Background: miRNAs are crucial in orchestrating the odontoblast differentiation through negatively regulating their target gene expression.
Results: miR-143, which is repressed by KLF4, can promote miR-145 expression, whereas the latter inhibits KLF4, which promotes odontoblast differentiation.
Conclusion: miR-143, miR-145, and KLF4 form a feedback loop in odontoblast.
Significance: This is the first report on feedback loop regulation of odontoblast differentiation in which miRNAs are involved.
Keywords: Dentin, Differentiation, Kruppel-like factor (KLF), MicroRNA, Tooth Development, Krüpple-like Factor 4 (Klf4), Osterix (Osx), MicroRNA-143, MicroRNA-145, Odontoblast
Abstract
Dentin tissue is derived from mesenchymal cells induced into the odontoblast lineage. The differentiation of odontoblasts is a complex process regulated by several transcriptional factor signaling transduction pathways. However, post-translational regulation of these factors during dentinogenesis remains unclear. To further explore the mechanisms, we investigated the role of microRNA (miRNA) during odontoblast differentiation. We profiled the miRNA expression pattern during mouse odontoblast differentiation using a microarray assay and identified that miR-145 and miR-143 were down-regulated during this process. In situ hybridization verified that the two miRNAs were gradually decreased during mouse odontoblast differentiation. Loss-of-function and gain-of-function experiments revealed that down-regulation of miR-145 and miR-143 could promote odontoblast differentiation and increased Dspp and Dmp1 expression in mouse primary dental pulp cells and vice versa. We found that miR-145 and miR-143 controlled odontoblast differentiation through several mechanisms. First, KLF4 and OSX bind to their motifs in Dspp and Dmp1 gene promoters and up-regulate their transcription thereby inducing odontoblast differentiation. The miR-145 binds to the 3′-UTRs of Klf4 and Osx genes, inhibiting their expression. Second, KLF4 repressed miR-143 transcription by binding to its motifs in miR-143 regulatory regions as detected by ChIP assay and dual luciferase reporter assay. Third, miR-143 regulates odontoblast differentiation in part through miR-145 pathway. Taken together, we for the first time showed that the miR-143 and miR-145 controlled odontoblast differentiation and dentin formation through KLF4 and OSX transcriptional factor signaling pathways.
Introduction
The mammalian tooth is a complex organ generated from sequential and reciprocal epithelium-mesenchyme interactions and has long been investigated as a model for organogenesis (1–3). The main hard content of a tooth is dentin, which is formed via the differentiation, secretion, and mineralization of odontoblasts, a kind of cranial neural crest derived cells (4). The first step in tooth development takes place when the signals from the early oral ectoderm lead to induction of the odontogenic potential of the adjacent neural crest cells. Subsequently, the odontoblast cell lineage commences dental papilla formation during the transition from the bud to the cap stage of tooth development, which is regulated by the epithelium signals from the enamel knot and epithelium bud (5). Then, the dental papilla cells located near the epithelium differentiate into pre-odontoblasts. The pre-odontoblasts continue to differentiate into polarizing odontoblasts, secretory odontoblast, and terminally differentiated odontoblasts lying in the most outer layer of the dental pulp (6, 7). Secretory odontoblasts synthesize and secrete dentin extracellular matrix proteins, including collagen and non-collagen proteins (7, 8). Many non-collagen proteins are essential for mineralized dentin formation (9). Among the non-collagen proteins in dentin, Dspp (dentin sialophosphoprotein) and Dmp1 (dentin matrix protein1) are regarded as important markers of odontoblast differentiation (7–10). Mutations of Dspp (11–14) and Dmp1 (15, 16) genes in humans and mice are associated with dentin genetic diseases.
Thus, several growth and transcription factors regulating the expression of Dspp and Dmp1 are regarded as being responsible for odontoblast differentiation (17). We previously reported that Krüpple-like factor 4 (KLF4) (18, 19) and osterix (OSX) (20) transcription factors were expressed within odontoprogenitor cells during development and played crucial roles in odontoblast differentiation and dentin formation at multiple levels. However, the post-transcriptional regulators of these genes during odontoblast differentiation still remain unknown.
MicroRNAs (miRNAs),4 a group of short (usually ∼22 nucleotides) non-coding RNAs, are formed through sequential maturation steps (21, 22). These RNAs regulate their target genes in a post-transcriptional manner. They mainly bind to the 3′-UTRs of target mRNAs in addition to the 5′-UTRs and coding regions and induce the degradation of mRNAs, eventually resulting in the down-regulation of their target gene expression (21–23). Many miRNAs have been detected in various tissues such as skin (24), hair (25), and teeth (26–28) and related to development and diseases (29, 30). A group of miRNAs was reported to be expressed in the murine molar tooth germs, showing their possible participation of tooth development. A further analysis showed that miRNAs are differentially expressed in the epithelium and mesenchyme of the teeth, indicating different roles of the miRNAs at the given stages of tooth development (27). DICER is an enzyme responsible for miRNA maturation. Conditional dicer-inactivated mice resulted in abnormal development of mouse teeth, indicating importance of miRNAs during tooth formation (20, 21). However, which miRNA regulates odontoblast differentiation and dentin formation remains unknown. Therefore, we hypothesize that certain specific miRNAs play an essential role during odontoblast differentiation.
In the present study, we performed miRNA profiling assays during mouse odontoblast differentiation and found miR-145 and miR-143 as two of the most down-regulated miRNAs during this process in vivo and in vitro. Thereafter, we investigated how these two miRNAs regulate odontoblast differentiation. We elucidated the inhibitory effects of miR-145 and miR-143 on odontoblast differentiation using both loss-of-function and gain-of-function approaches. Either miR-143 or miR-145 suppressed odontoblast differentiation as well as Dspp and Dmp1 gene expression. In contrast, knockdown of miR-143 and miR-145 promoted the odontoblast differentiation and enhanced these tooth-relate gene expression. We found that the miR-145 is able to bind to the 3′-UTRs of Klf4 and Osx genes and repress these two gene expressions. Both of KLF4 and OSX up-regulate Dspp and Dmp1 transcription via binding to their DNA binding sites in their promoters. In addition, we found that effect of miR-143 on odontoblast differentiation is in part through miR-145. KLF4 interacts with its DNA binding motifs in the miR-143 regulatory region and down-regulates the miR-143 expression.
EXPERIMENTAL PROCEDURES
Cell Line Generation and Von Kossa Assay
Protocols utilized for mouse experiments were approved by Animal Ethics Committee of Wuhan University. The mouse dental papilla mesenchymal cells (mDPCs) were isolated from the first molars of embryonic day 18.5 (E18.5) Swiss mice and washed with phosphate-buffered saline and then digested for 1 h at 37 °C in a solution of 3 mg/ml collagenase type I and 4 mg/ml of dispase (Invitrogen). The cells were maintained using Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal bovine serum (Invitrogen) plus antibiotic-antimycotic (Invitrogen,) cultured at 37 °C in a humidified air atmosphere containing 5% CO2. The medium was replaced every 2 days, and cells were spread after reaching confluence. The second passage of dental papilla cells was immortalized via transduction of lentivirus pLOX-tTagiresTK (a kind gift from Dr. Trono in the School of Life Science École Polytechnique Fédérale, Lausanne, Switzerland), following steps described previously (31). Thus, the pre-odontoblast cell line was obtained and named mDPC6T, which was maintained in the same condition as the primarily cultured mDPC. The 293FT cell line (human embryonic kidney cell line) was purchased from Invitrogen and was maintained in DMEM supplemented with 10% fetal bovine serum and antibiotic-antimycotic. For induction of odontoblast differentiation, 50 μg/ml ascorbic acid, 10 mmol/liter sodium β-glycerophosphate, and 10 nmol/liter dexamethasone (Sigma) were added to the medium for the duration of the experiment as described previously (18, 32). Von-Kossa staining was carried out following a protocol reported previously (33).
