supplemental Fig. S2B). These data indicate that regulation of ATG7 by HSF1 is independent of heat shock. Taken together, these results strongly indicate that carboplatin activates HSF1 which directly binds to the ATG7 promoter and transcriptionally activates ATG7 expression independent of heat shock.
FIGURE 4.
Chemotherapy activates HSF1 in breast cancer cells. A, immunoblot analysis of trimer formation of HSF1 and phosphorylation of Ser-326 residue in MDA-MB-231 cells treated with 75 μg/ml carboplatin. B, immunofluorescence staining of untreated MDA-MB-231 HSF1 scramble cells, carboplatin-treated, and stable HSF1-knockdown carboplatin-treated cells. HSF1, Alexa Fluor 594 (red); nucleus, DAPI. Merged image, nuclear localization of HSF1. Images shown are a representative panel. Magnification, ×60. Bars represent the mean intensity of HSF1 in the nucleus. Error bars, S.E. (*, p < 0.05). C, ChIP using HSF1 antibody in scramble untreated (white bars) and carboplatin-treated (black bars) and stable HSF1-knockdown carboplatin-treated (gray bars) cells. ATG7 (1) primers flank HSF1 binding region, ATG7 (4) primer set and β-globin are negative controls. **, p < 0.05. D, ChIP of acetylation of histone H3. The conditions are as described in C, except the antibody used is specific to acetyl-H3. Bars are mean of three independent experiments; error bars, S.E. E, HSF1 regulates ATG7 luciferase promoter activity. MDA-MB-231 scramble and stable HSF1-knockdown cells were transfected with ATG7, ATG7 mut1, ATG7 mut2. Control indicates untreated cells; carboplatin treatment was 75 μg/ml, 24 h. Results were normalized to a co-transfected Renilla. Bars are mean of three independent experiments; error bars, S.E. (*, p < 0.05; **, p < 0.02). β-Actin is the loading control.
Autophagy Confers HSF1-mediated Resistance to Chemotherapy-induced Cell Death
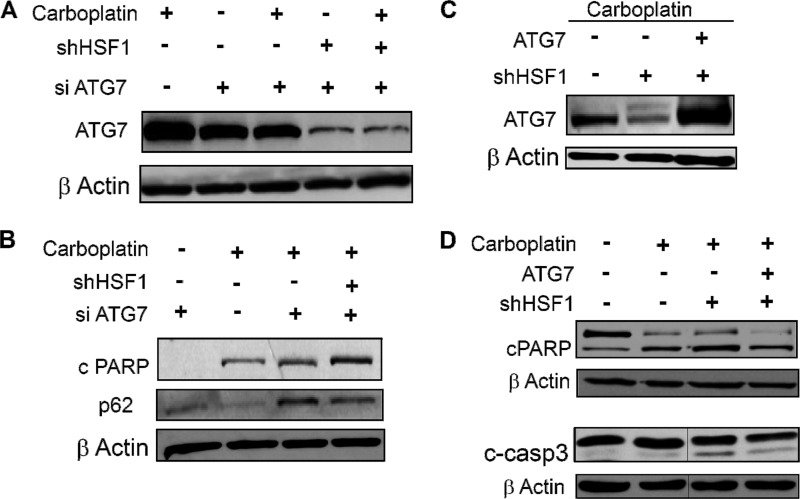
Next, to examine whether regulation of ATG7 by HSF1 plays a role in HSF1-mediated resistance to chemotherapy-induced cell death, we transiently knocked down ATG7 expression in MDA-MB-231 cells (Fig. 5A). Reduction of ATG7 expression reduced autophagy as indicated by the accumulation of p62 and also increased sensitivity to carboplatin treatment as shown by the increase in cleaved PARP (Fig. 5B). The transient knockdown efficiency of ATG7 was significantly higher in the HSF1-stable knockdown cells (Fig. 5A). This may explain the significantly higher apoptosis in the double-knockdown cells. To further confirm the role of ATG7 in chemosensitivity, ATG7 was overexpressed in HSF1-knockdown cells (Fig. 5C). Overexpression of ATG7 reduced carboplatin sensitivity, as indicated by the reduction of apoptosis in these cells detected by decrease in cleaved PARP and cleaved caspase 3 expression (Fig. 5D). These data indicate that autophagy regulated by ATG7 plays an important cytoprotective role in carboplatin-treated cancer cells.
