Background: Apoptotic nuclear morphology can occur independently of DFF40/CAD-mediated DNA fragmentation.
Results: DFF40/CAD induces 3′-OH single-strand DNA nicks/breaks and nuclear collapse during caspase-dependent apoptosis.
Conclusion: Caspase-dependent apoptotic nuclear collapse is prompted by DFF40/CAD-mediated single-strand DNA damage.
Significance: The knowledge of how apoptotic nuclear collapse occurs should be relevant to understand the final steps of cell demise and its influence on the cellular environment.
Keywords: Apoptosis, Caspase, DNA Damage, DNase, Neuroblastoma, DFF40/CAD, Apoptotic Nuclear Morphology, Oligonucleosomal DNA Degradation, Single-strand DNA Nicks/Breaks
Abstract
Apoptotic nuclear morphology and oligonucleosomal double-strand DNA fragments (also known as DNA ladder) are considered the hallmarks of apoptotic cell death. From a classic point of view, these two processes occur concomitantly. Once activated, DNA fragmentation factor, 40-kDa subunit (DFF40)/caspase-activated DNase (CAD) endonuclease hydrolyzes the DNA into oligonucleosomal-size pieces, facilitating the chromatin package. However, the dogma that the apoptotic nuclear morphology depends on DNA fragmentation has been questioned. Here, we use different cellular models, including MEF CAD−/− cells, to unravel the mechanism by which DFF40/CAD influences chromatin condensation and nuclear collapse during apoptosis. Upon apoptotic insult, SK-N-AS cells display caspase-dependent apoptotic nuclear alterations in the absence of internucleosomal DNA degradation. The overexpression of a wild-type form of DFF40/CAD endonuclease, but not of different catalytic-null mutants, restores the cellular ability to degrade the chromatin into oligonucleosomal-length fragments. We show that apoptotic nuclear collapse requires a 3′-OH endonucleolytic activity even though the internucleosomal DNA degradation is impaired. Moreover, alkaline unwinding electrophoresis and In Situ End-Labeling (ISEL)/In Situ Nick Translation (ISNT) assays reveal that the apoptotic DNA damage observed in the DNA ladder-deficient SK-N-AS cells is characterized by the presence of single-strand nicks/breaks. Apoptotic single-strand breaks can be impaired by DFF40/CAD knockdown, abrogating nuclear collapse and disassembly. In conclusion, the highest order of chromatin compaction observed in the later steps of caspase-dependent apoptosis relies on DFF40/CAD-mediated DNA damage by generating 3′-OH ends in single-strand rather than double-strand DNA nicks/breaks.
Introduction
Apoptosis is a type of regulated cell death that results in the orderly removal of unneeded, senescent, or defective cells that are, therefore, destined to die. This programmed cell death is an evolutionarily and genetically conserved biological process that is required during embryogenesis, tissue homeostasis, development of the nervous system, and regulation of the immune system (1, 2). An imbalance in this biological process plays an important role in several human pathological situations, including cancer and degenerative neuronal diseases (3). The key mediators of the apoptotic program are a family of cysteine peptidases called caspases, which remain inactive as zymogens in healthy cells. Once activated by cleavage, caspases will process a set of intracellular proteins in which limited proteolysis will determine the apoptotic features, yielding different morphological and biochemical alterations (4, 5). These alterations, considered as the hallmarks of canonical apoptotic cell death (6), include distinctive nuclear morphological modifications and internucleosomal double-strand cleavage of genomic DNA, yielding laddered patterns of oligonucleosomal-size fragments when resolved by conventional agarose gel electrophoresis (2, 7, 8). The main nuclease responsible for genomic DNA fragmentation during caspase-dependent apoptosis is known as DNA fragmentation factor, 40-kDa subunit (DFF40)3, caspase-activated nuclease in humans, and caspase-activated DNase (CAD) in mice (9–11). DFF40/CAD is a magnesium-dependent endonuclease specific for double-stranded DNA, which generates double-strand breaks (DSBs) with 3′-hydroxyl (3′-OH) ends in vitro (12). In growing non-apoptotic cells, DFF40/CAD is complexed with its chaperone-inhibitor, ICAD (13), also known as DNA fragmentation factor, 45-kDa subunit (DFF45) (11, 14). Two alternatively spliced isoforms of ICAD have been described, the long (ICADL) and the short (ICADS) variants. During apoptosis, caspase-3 cleaves and inhibits DFF45/ICADL, allowing the release and activation of DFF40/CAD endonuclease (11, 13, 14).
Besides DNA fragmentation, the nucleus adopts characteristic traits during caspase-dependent apoptosis, those being the other hallmark of apoptotic cell death (6). These changes include chromatin condensation (nuclear collapse) and shrinkage and fragmentation of the nucleus (nuclear disassembly). These apoptotic nuclear alterations have also been divided into early stage (stage I) (peripheral chromatin condensation) and late stage (stage II) (nuclear collapse and disassembly) (15). Both stages are caspase-dependent, and stage II nuclear morphology often arises concomitantly with DFF40/CAD-mediated DNA degradation (16). Indeed, the generation of oligonucleosomal double-strand DNA fragments by DFF40/CAD has been considered to be responsible for stage II but not for stage I nuclear morphology (15). Indeed, genetically modified CAD−/− DT40 chicken cells do not reach stage II chromatin condensation after apoptotic stimuli (17). Conversely, some studies indicate that stage II chromatin condensation and the oligonucleosomal DNA degradation processes can occur separately (18–23). Therefore, how DFF40/CAD endonuclease influences stage II chromatin condensation during caspase-dependent apoptotic cell death still remains elusive.
We have recently characterized the type of cell death that SK-N-AS cells suffer after apoptotic insult. They undergo an incomplete caspase-dependent apoptosis with highly compacted chromatin in the absence of DNA laddering (22). Finding such apoptotic behavior should provide new insights on how the final apoptotic chromatin compaction takes place and whether DFF40/CAD plays a role in this process. Here we report that the specific down-regulation of DFF40/CAD in SK-N-AS cells is sufficient to avoid nuclear collapse and disassembly (stage II nuclear morphology), thus reducing the number of apoptotic nuclei after STP treatment. The analysis of the nuclei in STP-treated MEFs from CAD knockout mice corroborates the relevance of endonuclease for stage II apoptotic nuclear morphology. In addition, the enzymatic activity of DFF40/CAD is necessary to reach stage II because the overexpression of different catalytic-null mutants of murine CAD in IMR-5 cells, a ladder- and stage II-deficient cellular model, does not promote apoptotic nuclear changes after treatment with STP. By TUNEL assay we have shown that STP induces a DFF40/CAD-dependent endonuclease activity. We also demonstrate that this endonuclease is responsible for single-strand break (SSB) generation during caspase-dependent cell death. Altogether, we demonstrate that apoptotic oligonucleosomal DNA degradation and stage II nuclear morphology both depend on DFF40/CAD activation. However, although the first process requires the classical nucleolytic action described for DFF40/CAD, i.e. generation of DSBs with 3′-OH ends, the occurrence of apoptotic chromatin collapse relies on 3′-OH SSBs in the DNA.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals were obtained from Sigma-Aldrich Quimica SA (Madrid, Spain) unless indicated otherwise. The pan-caspase inhibitor N-(2-Quinolyl)valyl-aspartyl-(2,6-difluorophenoxy)methyl ketone was from MP Biomedicals Europe (Illkirch, France). Anti-actin antibody (clone E361) (catalog no. BS1002, 1:5000) was from Bioworld Technology, Inc. (St. Louis Park, MN). Antibodies against DFF40/CAD (catalog no. AB16926, 1:500) and DNA, single strand-specific (clone F7-26) (catalog no. MAB3299, 1:50) were obtained from Millipore Iberica S.A.U. (Madrid, Spain). Antibody against DFF45/ICAD (clone 6B8) (catalog no. M037-3, 1:40,000) was from MBL (Naka-ku Nagoya, Japan). Peroxidase (POD)-conjugated secondary antibodies against mouse IgG (catalog no. A9044, 1:10,000) and rabbit IgG (catalog no. A0545, 1:20,000) were purchased from Sigma. The secondary antibody Alexa Fluor 594 goat anti-mouse IgM (catalog no. A21044, 1:1000) was from Molecular Probes (Barcelona, Spain).
Cell Lines and Culture Procedures
All cell lines used in this study were routinely grown in 100-mm culture dishes (BD Falcon, Madrid, Spain) containing 10 ml of DMEM supplemented with penicillin/streptomycin (100 units/ml and 100 μg/ml, respectively) and 10% heat-inactivated FBS (Invitrogen). Medium was routinely changed every 3 days. Cells were maintained at 37 °C in a saturating humidity atmosphere containing 95% air and 5% CO2. For the different experiments, cells were grown at the adequate cell densities in culture dishes or multiwell plates (BD Falcon) using the same culture conditions as described above.
Trypan Blue Exclusion Assay and Chromatin Staining with Hoechst 33258
A trypan blue exclusion assay as well as nuclear morphology staining by Hoechst 33258 were performed as established previously (24). Normal or apoptotic cell nuclei stained with Hoechst 33258 were visualized with a Nikon ECLIPSE TE2000-E microscope equipped with epifluorescence optics under UV illumination and a Hamamatsu ORCA-ER photographic camera. The altered nuclei were counted and scored as stage I (partial chromatin condensation) or stage II (high chromatin compaction). The y-axis title “apoptotic nuclei” in the corresponding graphs refers to stage II nuclear morphology.
Low Molecular Weight DNA Degradation Analysis
The DNA fragmentation assays were carried out as described previously (22). DNA was stained in ethidium bromide and then visualized using a Syngene Gene Genius UV transilluminator equipped with a photographic camera.
Total Protein Extractions and Western Blotting
Approximately 1 × 106 cells/condition were detached from the 35-mm culture dishes, pelleted at 600 × g for 5 min, and washed once with PBS. Then, cells were lysed with 100 μl of SET buffer (10 mm Tris-HCl (pH 6.8), 150 mm NaCl, 1 mm EDTA, 1% SDS) and heated at 95 °C for 10 min to get total protein extracts. The protein concentration in the supernatants was quantified by a modified Lowry assay (DC protein assay, Bio-Rad), and 15–30 μg of protein was loaded in SDS-polyacrylamide gels. The proteins were electrophoresed and electrotransferred onto a protran nitrocellulose transfer membrane (Whatman GmbH, Dassel, Germany). After blocking with TBS/0.1% Tween 20 containing 5% nonfat dry milk, the membranes were probed with the appropriate specific primary antibodies and incubated with the adequate secondary antibodies conjugated with horseradish peroxidase. Finally, immunoblots were developed with an EZ-ECL chemiluminescence detection kit (Biological Industries, Kibbutz Beit-Haemek, Israel).
Subcloning and Stable Transfection of mCAD Mutants
The pBluescript vectors carrying the open reading frames of mouse CAD (mCAD), the wild type (WT) form or different catalytically inactive mutants (H242A, H263A and H313A) were provided by Dr. Shigekazu Nagata (Kyoto University, Kyoto, Japan). (25). The mCAD H242A mutant abrogates Zn2+ incorporation, avoiding CAD dimerization and DNA binding (25). The mCAD H263A mutant lacks endonuclease activity because His-263 is the general base of the active site (26). The mCAD H313A mutant impedes Mg2+ binding, avoiding transition state stabilization without affecting CAD dimerization or DNA binding (27).