Microarray
The Agilent Mouse miRNA Microarray was employed to identify miRNAs expressed in primarily cultured mDPCs (undifferentiated odontoblasts). The miRNA expression profiles of differentiated odontoblasts at day 9 (day 9) after odontoblastic induction were compared with the profiles from mouse dental papilla cells of the same passage without odontoblastic induction (day 0). Total RNA from different samples was isolated using the mirVanaTM miRNA Isolation Kit (Am1560; Ambion, Austin, TX). Target labeling, hybridization, imaging, and data processing were performed in a facility at the ShanghaiBio Corp. National Engineering Center for Biochip using Gene Spring Software (version 11.0; Agilent Technologies).
In Situ Hybridization
Locked nucleic acid (LNA)-miRNA digoxin double-labeled probes were obtained from Exiqon (Vedbaek, Denmark), and in situ hybridization was performed as described previously (23). For tissue in situ hybridization analyses, mouse embryos were fixed in 4% paraformaldehyde at 4 °C overnight, and teeth at postnatal day 2 were decalcified in 10% EDTA after fixation for 2 days, followed by dehydration, paraffin embedding, and sectioning. Tissue sections were subjected to standard non-radioactive in situ hybridization as reported previously (34).
For cell fluorescent in situ hybridization, the second passage of primary mouse dental papilla cells were cultured on sterile glass slides in six-well plates and were induced in odontoblast differentiation medium at 0 day, 7 days, and 11 days. Cell slides were fixed with 4% paraformaldehyde for 30 min, and fluorescent in situ hybridization was performed as described previously (34, 35).
Total RNA Isolation and Quantitative Real-time PCR Assay
Total RNA was isolated with Qiagen miRNeasy® mini kit (217004; Qiagen, Valencia, CA) and treated with RNase-free DNase I (79254; Qiagen). For miRNA expression assays, total RNA was subjected to reverse transcription using the miSript Reverse Transcription Kit (218061; Qiagen). Then, quantitative real-time PCR (qRT-PCR) was carried out using the ABI PRISM 7500 real-time PCR System (Applied Biosystems) with miScript primer assay set and miScript SYBR Green PCR kit (218073; Qiagen). U6 snRNA level was used as an internal normalization control. For mRNA expression, total RNA was reverse transcribed with the SuperScriptTM III First-Strand Synthesis System (18080; Invitrogen) for qRT-PCR and assayed with TaqMan probes and primer sets in the ABI PRISM 7500 real-time PCR System (Applied Biosystems) with Platinum® Quantitative PCR SuperMix-UDG (11730-017; Invitrogen). The β-actin mRNA level was used as an internal normalization control. The sequences of primers and TaqMan probes employed in this study are listed in the supplementary information (supplemental Table S2).
Western Blotting
Western blotting was carried out as described previously (18), and membranes were blotted with antibodies for KLF4 (Abcam, Cambridge, UK), OSX (Santa Cruz Biotechonology, Inc., Santa Cruz, CA), DMP1 (a kind gift from Dr. Jerry Feng from the Baylor College of Dentistry), DSPP (a generous gift from Dr. Chunlin Qin from the Baylor College of Dentistry), and β-actin (Biomart, Shanghai, China).
Plasmid Construction
Mus musculus miR-145 and M. musculus miR-143 expression lentivectors were constructed by replacing the Homo sapiens miR-145 fragment in pLVTHM-has-miR-145 (kind gifts from Dr. Kosik of the University of California at Santa Barbara) (23) with the precursor of M. musculus miR-145/143 and a scrambled sequence. The miRNA sponges for M. musculus miR-145 and M. musculus miR-143 are listed in the supplemental data and synthesized by Invitrogen. The sponge sequences were then subcloned into the 3′-UTR of pcDNA5-d2eGFP (a kind gift from Dr. Philip Sharp, the Massachusetts Institute of Technology) as described previously (supplemental Table S3) (36). The lentivectors encoding the miRNA sponges for miR-145 and miR-143 were constructed by replacing the EGFP fragment in pLL3.7 (Addgene 11795) with d2eGFP together with the sponge sequence in the 3′-UTR of pcDNA5-d2eGFP, whereas d2eGFP-empty was used as a control.
For KLF4 and OSX expression lentivector constructs, we first replaced the EGFP fragment in pLL3.7 with the MCS-IRES-EGFP fragment from pIRES2-EGFP to obtain pLL-IG (control group). Using PCR, we subcloned the open reading frame (ORF) sequences of Klf4 (NM_010637.3; Genecopoeia) and Osx (a kind gift from Dr. Jerry Feng of the Baylor College of Dentistry) with/without their 3′-UTRs into AgeI and NheI sites in pLL-IG.
For Dual-Luciferase assays, we subcloned the 3′-UTRs of Klf4 and Osx into luciferase reporter construct, respectively, and deletion of the miR-145 binding site was carried out using overlapping PCR. The wild-type and mutant 3′-UTRs were inserted into the 3′-UTR of the pMIR-reporter (Ambion). A 3000-bp sequence of the 5′-UTR of miR-143 was amplified by PCR. The deletion of KLF4 binding sites in the miR-143 promoter were generated using overlapping PCR technique. The wild-type and mutant promoter regions of the miR-143 were subcloned into the pGL3-Basic (Promega, Madison, WI). All of the primers employed in plasmid constructs are listed in supplemental Table S2.
Oligonucleotide Transfection
M. musculus miR-145 and M. musculus miR-143 precursors and scrambled double-stranded RNAs as negative control (Ambion) were transfected into mDPC6T and 293FT cell lines using Lipofectamine 2000 (Invitrogen) at a final concentration of 50 nm. MiRCURY LNA-miRNA inhibitors for miR-145 and miR-143 and negative control (412385-00, 412356-00, 199004; Exiqon) were transfected into mDPC6T cells using Lipofectamine 2000 at a final concentration of 20 nm. siRNA for Osx and a negative control (s10061; Ambion) were used for transfection at a final concentration of 20 nm.
Lentivirus Production and Transduction
Lentivector was co-transfected with psPAX2 and pMD2.G (kind gifts from Dr. Trono) according to the protocol described previously (31). At 48- and 72-h post transfection, the culture medium was collected to be incubated with 293FT cell line in the presence of Protamine (Sigma) for titration. mDPC6T were infected with different lentivirus in the same multiplicity of infection of 7.
Dual-Luciferase Assay
For validation of the miR-145 binding site, mDPC6T cells were cotransfected with the pMIR reporter vector, phRL-TK (Promega), and pLVTHM-miR-145 or negative control. For validation of the KLF4 binding sites within the promoter region of miR-143, mDPC6T cells were co-transfected with wild-type or mutant miR-143 promoter-luciferase reporter vector, pRL-TK, and KLF4 expression plasmid or the negative control. At 48 h post transfection, cells were harvested and assayed with Dual-Luciferase assay (Promega) according to the manufacturer's instructions. All transfection assays were carried out three times independently.
ChIP Assay
A chromatin immunoprecipitation assay was carried out using EZ-ChIP (Millipore, Billerica, MA) with KLF4 antibody (Santa Cruz Biotechonology, Inc.). Primer sequences used in this assay are listed in supplemental Table S2.
Statistical Analysis
All data were presented as means ± S.D. Statistical analysis was performed with SPSS (version 15.0). Differences between individual groups were analyzed by Student's t test (two-tailed). p values of < 0.05 are considered statistically significant.
