Abstract
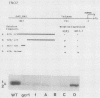
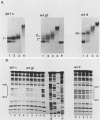
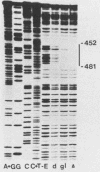
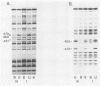
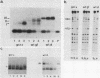
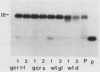
Transcription of the yeast enolase gene ENO2 is reduced 20- to 50-fold in strains carrying a null mutation in the positive regulatory gene GCR1. A small deletion mutation within one of two upstream activation sites (UAS elements) in the 5'-flanking region of ENO2 permitted wild-type levels of ENO2 gene expression in a strain carrying the gcr1 null mutation. These data show that sequences required for UAS element activity in GCR1 strains were required to repress ENO2 expression in a gcr1 strain. Protein factors that specifically bound to this UAS/repression site were identified. We show that the DNA-binding protein ABFI (autonomously replicating sequence-binding factor) is the major protein which binds the UAS/repression site. Minor DNA-binding activities that interact specifically with the UAS/repression site were also identified and may correspond to proteolytic breakdown products of ABFI. None of the observed binding activities were encoded by the GCR1 structural gene. A double-stranded oligonucleotide that included the UAS/repression site activated transcription of UAS-less ENO1 and ENO2 gene cassettes in vivo to wild-type levels in strains carrying the GCR1 allele as well as the gcr1 null mutation. These latter data show that the UAS/repression site is sufficient for transcriptional activation but is not sufficient to repress transcription of the enolase genes in a gcr1 genetic background.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker H. V. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986 Nov;6(11):3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Brindle P. K., Holland J. P., Willett C. E., Innis M. A., Holland M. J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol. 1990 Sep;10(9):4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R., Holland J. P., Yokoi T., Holland M. J. Identification of a regulatory region that mediates glucose-dependent induction of the Saccharomyces cerevisiae enolase gene ENO2. Mol Cell Biol. 1986 Jul;6(7):2287–2297. doi: 10.1128/mcb.6.7.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R., Yokoi T., Holland J. P., Pepper A. E., Holland M. J. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol. 1987 Aug;7(8):2753–2761. doi: 10.1128/mcb.7.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. J., Yokoi T., Holland J. P., Myambo K., Innis M. A. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):813–820. doi: 10.1128/mcb.7.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. G., Hershey J. W. A sensitive immunoblotting method for measuring protein synthesis initiation factor levels in lysates of Escherichia coli. J Biol Chem. 1981 Dec 25;256(24):12836–12839. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- Pavlović B., Hörz W. The chromatin structure at the promoter of a glyceraldehyde phosphate dehydrogenase gene from Saccharomyces cerevisiae reflects its functional state. Mol Cell Biol. 1988 Dec;8(12):5513–5520. doi: 10.1128/mcb.8.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweder K. S., Rhode P. R., Campbell J. L. Purification and characterization of proteins that bind to yeast ARSs. J Biol Chem. 1988 Nov 25;263(33):17270–17277. [PubMed] [Google Scholar]
- Uemura H., Jigami Y., Tanaka H., Toshimitsu N., Paterson M., Nakasato S. Nucleotide sequence of the 5' flanking region responsible for the enhancement of the expression of yeast enolase 1 gene. J Biochem. 1985 Sep;98(3):859–862. doi: 10.1093/oxfordjournals.jbchem.a135345. [DOI] [PubMed] [Google Scholar]
- Uemura H., Shiba T., Machida M., Matsui I., Jigami Y., Tanaka H. A positive regulatory sequence of the Saccharomyces cerevisiae ENO1 gene. J Biochem. 1987 Jul;102(1):181–189. doi: 10.1093/oxfordjournals.jbchem.a122031. [DOI] [PubMed] [Google Scholar]
- Uemura H., Shiba T., Paterson M., Jigami Y., Tanaka H. Identification of a sequence containing the positive regulatory region of Saccharomyces cerevisiae gene ENO1. Gene. 1986;45(1):67–75. doi: 10.1016/0378-1119(86)90133-2. [DOI] [PubMed] [Google Scholar]