The cDNA inserts described above were subcloned into the pcDNA3 mammalian expression vector (Invitrogen). The constructs were named pcDNA3/mCAD WT, pcDNA3/mCAD H242A, pcDNA3/mCAD H263A, and pcDNA3/mCAD H313A. Cells were transfected with the different constructs or pcDNA3 vector alone (Neo) using Lipofectamine transfection reagent (Invitrogen). Stably transfected cells were selected with 500 μg/ml G-418 (geneticin) (Invitrogen) and were used as a pool. The cDNA of mCAD WT was also subcloned into the pAG3 mammalian expression vector (provided by Dr. Carles Saura, Universitat Autònoma de Barcelona, Spain). The construct was named pAG3/mCAD WT. MEF CAD−/− cells were transfected with the pAG3 empty vector alone (Zeo) or the pAG3/mCAD WT construct by using Attractene transfection reagent (Qiagen) according to the instructions of the manufacturer. Stably transfected MEF cells were selected for a month in the presence of 0.4 mg/ml Zeocin (Invitrogen) and were used as a pool. The expression of mCAD was assessed by Western blot analysis in total protein extracts.
Design and Transfection of siRNAs
siRNA transfection was performed in SK-N-AS and SH-SY5Y cells using DharmaFECT 1 siRNA transfection reagent (Fisher Scientific, Madrid, Spain) in DMEM without serum and antibiotics according to the instructions of the manufacturer. Briefly, the day before transfection, 3 × 105 SK-N-AS cells (2 × 105 SH-SY5Y cells) were seeded in 60-mm culture dishes in DMEM with 10% FBS but without antibiotics. The final concentration of the selected siRNA (see below) or negative control (NR siRNA) was 140 nm, and 10 μl of Dharmafect 1 reagent was employed for each transfection. After 5 h of transfection, serum was added to a final concentration of 10%. Four days after transfection, cells were detached from culture dishes, reseeded in adequate plates, and treated the next day with STP. To evaluate the extent of DFF40/CAD, ICADL, or ICADS protein down-regulation, Western blot analyses of total protein lysates were performed 4 days after transfection. The sequences of the different siRNAs employed were as follows: 5′-GAGCUACAGAGGAGGACAU-3′ (human ICADS (hICADS) siRNA-1), 5′-UGUCAAUGUCACAGGAAAA-3′ (hICADS siRNA-2), 5′-GUGUAAAGAGGUAACAUAA-3′ (hICADS siRNA-3), 5′-CAGGAUUUGGAGUUGGUUA-3′ (hICADL siRNA), 5′-CCAGAGGGCUUGAGGACAU-3′ (CAD siRNA-1), 5′-GGAACAAGAUGGAAGAGAA-3′ (CAD siRNA-2), 5′-GGCUAAUGUUUGUAUUUUU-3′ (CAD siRNA-3), 5′-CCACCUUGCUUGAGGGACA-3′ (CAD siRNA-4), 5′-GAUUCAACCUGAUACAUUU-3′ (CAD siRNA-5), and 5′-GUAAGACACGACUUAUCGC-3′ (NR siRNA (28)).
TUNEL Assay
The TUNEL assays were performed as described previously with some modifications (29). Briefly, SH-SY5Y, SK-N-AS, and MEF CAD−/− cells were seeded onto 8-wells Lab-Tek chamber slides (Nalge Nunc International Corp., Rochester, NY) and treated with STP as indicated in the respective figure legends. Detection of DNA fragments carrying 3′-OH groups was carried out by fixing cells in freshly prepared 2% paraformaldehyde for 30 min at 4 °C and, after washing with PBS, fixed cells were permeabilized with 0.1% Triton X-100 and 0.1% sodium citrate for 90 min at 4 °C. Then, cells were rinsed with PBS containing 0.1% Triton X-100 and incubated with 100 μl of a reaction mixture containing 0.025 nmol fluorescein-12-dUTP, 0.25 nmol dATP, 2.5 mm CoCl2, 40 units of recombinant terminal deoxynucleotidyl transferase and 1X terminal deoxynucleotidyl transferase reaction buffer (Roche Applied Science) for 1 h at 37 °C. The reaction was ended by adding 20 mm EGTA. Cells were washed twice with PBS, counterstained with 0.05 μg/ml Hoechst 33258 in 20 mm EGTA, and microphotographs were taken under fluorescein-isothiocyanate or UV filters in an epifluorescence microscope (Nikon ECLIPSE TE2000-E) coupled to a Hamamatsu ORCA-ER camera.
Alkaline Agarose Gel Electrophoresis for SSB Detection
For alkaline agarose gel analysis, SH-SY5Y and SK-N-AS cells were seeded in 12 multiwell plates, and after 12 h of treatment with 1 μm STP, DNA from cells was extracted according to the low molecular weight DNA degradation analysis method. DNA was resuspended in 50 μl of 1 mm EDTA (pH 8.0) and 20 μg/ml DNase-free RNase A and heated at 50 °C for 20 min. Alkaline electrophoresis was carried out as described previously with minor modifications (30). Briefly, a 10-cm 0.9% agarose gel, prepared in 50 mm NaCl and 1 mm EDTA, was equilibrated for > 1 h at room temperature in freshly prepared alkaline gel running buffer (50 mm NaOH, 1 mm EDTA (pH 8.0)). Prior to loading, 10 μl of DNA were mixed with 2 μl of 6× alkaline gel loading buffer (300 mm NaOH, 6 mm EDTA, 60% glycerol). Gels were run at 20 V (2 V/cm) for 10–12 h at room temperature. After electrophoresis, the gel surface was rinsed in distilled water and incubated in neutralizing solution (0.1 m Tris-HCl (pH 8.0)) for 20 min. Neutralization continued for 40 min with fresh 0.1 m Tris-HCl (pH 8.0). Then, the gel was stained in ethidium bromide (1 μg/ml) for 15 min, rinsed twice with distilled water for 10 min each, and DNA was visualized using an UV transilluminator.
Single-strand DNA Damage Measurements with F7-26 Antibody by Flow Cytometry
For ssDNA damage measurements, cells were cultured in 35-mm culture dishes (1 × 106 cells/condition) in the presence or absence of STP as indicated in the respective figure legends. Cells were then fixed in ice-cold methanol-PBS (6:1) (−20 °C) and analyzed 24 h later. Fixed cells were centrifuged at 400 × g for 5 min, and the pellet obtained was resuspended in 0.25 ml of formamide and heated in a water bath at 75 °C for 10 min. The cells were then returned to room temperature for 5 min and incubated with 2 ml of 1% nonfat dry milk in PBS for 15 min, also at room temperature. Then, cells were centrifuged at 400 × g for 5 min, and the pellet was resuspended in 100 μl anti-ssDNA F7-26 antibody (1:10) diluted in PBS plus 5% FBS and incubated for 25 min at room temperature. 2 μl of Alexa Fluor 594-conjugated goat anti-mouse IgM (μ chain) antibody (1:50) (Invitrogen) was added, and the cell suspension was further incubated for 25 min, also at room temperature. Then, 900 μl of PBS was added to each tube, and cells were analyzed by flow cytometry. Flow cytometry analyses were conducted using a Cytomics FC 500 (Beckman Coulter). Red fluorescence in labeled and non-labeled cells was determined by excitation at 488 nm and emission at 620 nm (FL3 detector). Data were acquired and analyzed with CXP software. During the analyses, 10,000 events (cells) were analyzed. Density plots representing size (FCS, y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale) were plotted. Cell density (events) was shown on a pseudocolor scale from minimum density (blue) to maximum density (red).
Transmission Electron Microscopy
Transmission electron microscopy was used to examine the nuclear morphology from SK-N-AS cells transfected with NR or CAD siRNA and left untreated or treated with STP for 6 h. Pellets of cells from all conditions were fixed with 2.5% (v/v) glutaraldehyde and 2% (v/v) paraformaldehyde (EM grade, TAAB Laboratories, Berkshire, UK) in 100 mm PBS (pH 7.4) for 2 h and rinsed four times with 100 mm PBS. Pellets were then post-fixed in 1% (w/v) osmium tetroxide (TAAB Laboratories) containing 0.8% (w/v) potassium hexacyanoferrate (III) (Sigma) for 2 h and washed with 100 mm PBS. These steps were performed at 4 °C. Samples were dehydrated through a graded acetone series, embedded in Epon resin, and polymerized for 48 h at 60 °C. Ultrathin sections were mounted in copper grids (200 mesh), contrasted with uranyl acetate and lead citrate solutions, and evaluated with a transmission electron microscope (Jeol JEM-1400 equipped with a Gatan Ultrascan ES1000 charge-coupled device (CCD) camera).
Statistical Analysis
All experiments were repeated at least three times. Values are expressed as means ± S.E. Student's t-tests were used to determine the statistical significance in cell death measurements. p ≤ 0.01 was considered to be significant.
RESULTS
DFF40/CAD Is Required for Stage II Nuclear Morphology Even in the Absence of Oligonucleosomal DNA Fragmentation
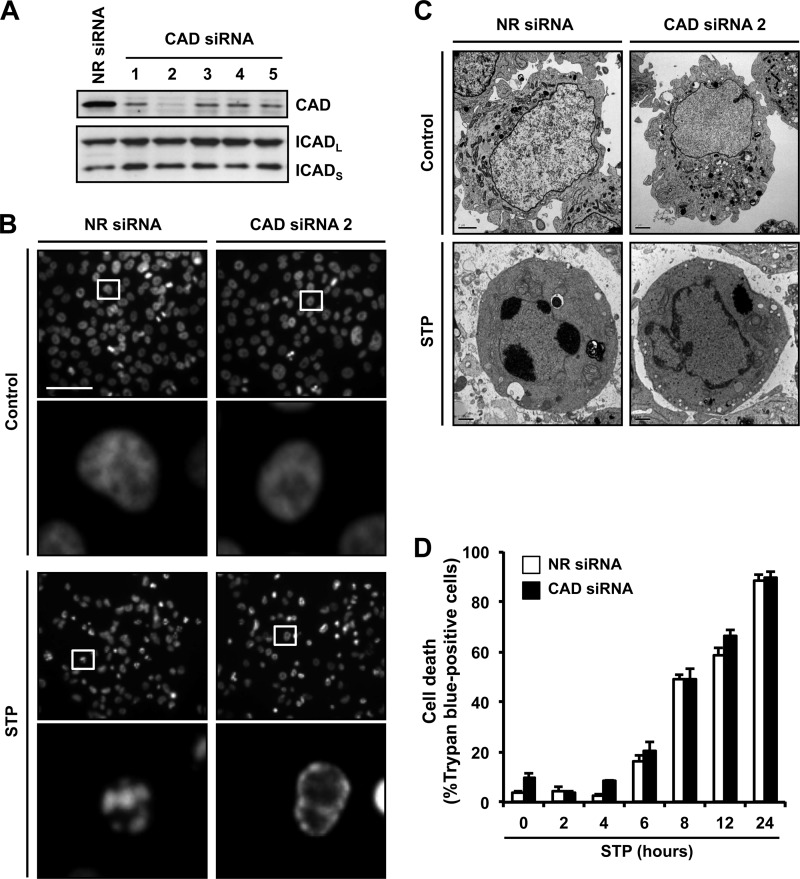
We have recently identified SK-N-AS cells, a human neuroblastoma-derived cell line, as a cellular model that undergoes incomplete apoptotic cell death after cytotoxic stimuli, characterized by the presence of apoptotic nuclei in the absence of DNA laddering (22). First, we established that the aspect of these nuclei depends on the activation of caspases (supplemental Fig. S1). Therefore, we wanted to ascertain whether DFF40/CAD played a role in the observed apoptotic nuclear morphology. For this, we designed five siRNAs against human DFF40/CAD mRNA (see “Experimental Procedures”). As shown in Fig. 1A, all siRNAs down-regulated the protein levels of the endonuclease without affecting the expression of either the long (ICADL) or the short (ICADS) isoforms of ICAD. Among the siRNAs tested, number 2 was the most effective at decreasing DFF40/CAD protein levels (Fig. 1A). When apoptotic nuclear morphology induced by STP was analyzed by Hoechst 33258 staining, we observed that the specific down-regulation of DFF40/CAD prevented nuclear collapse/disassembly (stage II nuclear morphology) but not chromatin condensation against the nuclear periphery (stage I) (Fig. 1B). Ultrastructurally, the cells transfected with an NR siRNA displayed aggregates of highly condensed granular chromatin, frequently associated with the appearance of dense semilunar caps after STP treatment (Fig. 1C). However, in CAD siRNA-transfected cells, the chromatin appeared marginal and homogenously distributed beneath the nuclear envelope, forming irregular and interconnected masses of heterochromatin in a coarse pattern (Fig. 1C). Irrespective of the morphological features observed, the profile of cell death was comparable between both transfected cells, establishing that the effect of DFF40/CAD down-regulation on the nuclear morphology was not due to a reduction in the indices of cell death (Fig. 1D).