RESULTS
miRNA Expression Profile during Murine Odontoblast Differentiation
To determine the key miRNA regulators of odontoblast differentiation, we first established an in vitro murine odontoblast differentiation model as described previously (18, 32). Mouse primary dental papilla cells were treated with odontoblast differentiation medium for 9 days and induced to odontoblast-like cells, whereas the primary dental papilla cells without induction were used as a control. To assess differential miRNA expression between the two groups, the miRNA profiles were performed using a RNA microarray. The results showed that 207 miRNAs among 657 candidate miRNAs were presented in both the induced odontoblast-like cells and uninduced dental papilla cells (supplemental Table S1 and supplemental Fig. S1). Six miRNAs were presented in the uninduced dental cells but were absent in the differentiated odontoblast-like cells. Only one miRNA was detected in the differentiated odontoblast-like cells but was absent in the uninduced dental papilla cells. Additionally, seven miRNAs were up-regulated, and 20 miRNAs were down-regulated in the induced cells compared with the uninduced cells, seven of which were listed in Fig. 1A (Fig. 1A, for more details please refer to supplemental Table S1). Notably, expressional levels of miR-134 and miR-145 were decreased in the induced cells.
FIGURE 1.
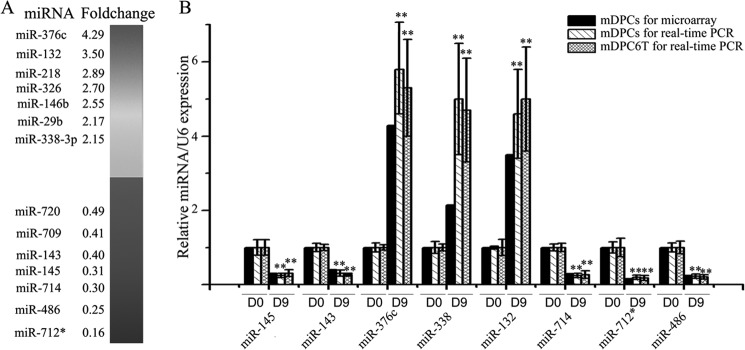
Expression change of miRNAs during mouse odontoblast differentiation. A, a random list of miRNAs significantly down-regulated or up-regulated (fold change >2, p < 0.01), 14 of 27 differentially expressed miRNAs were listed here. B, qRT-PCR validation of the miRNA microarray results, also the expression of these microRNAs during the differentiation of murine odontoblast cell line, mDPC6T. All data are presented as mean ± S.D. and are based on three independent experiments. Asterisks indicate statistical significance compared with the negative control groups; *, p < 0.05; **, p < 0.01.
To confirm the above results, we immortalized mouse dental papillary cells from the first lower molars at E18.5 using the method described previously (31) and named mDPC6T. This pre-odontoblast-like cell line exhibits the same traits as the mPDCs (supplemental Fig. S2). Thus, we employed these cells for our subsequent in vitro studies. Using the above method, the mDPC6T cells were induced with the differentiation medium for 9 days, and total RNAs were extracted. Using qRT-PCR, we examined miRNA expression levels in the mDPC6T cells treated with and without the differentiation medium. The results revealed the same trends of the qRT-PCR as that of microarray assay (Fig. 1B). Among them, miR-143 and miR-145 were greatly down-regulated in the in vitro odontoblast differentiation.
Down-regulation of miR-145 and miR-143 Expressions during Mouse Odontoblast Differentiation in Vivo
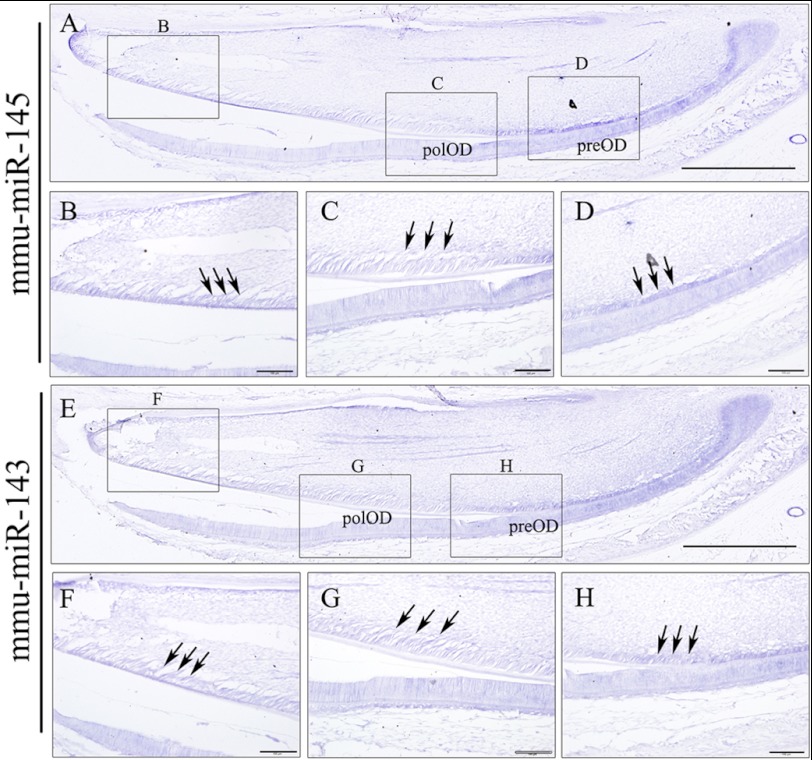
To investigate the miR-143 and miR-145 expression patterns in vivo, we first examined the expression of the miR-145 and miR-143 during odontoblast differentiation using postnatal day 2 mouse lower incisors because all stages of odontoblast development could be observed in one slide. In situ hybridization showed that these two miRNAs share the same expression patterns, and their high expression levels were found in pre-odontoblasts (Fig. 2, D and H). Notably, the two miRNA expression was gradually decreased with the odontoblast differentiation (Fig. 2, C and G). Also, the expression level of miR-145 and miR-143 in the odontoblast layer in the E18.5 mouse second lower molar were relatively higher than the first molar in which odontoblasts were better differentiated (supplemental Fig. S3). Furthermore, we specified the expression patterns of miR-145 and miR-143 during the differentiation of primarily cultured odontoblasts with fluorescent in situ hybridization. A wide spread signal of miR-145 and miR-143 could be detected in the nuclei and cytoplasm of dental papilla cells prior to odontoblastic induction. However, only very weak signals of miR-143 and miR-145 could be found within the nuclei at day 9 induction, which could be from the precursors of these two miRNAs (Fig. 3, A and B). Furthermore, we employed qRT-PCR to measure the expression levels of these miRNAs at different times during odontoblast differentiation. The results showed that the primarily cultured odontoblasts and mDPC6T cells shared the same miR-145 and miR-143 expression patterns, but miR-143 and miR-145 expressions in the induced cells were gradually decreased with time period induction. At day 9 induction, expressional levels of miR-143 and miR-145 were >4-fold lower than that of the uninduced cells (Fig. 3, C and D). We compared expression levels of miR-145 and miR-143 from E18.5 murine dental papilla, primary cultured odontoblasts from E18.5 murine dental papilla, and immortalized mouse mDPC6T cells. Data showed that there are no significant differences among these cells (supplemental Fig. S4). This indicates that both of miR-143 and miR-145 expressions are down-regulated during odontoblast differentiation in vitro and in vivo.
FIGURE 2.
Expression of miR-145 and miR-143 in the development incisor. miR-145 and miR-143 expression was detected by in situ hybridization mainly in the proximal end of incisor at postnatal day 2 (A and E). In the odontoblast layer of the incisor, the expression of both miR-145 and miR-143 was strongly localized in the preodontoblast (preOD) (D and H) and mature odontoblast (OD) (B and F), but weakly in the polarized odontoblast (polOD) (C and G). Arrows point out the odontoblast layer in the incisor. Scale bar for A and E, 500 μm; scale bar for B–D and F–H, 100 μm. mmu, M. musculus.
FIGURE 3.