FIGURE 5.
Autophagy influences drug sensitivity. A–C, immunoblot analysis of ATG7 in presence of siATG7 in MDA-MB-231 scramble control and HSF1-stable knockdown cells in the presence and absence of carboplatin (A); cPARP and p62 in transient ATG7-knockdown HSF1 scramble and HSF1-knockdown MDA-MB-231 cells (B); ATG7 in scramble control, stable HSF1-knockdown cells and stable HSF1-knockdown cells transiently overexpressing ATG7 in presence of carboplatin (C). D, total and cleaved PARP and cleaved caspase 3 levels in carboplatin-untreated and -treated scramble controls and in carboplatin-treated stable HSF1-knockdown and stable HSF1-knockdown cells transiently overexpressing ATG7. Vertical gray line in the blots indicates deleted lanes from the same blots.
HSF1 Expression Is Correlated with ATG7, and ATG7 Expression Is Associated with Poor Patient Survival in Breast Cancer Patients
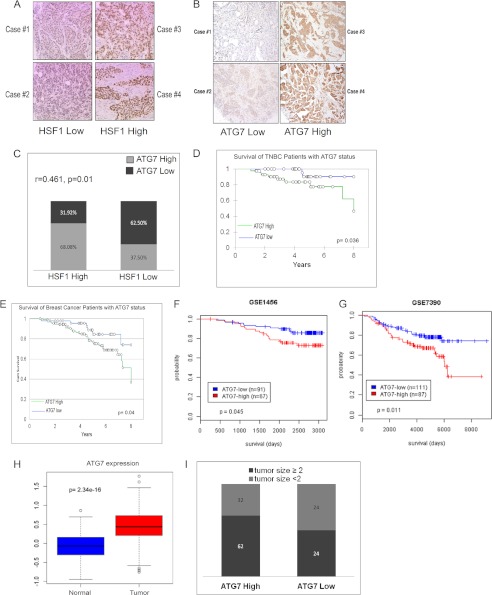
To study the clinical relevance of our findings, we carried out immunohistochemistry analysis for expression of HSF1 and ATG7 in breast cancer patient samples. A total of 142 patient samples were stained for HSF1 and ATG7 using commercially available antibodies (Fig. 6, A and B). Two pathologists scored the HSF1 and ATG7 staining by semiquantitative analysis of nuclear and cytoplasmic staining intensities, respectively. It has been reported that high levels of HSF1 are present in triple negative breast cancer (TNBC) (7) and are associated with poor prognosis in breast cancer (18). To explore further whether levels of ATG7 and HSF1 were correlated in TNBC we carried out tetrachoric correlation analysis. As shown in Fig. 6C there is a significant positive correlation between HSF1 expression and ATG7 expression levels in TNBC patient samples (r = 0.461, p = 0.01). Because the ATG7 and patient survival correlation has never been reported, we next focused our study on ATG7. Interestingly, Kaplan-Meier analysis suggests a significant association between ATG7 status and survival in TNBC, which is the more aggressive breast cancer subtype (Fig. 6D). Next, survival analysis of all 142 individual scored for ATG7 expression was obtained. The patients were divided into two groups, ATG7 high expressers and ATG7 low expressers. There were 94 (66.19%) patients for high ATG7 expression and 48 (33.80%) for low ATG7 expression. The Kaplan-Meier survival curve shows that patients with low ATG7 expression had significantly better survival compared with patients with high ATG7 expression (Fig. 6E, p = 0.04). Survival curves with similar results were obtained from analyzing survival of breast cancer patients using two different cancer patient databases (Fig. 6, F and G). Next, using The Cancer Genome Atlas (TCGA), we analyzed the expression of ATG7 in normal and breast tumors. There is a significant increase in expression level of ATG7 in tumors compared with normal tissues (Fig. 6H). We next determined whether ATG7 status correlates with tumor size in the breast cancer patient samples. As seen in Fig. 6I, high levels of ATG7 were associated with larger tumor size. These results support a possible role for ATG7 in clinical aggressiveness of the breast tumors. Taken together, these data indicate that ATG7 expression is associated with breast cancer patient survival and is significantly correlated with levels of HSF1, thus validating the clinical relevance of our findings.