FIGURE 1.
The apoptotic nuclear collapse and disassembly induced by STP in SK-N-AS cells requires the expression of DFF40/CAD endonuclease. A, SK-N-AS cells were transfected with an NR siRNA or five different CAD siRNAs (1–5). Total protein extracts of the different conditions were obtained, and DFF40/CAD protein levels (upper panel) were analyzed by Western blot analyses 5 days after transfection. The membrane was reprobed with anti-ICAD antibody (lower panel). B, SK-N-AS cells transfected with NR siRNA or CAD siRNA number 2 were treated with 1 μm STP for 6 h or left untreated (Control), and nuclear morphology was analyzed by staining the nuclei with Hoechst 33258. Scale bar = 40 μm. In each condition of treatment, the lower panels are magnifications of the insets in the upper panels. C, electron microscopy from SK-N-AS cells transfected and treated as in B. Scale bars = 40 μm. D, time course analysis of cell death by trypan blue exclusion assay. SK-N-AS cells transfected with NR (white bars) or DFF40/CAD (black bars) siRNA were treated with 1 μm STP.
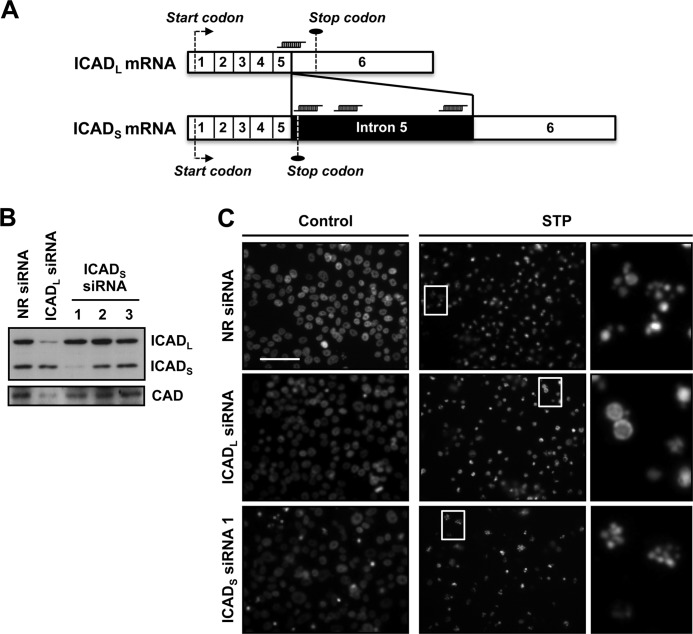
Apoptotic nuclear collapse is impaired in cells overexpressing several caspase-resistant ICAD mutants (31, 32). Therefore, we wanted to analyze the individual contribution of the ICADL and ICADS isoforms on STP-induced apoptotic nuclear morphology in SK-N-AS cells. Fig. 2A schematizes the ICADL and ICADS mRNAs and the position of each siRNAs employed. As shown in Fig. 2B, the siRNA against ICADL was effective at reducing its protein levels without affecting those of ICADS. On the other hand, among the three siRNAs chosen to down-regulate ICADS, only siRNA number 1 was effective at reducing the protein levels of this isoform without altering ICADL expression (Fig. 2B). Analysis of the nuclear morphology revealed that ICADL siRNA, but not ICADS siRNA, prevented stage II nuclear collapse in SK-N-AS cells treated with STP (Fig. 2C). The effect of ICADL siRNA on the apoptotic nuclear morphology is due to the decrease observed in the endogenous protein levels of DFF40/CAD (Fig. 2B). This fact is in agreement with previous data establishing the role of ICADL, but not ICADS, as a chaperone for DFF40/CAD (33) and, therefore, regulating the expression of the endonuclease (34). Regardless of the effect provoked on the aspect of the nuclei, none of the siRNAs designed avoided the STP-induced DNA degradation profile into high molecular weight (HMW) fragments (supplemental Fig. S2).
FIGURE 2.
Knockdown of ICADL, but not of ICADS, impedes stage II apoptotic nuclear morphology in SK-N-AS cells treated with STP. A, schematic illustrating ICADL and ICADS mRNAs showing the position against which the different siRNAs were designed. The numbers pictured in each mRNA (1-6, empty segments) represent the exons. The intron 5 (filled segment) is only preserved in ICADS mRNA. The specific siRNAs designed against ICADS are complementary to intron 5, whereas the ICADL siRNA is complementary to the exon-5/exon-6 junction. The start and stop codons are indicated in both mRNAs. B, SK-N-AS cells were transfected with NR siRNA, ICADL siRNA, or three different ICADS siRNAs (1-3). Total protein extracts of the different conditions were obtained, and ICADL and ICADS protein levels (upper panel) were analyzed by Western blot analysis 5 days after transfection. The membrane was reprobed with anti-DFF40/CAD antibody (lower panel). C, SK-N-AS cells transfected with NR siRNA, ICADL siRNA, or ICADS siRNA number 1 were treated with 1 μm STP for 24 h or left untreated (Control), and nuclear morphology was analyzed by staining the nuclei with Hoechst 33258. The right panels are magnifications of the insets in the center panels. Scale bars = 40 μm.
DFF40/CAD Catalytic Activity Is Required to Promote Apoptotic Nuclear Morphology
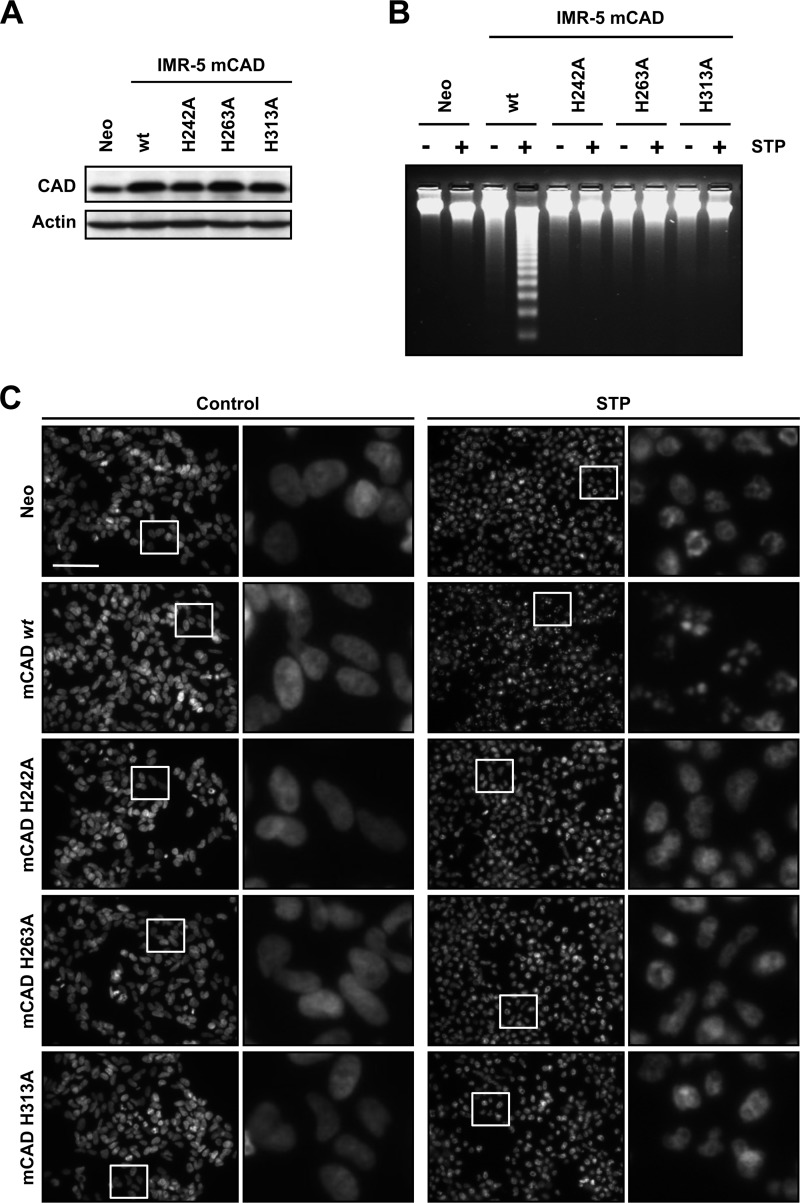
At this point, we demonstrated that SK-N-AS cells required DFF40/CAD to reach a high order of chromatin compaction during caspase-dependent apoptosis. Because apoptotic DNA degradation is a consequence of the endonucleolytic action of DFF40/CAD, we wanted to assess whether DFF40/CAD also required its enzymatic activity to promote stage II nuclear morphology after STP treatment. For this reason, we overexpressed several catalytically inactive mutants of mCAD (25) in human neuroblastoma-derived IMR-5 cells, a cellular model that, upon STP treatment, displays an incomplete apoptotic cell death characterized by the absence of both stage II nuclear morphology and oligonucleosomal DNA degradation (35). Therefore, we proceeded with the stable overexpression of mCAD H242A, H263A, or H313A mutants as well as the pcDNA3-empty vector (Neo) or wild-type mCAD in IMR-5 cells. As depicted in Fig. 3A, mCAD or the different mutants were efficiently overexpressed. As expected, only IMR-5 cells that overexpress the wild-type form of mCAD were able to degrade their chromatin into oligonucleosomal DNA fragments after treatment with STP (Fig. 3B). As shown in Fig. 3C, the analysis of the nuclear morphology revealed that only IMR-5 cells overexpressing the wild-type form of the endonuclease recovered their ability to display stage II chromatin condensation when challenged with STP. These results indicate that nuclear collapse and disassembly observed through apoptotic cell death, were strictly dependent on the enzymatic activity of mCAD. Therefore, the hydrolytic action of DFF40/CAD on chromatin is required to provoke both DNA laddering and nuclear morphological changes after apoptotic insult.
FIGURE 3.
The endonuclease activity of DFF40/CAD is necessary for apoptotic nuclear collapse and disassembly. IMR-5 cells stably transfected with empty pcDNA3 (Neo), pcDNA3-mCAD wild-type (mCAD WT), pcDNA3-mCADH242A (mCAD H242A), pcDNA3-mCADH263A (mCAD H263A), or pcDNA3-mCADH313A (mCAD H313A) plasmids were treated with 1 μm STP for 24 h or left untreated. A, total protein extracts from the different cell lines generated were analyzed by Western blot analysis to confirm the overexpression of the different mCAD variants. The membrane was reprobed with an antibody against actin as a loading control. B, genomic DNA analysis of the different stably transfected cell lines treated with STP (+) or left untreated (−). C, morphological aspect of the nuclei of the different cell lines stained with Hoechst 33258 after STP treatment or no treatment (Control). In each condition, the right panels are magnifications of the insets in the left panels. Scale bar = 40 μm.