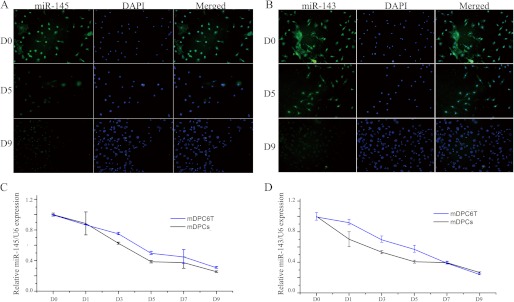
miR-145 and miR-143 were down-regulated during odontoblast differentiation. Fluorescent in situ hybridization results of miR-145 (A) and miR-143 (B) during the differentiation of primarily cultured odontoblasts at day 0 (D0), day 5 (D5), and day 9 (D9). Photos were taken at 20× magnification. Shown are qRT-PCR results for the expression of miR-145 (C) and miR-143 (D) during differentiation of primarily cultured odontoblasts and mDPC6T cells.
Down-regulation of miR-145 and miR-143 Promotes Odontoblast Differentiation
Because miR-145 and miR-143 were down-regulated during the differentiation of odontoblasts, we hypothesized that overexpression of miR-145 and miR-143 could have a negative effect on odontoblast differentiation. Thus, the pre-odontoblast cell line, mDPC6T, was infected with lentivirus encoding pre-miR-145, pre-miR-143 or scrambled miRNA (Fig. 4A). Four days after transduction, a significant increase of miR-145 or miR-143 was detected in cells infected with pLVTHM-miR-145 or pLVTHM-miR-143, respectively, compared with the negative control using qRT-PCR (Fig. 4B).
FIGURE 4.
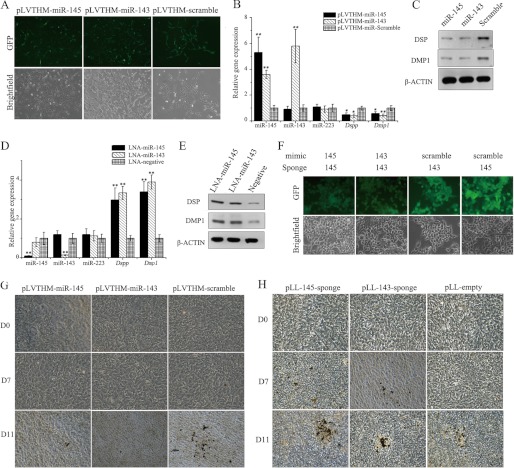
Enforced expression of miR-145 and miR-143 retarded odontoblast differentiation. A, lentivirus mediated miR-145 and miR-143 overexpression in mDPC6T. Shown are mRNA (B) and protein levels (C) of typical odontoblast markers after miR-145 and miR-143 overexpression. mRNA (D) and protein levels (E) of typical odontoblast markers 24 h after LNA-miR-145 and LNA-miR-143 inhibitor transfection are shown. F, validation of efficiency of lentivirus mediated miR-145 sponge and miR-143 sponge using 293FT cell line. G, Von-Kossa staining showing miR-145 and miR-143 overexpression inhibits mineralization of odontoblast. H, Von-Kossa staining showing down-regulation of miR-145 and miR-143 could accelerate or direct mineralization of odontoblast. Photos were taken at 200× magnification. Asterisks indicate statistical significance compared with the negative control groups. *, p < 0.05; **, p < 0.01.
To examine the effect of miR-145 and miR-143, the expression levels of the odontoblast markers Dmp1 and Dspp were measured using qRT-PCR and Western blotting assays. The results showed that enforced expression of miR-145 and miR-143 resulted in a significant decrease in Dmp1 and Dspp expression (Fig. 4, B and C). Four days following the transduction, the cells were cultured in odontoblastic induction medium for odontoblast differentiation assays. Von-Kossa staining was performed to identify the mineralization nodules on days 0, 7, and 11 post-induction. After day 7 induction, remarkable nodules were found in the negative control group, whereas scarce nodules were seen in the miR-145- or miR-143-treated groups (Fig. 4G). These results demonstrated that enforced expression of miR-145 and miR-143 significantly retarded cell differentiation. We then asked whether inhibition of the two miRNAs could promote odontoblast differentiation. Two approaches were employed to down-regulate the expression of the two miRNAs in odontoblasts: LNA-miR inhibitor transfection and lentivirus-mediated miRNA- sponge transduction (Fig. 4F). At 24 h after LNA-miR-145- or LNA-miR-143 inhibitor transfection, the expression levels of miR-145 and miR-143 were determined with qRT-PCR. The results showed that both the LNA-miR-145 and miR-143 inhibitors were able to repress the expression of miR-145 and miR-143, respectively, by >10-fold compared with cells transfected with the LNA-miR-negative inhibitor. Additionally, the down-regulation of miR-145 and miR-143 resulted in a significant up-regulation of the expression of the odontoblast differentiation markers Dmp1 and Dspp (Fig. 4, D and E). Furthermore, lentivirus-mediated miRNA sponges were employed to stably down-regulate the activity of miR-145 and miR-143. Four days following transduction of the miRNA sponges, the cells were treated with odontoblast differentiation medium and Von-Kossa staining was performed on days 0, 7, and 11 post-induction (Fig. 4H). The results showed that down-regulation of miR-145 or miR-143 greatly enhanced odontoblast mineralization: a large number of mineralization nodules were observed on day 7 using miR-145 and miR-143 sponge groups, whereas no nodules could be observed in the empty sponge group. More mineralization nodules were found on day 11 in the miR-145 and miR-143 sponge groups compared with the empty sponge group. The above results demonstrated that down-regulation of miR-145 and miR-143 could promote the differentiation of odontoblasts.
miR-143/miR-145 Axis Regulate Odontoblast Differentiation
In miR-143 gain-of-function experiments, we found that overexpression of miR-143 in the cells leads to dramatic increase expression of not only miR-143 but also miR-145 (Fig. 4B). This led us to investigate whether miR-143 was the upstream of miR-145 in the mDPC6T cell line. We then examined the expression level of miR-143 and miR-145 at 12, 24, and 48 h after LNA-miR-145 or 143 inhibitor transfection, respectively. qRT-PCR analysis showed that the miRNA inhibitors were able to repress the expression of their target genes at 12 h after transfection (Fig. 5A). In contrast, the miR-145 inhibitor had not any effect on miR-143 after transfection (Fig. 5B).
FIGURE 5.