FIGURE 6.
Atg7-high tumors associated with decreased survival in breast cancer patients. A and B, immunohistochemical staining of breast cancer patient samples for HSF1 (A) and ATG7 (B). C, correlation between expression of HSF1 and ATG7 in the TNBC patient samples. Tetrachoric correlation coefficient is shown along with log p value. D, Kaplan-Meier analysis of TNBC individuals scored for the ATG7 status in the study. E, Kaplan-Meier analysis of all individuals scored for the ATG7 status in the study. F and G, survival analysis for breast cancer patient based on ATG7 status using the GSE7390 database (F) and using the GSE1456 database (G). Log-rank p value is shown. H, box plot for expression of ATG7 in normal and tumor tissue using TCGA database. I, distribution of breast cancer patients with large tumor size (≥2) and small tumor size (<2) expressing different levels of ATG7.
DISCUSSION
HSF1, a master regulator of the heat shock response, has been shown to regulate expression of genes involved in cellular adaptation and survival (13). HSF1 has not only been associated with malignant transformation (14, 37), but high levels of nuclear HSF1 are also associated with poor prognosis in breast cancer (18). Trimer formation and subsequent nuclear translocation of HSF1 are associated with the active form of HSF1 that can bind to promoter of its target genes and regulate their expression (8). Here, for the first time, we show that treatment with a chemotherapeutic agent increases trimer formation and nuclear translocation of HSF1. Moreover, this translocation was accompanied by increased binding of HSF1 and higher H3 acetylation at ATG7 promoter region and enhanced ATG7 promoter activity. HSF1 has been shown to bind inducibly to a specific consensus motif that is associated with histone acetylation, H3K4 dimethylation, RNA polymerase II, and co-activators, which are markers of activated state of chromatin (38). Using stable HSF1-knockdown cells, we identified that binding of HSF1 was required to increase the H3 acetylation at the ATG7 promoter region. Because higher acetylation status of the ATG7 promoter region is associated with increased ATG7 expression (39), our data suggest that the absence of HSF1 at the ATG7 promoter prevented H3 acetylation, leading to repression of ATG7 and thus formation of autophagosomes leading to increased chemosensitivity. Furthermore, we identified that regulation of ATG7 by HSF1 is independent of heat shock. This observation is further supported by a recent report that shows that HSF1 drives a transcriptional program which is distinct from heat shock to support a variety of highly malignant cancers (7).
Defects in autophagy have been found in several human tumors, such as allelic loss of beclin1 observed often in human breast, ovarian, and prostate cancers (40–42). These observations suggest that autophagy may play a tumor suppressor role in cancer. However, a large number of recent reports have shown that autophagy can serve as a mechanism of adaptation in cancer cells to induce resistance to apoptosis for survival under conditions of metabolic stress (43). Furthermore, several cancer therapies including radiation therapy, chemotherapy, histone deacetylase inhibitors, anti-estrogen hormonal therapy as well as Herceptin treatment can induce autophagy as a protective and pro-survival mechanism in human cancer cells (26, 28, 29, 36, 44–46). Similarly, our data show that inhibition of autophagy increases sensitivity to carboplatin. Furthermore, knockdown of HSF1 along with inhibition of autophagy enhances the chemosensitivity of cancer cells. This indicates that inhibition of cytoprotective autophagy can lead to an increase in efficacy of chemotherapy.
This is the first report demonstrating that high ATG7 levels are associated with reduced survival in breast cancer patients. Unbiased bioinformatic study using breast cancer databases along with immunohistochemical analysis of 142 breast cancer patients supports the reverse correlation between ATG7 expression and patient survival, suggesting that ATG7 can be used as an independent marker to predict breast cancer disease outcome. A similar association between patient survival and nuclear HSF1 expression has been shown in breast cancer patients (18). Santagata et al. show that higher nuclear HSF1 levels in breast cancer patients are associated with poor patient survival. Similarly, our unbiased bioinformatic analysis of HSF1 expression levels supports this finding. In addition, we also found a significant correlation between ATG7 and HSF1 levels in the TNBC patient samples (r = 0.461). TNBC is a more aggressive and metastatic breast cancer subtype. It is characterized by lack of progesterone receptor, estrogen receptor, and HER2/ErbB2 receptor. The absence of these receptors for targeted therapy makes TNBC difficult to treat. We observed that higher ATG7 expression is associated with poor survival in TNBC patients, suggesting that ATG7 may have a strong prognostic value in these patients. Moreover, the positive correlation observed between levels of HSF1 and ATG7 in these patients further corroborates our in vitro findings.