Staurosporine Provokes DNA Damage in SK-N-AS Cells by Introducing Single-strand DNA Breaks with free 3′-OH Ends
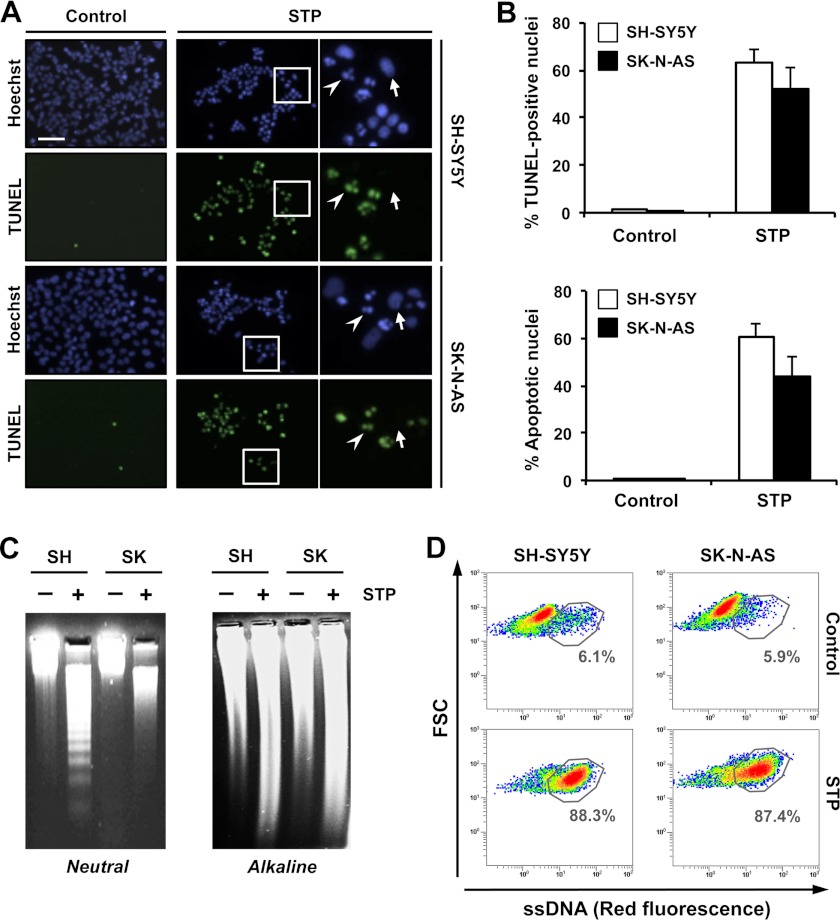
Taking into account the results obtained, and because the endonuclease activity of DFF40/CAD generates free 3′-OH group DNA ends (12), we wanted to explore whether the apoptotic nuclear morphology observed in SK-N-AS cells after STP treatment correlated with the generation of 3′-OH DNA ends. For this, we took advantage of the TUNEL assay, which allows the detection of free 3′-OH termini by labeling with modified nucleotides, e.g. FITC-dUTP. Interestingly, as shown in Fig. 4A, TUNEL reactivity was detected in both SK-N-AS and SH-SY5Y cells treated with STP for 12 h. Moreover, the percentage of TUNEL-positive nuclei was comparable between both cell lines, a result that directly correlated with the number of cells displaying stage II apoptotic nuclear morphology (Fig. 4B). Subsequently, we wanted to determine the DNA damage occurrence in SK-N-AS cells challenged with STP. Isolated DNA from untreated or STP-treated cells was electrophoresed under alkaline/denaturing conditions to unwind strand breaks or labile sites present in double-stranded DNA, yielding ssDNA (36). As shown in Fig. 4C, genomic DNA isolated from STP-treated SK-N-AS or SH-SY5Y cells migrated faster on alkaline agarose gels than their respective untreated controls, evidencing that, regardless of the extent of DSBs, the alkaloid provoked DNA damage. Although SSBs revealed by the alkaline unwinding gel electrophoresis assay are usually interpreted as an evidence of DNA damage, we took advantage of the F7-26 monoclonal antibody, which does not recognize DNA in double-stranded conformations but specifically reveals exogenously induced ssDNA damage (37). Flow cytometry analysis demonstrated that after STP treatment, either SK-N-AS or SH-SY5Y cells showed a new cell population with higher reactivity to the F7-26 antibody, indicating the presence of ssDNA stretches (Fig. 4D). The generation of ssDNA nicks/breaks with 3′-OH ends was evidenced in SK-N-AS and SH-SY5Y cells by employing in vitro polymerization assays using either DNA polymerase I or its (large) Klenow fragment (supplemental Figs. S3 and S4).
FIGURE 4.
STP induces endonuclease activity in SK-N-AS cells that is evidenced by the generation of SSBs and free 3′-OH ends in the DNA. A, TUNEL reactivity and Hoechst nuclear staining in SH-SY5Y or SK-N-AS cells treated (STP) or not treated (Control) with 1 μm STP for 12 h. The right panels are magnifications of the insets in the center panels. The arrows show non-apoptotic nuclei that are negative for TUNEL reactivity. The arrowheads indicate apoptotic and TUNEL-positive nuclei. Scale bar = 40 μm. B, quantification of the data presented in A representing TUNEL-positive nuclei (upper panel) and apoptotic nuclei (lower panel) in SH-SY5Y (white bars) and SK-N-AS (black bars) cells upon STP treatment. The means ± S.E. of three independent experiments are shown. C, SH-SY5Y (SH) and SK-N-AS (SK) cells were left untreated (-) or treated (+) with 1 μm STP for 12 h. Genomic DNA was extracted and analyzed by neutral conventional (left panel) or alkaline (right panel) agarose gel electrophoresis and subsequent ethidium bromide staining (see “Experimental Procedures”). D, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26 in SK-N-AS and SH-SY5Y cells left untreated (Control) or treated with 1 μm STP for 12 h and fixed immediately (see “Experimental Procedures”). Flow cytometry data are shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 5%).
DNA SSBs Produced during Apoptotic Cell Death Correlate with a Proper Condensation of the Chromatin, Leading to Nuclear Collapse
With the aim to establish a correlation between the occurrence of DNA damage by SSBs and the presence of apoptotic collapsed nuclei, we took advantage of the intrinsic properties of IMR-5 cells (35). SK-N-AS and IMR-5 cells were cultured in the presence of STP for 6 h, and ssDNA damage was assessed by flow cytometry analysis using the F7-26 monoclonal antibody. As depicted in Fig. 5A, the presence of STP in the culture medium provoked a minor percentage of IMR-5 cells to display ssDNA damage when compared with SK-N-AS cells. These results correlated with the limited number of TUNEL-positive and apoptotic nuclei observed in STP-treated IMR-5 cells compared with those obtained in STP-treated SK-N-AS cells (Fig. 5, B and C). IMR-5 cells, which fail to activate DFF40/CAD at the final steps of caspase-dependent cell death (35), displayed stage II nuclear morphology and degradation of their chromatin into oligonucleosomal-size pieces after apoptotic insult only when DFF40/CAD wild-type was overexpressed (Fig. 3). As shown in Fig. 6A, the overexpression of mCAD provoked a marked increase in the number of cells displaying ssDNA damage after STP treatment when compared with empty-pcDNA3 IMR-5 (IMR-5 Neo) cells. These results directly correlated with the data obtained by the TUNEL assay. In this sense, the number of TUNEL-positive nuclei increased in IMR-5 mCAD cells compared with IMR-5 Neo cells (Fig. 6, B and C). As observed in Fig. 6C, the percentage of TUNEL-positive nuclei in both IMR-5-transfected cells correlated directly with the percentage of apoptotic nuclei afforded upon STP treatment (Fig. 6, B and C). Therefore, the results obtained support the concept that DFF40/CAD activity might be necessary to induce DNA damage during caspase-dependent cell death.
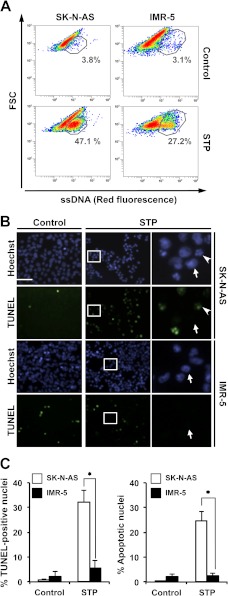
FIGURE 5.
The absence of stage II nuclear morphology in injured IMR-5 cells correlates with reduced DNA damage in 3′-OH SSBs. SK-N-AS and IMR-5 cells were left untreated (Control) or treated with 1 μm STP for 6 h. A, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26. Data are shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 10%). B, TUNEL reactivity and Hoechst nuclear staining in SK-N-AS or IMR-5 cells untreated (Control) or treated with STP. The right panels are magnifications of the insets in the center panels. The arrows show stage I nuclei negative for TUNEL reactivity. The arrowheads indicate representative apoptotic stage II and TUNEL-positive nuclei. Scale bar = 40 μm. C, quantification of the data presented in B representing TUNEL-positive nuclei (left panel) and apoptotic nuclei (right panel) in SK-N-AS (white bars) and IMR-5 (black bars) cells upon STP treatment. The means ± S.E. of three independent experiments are represented. Asterisks indicate p ≤ 0.01.
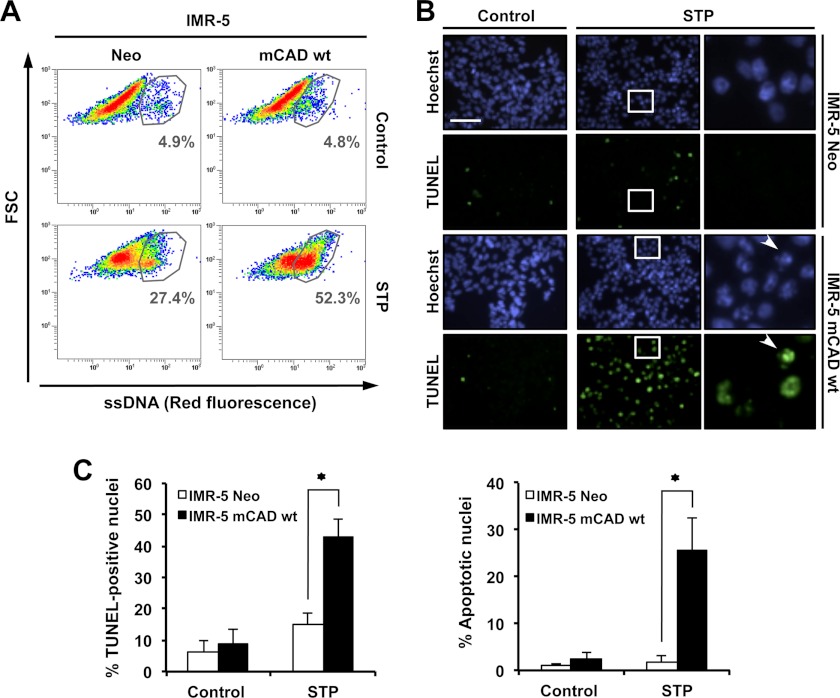
FIGURE 6.
The apoptotic nuclear morphology observed in STP-treated IMR-5 cells overexpressing mCAD correlates with a higher percentage of cells carrying 3′-OH SSBs and DNA damage. IMR-5 cells stably transfected with empty pcDNA3 (Neo) or pcDNA3-mCAD wild-type (mCAD WT) were left untreated (Control) or treated with 1 μm STP for 4 h. A, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26. Data are shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 10%). B, TUNEL reactivity and Hoechst nuclear staining of IMR-5 Neo or IMR-5 mCAD WT cells left untreated (Control) or treated with STP. The right panels are magnifications of the insets in the center panels. The arrowheads indicate representative apoptotic stage II and TUNEL-positive nuclei. Scale bar = 40 μm. C, quantification of the data presented in B representing TUNEL-positive nuclei (left panel) and apoptotic nuclei (right panel) in IMR-5 Neo (white bars) or IMR-5 mCAD WT (black bars) cells upon STP treatment. The means ± S.E. of three independent experiments are represented. Asterisks indicate p ≤ 0.01.