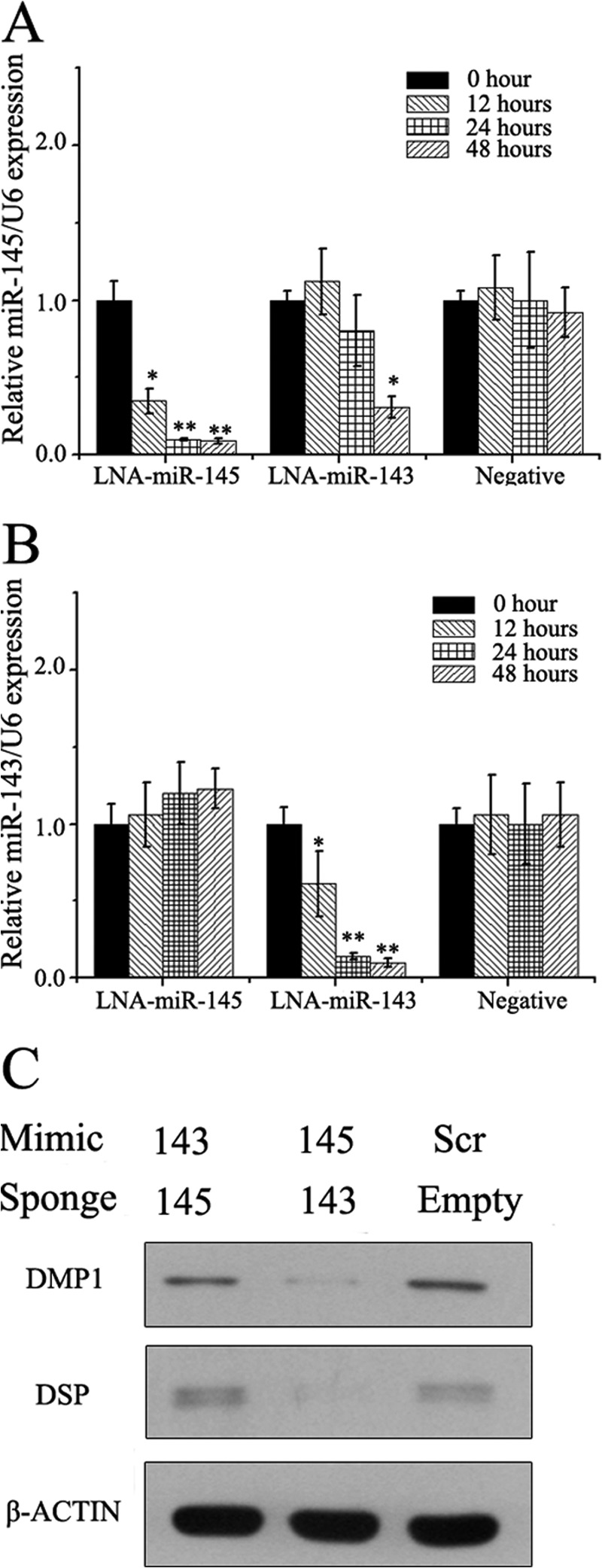
miR-143 functions partially through miR-145 in odontoblasts. A, expression change of miR-145 after transfection of LNA-miR-145, LNA-miR-143, or negative inhibitor. B, expression change of miR-143 after transfection of LNA-miR-145, LNA-miR-143, or negative inhibitor. *, p < 0.05; **, p < 0.01. C, modulation of odontoblast markers protein level by miR-145 mimics + miR-143 sponge and miR-143 mimics + miR-145 sponge. Scr, scrambled.
We then asked whether miR-143 retarded the differentiation of odontoblast through miR-145 in the mDPC6T cell line. Thus, we blocked the activity of miR-145 by stably expressing the miR-145 sponge in mDPC6T cells and overexpressed miR-143. 48 h after miR-143 transfection, proteins were extracted. Western blotting analysis showed that when miR-145 was blocked, miR-143 could not depress the expression levels of DMP1 and DSPP compared with the miR-143 overexpressed without miR-145-sponge, though the levels were still lower than in the miR scramble transfection group (Fig. 5C) These results indicated that miR-143 inhibited the differentiation of odontoblasts in part through miR-145.
miR-145 Targets Klf4 and Osx in Pre-odontoblast Cells
In the previous experiment, we found that miR-145 was the downstream of miR-143 in the mDPC6T cell line and that miR-143 retarded the differentiation of odontoblasts in part through miR-145. We then asked in what way miR-145 inhibited odontoblast differentiation. It has been reported that miRNA functions mainly through binding to the 3′-UTRs of its target genes and thereby inhibiting their expression levels (37). Therefore, we performed a bioinformatic screen for mRNAs whose 3′-UTRs contained sequences complementary to miR-145. Screening was carried out using two different algorithms (TargetScan and miRanda) (38), and the results were confirmed using RNA22 (39). We considered all of the candidate transcription factors associated with bone/tooth development from the predictions. Among the predictions, Klf4 and Osx were chosen for further analysis (Fig. 6D). Based on our previous studies, expression of Klf4 and Osx genes were gradually increased during odontoblast differentiation (18–20). In the present study, we found that both of miR-143 and miR-145 were decreased with odontoblast differentiation in vitro and in vivo. It is possible that Klf4 and Osx expression is relevant to expression of the miR-143 and miR-145 during odontoblast differentiation.
FIGURE 6.
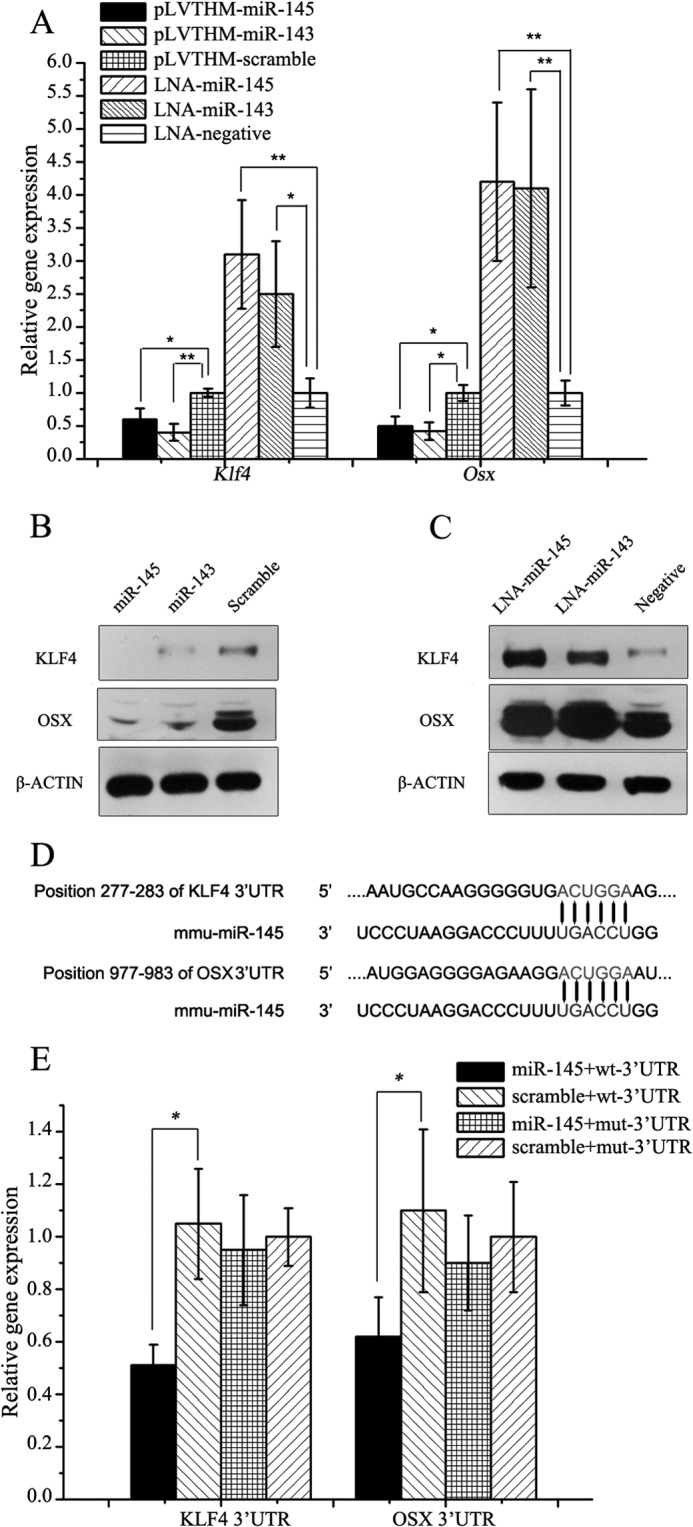
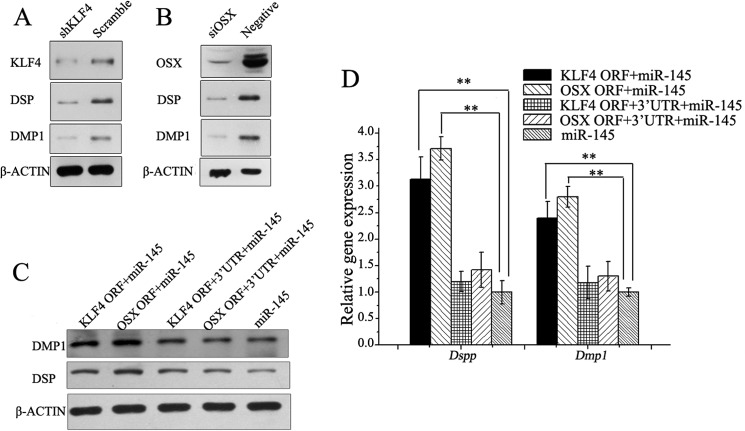
miR-145 directly targeted Klf4 and Osx in odontoblasts. A, mRNA of Klf4 and Osx when miR-145 and miR-143 were up-regulated and down-regulated. B, protein levels of KLF4 and OSX when miR-145 and miR-143 were up-regulated. C, protein levels of KLF4 and OSX when miR-145 and miR-143 were down-regulated. D, in silico assay reveals binding sites for miR-145 with 3′-UTR of Klf4 and Osx. E, validation of miR-145 binding sites using Dual-Luciferase assay, miR-145 can depress the luciferase activity of pMIR-reporter with the 3′-UTR from Klf4 and Osx compared with their mutant 3′-UTRs. All data are presented as mean ± S.D. and are based on triplicate independent experiments. Asterisks indicate statistical significance compared with the negative control groups: *, p < 0.05; **, p < 0.01. mmu, M. musculus.