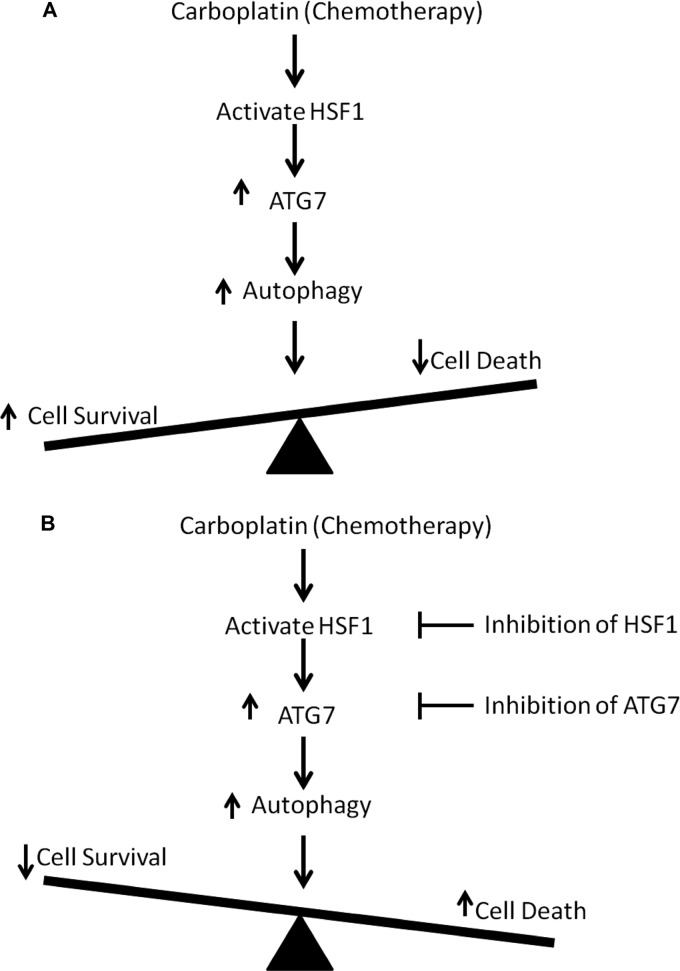
In conclusion, this is the first study to identify the critical function of HSF1 in regulating drug-mediated autophagy in breast cancer cells. We identified a novel target gene of HSF1, ATG7. We propose a mechanism of action in which stress induced by chemotherapeutic agent carboplatin activates HSF1, leading to continued expression of ATG7 and induction of autophagy (Fig. 7A). This autophagy has a cytoprotective role leading to drug resistance in the cells. Therefore, knockdown of HSF1 inhibits induction of autophagy in chemotherapy-treated breast cancer cells, increasing drug sensitivity to carboplatin in the cells. Our data from in vitro study along with the patient data analyses clearly support the rationale to design novel drugs or combination therapies to target HSF1 as well as its target gene ATG7, leading to inhibition of autophagy (Fig. 7B). These findings provide insights into novel mechanisms for HSF1-mediated chemoresistance and have significant implications in the development of strategies to overcome cancer cell resistance to chemotherapy.
FIGURE 7.
Proposed model. A, carboplatin treatment activates HSF1 (increasing trimer formation, nuclear translocation, and phosphorylation) which allows HSF1 to bind to and up-regulate transcription of ATG7 leading to an increase in autophagy. An increase in autophagy leads to an increase in cell survival and a reduction in amount of cell death. B, inhibition of HSF1 activity or reduction in ATG7 levels may lead to a reduction in autophagy and improve the cellular response to carboplatin.
Acknowledgment
We thank Dr. Richard Voellmy for providing HSF1 and HSF1-DN constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA149646 (to M. T.). This work was also supported by Radiumhospitalets Legater Award Project 334003 (to M. T. and O. F.) and by The Vincent F. Kilborn, Jr. Cancer Research Foundation (to M. T.).