DFF40/CAD Is the Main Endonuclease Responsible for Single-strand DNA Damage during Apoptosis
The results obtained indicated that apoptotic nuclear changes strongly correlated with the generation of DNA SSBs carrying 3′-OH-free ends. Moreover, the results shown in Fig. 6, A and B, strongly suggest that DFF40/CAD could be the main endonuclease implicated in the DNA damage process observed. First, we sought to analyze whether the overexpression of mCAD (Fig. 7A) could increase the percentage of SK-N-AS cells generating ssDNA damage after STP treatment. Although DFF40/CAD-overexpressed SK-N-AS cells digested their chromatin into oligonucleosomal double-stranded fragments (Fig. 7B), the number of cells carrying ssDNA damage did not increase when compared with empty-pcDNA3-transfected (Neo) cells (C). Moreover, neither the number of TUNEL-positive nor of apoptotic nuclei was altered by the overexpression of endonuclease (Fig. 7, D and E). Altogether, these results pointed out that both 3′-OH DNA breaks and stage II nuclear morphology observed during caspase-dependent apoptosis relied on DNA SSB generation rather than on DSB oligonucleosomal DNA fragmentation. Moreover, as shown in Fig. 7, the overexpression of mCAD in SK-N-AS cells failed to demonstrate a direct implication of endonuclease on apoptotic ssDNA damage. Consequently, to confirm whether DFF40/CAD played a role in the generation of SSBs during STP-induced DNA damage, we proceeded with its specific knockdown in SK-N-AS cells (Fig. 8A). As shown in Fig. 8B, the ssDNA damage induced by STP in SK-N-AS cells transfected with an NR siRNA was almost completely abolished in cells in which DFF40/CAD protein levels were previously down-regulated (CAD siRNA) (Fig. 8A). Accordingly, with this result, the generation of 3′-OH ends in DFF40/CAD knockdown cells was also impeded (Fig. 8, C and D). As expected, and contrary to NR siRNA-transfected SK-N-AS cells, most of CAD siRNA-transfected cells displayed stage I instead of stage II nuclear morphology after STP treatment (Fig. 8, C and E). The same results were obtained in CAD siRNA-transfected and STP-treated SH-SY5Y cells (supplemental Fig. S5).
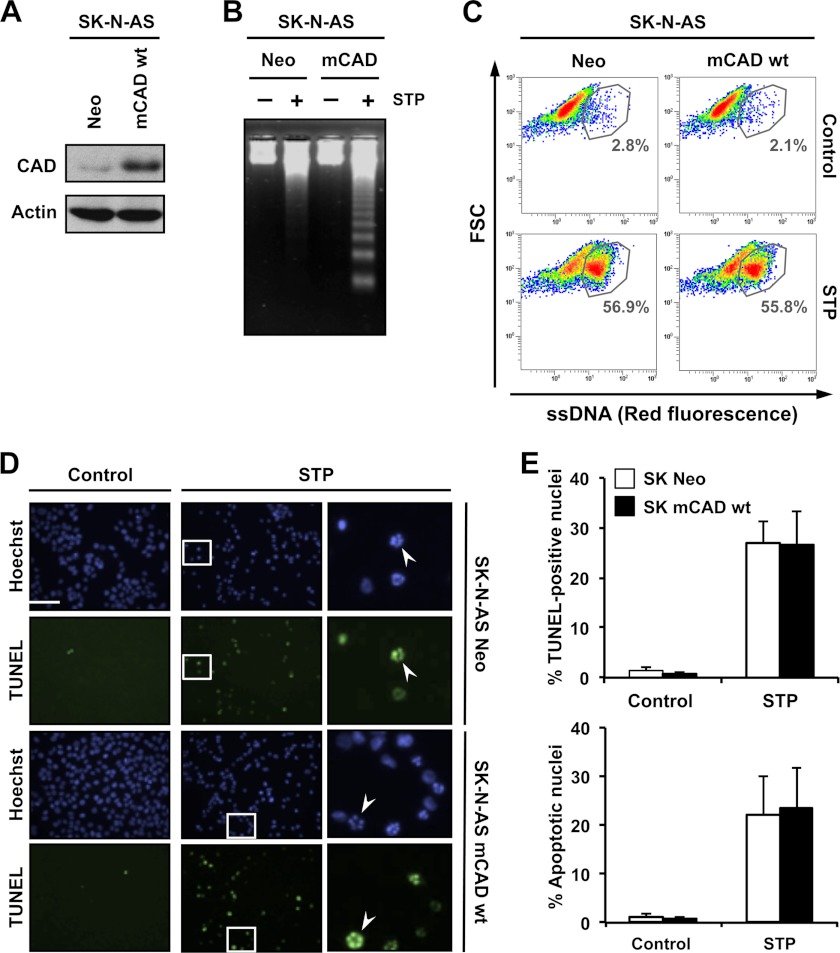
FIGURE 7.
mCAD overexpression in SK-N-AS cells does not alter the percentage of STP-treated cells displaying ssDNA damage even though oligonucleosomal DNA degradation is observed. SK-N-AS cells stably transfected with empty pcDNA3 (Neo) or pcDNA3-mCAD wild-type (mCAD WT) were left untreated (Control) or treated with 1 μm STP for 4 h. A, total protein extracts from untreated cells were analyzed by Western blot analysis to confirm the overexpression of mCAD. The membrane was reprobed with an antibody against actin as a loading control. B, genomic DNA analysis of both stably transfected cell lines after treatment with STP (+) or no treatment (−). C, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26. Data is shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 5%). D, TUNEL reactivity and Hoechst nuclear staining from control and STP-treated cells were assessed. Representative images are indicated. The right panels are magnifications of the insets in the center panels. The arrowheads indicate apoptotic nuclei, which are also positive for TUNEL reactivity. Scale bar = 40 μm. E, quantification of TUNEL-positive nuclei (upper panel) and apoptotic nuclei (lower panel) in SK-N-AS Neo (white bars) and SK-N-AS mCAD WT (black bars) cells from D. The means ± S.E. of three independent experiments are represented.
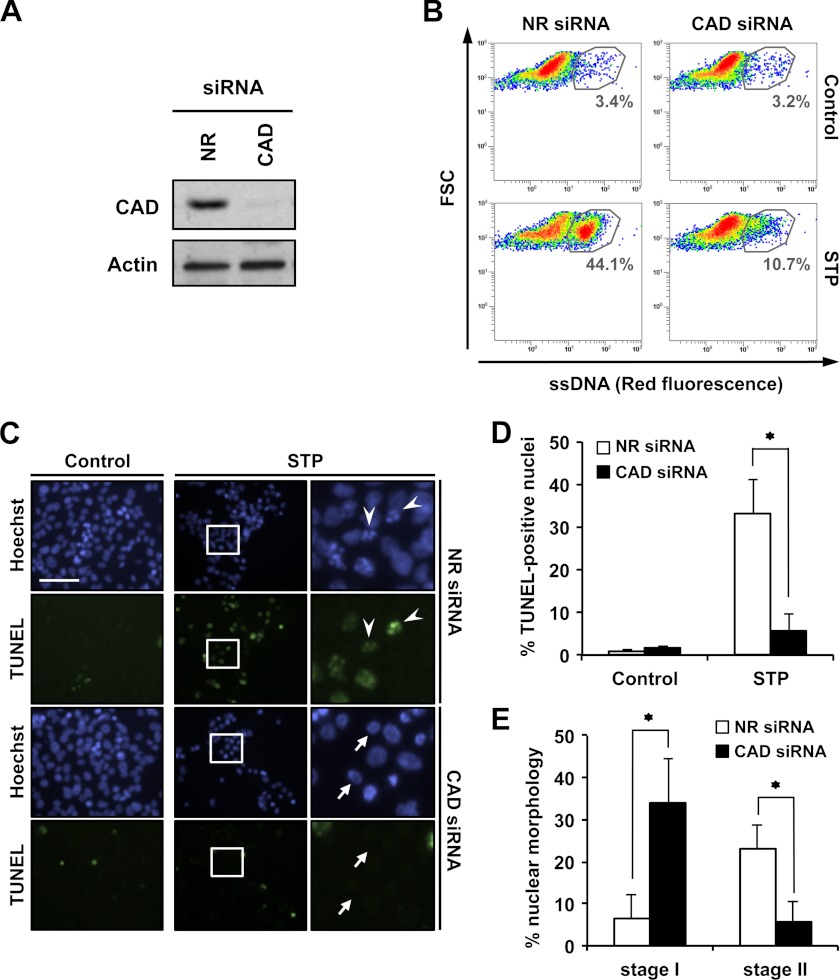
FIGURE 8.
DFF40/CAD is a key effector for 3′-OH SSBs DNA damage occurrence and nuclear collapse during apoptosis. SK-N-AS cells were transfected with NR siRNA or CAD siRNA (see “Experimental Procedures”). Then, cells were left untreated (Control) or treated with 1 μm STP for 4 h. A, total protein extracts from untreated cells were analyzed by Western blot analysis to confirm the down-regulation of DFF40/CAD. The membrane was reprobed with an antibody against actin as a loading control. B, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26. Data are shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 5%). C, TUNEL reactivity and Hoechst nuclear staining were performed. The right panels are magnifications of the insets in the center panels. The arrowheads indicate apoptotic nuclei that are positive for TUNEL. The arrows show stage I apoptotic nuclei that are negative for TUNEL reactivity. Scale bar = 40 μm. D, quantification of TUNEL-positive nuclei in SK-N-AS NR siRNA (white bars) and SK-N-AS CAD siRNA (black bars) cells treated with STP or left untreated (Control). E, stage I and stage II nuclear morphology of untreated (Control) or STP-treated SK-N-AS NR siRNA (white bars) or SK-N-AS CAD siRNA (black bars) cells were quantified. In D and E, the means ± S.E. of three independent experiments are shown. Asterisks indicate p ≤ 0.01.
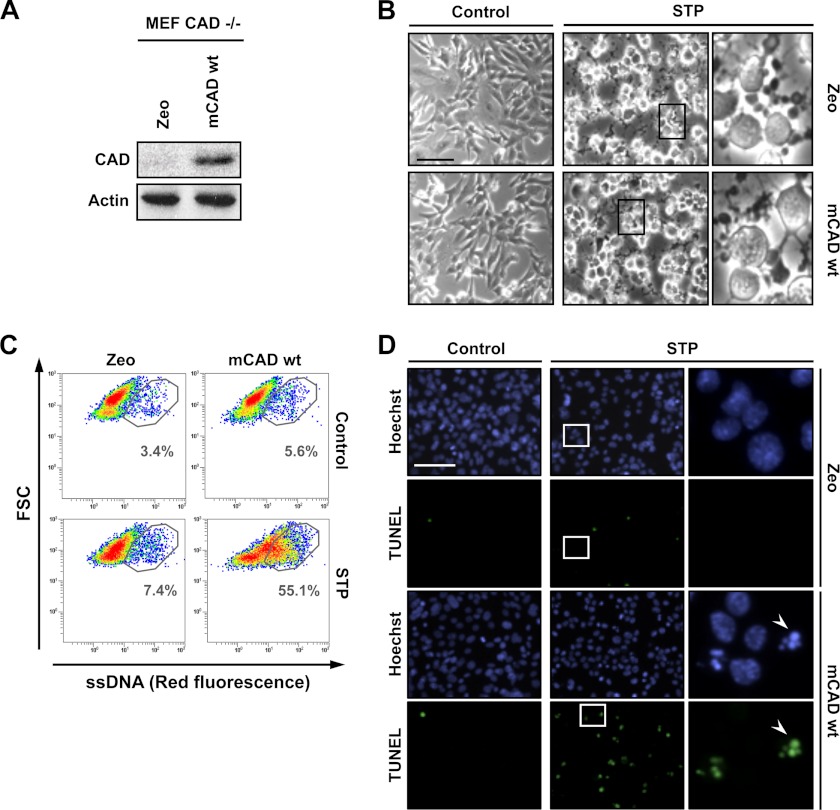
To expand the results obtained we used MEF cells from CAD knockout mice. For this purpose, we proceeded to stably transfect the cells with the pAG3-mCAD or pAG3-empty (Zeo) vectors. The expression of mCAD was assessed by Western blot analysis in total protein extracts (Fig. 9A). As shown in Fig. 9B, mCAD and Zeo cells underwent comparable morphological changes upon treatment with 1 μm STP for 10 h. The analysis of ssDNA damage by flow cytometry revealed that STP did not induce ssDNA breaks in cells lacking DFF40/CAD expression (Zeo cells) (Fig. 9C). This result correlated with a minimal detection of free 3′-OH DNA ends and the absence of chromatin collapse in MEF CAD−/− Zeo cells challenged with STP (Fig. 9D). On the other hand, upon STP insult, the overexpression of mCAD made MEF CAD−/− cells competent to generate ssDNA damage (Fig. 9C) and to display stage II nuclear morphology with free 3′-OH DNA ends (D).
