Background: EXTL2 is one of three EXT-like genes homologous to the tumor suppressor EXT gene family members and encodes an N-acetylhexosaminyltransferase.
Results: EXTL2 terminated polymerization of glycosaminoglycan (GAG) chains by transferring an N-acetylglucosamine residue to the phosphorylated tetrasaccharide linkage region.
Conclusion: EXTL2 controlled GAG biosynthesis in a xylose kinase-dependent manner.
Significance: Lack of EXTL2 causes GAG overproduction associated with pathological processes.
Keywords: Glycobiology, Glycosaminoglycan, Glycosyltransferases, Proteoglycan, Proteoglycan Synthesis
Abstract
Mutant alleles of EXT1 or EXT2, two members of the EXT gene family, are causative agents in hereditary multiple exostoses, and their gene products function together as a polymerase in the biosynthesis of heparan sulfate. EXTL2, one of three EXT-like genes in the human genome that are homologous to EXT1 and EXT2, encodes a transferase that adds not only GlcNAc but also N-acetylgalactosamine to the glycosaminoglycan (GAG)-protein linkage region via an α1,4-linkage. However, both the role of EXTL2 in the biosynthesis of GAGs and the biological significance of EXTL2 remain unclear. Here we show that EXTL2 transfers a GlcNAc residue to the tetrasaccharide linkage region that is phosphorylated by a xylose kinase 1 (FAM20B) and thereby terminates chain elongation. We isolated an oligosaccharide from the mouse liver, which was not detected in EXTL2 knock-out mice. Based on structural analysis by a combination of glycosidase digestion and 500-MHz 1H NMR spectroscopy, the oligosaccharide was found to be GlcNAcα1-4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate), which was considered to be a biosynthetic intermediate of an immature GAG chain. Indeed, EXTL2 specifically transferred a GlcNAc residue to a phosphorylated linkage tetrasaccharide, GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate). Remarkably, the phosphorylated linkage pentasaccharide generated by EXTL2 was not used as an acceptor for heparan sulfate or chondroitin sulfate polymerases. Moreover, production of GAGs was significantly higher in EXTL2 knock-out mice than in wild-type mice. These results indicate that EXTL2 functions to suppress GAG biosynthesis that is enhanced by a xylose kinase and that the EXTL2-dependent mechanism that regulates GAG biosynthesis might be a “quality control system” for proteoglycans.
Introduction
Glycosaminoglycans (GAGs)3 are abundant on the surfaces of most cells and in extracellular matrices as components of proteoglycans (PGs) (1). PGs are known to function as cofactors in a variety of biological processes, including cell adhesion, angiogenesis, morphogenesis, and regulating the effects of growth factors and cytokines (2). Most biological activities of GAGs are attributable to the GAG side chains that interact with diverse protein ligands via specific saccharide sequences (3). Linear, sulfated GAGs are loosely classified as either heparan sulfates (HSs) or chondroitin sulfates (CSs). Both types of GAG chains are covalently attached to their respective core proteins via the so-called GAG-protein linkage region, GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser, which is formed via the stepwise addition of individual monosaccharide residues by a respective specific glycosyltransferase. In addition, a xylose residue is transiently phosphorylated during formation of the linkage region (4, 5). Once N-acetylglucosamine (GlcNAc) is transferred to the linkage region, the backbone of an HS is formed by alternating the addition of glucuronic acid (GlcUA) and GlcNAc via the actions of GlcNAc transferase and GlcUA transferase, respectively. In contrast, if an N-acetylgalactosamine (GalNAc), rather than a GlcNAc, is the first residue to be added to the linkage region, the backbone of a CS, rather than an HS, is synthesized; this CS backbone is formed via alternating the addition of GlcUA and GalNAc by the concerted actions of GalNAc transferase and GlcUA transferase. It seems that almost all of the glycosyltransferases involved in GAG biosynthesis (1) and the only kinase responsible for the phosphorylation of xylose (5) have been identified and that the molecular mechanisms mediating GAG biosynthesis have been largely solved.
The genes EXT1 and EXT2, members of the tumor suppressor EXT gene family, are associated with hereditary multiple exostoses, an autosomal dominant disorder characterized by aberrant bone formation that most commonly occurs in the juxtaepiphyseal regions of the long bones (6–8). Previous studies have shown that as recombinant proteins EXT1 and EXT2 have both GlcNAc and GlcUA transferase activity and form an EXT1·EXT2 heterooligomeric complex in the Golgi apparatus to yield a more active, biologically relevant enzyme form (9, 10). Notably, a deficiency of either EXT1 or EXT2 causes hereditary multiple exostoses (11–13). These findings indicate that both EXT1 and EXT2 are essential glycosyltransferases for HS biosynthesis. Three highly homologous EXT-like genes, EXTL1–EXTL3, have been cloned (6–8), and based on their chromosomal locations, they may each be a tumor suppressor (6, 7, 13–15); however, none is linked with hereditary multiple exostoses. EXTL2 protein is an N-acetylhexosaminyltransferase that transfers not only GalNAc, but also GlcNAc, to the common linkage region core tetrasaccharide via an α1,4-linkage (16). However, the function of EXTL2 in GAG biosynthesis and the biological significance of EXTL2 have not been established. Here, we discovered a novel regulation mechanism; specifically, EXTL2 controls GAG biosynthesis in a coordinated manner with FAM20B, which catalyzes the phosphorylation of a xylose residue.
Although 2-O-phosphorylation of xylose has been detected in the GAG-protein linkage region (GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser) of PGs, the enzyme responsible for the phosphorylation of xylose remained unclear. We have recently identified FAM20B as a kinase that phosphorylates the xylose residue in the linkage region (5). FAM20B regulates the amount of GAG chain synthesis by controlling the number of GAG chains and plays an important role in the biosynthesis of GAG (5). We hypothesized that the phosphorylation of xylose by FAM20B facilitates the formation of the linkage region and thereby leads to enhanced GAG biosynthesis. Here we report that EXTL2 controls GAG biosynthesis in a FAM20B-dependent manner.
EXPERIMENTAL PROCEDURES
Isolation of Mouse EXTL2 Genomic Clones and Analysis of the Mouse EXTL2 Gene
A mouse 129SVJ λ FIX II genomic DNA library (Stratagene) was screened with a cloned human EXTL2 cDNA fragment of 1.1 kb (16) using the Gene Images random prime labeling module (GE Healthcare). Positive clones obtained by screening ∼1 million plaques were further rescreened at least three times. Insert DNA fragments were initially characterized by restriction digestion and Southern blot analysis. Mouse genomic DNA fragments that hybridized to the human EXTL2 cDNA probe were subcloned into Bluescript plasmid vectors and sequenced. Two positive clones were isolated, which each contained full-length mouse EXTL2 cDNA. PCR-based sequencing and primers derived from mouse EXTL2 cDNA sequences were used to determine the genomic structure of EXTL2 (Fig. 1A) and the complete sequence of a full-length EXTL2 cDNA. The site of transcription initiation was determined by using a Cap site cDNATM kit (Wako, Osaka, Japan) as described previously (Fig. 1A) (17).