This article contains supplemental Figs. S1 and S2.
- HSF1
- heat shock factor 1
- 7-AAD
- 7-aminoactinomycin D
- ATG7
- Autophagy-related Protein 7
- AVO
- acidic vesicular organelle
- cPARP
- cleaved PARP
- DN
- dominant negative
- 3-MA
- 3-methyladinine
- PARP
- poly(ADP-ribose) polymerase
- TCGA
- The Cancer Genome Atlas
- TNBC
- triple negative breast cancer.
REFERENCES
- 1. Perez E. A. (2004) Carboplatin in combination therapy for metastatic breast cancer. Oncologist 9, 518–527 [DOI] [PubMed] [Google Scholar]
- 2. Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. (2004) DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc. Natl. Acad. Sci. U.S.A. 101, 4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) ChIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Inouye S., Fujimoto M., Nakamura T., Takaki E., Hayashida N., Hai T., Nakai A. (2007) Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J. Biol. Chem. 282, 33210–33217 [DOI] [PubMed] [Google Scholar]
- 5. Lee Y. J., Kim E. H., Lee J. S., Jeoung D., Bae S., Kwon S. H., Lee Y. S. (2008) HSF1 as a mitotic regulator: phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 68, 7550–7560 [DOI] [PubMed] [Google Scholar]
- 6. Khaleque M. A., Bharti A., Gong J., Gray P. J., Sachdev V., Ciocca D. R., Stati A., Fanelli M., Calderwood S. K. (2008) Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene 27, 1886–1893 [DOI] [PubMed] [Google Scholar]
- 7. Mendillo M. L., Santagata S., Koeva M., Bell G. W., Hu R., Tamimi R. M., Fraenkel E., Ince T. A., Whitesell L., Lindquist S. (2012) HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baler R., Dahl G., Voellmy R. (1993) Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor HSF1. Mol. Cell. Biol. 13, 2486–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sarge K. D., Murphy S. P., Morimoto R. I. (1993) Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13, 1392–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akerfelt M., Morimoto R. I., Sistonen L. (2010) Heat shock factors: integrators of cell stress, development, and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christians E. S., Yan L. J., Benjamin I. J. (2002) Heat shock factor 1 and heat shock proteins: critical partners in protection against acute cell injury. Crit. Care Med. 30, S43–50 [PubMed] [Google Scholar]
- 12. Westerheide S. D., Morimoto R. I. (2005) Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 280, 33097–33100 [DOI] [PubMed] [Google Scholar]
- 13. Page T. J., Sikder D., Yang L., Pluta L., Wolfinger R. D., Kodadek T., Thomas R. S. (2006) Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol. Biosyst. 2, 627–639 [DOI] [PubMed] [Google Scholar]
- 14. Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y. H., Zhou M., Liu H., Ding Y., Khong H. T., Yu D., Fodstad O., Tan M. (2009) Up-regulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene 28, 3689–3701 [DOI] [PubMed] [Google Scholar]
- 16. Cen H., Zheng S., Fang Y. M., Tang X. P., Dong Q. (2004) Induction of HSF1 expression is associated with sporadic colorectal cancer. World J. Gastroenterol. 10, 3122–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoang A. T., Huang J., Rudra-Ganguly N., Zheng J., Powell W. C., Rabindran S. K., Wu C., Roy-Burman P. (2000) A novel association between the human heat shock transcription factor 1 (HSF1) and prostate adenocarcinoma. Am. J. Pathol. 156, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santagata S., Hu R., Lin N. U., Mendillo M. L., Collins L. C., Hankinson S. E., Schnitt S. J., Whitesell L., Tamimi R. M., Lindquist S., Ince T. A. (2011) High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 18378–18383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., Thompson C. B. (2005) Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 [DOI] [PubMed] [Google Scholar]
- 20. Karantza-Wadsworth V., Patel S., Kravchuk O., Chen G., Mathew R., Jin S., White E. (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mathew R., Karantza-Wadsworth V., White E. (2007) Role of autophagy in cancer. Nat. Rev. Cancer 7, 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yorimitsu T., Klionsky D. J. (2005) Autophagy: molecular machinery for self-eating. Cell Death Differ. 12, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 24. Vazquez-Martin A., Oliveras-Ferraros C., Menendez J. A. (2009) Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One 4, e6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abedin M. J., Wang D., McDonnell M. A., Lehmann U., Kelekar A. (2007) Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 14, 500–510 [DOI] [PubMed] [Google Scholar]