FIGURE 9.
MEF CAD−/− cells develop the ability to display 3′-OH ssDNA damage and stage II apoptotic nuclear morphology after STP treatment when mCAD is overexpressed. A–D, MEF CAD−/− cells stably transfected with empty pAG3 (Zeo) or pAG3-mCAD wild-type (mCAD WT) were left untreated (Control) or treated with 1 μm STP for 10 h. A, total protein extracts from untreated cells were analyzed by Western blot analysis to confirm the overexpression of mCAD WT. The membrane was reprobed with an antibody against actin as a loading control. B, microphotographs using a phase contrast microscopy showing the morphological appearance of MEF CAD−/− Zeo or mCAD WT cells after the indicated treatment. The right panels are magnifications of the insets in the center panels. C, flow cytometry analysis of ssDNA damage using the monoclonal antibody F7-26. The data are shown as density plots representing size (y axis, in a logarithm scale) versus intensity of red fluorescence (x axis, in a logarithm scale). FSC, forward scatter of light. Cell density (events) is shown on a pseudocolor scale from minimum density (blue) to maximum density (red). The circled populations correspond to cells presenting ssDNA damage. The percentage of gated cells in each condition is indicated. The experiment was repeated three times with a low variability (< 5%). D, TUNEL reactivity and Hoechst nuclear staining were performed. The right panels are magnifications of the insets in the center panels. The arrowheads indicate an apoptotic nucleus that is positive for TUNEL. Scale bars = 40 μm.
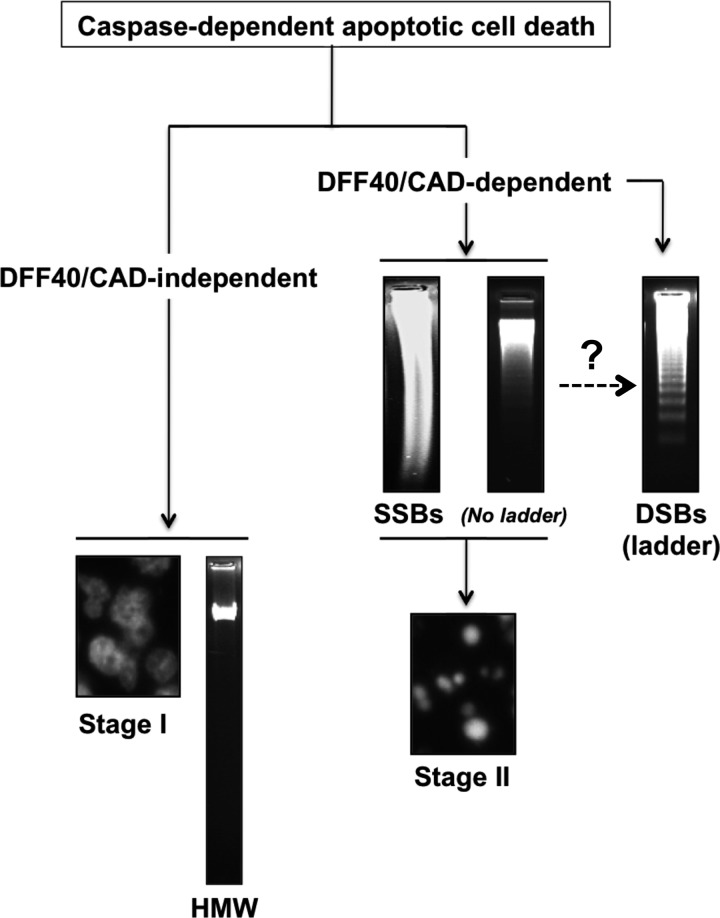
The data presented indicate that the DNA damage observed during caspase-dependent apoptosis is mainly due to the activation of DFF40/CAD endonuclease, which introduces 3′-OH SSBs into the DNA facilitating nuclear collapse (stage II). A major degree of DFF40/CAD activity should lead to a more advanced step in the apoptotic DNA fragmentation, i.e. DSBs into oligonucleosomal DNA fragments or DNA laddering. On the other hand, DFF40/CAD seems to not be involved in peripheral chromatin condensation (stage I) or HMW DNA degradation, at least in our human cellular models (Fig. 10). Of note, it has been reported that HMW DNA degradation does not take place in cells from CAD knockout mice (38). This difference could be explained by an exclusive role of DFF40/CAD during embryogenesis, being the sole endonuclease able to carry out HMW DNA degradation. This function seems to be replaced by other non-characterized nuclease/s in malignant (adult) cells. In any case, our data point to a non-redundant cellular function of DFF40/CAD for the generation of 3′-OH SSBs, chromatin compaction, and LMW DNA degradation during caspase-dependent apoptotic cell death, at least under our experimental conditions.
FIGURE 10.
Proposed model of DFF40/CAD involvement in both apoptotic hallmarks during caspase-dependent cell death in tumor-derived cells. After apoptotic insult, DFF40/CAD is released from its inhibitor ICAD and, therefore, activated. On one hand, the endonuclease will induce SSBs, promoting nuclear collapse/disassembly (stage II). On the other hand, possibly in a second step, DFF40/CAD will degrade the chromatin into oligonucleosomal-size DSBs, generating the DNA ladder. The stage I apoptotic nuclear morphology and HMW DNA fragmentation are DFF40/CAD-independent processes.
DISCUSSION
Here, we have taken advantage of the responsiveness of MEF CAD−/− cells and three human-derived neuroblastoma cells toward the same apoptotic stimulus to unravel the mechanism by which DFF40/CAD influences the nuclear morphological changes observed throughout caspase-dependent apoptotic cell death. We have previously characterized SH-SY5Y cells that undergo canonical apoptotic cell death after different external insults (39–41). On the other hand, IMR-5 cells display caspase-dependent cell death in the absence of oligonucleosomal DNA degradation and stage II nuclear morphology, only showing marginal chromatin condensation inside of the nucleus (stage I) after apoptotic stimuli (32, 35, 39). More recently, we have identified SK-N-AS cells as an interesting cellular model to elucidate how stage II apoptotic nuclear morphology takes place. Indeed, these cells hold a defect in the hydrolysis of their chromatin into oligonucleosomal-size fragments, albeit the presence of stage II apoptotic nuclei upon cytotoxic insult (22). We show here that DFF40/CAD knockdown in SK-N-AS cells completely avoids caspase-dependent nuclear collapse and disassembly without affecting the initial ring-like marginalization of the chromatin inside the nucleus (stage I). Accordingly, MEF CAD−/− cells only display stage I nuclear morphology upon apoptotic stimulus, and the overexpression of a wild-type form of DFF40/CAD restores stage II. In fact, apoptotic nuclear collapse/disassembly requires a fully functional wild-type form of DFF40/CAD because the overexpression of different catalytic-null mutants in IMR-5 cells do not promote stage II nor DNA laddering after apoptotic insult. Hence, caspase-dependent stage II apoptotic nuclear morphology needs the catalytic activity of DFF40/CAD.
The nuclear changes observed during apoptosis have attracted many researchers in this field. As a consequence, besides DFF40/CAD, other factors coupling to stage II chromatin condensation have been identified, e.g. the caspase-generated N-terminal fragment of gelsolin (42) and acinus (43) and the release of nuclear actin (44). However, the exact contribution of each factor still remains controversial. The different stimuli employed, the metabolic state of each individual in a population of cells, or even the particular subcellular contexts could be determinants for the player/s that will trigger chromatin condensation upon cytotoxic insult. However, we can also speculate that the abovementioned molecules (and probably others that remain to be identified) could be parts of intracellular pathways that are somehow interconnected. In this context, a correlation between DFF40/CAD activity and other caspase-dependent stage II-mediating factors, such as acinus, gelsolin, or even nuclear actin, should be plausible. Therefore, it would be of outstanding interest for the field to know whether there exists any molecular relationship between DFF40/CAD and the abovementioned condensing factors to mediate stage II chromatin compaction.
Although the presence of multiple single-strand cuts in the double helix of DNA of apoptotic cells has been observed previously (45), the nuclease implicated and, more importantly, its biological significance, remained largely unsolved. During STP-induced apoptotic cell death, SK-N-AS and SH-SY5Y cells undergo a process of DNA hydrolysis characterized by the presence of SSBs with 3′-OH termini. The fact that SK-N-AS cells are defective in degrading their chromatin into oligonucleosomal-length DSBs fragments but effective in generating stage II chromatin condensation after caspase activation demonstrates that stage II nuclear morphology can take place independently of internucleosomal DNA degradation. This fact is further supported by the specific down-regulation of DFF40/CAD in SK-N-AS cells that precludes the occurrence of SSBs with 3′-OH DNA ends. The absence of 3′-OH DNA SSBs avoids the progression toward chromatin collapse of the nuclei during apoptotic cell death. Therefore, the results presented here point to DFF40/CAD endonuclease as the key molecule controlling stage II nuclear morphology by introducing single-strand cuts in the DNA of preapoptotic cells. The observations reported here are in agreement with the finding that DFF40/CAD can indeed introduce single-strand nicks in DNA in vitro (46).
How the endonuclease independently controls both apoptotic hallmarks should be linked to the degree of its activation/activity or its protein levels. The degree of DFF40/CAD activation is directly related to the proteolytic action of caspases, and its enzymatic activity is further regulated by other factors, including histone H1 (11, 47), HMG-1 (11), CIIA (48), or nucleophosmin/B23 (49). On the other hand, we have recently described that SK-N-AS cells show higher DFF40/CAD protein amounts than IMR-5 cells but less than SH-SY5Y cells (22). Therefore, it should be plausible that stage II nuclear morphology (mediated by the induction of ssDNA nicks/breaks) or DNA laddering require different amounts of DFF40/CAD protein for it to take place. In this regard, cells with a high protein content of DFF40/CAD (e.g. SH-SY5Y cells) display ssDNA nicks/breaks, stage II nuclear morphology, and DNA laddering. Cells showing a medium content of the endonuclease (e.g. SK-N-AS cells) are able to generate ssDNA nicks/breaks and chromatin collapse but not DNA laddering. Finally, cells containing low levels of DFF40/CAD (e.g. IMR-5 cells) do not display ssDNA nicks/breaks, stage II nuclear morphology, or DNA laddering.