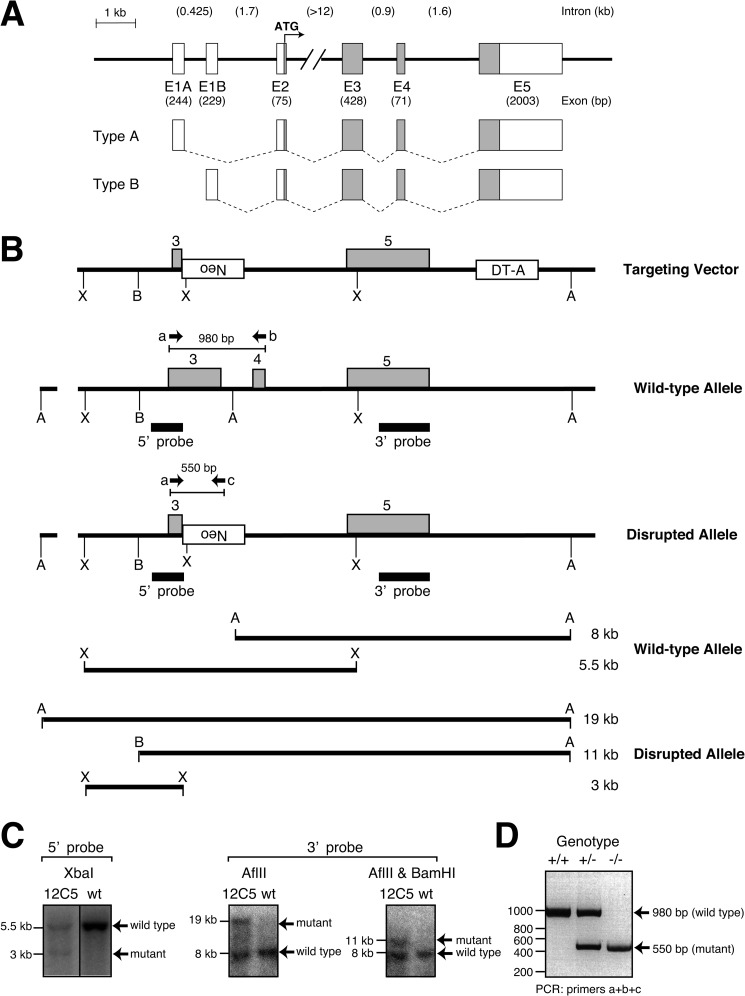
FIGURE 1.
Genomic map and targeted disruption of the mouse EXTL2 gene. A, exons are labeled E1–E5 and denoted by boxes. Gray boxes represent coding regions, and open boxes denote 5′- and 3′-untranslated regions. Shown below are the splicing patterns for each type message described here. B, a targeting vector of the replacement type was constructed as shown in supplemental Fig. 1. The diptheria toxin A (DT-A) gene in the targeting vector was used as a negative selectable marker. The neomycin (Neo) gene was driven by the phosphoglycerate kinase I promoter. The transcriptional direction of the Neo gene was anti-parallel to that of the EXTL2. In the event of homologous recombination, the disrupted allele will have acquired additional sites for the restriction endonucleases XbaI (X) and lost the AflII (A) site located between exons 3 and 4. The expected digestion patterns with XbaI, AflII, and AflII and BamHI (B) are shown at the bottom. The DNA probes used for Southern blotting are depicted as black boxes. The primers used for genotyping are represented by arrows (a, b, and c). C, genomic DNA prepared from wild-type embryonic stem cells and clone 12C5 were each used for Southern blot analysis after digestion with XbaI (left blot), AflII (middle blot) and double digestion with AflII and BamHI (right blot). Filters were probed with the 5′ probe or the 3′ probe indicated in B. D, RT-PCR analysis of RNA prepared from tail tissue was carried out using primers a, b, and c. A 980-bp fragment was generated from the wild-type allele (primers a and b), and a 550-bp fragment was amplified from the disrupted allele (primers a and c). ATG, start methionine; A, AflII site; B, BamHI site; X, XbaI site; +/+, wild type; +/−, heterozygous; −/−, homozygous knock-out.
Generation of EXTL2 Knock-out Mice
The strategy of construction of the EXTL2 targeting vector is shown in supplemental Fig. 1. The linearized targeting vector (20 μg) was introduced into 107 E14-1 mouse embryonic stem cells (18) via electroporation (250 V, 500 microfarads); cells were then subjected to selection with 250 μg (active form)/ml G418 (Invitrogen) for 7–10 days. Homologous recombinants were screened by PCR and confirmed by Southern blot hybridization with an external 5′ and 3′ probe (see Fig. 1B). Chimeric mice were generated using a modified version of an aggregation method described previously (19). Male chimeras were mated with C57BL/6J females, and homozygous mutant mice were generated by intercrossing of heterozygotes. Genotypes were determined by amplification of tail DNA via PCR with allele-specific primers with wild-type sequences (primer a, 5′-TTCACCCTCATCATGCAGAC-3′; primer b, 5′-CTGCCAAATTGAGAAAGCG-3′) or mutant sequences derived from the recombinant neo gene (primer c, 5′-GCTCGCTGATCAGCCTCGACTGTGC-3′). EXTL2+/− mice were back-crossed to C57BL/6J mice for 11 generations. Mice were kept under specific pathogen-free conditions in an environmentally controlled, clean room at the Institute of Laboratory Animals, Kobe Pharmaceutical University; animals were maintained on standard mouse/rat food and on a 12-h light/12-h dark cycle. All experiments were conducted according to institutional ethics guidelines for animal experiments and safety guidelines for gene manipulation experiments. All animal procedures were approved by the Kobe Pharmaceutical University Committee on Animal Research and Ethics.
Northern Blots and in Situ Hybridization
The mouse multiple tissue Northern (MTNTM) blot with 2 μg of poly(A)+ RNAs (Clontech, catalog no. 7762-1) was hybridized with a cDNA probe specific to mEXTL2. This specific cDNA probe to EXTL2 was prepared from pGEM-T(Easy)-mEXTL2 (open reading frame) by digestion with NotI; cDNA probes (20 ng) were labeled with [α-32P]dCTP (3,000 Ci/mmol) (Muromachi Technos Co., Ltd., Japan) using an oligolabeling kit (GE Healthcare). MTNTM blots were hybridized with radiolabeled cDNA probe in ExpressHyb hybridization solution (Clontech) overnight at 68 °C according to the manufacturer's protocols. Sections of mouse embryos on slides (embryonic days 8–16) (Novagen) were subjected to in situ hybridization according to the manufacturer's instructions. Briefly, after wax was removed from the sections, the sections were treated with proteinase K (10 μg/ml) at room temperature for 20 min, washed, and prehybridized for 1 h at 42 °C. Hybridization with digoxygenin-labeled RNA probes prepared using a DNA digoxygenin RNA labeling kit (SP6/T7) (Roche Applied Science) was performed overnight at 50 °C. Slides were then washed at 50 °C and incubated with alkaline phosphatase-conjugated sheep anti-digoxygenin Fab fragments (1:500 (Roche Applied Science)) for 30 min at room temperature. Immunostaining was visualized by adding 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium alkaline phosphatase substrate.
Preparation of Embryonic Fibroblasts
Mouse embryonic fibroblasts (MEFs) were generated from littermates of one of two genotypes, EXTL2+/+ or EXTL2−/−. Primary MEFs were harvested from embryonic day 14 embryos. Briefly, pregnant female mice were anesthetized using pentobarbital; each uterus was opened up; each embryo was placed into a separate 10-cm Petri dish; and then the head, limbs, and liver were removed from each embryo. Embryos were subsequently minced and then incubated at 37 °C in the presence of 6 ml of 0.05% trypsin, 0.02% EDTA for 20 min in a humidified incubator. Trypsin-treated embryos were homogenized by pipetting each mixture up and down until a viscous fluid was obtained and only a few tissue clumps remained. Each homogenized embryo was incubated again in the presence of 6 ml of 0.05% trypsin, 0.02% EDTA for 20 min. After the addition of 2 ml of fetal bovine serum, homogenized embryos were centrifuged at 100 × g for 5 min. Cell pellets were suspended in fresh DMEM (Wako Pure Chemical Industries, Ltd.) containing 10% FBS (Biowest), 100 units/ml penicillin, and 100 μg/ml streptomycin; each cell suspension was then transferred to a 10-cm dish.