To determine whether miR-145 controls odontoblast differentiation through KLF4 and OSX signaling pathways, we performed both of gain-of-function and loss-of-function approaches. The results showed that Klf4 and Osx expression was depressed when miR-145 was overexpressed (Fig. 6, A and B), whereas their expression was induced when miR-145 was knocked down by the LNA-miRNA inhibitor (Fig. 6, A and C) using qRT-PCR and Western blot analyses. Similar to miR-145, miR-143 had the similar effects on KLF4 and OSX (Fig. 6, A–C).
To further determine whether miR-145 controls the expression of Klf4 and Osx by binding to the 3′-UTRs in both genes, we performed a Dual-Luciferase assay in the mDPC6T cells. The wild-type and mutant 3′-UTRs with and without the miR-145 binding sites were subcloned into downstream of the firefly luciferase gene, respectively. The reporter constructs were transfected into mDPC6T with phRLuc-TK (Promega) and pLVTHM-miR-145. As expected, co-transfection of the miR-145 dramatically reduced the wild-type reporter activity, whereas the mutant construct had no effect (Fig. 6E). These data demonstrate that miR-145 binds to the 3′-UTR of the Klf4 and Osx genes, resulting in inhibition of both the two gene expression. This indicates that miR-143 and miR-145 regulate the cell differentiation via KLF4 and OSX pathways.
miR-145, KLF4/OSX Axes Are Necessary to Tooth-related Gene Expression and Odontoblast Differentiation
The above results revealed that miR-145 and miR-143 inhibit Klf4 and Osx expression. Next, we tested whether KLF4 and OSX regulated odontoblast differentiation and tooth-related gene activity. We employed an RNA interference approach to knock down KLF4 and OSX. The results showed that a lentivirus encoding an shRNA specific for Klf4 (40) and siRNA for Osx caused the down-regulation of DMP1 and DSPP expression in mDPC6T cells (Fig. 7, A and B), indicating that the role of the two miRNAs in odontoblast differentiation is via KLF4 and OSX pathways.
FIGURE 7.
KLF4 and OSX can promote odontoblast differentiation. A, down-regulation of KLF4 inhibits odontoblast differentiation. B, down-regulation of Osx inhibits odontoblast differentiation. mRNA (C) and protein levels (D) of odontoblast markers could be rescued by Klf4 and Osx without 3′-UTR in miR-145-overexpressed odontoblasts. Asterisks indicate statistical significance compared with the negative control groups. *, p < 0.05; **, p < 0.01.
Although the functions of miR-145, KLF4, and OSX with odontoblast differentiation have been clarified, the direct relationships between miR-145, KLF4, OSX, and cell phenotype remain unclear. Thus, we adopted a “rescue” method to validate whether miR-145 arrests odontoblast differentiation via the direct regulation of its targets, Klf4 and Osx. We constructed four lentivector constructs: with two encoding the full-length sequence of Klf4 with (pLL-Klf4) or without its 3′-UTR (pLL-Klf4–3′-UTR) and another two encoding the full length of Osx either with (pLL-Osx) or without its 3′-UTR (pLL-Osx-3′-UTR). The mDPC6T cells were transduced with pLVTHM-miR-145 and one of the lentivirus constructs described above, respectively. We observed that Klf4 or Osx without the 3′-UTRs could partially rescue the odontoblast phenotype, whereas those with the wild-type 3′-UTRs had no impact on the odontoblast phenotype (Fig. 7, C and D). These results indicate that miR-145 and miR-143 arrest odontoblast differentiation by down-regulating Klf4 and Osx expression directly and that the down-regulation of miR-145 and miR-143 during odontoblast differentiation tunes down the repression of Klf4 and Osx, which in turn promotes the odontoblast differentiation.
KLF4 Depresses miR-143 Expression
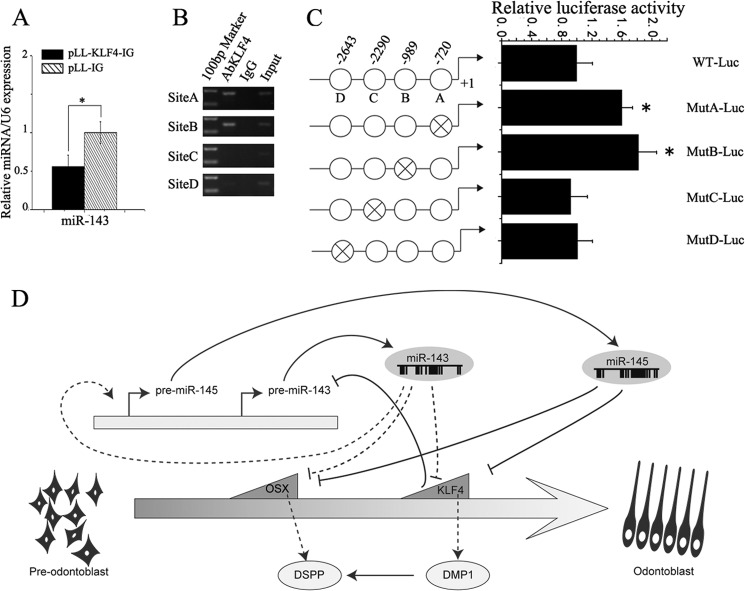
In the previous part of this study, we demonstrated that overexpression of full-length KLF4 induced odontoblast marker gene expression. Interestingly, we also found that overexpression of KLF4 can decrease the miR-143 expression (Fig. 8A). It led us to conjecture what mechanism regulates miR-143 in odontoblasts. Therefore, we subcloned the 3-kb upstream region of M. musculus miR-143 into pGL3-basic to construct pGL3-miR-143 promoter. The luciferase assay revealed that this region was sufficient to promote firefly luciferase activity. To identify the potential regulatory elements within the 3-kb miR-143 promoter, we analyzed this region using the online TESS computational program. In silico assays revealed that there are four potential KLF4-binding motifs (CACCC) (Fig. 7C). To determine whether KLF4 can interact with the miR-143 promoter in vivo, we performed ChIP assay. ChIP assay revealed that KLF4 was recruited to the three binding sites (Fig. 8B), in particular site A (−720) and site B (−989). To test whether these predicted binding sites were functional, we mutated each binding site and ligated them into upstream of firefly luciferase construct. Then, each of these reporter constructs was co-transfected with the KLF4 expression plasmid into the mDPC6T cells. The results showed that only mutations of the sites A and B had an increase of the luciferase-report activity in mDPC6T cells (Fig. 8C). It demonstrates that KLF4 negatively regulates the miR-143 gene via feedback loop signaling.