However, another interesting aspect is the subcellular localization of this endonuclease. In this regard, we also described that apoptotic oligonucleosomal DNA degradation relies on a cytosolic, rather than a nucleoplasmic, pool of DFF40/CAD (22). Although the DFF40/CAD cytosolic pool is highly reduced in SK-N-AS cells when compared with SH-SY5Y cells (22), it might still be enough to mediate ssDNA nicks/breaks and, subsequently, the topological changes in the chromatin of apoptotic SK-N-AS cells. However, the amount of DFF40/CAD in the cytosolic fraction of IMR-5 cells is similar to that observed in SK-N-AS cells (22). Therefore, an interesting question arises from these observations regarding the potential role of the nucleoplasmic pool of DFF40/CAD. In this regard, the SK-N-AS cells present nucleoplasmic amounts of DFF40/CAD comparable with that observed in SH-SY5Y cells (22). Interestingly, DFF40/CAD is not detected in the nucleoplasmic fraction of healthy IMR-5 cells.4 Therefore, the nucleoplasmic pool of DFF40/CAD could also play a role in the generation of the apoptotic hallmarks, at least for the highly chromatin compaction occurrence induced by the generation of ssDNA nicks/breaks. Another interesting open question is whether the DNA laddering comes from previous DFF40/CAD-mediated ssDNA breaks. In this sense, Widlak and Garrard (46) reported that, depending on the in vitro conditions employed, DFF40/CAD may also cleave each strand of DNA stepwise. Therefore, the results presented here support the notion that DFF40/CAD acts similarly in cellula, first by introducing single-strand nicks/breaks and later by cleaving the adjacent second strand, yielding laddered patterns of oligonucleosomal-size fragments (see Fig. 10).
The presence of single-strand (transient) DNA lesions that permit the rearrangement of genetic material has also been observed during cell differentiation (50–53). More recently, the SSBs generated during terminal differentiation of skeletal muscle cells have been attributed to DFF40/CAD (54, 55). These DNA strand breaks commonly accumulate in specific DNA sequences placed at the nuclear matrix attachment regions (56). Interestingly, during apoptosis, and once activated by caspases, DFF40/CAD also associates with matrix attachment regions (57), promoting the detachment of interchromatin granule clusters via cleavage of susceptible A/T-rich matrix attachment regions of chromatin (58). Thus, DFF40/CAD nucleolytic action should apparently take place in matrix attachment regions, either in cell differentiation or apoptotic cell death processes. Therefore, the role of DFF40/CAD in apoptosis or differentiation should rely directly on its catalytic function. Although DNA damage observed during cell differentiation is a reversible controlled process (59), we show here that SSBs generated during apoptosis persist over time. The transient nature of the SSBs generated during cytodifferentiation probably relies on an efficient DNA repair system (reviewed in Refs. 56 and 60). In contrast, many key components of the DNA repair machinery are processed and inhibited during apoptosis by the proteolytic action of caspases (reviewed in Ref. 61). Alternatively, DFF40/CAD could be regulated by the negative (21, 48, 49, 62–64) or positive (65) modulators explained above to reach the required enzymatic activity for cell differentiation or apoptotic cell death. Besides, the amount of DFF40/CAD bound to the chromatin should also be crucial for the generation of SSBs or DSBs upon apoptotic insult. In this sense, we have recently demonstrated that DFF40/CAD binds to the chromatin in both SK-N-AS and SH-SY5Y cells after apoptotic stimuli. However, a minor amount of the endonuclease associated with the chromatin in SK-N-AS cells leads to a defect in oligonucleosomal DSBs generation upon apoptotic challenge (22). Nevertheless, we demonstrate here that this amount of chromatin-associated DFF40/CAD is enough to generate a percentage of cells displaying DNA SSBs comparable with that observed in SH-SY5Y cells.
From a more functional point of view, although caspase-mediated activation of DFF40/CAD plays an active and crucial role during skeletal muscle differentiation (54), the endonuclease seems relegated to a more passive function during apoptosis. Indeed, it is well known that DFF40/CAD action is dispensable for caspase-dependent cell death (38, 66), whereas DFF40/CAD-mediated apoptotic DNA degradation has been classically considered as the point of no return of apoptosis. Taking into account the role of DFF40/CAD in cell differentiation, and on theoretical grounds, we could envisage a more active role of this endonuclease during apoptotic cell death, mainly when macromolecular synthesis is involved (7, 67). Indeed, because many of the proteins implicated in macromolecular synthesis are neither cleaved nor inhibited during apoptosis (61), it seems reasonable to think that the DNA stretches in SK-N-AS cells highlighted by DNA polymerase I could also serve as templates for non-inhibited endogenous polymerases, at the beginning of the apoptotic process. Irrespective of these theoretical considerations, the detailed study of the molecular mechanisms controlling nuclear collapse/disassembly during caspase-dependent apoptosis should be of outstanding interest to comprehend how chromatin is being compacted inside the nucleus of a cell committed to die. The physiopathological relevance relies on the fact that accurate packaging of the DNA (fragmented or not) would potentially minimize the spreading of harmful genes, such as oncogenes or viral DNA, through horizontal gene transfer (68).
Acknowledgments
We thank Dr. Shigekazu Nagata from the Department of Medical Chemistry, Graduate School of Medicine, Kyoto University, Kyoto, Japan, for the MEF CAD−/− cells and the pBluescript vectors carrying mCAD or the different mutants employed in this study. We also thank Dr. Carles Saura from Institut de Neurociències, Facultat de Medicina, Universitat Autònoma de Barcelona, Barcelona, Spain, for the pAG3 plasmid; the personnel of Servei de Microscopia from Universitat Autonoma de Barcelona; A. Sanchez-Chardi; and Francisca Cardoso for technical support. We also thank Dr. José R. Bayascas and Dr. Néstor Gómez for many discussions and the other members from both laboratories for helpful criticism.
This work was supported by Ministerio de Ciencia e Innovación/Fondo Europeo de Desarrollo Regional (FEDER) Grants SAF2011-24081 and SAF2012-31485 (to V. J. Y.) and SAF2010-19953 (to J. X. C.), by Centro de Investigacion Biomedica en Red sobre Enfermedades Neurodegenerativas (CIBERNED) Grant CB06/05/0042, and by Generalitat de Catalunya Grant SGR2009-346. This work was also supported by a fellowship from Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR, Generalitat de Catalunya) (to V. I. G.), by Postdoctoral Contract TRA2009-0185 from the Ministerio de Ciencia e Innovación (to E. G. G.), by Formación de Personal Investigador (FPI) Fellowship BES-2099-028572 from the Ministerio de Ciencia e Innovación (to M. G. B.), and by the Ramón y Cajal Program (Spanish Ministerio de Educación y Ciencia) (to V. J. Y.).

This article contains supplemental Figs. S1–S5 and Experimental Procedures.
V. Iglesias-Guimarais and V. J. Yuste, unpublished results.
- DFF40
- DNA fragmentation factor, 40-kDa subunit
- CAD
- caspase-activated DNase
- DSB
- double-strand break
- ICAD
- inhibitor of caspase-activated DNase
- ICADL
- inhibitor of caspase-activated DNase, long variant
- ICADS
- inhibitor of caspase-activated DNase, short variant
- STP
- staurosporine
- MEF
- mouse embryonic fibroblasts
- SSB
- single-strand break
- mCAD
- mouse caspase-activated DNase
- NR
- non-relevant
- ssDNA
- single-stranded DNA
- HMW
- high molecular weight.
REFERENCES
- 1. Jacobson M. D., Weil M., Raff M. C. (1997) Programmed cell death in animal development. Cell 88, 347–354 [DOI] [PubMed] [Google Scholar]
- 2. Hipfner D. R., Cohen S. M. (2004) Connecting proliferation and apoptosis in development and disease. Nat. Rev. Mol. Cell Biol. 5, 805–815 [DOI] [PubMed] [Google Scholar]
- 3. Thompson C. B. (1995) Apoptosis in the pathogenesis and treatment of disease. Science 267, 1456–1462 [DOI] [PubMed] [Google Scholar]
- 4. Timmer J. C., Salvesen G. S. (2007) Caspase substrates. Cell Death Differ. 14, 66–72 [DOI] [PubMed] [Google Scholar]
- 5. Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wlodkowic D., Telford W., Skommer J., Darzynkiewicz Z. (2011) Apoptosis and beyond. Cytometry in studies of programmed cell death. Methods Cell Biol. 103, 55–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wyllie A. H., Morris R. G., Smith A. L., Dunlop D. (1984) Chromatin cleavage in apoptosis. Association with condensed chromatin morphology and dependence on macromolecular synthesis. J. Pathol. 142, 67–77 [DOI] [PubMed] [Google Scholar]
- 8. Zamzami N., Kroemer G. (1999) Condensed matter in cell death. Nature 401, 127–128 [DOI] [PubMed] [Google Scholar]
- 9. Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. (1998) A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature 391, 43–50 [DOI] [PubMed] [Google Scholar]
- 10. Halenbeck R., MacDonald H., Roulston A., Chen T. T., Conroy L., Williams L. T. (1998) CPAN, a human nuclease regulated by the caspase-sensitive inhibitor DFF45. Curr. Biol. 8, 537–540 [DOI] [PubMed] [Google Scholar]
- 11. Liu X., Li P., Widlak P., Zou H., Luo X., Garrard W. T., Wang X. (1998) The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. U.S.A. 95, 8461–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Widlak P., Li P., Wang X., Garrard W. T. (2000) Cleavage preferences of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease) on naked DNA and chromatin substrates. J. Biol. Chem. 275, 8226–8232 [DOI] [PubMed] [Google Scholar]
- 13. Sakahira H., Enari M., Nagata S. (1998) Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis. Nature 391, 96–99 [DOI] [PubMed] [Google Scholar]
- 14. Liu X., Zou H., Slaughter C., Wang X. (1997) DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell 89, 175–184 [DOI] [PubMed] [Google Scholar]
- 15. Susin S. A., Daugas E., Ravagnan L., Samejima K., Zamzami N., Loeffler M., Costantini P., Ferri K. F., Irinopoulou T., Prévost M. C., Brothers G., Mak T. W., Penninger J., Earnshaw W. C., Kroemer G. (2000) Two distinct pathways leading to nuclear apoptosis. J. Exp. Med. 192, 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bortner C. D., Oldenburg N. B., Cidlowski J. A. (1995) The role of DNA fragmentation in apoptosis. Trends Cell Biol. 5, 21–26 [DOI] [PubMed] [Google Scholar]
- 17. Samejima K., Tone S., Earnshaw W. C. (2001) CAD/DFF40 nuclease is dispensable for high molecular weight DNA cleavage and stage I chromatin condensation in apoptosis. J. Biol. Chem. 276, 45427–45432 [DOI] [PubMed] [Google Scholar]
- 18. Fournel S., Genestier L., Rouault J. P., Lizard G., Flacher M., Assossou O., Revillard J. P. (1995) Apoptosis without decrease of cell DNA content. FEBS Lett. 367, 188–192 [DOI] [PubMed] [Google Scholar]
- 19. Oberhammer F., Wilson J. W., Dive C., Morris I. D., Hickman J. A., Wakeling A. E., Walker P. R., Sikorska M. (1993) Apoptotic death in epithelial cells. Cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 12, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamaguchi K., Uzzo R., Dulin N., Finke J. H., Kolenko V. (2004) Renal carcinoma cells undergo apoptosis without oligonucleosomal DNA fragmentation. Biochem. Biophys. Res. Commun. 318, 710–713 [DOI] [PubMed] [Google Scholar]