Real-time PCR Analysis
Total RNA was extracted from cells by the guanidine phenol method using TRIzol reagent (Invitrogen) according to the manufacturer's protocols. Aliquots (1 μg) of total RNA were digested with 2 IU of RQ1 RNase-free DNase (Promega) for 30 min at 37 °C and then incubated for 10 min at 65 °C with stop solution (Promega). For reverse transcription, total RNA (0.75 μg) was treated with Moloney murine leukemia virus reverse transcriptase (Invitrogen) using random primers (nonadeoxyribonucleotide mixture; pd(N)9) (Takara bio Inc., Shiga, Japan). Quantitative real-time PCR was conducted using FastStart DNA Master plus SYBR Green I and a LightCycler 1.5 (Roche Applied Science) according to the manufacturer's protocols. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for quantification. The primers used for real-time PCR are shown in supplemental Table 1.
Disaccharide Analysis of GAGs from Mouse Tissues and Cells
GAGs were isolated and purified from mouse tissues and cells as described previously (20). Briefly, tissues or cells were homogenized and extracted with acetone three times and air-dried thoroughly. The dried materials were digested with heat-activated actinase E (10% by weight of dried materials) in 0.1 m borate sodium, pH 8.0, containing 10 mm CaCl2 at 55 °C for 48 h. The samples were adjusted to 5% trichloroacetic acid and centrifuged. The resultant supernatants were extracted with diethyl ether three times to remove trichloroacetic acid and then neutralized using 20% NH4HCO3. The aqueous phase containing 5% sodium acetate was adjusted to 80% ethanol and left overnight at −30 °C. The resultant precipitate was dissolved in 50 mm pyridine acetate, pH 5.0, and subjected to gel filtration on a PD-10 column (GE Healthcare) using 50 mm pyridine acetate, pH 5.0, as an eluent. The flow-through fractions were collected and evaporated to dryness. Purified GAGs were digested with chondroitinase ABC from Proteus vulgaris (EC 4.2.2.4) (10 mIU) or with a mixture of heparinase from Flavobacterium heparinum (EC 4.2.2.7) (1 mIU) and heparitinase from F. heparinum (EC 4.2.2.8) (1 mIU) at 37 °C for 4 h. The digests were derivatized with a fluorophore 2-aminobenzamide and then analyzed by high performance liquid chromatography (HPLC) as reported previously (21).
Isolation of Linkage Region Oligosaccharides from Mouse Liver
Mouse livers were homogenized in acetone; tissue samples were treated with acetone three times and air-dried thoroughly. The dried materials (120–140 mg) were dissolved in 0.5 m LiOH and kept at 4 °C on a rotator for 16 h to release O-linked saccharides from core proteins (22). After neutralization, the samples were adjusted to 5% (v/v) in trichloroacetic acid, incubated on ice for 1 h, and centrifuged. The resulting supernatants were extracted with diethyl ether three times to remove the trichloroacetic acid, adjusted to pH 5–6 with 10% NaHCO3, and subjected to ion exchange chromatography on a column (2-ml bed volume) of AG 50W-X2 (H+ form; Bio-Rad) using H2O as an eluent. The flow-through fraction containing the O-linked oligosaccharide components was pooled and neutralized with 10% NaHCO3.
Derivatization of the Isolated Oligosaccharide with 2-Aminobenzamide (2AB)
Derivatization with 2AB of the oligosaccharides was performed as described (23, 24). The labeled oligosaccharides were subjected to gel filtration HPLC on a Shodex Asahipak column, GS320 7G (7.6 × 500 mm) (Showa Denko), using a flow rate of 0.5 ml/min and 5 mm NH4HCO3 containing 50% acetonitrile as the mobile phase. Fluorescence of the eluates was monitored at an excitation wavelength of 330 nm and an emission wavelength of 420 nm. The fractions containing 2AB-labeled oligosaccharide were pooled, evaporated to dryness, and used for the structural analysis.
Enzyme Digestion
Enzyme digestion with heparitinase (EC 4.2.2.8) from F. heparinum (3 mIU) (Seikagaku Biobusiness Corp.), alkaline phosphatase (1 unit) (Roche Applied Science), chondroitinase AC-II (EC 4.2.2.5) from Arthrobacter aurescens (10 mIU), or α-N-acetylgalactosaminidase (EC 3.2.1.49) from Acremonium sp. (20 mIU) was carried out in a total volume of 20 μl of appropriate buffer at 37 °C overnight (25).
500-MHz 1H NMR Spectroscopy
The 2AB-oligosaccharide was repeatedly exchanged in D2O with intermittent lyophilization. The 1H NMR spectrum was measured in a Varian VXR-500 using a nanoprobe at 26 °C. Chemical shifts were given relative to sodium 4,4-dimethyl-4-silapentane-1-sulfonate but were actually measured indirectly relative to acetone (δ2.225) in D2O (26).
Expression of Soluble Forms of EXTL2, FAM20B, and EXT1·EXT2
Soluble forms of EXTL2, FAM20B, EXT1, and EXT2 each fused with the cleavable insulin signal sequence and the protein A IgG-binding domain were constructed as described (5, 16, 27). Each expression plasmid (6.0 μg) was transfected into COS-1 cells on 100-mm plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For co-transfection experiments, pairs of expression plasmids (3.0 μg each) were co-transfected into COS-1 cells on 100-mm plates using Lipofectamine 2000 as above. After 2 days of culture at 37 °C, 4 ml of the culture medium was collected and incubated with 50 μl of IgG-Sepharose (GE Healthcare) at 4 °C overnight. The beads were recovered by centrifugation, washed with, and then resuspended in each of the assay buffers described below. FAM20B kinase reactions (5) were incubated in reaction mixtures containing the following constituents in a total volume of 20 μl: 0.5 nmol of α-thrombomodulin with a truncated linkage region tetrasaccharide (GlcUAβ1–3Galβ1–3Galβ1–4Xyl), 10 μm [γ-32P]ATP (1.0 × 104 dpm), 50 mm Tris-HCl (pH 7.0), 10 mm MnCl2, 10 mm CaCl2, 0.1% BSA, and 10 μl of the resuspended beads. These mixtures were incubated for 4 h at 37 °C. The labeled products were isolated using a syringe column (1 ml) packed with Sephadex G-25 (28). Enzyme assays for α-GlcNAc transferase (29) and α-GalNAc transferase (30) were carried out as described previously (27, 28). The resultant products were isolated using a syringe column method and were then subjected to an HS chain polymerization reaction using an EXT1·EXT2 complex as the enzyme source (27). The reaction products were analyzed by gel filtration on a Superdex 75 HR 10/30 column, and the radioactivity in each collected fraction (1.2 ml each) was measured in a liquid scintillation counter (TRI-CARB 2900TR, Packard Instrument Co.).
Plasmid Construction and Establishment of FAM20B and EXTL2-expressing HeLa Clones
Human EXTL2 cDNA was amplified from human fetal placenta cDNA library via a reverse transcription-coupled polymerase chain reaction that included the following primers: 5′-GCTCTAGAACAAGTGAGCCTGCTT-3′ (XbaI site underlined) and 5′-CGGGATCCAAGCTACTCAAATGCCAAGCAG-3′ (BamHI site underlined). The resultant amplification products were inserted into the SrfI sites of the pCMVscript vector (Stratagene). The recombinant pCMV-hEXTL2 plasmid was double-digested with NotI and KpnI, and resulting DNA fragments were subcloned into NotI and KpnI sites of the pcDNA3.1-zeo(−) expression vector (Invitrogen). Integrity of the resulting vector, pcDNA3.1-zeo(−)-hEXTL2, was confirmed by sequencing the entire coding region and the ligation joints. The expression plasmid, pcDNA3.1-zeo(−)-hEXTL2, was transfected into HeLa cells that carried the pCMV empty vector (HeLa mock) or the pCMV-FAM20B vector (HeLa-FAM20B) using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions; both pCMV-HeLa cell lines had been established previously (5). Four kinds of transfectants (HeLa-mock-empty, HeLa-mock-hEXTL2, HeLa-FAM20B-empty, and HeLa-FAM20B-hEXTL2) were cultured in the presence of 800 μg/ml G418 and 150 μg/ml Zeocin (Invitrogen). G418 and Zeocin-resistant clones were then picked and propagated for experiments.