FIGURE 8.
KLF4 down-regulated the expression of miR-143. A, qRT-PCR shows overexpression of KLF4 can down-regulate the expression of miR-143. C, predicted potential binding sites of KLF4 in the 3 kb upstream region of miR-143. B, ChIP analysis of KLF4 binding sites in the 3 kb within the upstream of miR-143. C, Dual-Luciferase assay showing different luciferase (Luc) activity of the mutated promoter of miR-143. Luciferase activities were normalized with the relative activity of pGL-WT-Luc co-transfected with KLF4. Asterisks indicate statistical significance compared with the negative control groups. *, p < 0.05; **, p < 0.01. D, schematic diagram showing the regulatory relationships among miR-143, miR-145, Klf4 and Osx during odontoblast differentiation.
DISCUSSION
The development of a tooth, particularly in odontoblast differentiation, is a masterpiece of orchestration of growth and transcription factors. Previous studies demonstrated that transcription factors are involved in controlling the expression of numerous genes, executing cellular programs and tooth development. However, post-transcriptional regulations of these factors by miRNAs remain unknown. In this study, we employed miRNA microarray to profile the miRNA expression pattern during odontoblast differentiation. Our study found that the miR-143 and miR-145 act as important factors in this process. Effect of the miR-143 and miR-145 on odontoblast differentiation and tooth-relate gene expression is involved in KLF4 and OSX signaling transduction pathways.
Using miRNA microarray, fluorescent in situ hybridization and qRT-PCR analyses, we showed that miR-143 and miR-145 expression is down-regulated during murine odontoblast differentiation in vitro. Using mouse incisor tooth sections, in vivo study further demonstrated that the two miRNA expression is gradually decreased with odontoblast cytodifferentiation by using in situ hybridization assay, whereas expression of Klf4 and Osx is increased during odontoblast differentiation of mouse tooth development (21, 22). Previous works on miR-145 and miR-143 have mostly reported their up-regulation during cell differentiation such as embryonic stem and smooth muscle cells (23, 41–43). They mainly function as promoters of differentiation. In this study, our data showed that both the miR-145 and miR-143 serve as repressors and inhibit odontoblast differentiation during tooth development. These findings demonstrate the dual effects of these multifunctional miRNAs in different biological roles, and these different functions of the two miRNAs may depend on the different cell types and tissues.
As there are different expressional levels between the miR-143, miR-145, and Klf4 and Osx in odontoblast differentiation during murine tooth development, we investigated the relationship between the two miRNAs and the two transcription factors and found that the miR-145 directly represses their gene expression through binding to their 3′-UTRs of Klf4 and Osx. We failed to find the binding site of miR-143 within the 3′-UTRs of Klf4 or Osx (44). However, the miR-143 inhibited Klf4 and Osx expression through the miR-145. When the activity of miR-145 was blocked, miR-143 failed to down-regulate the expression of DMP1 and DSPP. All of these results further indicated that miR-143 regulates odontoblast differentiation through miR-145 pathway.
Klf4 has been extensively investigated for its roles in tumorigenesis, cell proliferation, and differentiation. Studies showed that Klf4 is responsible for promoting the differentiation of many cell types such as adipocytes (45), monocytes (46), and goblet cells (47). However, an inhibitory effect of Klf4 on cytodifferentiation has also been demonstrated based on its ability to maintain the stemness of embryonic stem cells (23, 48). Our previous study found Klf4 exhibited a distinct expression pattern during tooth development, especially with respect to odontoblast differentiation. In situ hybridization and immunochemistry analyses demonstrated that Klf4 first appeared in the polarizing odontoblast in E18.5 of murine first molar, indicating its role in odontoblast differentiation (19). When Klf4 was overexpressed in human dental pulp cells, the expression level of Dmp1 increased dramatically, with minor up-regulation of Dspp detected, revealing that Klf4 can induce these cells into odontoblast-like cells (18). As a transcription factor, KLF4 can specifically bind to the CACCC motif in the promoter regions of its downstream targets and initiate transcription such as Dmp1 (46). We have shown that in human dental pulp cells and murine odontoblasts, enforced expression of KLF4 mainly up-regulate the expression of Dmp1, whereas DMP1 can act as transcription factor promoting the Dspp expression (49), thus explaining the down-regulation of both Dmp1 and Dspp after the knockdown of Klf4. However, when KLF4 was overexpressed, the expression of miR-143 was depressed. ChIP and Dual-Luciferase assay showed that KLF4 could bind to the promoter region of miR-143, thus repressed its expression. The mechanisms that KLF4 up- and down-regulates the Dmp1, Dspp, and miR-143 gene expression need to be further investigated.
On the basis of the results presented in this study and all of the above considerations, we propose a model to further explain the role of miR-143, miR-145, Klf4, and Osx during odontoblast differentiation, schematically depicted in Fig. 8D. Our current study demonstrated that in vitro induction of odontoblast differentiation decreases the expression of miR-145 and miR-143. The decrease of miR-143 can partially down-regulate miR-145 expression and thus release the expression of the target genes Klf4 and Osx. Both KLF4 and OSX up-regulate odontoblast maker genes, Dspp and Dmp1, thereafter inducing odontoblast differentiation. Moreover, KLF4 binds to its DNA binding sites in miR-143 regulatory regions and inhibits its transcription in mDPC6T cells. This feedback loop signaling illustrates the orchestration between miRNAs and their targets during odontoblast differentiation.
This work was supported by National Natural Science Foundation of China Grants 81070797 and 81271099 and National 973 Project of China Grant 2010CB534915.