- 21. Li X., Wang J., Manley J. L. (2005) Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 19, 2705–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iglesias-Guimarais V., Gil-Guiñon E., Gabernet G., García-Belinchón M., Sánchez-Osuna M., Casanelles E., Comella J. X., Yuste V. J. (2012) Apoptotic DNA degradation into oligonucleosomal fragments, but not apoptotic nuclear morphology, relies on a cytosolic pool of DFF40/CAD endonuclease. J. Biol. Chem. 287, 7766–7779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takahashi M., Ozaki T., Takahashi A., Miyauchi M., Ono S., Takada N., Koda T., Todo S., Kamijo T., Nakagawara A. (2007) DFF45/ICAD restores cisplatin-induced nuclear fragmentation but not DNA cleavage in DFF45-deficient neuroblastoma cells. Oncogene 26, 5669–5673 [DOI] [PubMed] [Google Scholar]
- 24. Bayascas J. R., Yuste V. J., Solé C., Sánchez-López I., Segura M. F., Perera R., Comella J. X. (2004) Characterization of splice variants of human caspase-activated DNase with CIDE-N structure and function. FEBS Lett. 566, 234–240 [DOI] [PubMed] [Google Scholar]
- 25. Sakahira H., Takemura Y., Nagata S. (2001) Enzymatic active site of caspase-activated DNase (CAD) and its inhibition by inhibitor of CAD. Arch. Biochem. Biophys. 388, 91–99 [DOI] [PubMed] [Google Scholar]
- 26. Korn C., Scholz S. R., Gimadutdinow O., Pingoud A., Meiss G. (2002) Involvement of conserved histidine, lysine and tyrosine residues in the mechanism of DNA cleavage by the caspase-3 activated DNase CAD. Nucleic Acids Res. 30, 1325–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Woo E. J., Kim Y. G., Kim M. S., Han W. D., Shin S., Robinson H., Park S. Y., Oh B. H. (2004) Structural mechanism for inactivation and activation of CAD/DFF40 in the apoptotic pathway. Mol. Cell 14, 531–539 [DOI] [PubMed] [Google Scholar]
- 28. Sancho P., Fabregat I. (2010) NADPH oxidase NOX1 controls autocrine growth of liver tumor cells through up-regulation of the epidermal growth factor receptor pathway. J. Biol. Chem. 285, 24815–24824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bayascas J. R., Yuste V. J., Benito E., Garcia-Fernàndez J., Comella J. X. (2002) Isolation of AmphiCASP-3/7, an ancestral caspase from amphioxus (Branchiostoma floridae). Evolutionary considerations for vertebrate caspases. Cell Death Differ. 9, 1078–1089 [DOI] [PubMed] [Google Scholar]
- 30. Chalmers C. R., Fenwick S. W., Toogood G. J., Hull M. A. (2006) Immunohistochemical measurement of endothelial cell apoptosis and proliferation in formalin-fixed, paraffin-embedded human cancer tissue. Angiogenesis 9, 193–200 [DOI] [PubMed] [Google Scholar]
- 31. Sakahira H., Enari M., Ohsawa Y., Uchiyama Y., Nagata S. (1999) Apoptotic nuclear morphological change without DNA fragmentation. Curr. Biol. 9, 543–546 [DOI] [PubMed] [Google Scholar]
- 32. Yuste V. J., Sánchez-López I., Solé C., Moubarak R. S., Bayascas J. R., Dolcet X., Encinas M., Susin S. A., Comella J. X. (2005) The contribution of apoptosis-inducing factor, caspase-activated DNase, and inhibitor of caspase-activated DNase to the nuclear phenotype and DNA degradation during apoptosis. J. Biol. Chem. 280, 35670–35683 [DOI] [PubMed] [Google Scholar]
- 33. Ageichik A. V., Samejima K., Kaufmann S. H., Earnshaw W. C. (2007) Genetic analysis of the short splice variant of the inhibitor of caspase-activated DNase (ICAD-S) in chicken DT40 cells. J. Biol. Chem. 282, 27374–27382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagase H., Fukuyama H., Tanaka M., Kawane K., Nagata S. (2003) Mutually regulated expression of caspase-activated DNase and its inhibitor for apoptotic DNA fragmentation. Cell Death Differ. 10, 142–143 [DOI] [PubMed] [Google Scholar]
- 35. Yuste V. J., Bayascas J. R., Llecha N., Sánchez-López I., Boix J., Comella J. X. (2001) The absence of oligonucleosomal DNA fragmentation during apoptosis of IMR-5 neuroblastoma cells. Disappearance of the caspase-activated DNase. J. Biol. Chem. 276, 22323–22331 [DOI] [PubMed] [Google Scholar]
- 36. Fornace A. J., Jr., Dobson P. P., Kinsella T. J. (1986) Analysis of the effect of DNA alkylation on alkaline elution. Carcinogenesis 7, 927–932 [DOI] [PubMed] [Google Scholar]
- 37. Frankfurt O. S. (1987) Detection of DNA damage in individual cells by flow cytometric analysis using anti-DNA monoclonal antibody. Exp. Cell Res. 170, 369–380 [DOI] [PubMed] [Google Scholar]
- 38. Kawane K., Fukuyama H., Yoshida H., Nagase H., Ohsawa Y., Uchiyama Y., Okada K., Iida T., Nagata S. (2003) Impaired thymic development in mouse embryos deficient in apoptotic DNA degradation. Nat. Immunol. 4, 138–144 [DOI] [PubMed] [Google Scholar]
- 39. Boix J., Llecha N., Yuste V. J., Comella J. X. (1997) Characterization of the cell death process induced by staurosporine in human neuroblastoma cell lines. Neuropharmacology 36, 811–821 [DOI] [PubMed] [Google Scholar]
- 40. Ribas J., Yuste V. J., Garrofé-Ochoa X., Meijer L., Esquerda J. E., Boix J. (2008) 7-Bromoindirubin-3′-oxime uncovers a serine protease-mediated paradigm of necrotic cell death. Biochem. Pharmacol. 76, 39–52 [DOI] [PubMed] [Google Scholar]
- 41. Perez-Alvarez S., Iglesias-Guimarais V., Solesio M. E., Melero-Fernandez de Mera R. M., Yuste V. J., Galindo M. F., Jordán J. (2011) Methadone induces CAD degradation and AIF-mediated necrotic-like cell death in neuroblastoma cells. Pharmacol. Res. 63, 352–360 [DOI] [PubMed] [Google Scholar]
- 42. Kothakota S., Azuma T., Reinhard C., Klippel A., Tang J., Chu K., McGarry T. J., Kirschner M. W., Koths K., Kwiatkowski D. J., Williams L. T. (1997) Caspase-3-generated fragment of gelsolin. Effector of morphological change in apoptosis. Science 278, 294–298 [DOI] [PubMed] [Google Scholar]
- 43. Sahara S., Aoto M., Eguchi Y., Imamoto N., Yoneda Y., Tsujimoto Y. (1999) Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature 401, 168–173 [DOI] [PubMed] [Google Scholar]
- 44. Widlak P., Palyvoda O., Kumala S., Garrard W. T. (2002) Modeling apoptotic chromatin condensation in normal cell nuclei. Requirement for intranuclear mobility and actin involvement. J. Biol. Chem. 277, 21683–21690 [DOI] [PubMed] [Google Scholar]
- 45. Peitsch M. C., Müller C., Tschopp J. (1993) DNA fragmentation during apoptosis is caused by frequent single-strand cuts. Nucleic Acids Res. 21, 4206–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Widlak P., Garrard W. T. (2001) Ionic and cofactor requirements for the activity of the apoptotic endonuclease DFF40/CAD. Mol. Cell Biochem. 218, 125–130 [DOI] [PubMed] [Google Scholar]
- 47. Liu X., Zou H., Widlak P., Garrard W., Wang X. (1999) Activation of the apoptotic endonuclease DFF40 (caspase-activated DNase or nuclease). Oligomerization and direct interaction with histone H1. J. Biol. Chem. 274, 13836–13840 [DOI] [PubMed] [Google Scholar]
- 48. Cho S. G., Kim J. W., Lee Y. H., Hwang H. S., Kim M. S., Ryoo K., Kim M. J., Noh K. T., Kim E. K., Cho J. H., Yoon K. W., Cho E. G., Park H. S., Chi S. W., Lee M. J., Kang S. S., Ichijo H., Choi E. J. (2003) Identification of a novel antiapoptotic protein that antagonizes ASK1 and CAD activities. J. Cell Biol. 163, 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahn J. Y., Liu X., Cheng D., Peng J., Chan P. K., Wade P. A., Ye K. (2005) Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol. Cell 18, 435–445 [DOI] [PubMed] [Google Scholar]
- 50. Modak S. P., Bollum F. J. (1972) Detection and measurement of single-strand breaks in nuclear DNA in fixed lens sections. Exp. Cell Res. 75, 307–313 [DOI] [PubMed] [Google Scholar]
- 51. Williams G. T., Johnstone A. P. (1983) ADP-ribosyl transferase, rearrangement of DNA, and cell differentiation. Biosci. Rep. 3, 815–830 [DOI] [PubMed] [Google Scholar]
- 52. Coulton G. R., Rogers B., Strutt P., Skynner M. J., Watt D. J. (1992) In situ localisation of single-stranded DNA breaks in nuclei of a subpopulation of cells within regenerating skeletal muscle of the dystrophic mdx mouse. J. Cell Sci. 102, 653–662 [DOI] [PubMed] [Google Scholar]
- 53. Dawson B. A., Lough J. (1988) Immunocytochemical localization of transient DNA strand breaks in differentiating myotubes using in situ nick-translation. Dev. Biol. 127, 362–367 [DOI] [PubMed] [Google Scholar]
- 54. Larsen B. D., Rampalli S., Burns L. E., Brunette S., Dilworth F. J., Megeney L. A. (2010) Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc. Natl. Acad. Sci. U.S.A. 107, 4230–4235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larsen B. D., Megeney L. A. (2010) Parole terms for a killer. Directing caspase3/CAD induced DNA strand breaks to coordinate changes in gene expression. Cell Cycle 9, 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sjakste N., Sjakste T. (2007) Possible involvement of DNA strand breaks in regulation of cell differentiation. Eur. J. Histochem. 51, 81–94 [PubMed] [Google Scholar]
- 57. Lechardeur D., Xu M., Lukacs G. L. (2004) Contrasting nuclear dynamics of the caspase-activated DNase (CAD) in dividing and apoptotic cells. J. Cell Biol. 167, 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dieker J., Iglesias-Guimarais V., Décossas M., Stevenin J., van der Vlag J., Yuste V. J., Muller S. (2012) Early apoptotic reorganization of spliceosomal proteins involves caspases, CAD and rearrangement of NuMA. Traffic 13, 257–272 [DOI] [PubMed] [Google Scholar]
- 59. Farzaneh F., Zalin R., Brill D., Shall S. (1982) DNA strand breaks and ADP-ribosyl transferase activation during cell differentiation. Nature 300, 362–366 [DOI] [PubMed] [Google Scholar]
- 60. Sherman M. H., Bassing C. H., Teitell M. A. (2011) Regulation of cell differentiation by the DNA damage response. Trends Cell Biol. 21, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fischer U., Jänicke R. U., Schulze-Osthoff K. (2003) Many cuts to ruin. A comprehensive update of caspase substrates. Cell Death Differ. 10, 76–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boulares A. H., Zoltoski A. J., Yakovlev A., Xu M., Smulson M. E. (2001) Roles of DNA fragmentation factor and poly(ADP-ribose) polymerase in an amplification phase of tumor necrosis factor-induced apoptosis. J. Biol. Chem. 276, 38185–38192 [DOI] [PubMed] [Google Scholar]
- 63. Ahn J. Y., Liu X., Liu Z., Pereira L., Cheng D., Peng J., Wade P. A., Hamburger A. W., Ye K. (2006) Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 25, 2083–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Widlak P., Garrard W. T. (2006) The apoptotic endonuclease DFF40/CAD is inhibited by RNA, heparin and other polyanions. Apoptosis 11, 1331–1337 [DOI] [PubMed] [Google Scholar]
- 65. Lu C., Zhu F., Cho Y. Y., Tang F., Zykova T., Ma W. Y., Bode A. M., Dong Z. (2006) Cell apoptosis. Requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell 23, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schulze-Osthoff K., Walczak H., Dröge W., Krammer P. H. (1994) Cell nucleus and DNA fragmentation are not required for apoptosis. J. Cell Biol. 127, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sen S. (1992) Programmed cell death. Concept, mechanism and control. Biol. Rev. Camb. Philos. Soc. 67, 287–319 [DOI] [PubMed] [Google Scholar]
- 68. Widlak P., Garrard W. T. (2009) Roles of the major apoptotic nuclease-DNA fragmentation factor-in biology and disease. Cell. Mol. Life Sci. 66, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]