Statistical Analysis
Data are expressed as mean ± S.D. Statistical significance was determined by Student's t test.
RESULTS
Expression of EXTL2 mRNA during Embryogenesis and Expression of EXTL2 in Adult Tissues
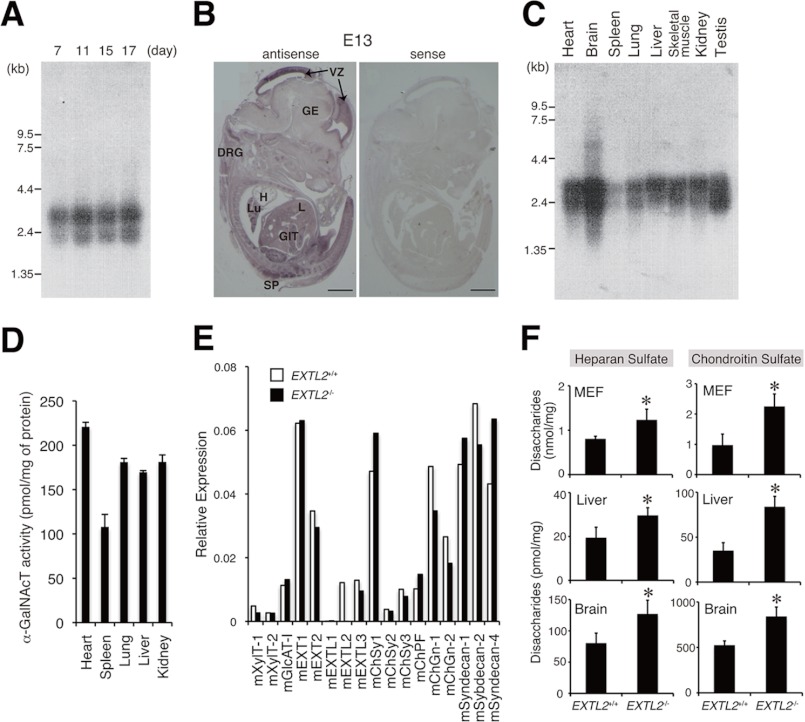
Findings from Northern blot analyses indicated that there were two transcript variants (∼2.8 and 2.2 kb) derived from mouse EXTL2 (Fig. 2, A and C). Both variants were detected at embryonic day 7, increased from embryonic day 7 to 11, and subsequently maintained a constant level (Fig. 2A). Based on in situ hybridization results, EXTL2 mRNA was ubiquitously expressed in embryonic tissues but was especially abundant in embryonic brain, spinal cord, lung, liver and gastrointestinal tract (Fig. 2B). In adult tissues, the strongest expression was evident in the brain, heart, and testis, where both transcript variants were expressed at comparable levels (Fig. 2C). EXTL2 mRNA was expressed at moderate levels in the lung, liver, skeletal muscle, and kidney, and the 2.8-kb signal was more intense than the 2.2-kb signal in each of these tissues (Fig. 2C). We measured α1,4-N-acetylgalactosaminyltransferase (α-GalNAcT) activities in several tissues as an indicator of the expression level of EXTL2 proteins (Fig. 2D). α-GalNAcT catalyzes the transfer of a GalNAc residue to the tetrasaccharide linkage region, GlcUAβ1–3Galβ1–3Galβ1–4Xyl, through α1,4-linkage. Each α-GalNAcT activity value represents the expression level of EXTL2 protein because all glycosyltransferases other than EXTL2 did not exhibit α-GalNAcT activities in vivo (supplemental Fig. 2). Consistent with the expression pattern of EXTL2 mRNA, EXTL2 enzyme activities were very highly expressed in the heart, whereas considerably higher expression of EXTL2 enzyme activities was observed in the liver, lung, and kidney (Fig. 2D).
FIGURE 2.
Analysis of the expression patterns of EXTL2 during embryogenesis and in adult tissues, the expression profile of GAG biosynthetic enzymes, and GAG synthesis in EXTL2+/+ or EXTL2−/− mice. A, mouse EXTL2 transcripts expressing during embryogenesis were detected on Northern blots using a radiolabeled mouse EXTL2 cDNA as a probe. Two transcripts of 2.2 and 2.8 kb were detected. B, tissue distribution of EXTL2 mRNA was examined using in situ hybridization. Sagittal sections of whole embryos at embryonic day 13 were hybridized with antisense probe and sense probes. VZ, ventricular zone; GE, ganglionic eminence; DRG, dorsal root ganglia; H, heart; Lu, lung; L, liver; GIT, gastrointestinal tract; SP, spinal cord. C, mouse EXTL2 transcripts in adult tissues were examined in Northern blots. D, α-GalNAcT activities in various adult mouse tissues were measured. E, the expression of genes encoding glycosyltransferases involved in GAG biosynthesis or the syndecans was assayed in embryonic fibroblasts (MEF) using a real-time PCR method. F, the amounts of HS and CS produced in MEF, liver, and brain were analyzed as described under “Experimental Procedures.” Data represent mean ± S.D. (error bars) of values from three specimens. *, p < 0.05.
HS and CS Levels Were Higher in EXTL2−/− Cells and Tissues than in EXTL2+/+ Cells and Tissues
We first checked levels of mRNA expression in MEFs derived from EXTL2+/+ and EXTL2−/− mice for genes encoding GAG biosynthetic enzymes or core proteins within some proteoglycans. The expression of mRNAs encoding GAG biosynthetic enzymes involved in the formation of linkage region (XylT-1, XylT-2, and GlcAT-I) or in the synthesis of the disaccharide repeating region of HS chains (EXT1, EXT2, EXTL1, and EXTL3) and CS chains (ChSy1, ChSy2, ChSy3, ChPF, ChGn-1, and ChGn-2) was similar in EXTL2−/− MEFs and EXTL2+/+ MEFs (Fig. 2E). In addition, the expression of mRNAs encoding core proteins of the syndecan family, which are relatively highly expressed in fibroblasts, were expressed at similar levels in EXTL2−/− MEFs and EXTL2+/+ MEFs. These results indicated that there were no differences between EXTL2+/+ and EXTL2−/− MEFs in the expression of mRNAs encoding GAG biosynthetic enzymes or syndecans. Next, we analyzed the amounts of HS and CS synthesized in MEFs, liver samples, and brain samples derived from EXTL2+/+ or EXTL2−/− mice (Fig. 2F). Lack of EXTL2 significantly increased the amount of HS and of CS in MEFs, in the liver, and in the brain (Fig. 2F). The EXTL2 deficiency hardly affected the sulfation patterns of HSs or CSs in samples of liver or brain from adult mice (8 weeks old) (Tables 1 and 2). In contrast, the EXTL2 deficiency greatly affected 6-O-sulfation of HSs and of CSs; sulfation levels were significantly and substantially lower in EXTL2−/− MEFs than in EXTL2+/+ MEFs (Tables 1 and 2). However, the mechanism by which EXTL2 affects 6-O-sulfation of HSs or CSs in MEFs remains unclear (see “Discussion”).
TABLE 1.