- 26. Qadir M. A., Kwok B., Dragowska W. H., To K. H., Le D., Bally M. B., Gorski S. M. (2008) Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res. Treat. 112, 389–403 [DOI] [PubMed] [Google Scholar]
- 27. Zhu K., Dunner K., Jr., McConkey D. J. (2010) Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 29, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Apel A., Herr I., Schwarz H., Rodemann H. P., Mayer A. (2008) Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy. Cancer Res. 68, 1485–1494 [DOI] [PubMed] [Google Scholar]
- 29. O'Donovan T. R., O'Sullivan G. C., McKenna S. L. (2011) Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy 7, 509–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan M., Jing T., Lan K. H., Neal C. L., Li P., Lee S., Fang D., Nagata Y., Liu J., Arlinghaus R., Hung M. C., Yu D. (2002) Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to Taxol-induced apoptosis. Mol. Cell 9, 993–1004 [DOI] [PubMed] [Google Scholar]
- 31. Jackson W. T., Giddings T. H., Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., Kirkegaard K. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desai S., Maurin M., Smith M. A., Bolick S. C., Dessureault S., Tao J., Sotomayor E., Wright K. L. (2010) PRDM1 is required for mantle cell lymphoma response to bortezomib. Mol. Cancer Res. 8, 907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shousha S. (2008) Oestrogen receptor status of breast carcinoma: Allred/H score conversion table. Histopathology 53, 346–347 [DOI] [PubMed] [Google Scholar]
- 34. Uebersax J. S. (2006) TetMat User Guide (v 1.0) Computer program documentation [Google Scholar]
- 35. Zhao Y., Liu H., Liu Z., Ding Y., Ledoux S. P., Wilson G. L., Voellmy R., Lin Y., Lin W., Nahta R., Liu B., Fodstad O., Chen J., Wu Y., Price J. E., Tan M. (2011) Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res. 71, 4585–4597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu D., Yang Y., Liu Q., Wang J. (2011) Inhibition of autophagy by 3-MA potentiates cisplatin-induced apoptosis in esophageal squamous cell carcinoma cells. Med. Oncol. 28, 105–111 [DOI] [PubMed] [Google Scholar]
- 37. Meng L., Gabai V. L., Sherman M. Y. (2010) Heat-shock transcription factor HSF1 has a critical role in human epidermal growth factor receptor-2-induced cellular transformation and tumorigenesis. Oncogene 29, 5204–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guertin M. J., Lis J. T. (2010) Chromatin landscape dictates HSF binding to target DNA elements. PLoS Genet. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eisenberg T., Knauer H., Schauer A., Büttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L., Fussi H., Deszcz L., Hartl R., Schraml E., Criollo A., Megalou E., Weiskopf D., Laun P., Heeren G., Breitenbach M., Grubeck-Loebenstein B., Herker E., Fahrenkrog B., Fröhlich K. U., Sinner F., Tavernarakis N., Minois N., Kroemer G., Madeo F. (2009) Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314 [DOI] [PubMed] [Google Scholar]
- 40. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 41. Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E. L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. (2003) Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yue Z., Jin S., Yang C., Levine A. J., Heintz N. (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paglin S., Hollister T., Delohery T., Hackett N., McMahill M., Sphicas E., Domingo D., Yahalom J. (2001) A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 61, 439–444 [PubMed] [Google Scholar]
- 45. Xi G., Hu X., Wu B., Jiang H., Young C. Y., Pang Y., Yuan H. (2011) Autophagy inhibition promotes paclitaxel-induced apoptosis in cancer cells. Cancer Lett. 307, 141–148 [DOI] [PubMed] [Google Scholar]
- 46. Lopez G., Torres K., Liu J., Hernandez B., Young E., Belousov R., Bolshakov S., Lazar A. J., Slopis J. M., McCutcheon I. E., McConkey D., Lev D. (2011) Autophagic survival in resistance to histone deacetylase inhibitors: novel strategies to treat malignant peripheral nerve sheath tumors. Cancer Res. 71, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]