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
- miRNA
- microRNA
- mDPC
- mouse dental papilla mesenchymal cell
- E18.5
- embryonic day 18.5
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1. Thesleff I. (2003) Epithelial-mesenchymal signalling regulating tooth morphogenesis. J. Cell Sci. 116, 1647–1648 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Y. D., Chen Z., Song Y. Q., Liu C., Chen Y. P. (2005) Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 15, 301–316 [DOI] [PubMed] [Google Scholar]
- 3. Tummers M., Thesleff I. (2009) J. Exp. Zool. B Mol. Dev. Evol. 312B, 309–319 [DOI] [PubMed] [Google Scholar]
- 4. Arana-Chavez V. E., Massa L. F. (2004) Odontoblasts: the cells forming and maintaining dentine. Int. J. Biochem. Cell Biol. 36, 1367–1373 [DOI] [PubMed] [Google Scholar]
- 5. Liu F., Chu E. Y., Watt B., Zhang Y., Gallant N. M., Andl T., Yang S. H., Lu M. M., Piccolo S., Schmidt-Ullrich R., Taketo M. M., Morrisey E. E., Atit R., Dlugosz A. A., Millar S. E. (2008) Wnt/β-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 313, 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thesleff I., Keränen S., Jernvall J. (2001) Enamel knots as signaling centers linking tooth morphogenesis and odontoblast differentiation. Adv. Dent. Res. 15, 14–18 [DOI] [PubMed] [Google Scholar]
- 7. Balic A., Aguila H. L., Mina M. (2010) Identification of cells at early and late stages of polarization during odontoblast differentiation using pOBCol3.6GFP and pOBCol2.3GFP transgenic mice. Bone 47, 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Qin C., D'Souza R., Feng J. Q. (2007) Dentin matrix protein 1 (DMP1): new and important roles for biomineralization and phosphate homeostasis. J. Dent. Res. 86, 1134–1141 [DOI] [PubMed] [Google Scholar]
- 9. Qin C., Baba O., Butler W. T. (2004) Post-translational modifications of sibling proteins and their roles in osteogenesis and dentinogenesis. Crit. Rev. Oral Biol. Med. 15, 126–136 [DOI] [PubMed] [Google Scholar]
- 10. Janebodin K., Horst O. V., Ieronimakis N., Balasundaram G., Reesukumal K., Pratumvinit B., Reyes M. (2011) Isolation and characterization of neural crest-derived stem cells from dental pulp of neonatal mice. PLoS One 6, e27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J. W., Simmer J. P. (2007) Hereditary dentin defects. J. Dent. Res. 86, 392–399 [DOI] [PubMed] [Google Scholar]
- 12. Kim J. W., Nam S. H., Jang K. T., Lee S. H., Kim C. C., Hahn S. H., Hu J. C., Simmer J. P. (2004) A novel splice acceptor mutation in the DSPP gene causing dentinogenesis imperfecta type II. Hum. Genet. 115, 248–254 [DOI] [PubMed] [Google Scholar]
- 13. Zhang X., Zhao J., Li C., Gao S., Qiu C., Liu P., Wu G., Qiang B., Lo W. H., Shen Y. (2001) DSPP mutation in dentinogenesis imperfecta Shields type II. Nat. Genet. 27, 151–152 [DOI] [PubMed] [Google Scholar]
- 14. Xiao S., Yu C., Chou X., Yuan W., Wang Y., Bu L., Fu G., Qian M., Yang J., Shi Y., Hu L., Han B., Wang Z., Huang W., Liu J., Chen Z., Zhao G., Kong X. (2001) Dentinogenesis imperfecta 1 with or without progressive hearing loss is associated with distinct mutations in DSPP. Nat. Genet. 27, 201–204 [DOI] [PubMed] [Google Scholar]
- 15. Turan S., Aydin C., Bereket A., Akcay T., Güran T., Yaralioglu B. A., Bastepe M., Jüppner H. (2010) Identification of a novel dentin matrix protein-1 (DMP-1) mutation and dental anomalies in a kindred with autosomal recessive hypophosphatemia. Bone 46, 402–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Y., Chen L., Ma S., Zhou J., Zhang H., Feng J. Q., Qin C. (2011) Roles of DMP1 processing in osteogenesis, dentinogenesis and chondrogenesis. Cells Tissues Organs 194, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ivey K. N., Srivastava D. (2010) MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem cell 7, 36–41 [DOI] [PubMed] [Google Scholar]
- 18. Lin H., Xu L., Liu H., Sun Q., Chen Z., Yuan G., Chen Z. (2011) KLF4 promotes the odontoblastic differentiation of human dental pulp cells. J. Endod. 37, 948–954 [DOI] [PubMed] [Google Scholar]
- 19. Chen Z., Couble M. L., Mouterfi N., Magloire H., Chen Z., Bleicher F. (2009) Spatial and temporal expression of KLF4 and KLF5 during murine tooth development. Arch. Oral Biol. 54, 403–411 [DOI] [PubMed] [Google Scholar]
- 20. Chen S., Gluhak-Heinrich J., Wang Y. H., Wu Y. M., Chuang H. H., Chen L., Yuan G. H., Dong J., Gay I., MacDougall M. (2009) Runx2, osx, and dspp in tooth development. J. Dent. Res. 88, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. He L., Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 22. Bushati N., Cohen S. M. (2007) MicroRNA functions in Annual Review of Cell and Developmental Biology, pp. 175–205, Annual Reviews, Palo Alto, CA: [DOI] [PubMed] [Google Scholar]
- 23. Li H., Xie H., Liu W., Hu R., Huang B., Tan Y. F., Xu K., Sheng Z. F., Zhou H. D., Wu X. P., Luo X. H. (2009) A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Invest. 119, 3666–3677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yi R., O'Carroll D., Pasolli H. A., Zhang Z., Dietrich F. S., Tarakhovsky A., Fuchs E. (2006) Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 38, 356–362 [DOI] [PubMed] [Google Scholar]
- 25. Andl T., Murchison E. P., Liu F., Zhang Y., Yunta-Gonzalez M., Tobias J. W., Andl C. D., Seykora J. T., Hannon G. J., Millar S. E. (2006) The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 16, 1041–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jevnaker A. M., Osmundsen H. (2008) MicroRNA expression profiling of the developing murine molar tooth germ and the developing murine submandibular salivary gland. Arch. Oral Biol. 53, 629–645 [DOI] [PubMed] [Google Scholar]
- 27. Cao H., Wang J., Li X., Florez S., Huang Z., Venugopalan S. R., Elangovan S., Skobe Z., Margolis H. C., Martin J. F., Amendt B. A. (2010) MicroRNAs play a critical role in tooth development. J. Dent. Res. 89, 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Michon F., Tummers M., Kyyrönen M., Frilander M. J., Thesleff I. (2010) Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 340, 355–368 [DOI] [PubMed] [Google Scholar]
- 29. Park C. Y., Choi Y. S., McManus M. T. (2010) Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 19, R169–R175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michon F. (2011) Birth defects. Res. A Clin. Mol. Teratol. 91, 763–769 [DOI] [PubMed] [Google Scholar]
- 31. Salmon P., Oberholzer J., Occhiodoro T., Morel P., Lou J., Trono D. (2000) Reversible immortalization of human primary cells by lentivector-mediated transfer of specific genes. Mol. Ther. 2, 404–414 [DOI] [PubMed] [Google Scholar]
- 32. Chen S., Rani S., Wu Y., Unterbrink A., Gu T. T., Gluhak-Heinrich J., Chuang H. H., Macdougall M. (2005) Differential regulation of dentin sialophosphoprotein expression by Runx2 during odontoblast cytodifferentiation. J. Biol. Chem. 280, 29717–29727 [DOI] [PubMed] [Google Scholar]
- 33. Wu X., Zeng L. H., Taniguchi T., Xie Q. M. (2007) Activation of PKA and phosphorylation of sodium-dependent vitamin C transporter 2 by prostaglandin E2 promote osteoblast-like differentiation in MC3T3-E1 cells. Cell Death Differ. 14, 1792–1801 [DOI] [PubMed] [Google Scholar]
- 34. Pena J. T., Sohn-Lee C., Rouhanifard S. H., Ludwig J., Hafner M., Mihailovic A., Lim C., Holoch D., Berninger P., Zavolan M., Tuschl T. (2009) miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods 6, 139–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Silahtaroglu A. N., Nolting D., Dyrskjøt L., Berezikov E., Møller M., Tommerup N., Kauppinen S. (2007) Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2, 2520–2528 [DOI] [PubMed] [Google Scholar]
- 36. Ebert M. S., Neilson J. R., Sharp P. A. (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 39. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 40. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. (2010) Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 285, 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ono K. (2011) MicroRNA links obesity and impaired glucose metabolism. Cell Res. 21, 864–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yi C., Xie W. D., Li F., Lv Q., He J., Wu J., Gu D., Xu N., Zhang Y. (2011) MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin. FEBS Lett. 585, 3303–3309 [DOI] [PubMed] [Google Scholar]
- 44. Liu H., Zhang S., Lin H., Jia R., Chen Z. (2012) Identification of microRNA-RNA interactions using tethered RNAs and streptavidin aptamers. Biochem. Biophys. Res. Commun. 422, 405–410 [DOI] [PubMed] [Google Scholar]
- 45. Birsoy K., Chen Z., Friedman J. (2008) Transcriptional regulation of adipogenesis by KLF4. Cell Metab. 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Feinberg M. W., Wara A. K., Cao Z., Lebedeva M. A., Rosenbauer F., Iwasaki H., Hirai H., Katz J. P., Haspel R. L., Gray S., Akashi K., Segre J., Kaestner K. H., Tenen D. G., Jain M. K. (2007) The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 26, 4138–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Katz J. P., Perreault N., Goldstein B. G., Lee C. S., Labosky P. A., Yang V. W., Kaestner K. H. (2002) The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129, 2619–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 49. Narayanan K., Gajjeraman S., Ramachandran A., Hao J., George A. (2006) Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J. Biol. Chem. 281, 19064–19071 [DOI] [PubMed] [Google Scholar]