Disaccharide composition of HS from embryonic fibroblasts, livers, or brains of EXTL2+/+ or EXTL2−/− mice
Values are expressed as pmol of disaccharide per mg of dried homogenate of cells or tissues and the means ± S.E. of three (embryonic fibroblasts) or four determinations (livers and brains). Embryonic fibroblasts were isolated from EXTL2+/+ or EXTL2−/− embryos at embryonic day 18.5. Livers and brains were extirpated from mice (approximately 8 weeks old). ΔDiHS-0S, ΔHexUAα1–4GlcNAc; ΔDiHS-6S, ΔHexUAα1–4GlcNAc(6S); ΔDiHS-NS, ΔHexUAα1–4GlcNS; ΔDiHS-diS1, ΔHexUAα1–4Glc(NS,6S); ΔDi-diS2, ΔHexUA(2S)α1–4GlcNS; ΔDi-triS, ΔHexUA(2S)α1–4Glc(NS,6S). ND, not detected.
| Composition | Embryonic fibroblasts |
Liver |
Brain |
|||
|---|---|---|---|---|---|---|
| EXTL2+/+ | EXTL2−/− | EXTL2+/+ | EXTL2−/− | EXTL2+/+ | EXTL2−/− | |
| pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | ||||
| ΔDiHS-0S | 342.9 ± 41.5 (42.6) | 801.5 ± 267.6 (65.0) | 12.3 ± 1.8 (35.1) | 26.3 ± 4.6 (31.3) | 33.9 ± 5.5 (42.0) | 49.3 ± 5.2 (36.0) |
| ΔDiHS-6S | 123.3 ± 29.6 (15.3) | 56.8 ± 11.8 (4.6) | 1.6 ± 0.9 (4.7) | 4.5 ± 0.7 (5.4) | 9.3 ± 1.7 (10.9) | 15.1 ± 3.0 (12.7) |
| ΔDiHS-NS | 218.4 ± 12.2 (27.1) | 306.6 ± 70.0 (24.9) | 9.5 ± 1.0 (27.1) | 24.7 ± 3.3 (29.5) | 29.0 ± 8.6 (39.4) | 51.3 ± 12.3 (40.4) |
| ΔDiHS-diS1 | 60.9 ± 13.0 (7.6) | 9.9 ± 8.6 (0.8) | 3.5 ± 3.5 (9.9) | 4.5 ± 2.1 (5.4) | ND | ND |
| ΔDiHS-diS2 | 17.6 ± 24.9 (2.2) | 24.9 ± 11.5 (2.0) | 3.7 ± 0.9 (10.5) | 9.2 ± 3.5 (11.0) | 6.7 ± 1.4 (6.2) | 9.1 ± 2.0 (8.8) |
| ΔDiHS-triS | 41.8 ± 11.2 (5.2) | 32.9 ± 28.4 (2.7) | 4.5 ± 1.7 (12.7) | 10.1 ± 4.3 (12.1) | 1.5 ± 0.8 (1.6) | 2.3 ± 0.4 (2.1) |
| Total | 804.9 ± 58.3 | 1232.7 ± 239.8 | 35.1 ± 8.8 | 83.8 ± 11.6 | 80.4 ± 16.0 | 127.0 ± 22.0 |
TABLE 2.
Disaccharide composition of CS from embryonic fibroblasts, livers, or brains of EXTL2+/+ or EXTL2−/− mice
Values are expressed as pmol of disaccharide per mg of dried homogenate of cells or tissues and the means ± S.E. of three (embryonic fibroblasts) or four determinations (livers and brains). Embryonic fibroblasts were isolated from EXTL2+/+ or EXTL2−/− embryos at embryonic day 18.5. Livers and brains were extirpated from mice (approximately 8 weeks old). ΔDi-0S, ΔHexUAα1–3GalNAc; ΔDi-6S, ΔHexUAα1–3GalNAc (6S); ΔDi-4S, ΔHexUAα1–3GalNAc(4S); ΔDi-diSD, ΔHexUA(2S)α1–3GalNAc(6S); ΔDi-diSE, ΔHexUAα1–3GalNAc(4S,6S); ΔDi-triS, ΔHexUA(2S)α1–3GalNAc(4S,6S). ND, not detected.
| Composition | Embryonic fibroblasts |
Liver |
Brain |
|||
|---|---|---|---|---|---|---|
| EXTL2+/+ | EXTL2−/− | EXTL2+/+ | EXTL2−/− | EXTL2+/+ | EXTL2−/− | |
| pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | ||||
| ΔDi-0S | 51.5 ± 89.2 (5.3) | 194.6 ± 41.3 (8.6) | 2.7 ± 0.5 (13.6) | 2.6 ± 0.4 (8.8) | 49.8 ± 10.6 (9.5) | 65.5 ± 3.6 (7.8) |
| ΔDi-6S | 17.5 ± 17.6 (1.8) | 11.3 ± 10.4 (0.5) | ND | ND | 30.7 ± 1.5 (5.9) | 33.9 ± 1.9 (4.0) |
| ΔDi-4S | 717.3 ± 334.1 (73.7) | 2001.7 ± 354.7 (89.0) | 11.9 ± 4.1 (61.2) | 17.8 ± 2.4 (60.1) | 436.4 ± 35.9 (83.2) | 723.8 ± 99.4 (87.1) |
| ΔDi-diSD | ND | ND | ND | ND | 7.7 ± 5.9 (1.5) | 15.9 ± 2.8 (1.9) |
| ΔDi-diSE | 187.4 ± 112.9 (19.2) | 42.5 ± 11.3 (1.9) | 4.9 ± 2.4 (25.4) | 9.2 ± 1.2 (30.9) | ND | ND |
| ΔDi-triS | ND | ND | ND | ND | ND | ND |
| Total | 973.8 ± 362.8 | 2250.0 ± 414.0 | 19.5 ± 4.8 | 29.6 ± 3.5 | 524.6 ± 46.8 | 841.5 ± 102.5 |
Accumulation of Phosphorylated Pentasaccharide Linkages That Terminate in α-GlcNAc in EXTL2+/+ Liver
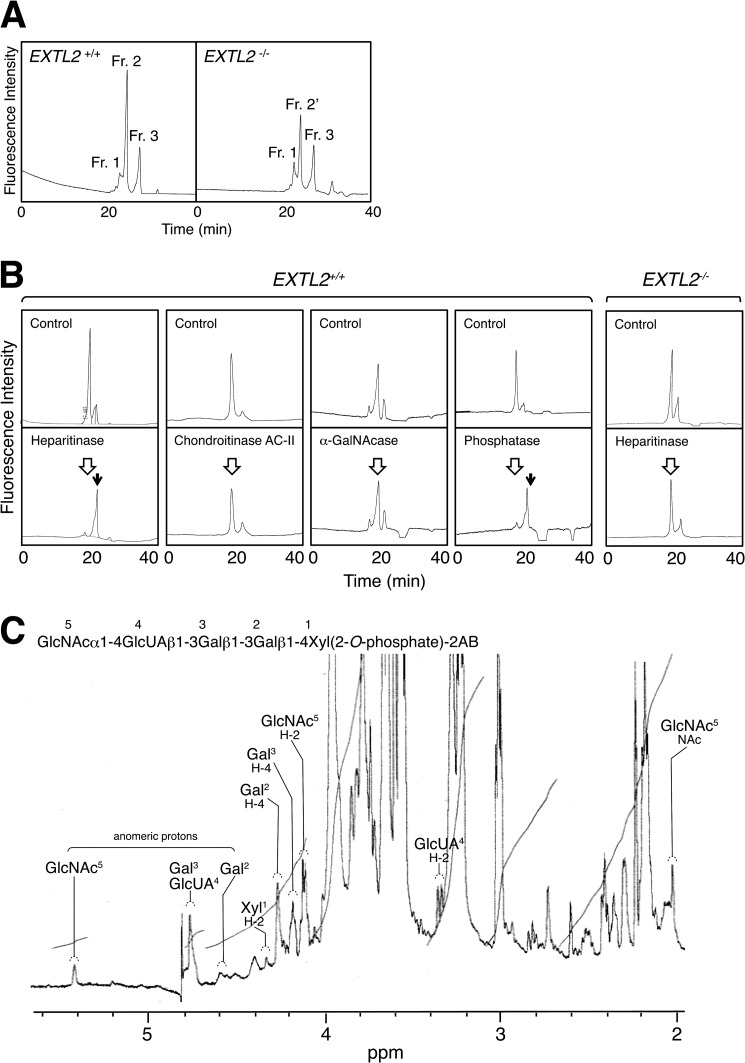
We previously demonstrated that EXTL2 encodes an enzyme with two catalytic activities, an α-GalNAcT activity and an N-acetylglucosaminyltransferase-I (GlcNAcT-I) activity, namely an α1,4-N-acetylhexosaminyltransferase activity that transfers GalNAc/GlcNAc to a tetrasaccharide that represents the common GAG-protein linkage region (16). In addition, the α-GalNAc-capped pentasaccharide serine (GalNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser), which is a reaction product of α-GalNAcT, was not utilized as an acceptor for polymerization; this finding indicated that the addition of an α-GalNAc residue may serve as a stop signal that precludes further elongation of HS chains and of CS chains (25). Based upon these results, we speculated that EXTL2 negatively regulates GAG biosynthesis by adding a stop signal to a growing chain; thus, lack of EXTL2 might increase the amount of HS and CS. Here, we investigated α-GalNAc-capped pentasaccharide linkage structure on naturally occurring proteoglycans in EXTL2+/+ liver. O-Glycosidically linked oligosaccharides were liberated by mild alkaline treatment from powder derived from acetone-treated liver; these oligosaccharides were then purified from the slurry by passage through a cation exchange column as described under “Experimental Procedures.” The isolated oligosaccharide fraction was derivatized with a fluorophore, 2AB, and fractionated by gel filtration HPLC; this process resulted in three fluorescent components (Fig. 3A). The fraction 2 peak was smaller in samples from EXTL2−/− liver than in samples from EXTL2+/+ liver (Fig. 3A); therefore, fraction 2 may have contained the target oligosaccharides. Consequently, samples of fraction 2 were subjected to further analyses, but fractions 1 and 3 were not analyzed further. The oligosaccharides in fraction 2 isolated from EXTL2+/+ liver were digested with heparitinase, which can cleave the α1–4 linkage of GlcNAcα1-4GlcUA. Interestingly, these heparitinase-treated oligosaccharides shifted to the elution position of GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB (Fig. 3B). In contrast, the oligosaccharides in fraction 2 were resistant to chondroitinase AC-II, which can cleave the β1–4 linkage of GalNAcβ1–4GlcUA, and to α-N-acetylgalactosaminidase (α-GalNAcase), which removes an α-linked GalNAc residue located at the non-reducing terminus of sugar chains (Fig. 3B). Notably, the oligosaccharides in fraction 2 were sensitive to alkaline phosphatase. To date, structural studies have shown that a Xyl residue at position 2 in the GAG-protein linkage region is phosphorylated (1). Based on these findings, we hypothesized that the structure of the oligosaccharides in fraction 2 was GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1-4Xyl(2-O-phosphate)-2AB. Furthermore, the oligosaccharides in fraction 2′ isolated from EXTL2−/− liver were resistant to heparitinase and chondroitinase AC-II, suggesting that the oligosaccharides had neither GlcNAcα1–4GlcUA nor GalNAcβ1–4GlcUA structure at the non-reducing terminus. Thus, we concluded that components containing fraction 2′ were not GAG-derived oligosaccharides. To test our hypothesis regarding the oligosaccharides in fraction 2 that were isolated from EXTL2+/+ liver, we used 1H NMR spectroscopy to characterize the oligosaccharides in fraction 2. The one-dimensional spectrum of the isolated oligosaccharides was recorded at 26 °C (Fig. 3C). Signals at δ4.4–5.5 ppm were identified as H-1 resonances of the constituent saccharide residues by comparison with the NMR spectra of the previously reported linkage oligosaccharides (31). NMR data from the analysis of the saccharides are summarized in Table 3. Based on these data, we concluded that the elongation of some GAG chains was terminated after a α-GlcNAc residue was added, via an α1–4 linkage, to a linkage tetrasaccharide that harbored a phosphorylated xylose residue; the structure of these phosphorylated linkers was GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate).
FIGURE 3.
Characterization of the truncated oligosaccharides isolated from wild-type mouse liver. A, the 2AB-labeled oligosaccharides were fractionated by gel filtration HPLC on a GS-320 column. B, fraction 2 isolated from the liver was digested with various glycosidases, and digests were analyzed by gel filtration HPLC on a column of GS-320. The open arrows and the filled arrows denote the elution positions of undigested compound and the authentic compound, GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-2AB, respectively. C, 500-MHz 1H NMR one-dimensional spectrum of fraction 2. The letters and numbers refer to the corresponding residues in the structure.
TABLE 3.
1H chemical shifts of the constituent monosaccharides within linkage oligosaccharides
Chemical shifts are given in ppm relative to sodium 4,4-dimethyl-4-silapentane-1-sulfonate (26) but were actually measured indirectly relative to the values to acetone in D2O at 26 °C. 1H chemical shifts of the constituent monosaccharides of fraction 2 are shown together with those of the reference compounds: R1, GlcUAβ1–3Galβ1–3Galβ1–4Xyl-2AB (23); R2, GalNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl-O-Ser (31); R3, ΔHexUAα1–3GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate)-ol (36). ND, not determined.
| Residue | Fraction 2 | R1 (GlcUA-Gal-Gal-Xyl-2AB) | R2 (GalNAcα1–4GlcUA-Gal-Gal-Xyl-Ser) | R3 (ΔHexA-GalNAc-GlcUA-Gal-Gal-Xyl(2P)-ol) |
|---|---|---|---|---|
| Xyl-1 or Xyl-ol | ||||
| H-1α | 3.785 | |||
| H-1β | 4.431 | 3.872 | ||
| H-2 | 4.35 | 3.94 | 3.358 | 4.267 |
| H-3 | NDa | 3.80 | ND | 3.89 |
| H-4 | 4.04 | 4.02 | ND | 4.034 |
| H-5 | ND | ND | ND | 3.89 |
| Gal-2 | ||||
| H-1 | 4.56 | 4.618 | 4.53 | 4.628 |
| H-2 | ND | 3.73 | ND | 3.736 |
| H-3 | ND | 3.80 | ND | 3.849 |
| H-4 | 4.28 | 4.188 | 4.171 | 4.186 |
| H-5 | ND | ND | ND | ND |
| H-6 | ND | ND | ND | ND |
| Gal-3 | ||||
| H-1 | 4.78 | 4.670 | 4.652 | 4.666 |
| H-2 | ND | 3.75 | ND | 3.744 |
| H-3 | ND | 3.80 | ND | 3.806 |
| H-4 | 4.20 | 4.188 | 4.171 | 4.161 |
| H-5 | ND | ND | ND | ND |
| H-6 | ND | ND | ND | ND |
| GlcUA-4 | ||||
| H-1 | 4.78 | 4.672 | 4.667 | 4.666 |
| H-2 | 3.37 | 3.42 | 3.436 | 3.455 |
| H-3 | ND | ND | 3.69 | 3.625 |
| H-4 | ND | ND | ND | 3.781 |
| H-5 | ND | ND | ND | 3.722 |
| GlcNAc-5 or GalNAc-5 | ||||
| H-1 | 5.44 | 5.437 | 4.532 | |
| H-2 | 4.14 | 4.153 | 4.003 | |
| H-3 | ND | ND | 3.905 | |
| H-4 | ND | ND | 4.098 | |
| H-5 | ND | ND | ND | |
| H-6 | ND | ND | ND | |
| NAc | 2.045 | 2.049 | 2.059 | |
Transfer of GlcNAc through α1,4-Linkage to GlcUA-Gal-Gal-Xyl(2-O-phosphate) on α-Thrombomodulin by EXTL2
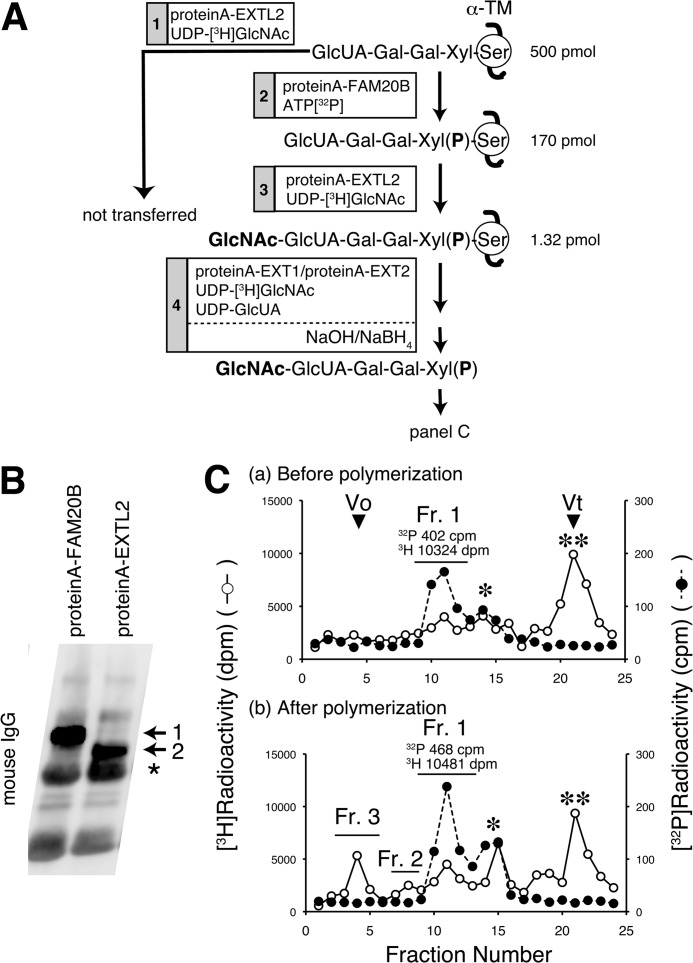
We next used an in vitro approach to examine whether transfer of a GlcNAc residue to the phosphorylated linkage tetrasaccharide was catalyzed by EXTL2. In a preliminary experiment, we had found that EXTL2 could not transfer α-GlcNAc to an unmodified tetrasaccharide linkage region, GlcUAβ1–3Galβ1–3Galβ1–4Xyl. Therefore, we investigated the significance of phosphorylation of the xylose residue in the EXTL2-catalyzed transfer of α-GlcNAc to the tetrasaccharide linkage region. We used α-thrombomodulin (α-TM) carrying a truncated tetrasaccharide linkage structure, GlcUA-Gal-Gal-Xyl (23), as an acceptor substrate. We recently identified FAM20B as a kinase involved in the phosphorylation of the xylose residue (5). After FAM20B-catalyzed phosphorylation of the xylose residue, EXTL2-catalyzed addition of GlcNAc onto the phosphorylated tetrasaccharide structure, GlcUA-Gal-Gal-Xyl(2-O-phosphate), on an α-TM was examined. The experimental design is illustrated in Fig. 4A. Functional, soluble forms of the enzymes were expressed in COS-1 cells as protein A-tagged fusion proteins. Cells secreted the protein A-tagged enzymes into medium, and these tagged enzymes were purified using IgG-Sepharose beads. After checking the expression level of each tagged enzyme (Fig. 4B), we used the purified proteins as an enzyme source. EXTL2 could not transfer [3H]GlcNAc to α-TM before FAM20B-catalyzed phosphorylation (reaction 1 in Fig. 4A). However, [3H]GlcNAc (1.32 pmol) was readily added onto 32P-labeled tetrasaccharides on α-TM (170 pmol) via EXTL2 (reaction 3 in Fig. 4A) following FAM20B-catalyzed phosphorylation of this tetrasaccharide (reaction 2 in Fig. 4A). These results indicated that EXTL2 specifically transferred a GlcNAc residue to the phosphorylated tetrasaccharide.
FIGURE 4.
Analysis of the termination mechanism of GAG chains by EXTL2. A, the experimental design is shown. Reaction 1, α-TM (500 pmol) was used as an acceptor for the GlcNAc transfer reaction catalyzed by recombinant protein A-EXTL2. Recombinant EXTL2 could not transfer a GlcNAc to unmodified linkage tetrasaccharide on α-TM. Reaction 2, 170 pmol of 500 pmol of α-TM was phosphorylated by protein A-FAM20B. Reaction 3, phosphorylated linkage tetrasaccharide was used as an acceptor for the GlcNAc transfer reaction catalyzed by protein A-EXTL2, and consequently 1.32 pmol of phosphorylated linkage pentasaccharide was produced. Reaction 4, after the polymerization reaction was carried out using protein A-EXT1·protein A-EXT2 complexes as an enzyme source, sugar chains released from core proteins by alkaline β-elimination with NaOH/NaB3H4 were analyzed by gel filtration chromatography (C). B, protein A-FAM20B and protein A-EXTL2 expressed in COS-1 cells were collected via IgG-Sepharose and analyzed on immunoblots. 1, protein A-FAM20B; 2, protein A-EXTL2; *, IgG. C, the products obtained from reactions 3 and 4 described in A were subjected to gel filtration chromatography using a Superdex 75 column, and the elution profiles of the products from reactions 3 and 4 are shown in a and b, respectively. Fr. 1, 5–6-mer; Fr. 2, 8–12-mer; Fr. 3, larger molecules; *, UDP-[3H]GlcNAc, free [3H]H2PO4, or free [3H]GlcNAc; **, [3H]HO.
The EXT1·EXT2 Polymerase Complex Could Not Use a Phosphorylated Pentasaccharide, GlcNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate) as a Primer for Polymerization
We previously demonstrated that a non-phosphorylated pentasaccharide linkage structure GlcNAc-GlcUA-Gal-Gal-Xyl functions as a good acceptor in the polymerization reaction (21). Here we investigated whether polymerization could occur on a phosphorylated linkage pentasaccharide GlcNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate). A phosphorylated pentasaccharide GlcNAc-GlcUA-Gal-Gal-Xyl(2-O-phosphate) attached to α-TM was generated using the tetrasaccharide GlcUA-Gal-Gal-Xyl attached to α-TM as a primer and recombinant FAM20B and EXTL2 as enzyme sources (reactions 2 and 3 in Fig. 4A). This pentasaccharide-protein was incubated with the HS polymerase (i.e. functional EXT1·EXT2 complexes) in the presence of UDP-[3H]GlcNAc and UDP-GlcUA (reaction 4 in Fig. 4A); the EXT1 and EXT2 proteins had been co-expressed as protein A fusion proteins. After the polymerization reaction, NaB3H4-mediated alkaline β-elimination was used to release the O-linked sugar chains from the core α-TM proteins; these sugar chains were then subjected to gel filtration analysis using a Superdex 75 column. Incorporation of [3H]radioactivity into phosphorylated pentasaccharides (fraction 1) did not differ between prepolymerization and postpolymerization reaction mixtures (Fig. 4C). Octa- or decasaccharides (fraction 2) and polymerized chains (fraction 3) were not labeled with 32P, but they were labeled with 3H, probably because residual non-phosphorylated linkage structures would be used as an acceptor (Fig. 4C). These results indicated that polymerization and transfer of a sugar residue never occurred on phosphorylated pentasaccharides.
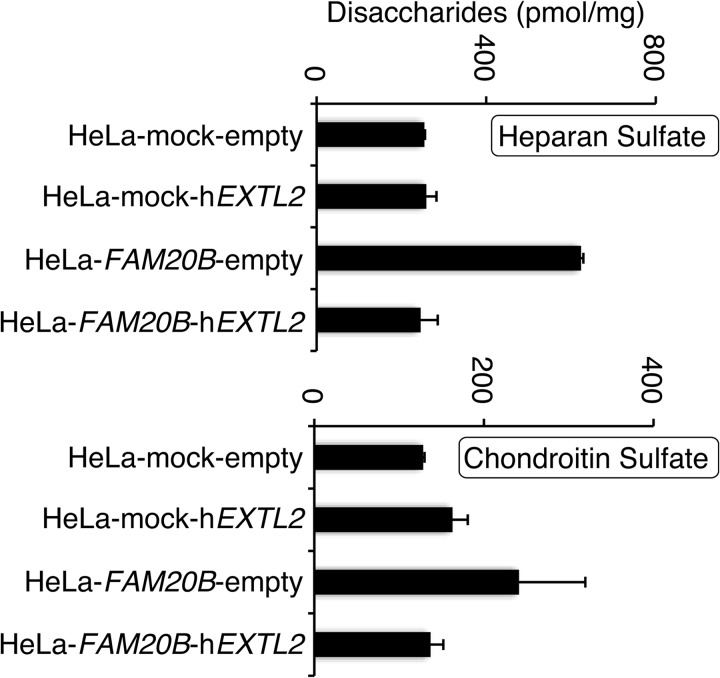
To investigate whether EXTL2 could suppress the GAG biosynthesis that is augmented by FAM20B, we generated HeLa cells that expressed either FAM20B or EXTL2 or both FAM20B and EXTL2. FAM20B-expressing HeLa cells produced many more GAG chains than did HeLa cells carrying an empty vector. Interestingly, introduction of EXTL2 into FAM20B-expressing HeLa cells attenuated the GAG synthesis; the levels of GAG synthesis were similar in HeLa cells expressing both FAM20B and EXTL2 and HeLa cells carrying both empty vectors, pCMV and pcDNA3.1-zeo(−) (Fig. 5 and Tables 4 and 5). These results indicated that EXTL2 could control the GAG biosynthesis that was accelerated by FAM20B.
FIGURE 5.
The analysis of GAGs synthesized by HeLa clones that expressed FAM20B, EXTL2, both, or neither. The expression plasmid, pcDNA3.1-zeo(−)-hEXTL2, was transfected into HeLa cells carrying pCMV empty vector (HeLa mock) or pCMV-FAM20B vector (HeLa-FAM20B). Four kinds of transfectants (HeLa-mock-empty, HeLa-mock-hEXTL2, HeLa-FAM20B-empty, and HeLa-FAM20B-hEXTL2) were subjected to GAG disaccharide composition analysis. Error bars, S.D.
TABLE 4.
Disaccharide composition of HS from HeLa clones
Values are expressed as pmol of disaccharide/mg of dried homogenate of cells and the means ± S.E. of two determinations. ΔDiHS-0S, ΔHexUAα1–4GlcNAc; ΔDiHS-6S, ΔHexUAα1–4GlcNAc(6S); ΔDiHS-NS, ΔHexUAα1–4GlN(NS); ΔDiHS-diS1, ΔHexUAα1–4GlcN(NS,6S); ΔDiHS-diS2, ΔHexUA(2S)α1–4GlcN(NS); ΔDiHS-triS, ΔHexUA(2S)α1–4GlcN(NS,6S). ND, not detected.
| Composition | HeLa-mock | HeLa-mock-hEXTL2 | HeLa-FAM20B-empty | HeLa-FAM20B-hEXTL2 |
|---|---|---|---|---|
| pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | |
| ΔDiHS-0Sa | 104.9 ± 8.3 (41.5) | 123.3 ± 41.6 (47.7) | 276.9 ± 18.3 (44.4) | 113.3 ± 5.0 (46.2) |
| ΔDiHS-6S | 7.9 ± 11.1 (2.8) | 8.6 ± 1.1 (3.6) | 12.1 ± 3.3 (2.0) | 7.8 ± 5.0 (3.1) |
| ΔDiHS-NS | 91.8 ± 16.0 (36.1) | 82.3 ± 22.8 (32.2) | 225.0 ± 17.7 (36.2) | 73.8 ± 2.2 (30.1) |
| ΔDiHS-diS1 | ND | ND | ND | ND |
| ΔDiHS-diS2 | 23.3 ± 2.8 (9.2) | 21.1 ± 13.2 (7.7) | 48.6 ± 8.3 (7.8) | 19.6 ± 6.8 (7.9) |
| ΔDiHS-triS | 26.3 ± 2.7 (10.4) | 23.5 ± 11.8 (8.8) | 61.2 ± 22.8 (9.7) | 30.9 ± 2.3 (12.7) |
| Total | 254.2 ± 41.0 | 258.8 ± 88.3 | 623.8 ± 28.5 | 245.4 ± 16.7 |
TABLE 5.
Disaccharide composition of CS from HeLa clones
Values are expressed as pmol of disaccharide/mg of dried homogenate of cells and the means ± S.E. of two determinations. ΔDi-0S, ΔHexUAα1–3GalNAc; ΔDi-6S, ΔHexUAα1–3GalNAc (6S); ΔDi-4S, ΔHexUAα1–3GalNAc(4S); ΔDi-diSD, ΔHexUA(2S)α1–3GalNAc(6S); ΔDi-diSE, ΔHexUAα1–3GalNAc(4S,6S); ΔDi-triS, ΔHexUA(2S)α1–3GalNAc(4S,6S). ND, not detected.
| Composition | HeLa-mock | HeLa-mock-hEXTL2 | HeLa-FAM20B-empty | HeLa-FAM20B-hEXTL2 |
|---|---|---|---|---|
| pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | pmol/mg (mol %) | |
| ΔDi-0S | 3.2 ± 0.02 (2.5) | 2.8 ± 0.2 (1.8) | 3.4 ± 1.4 (1.4) | 3.8 ± 2.3 (2.8) |
| ΔDi-6S | 17.7 ± 0.9 (13.8) | 36.1 ± 8.9 (22.0) | 77.3 ± 27.2 (31.9) | 31.5 ± 3.4 (23.2) |
| ΔDi-4S | 107.4 ± 0.7 (83.7) | 124.1 ± 8.9 (76.3) | 160.6 ± 49.6 (66.7) | 101.9 ± 20.8 (74.0) |
| ΔDi-diSD | ND | ND | ND | ND |
| ΔDi-diSE | ND | ND | ND | ND |
| ΔDi-triS | ND | ND | ND | ND |
| Total | 128.4 ± 1.6 | 163.0 ± 18.0 | 241.4 ± 78.2 | 137.1 ± 15.1 |
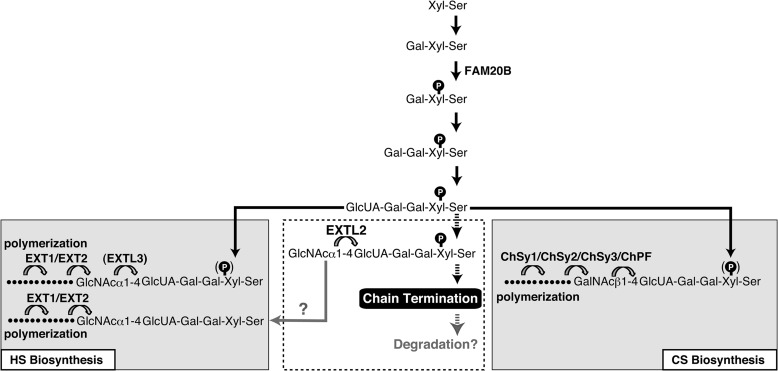
These results are summarized in Fig. 6. During formation of the tetrasaccharide linkage structure, Xyl residues are transiently phosphorylated. Transient phosphorylation of Xyl residues is thought to enhance the formation of linkage region (4, 5, 32). Subsequently, rapid dephosphorylation is induced just before initiation of polymerization; however, the enzyme responsible for the dephosphorylation of xylose remains unknown. Notably, the formation of the disaccharide repeat region of HS or CS chains is initiated on a dephosphorylated tetrasaccharide linkage structure. If there is a time lag before dephosphorylation, EXTL2 can transfer GlcNAc to phosphorylated linkage tetrasaccharides, leading to termination of GAG synthesis (Fig. 6). Based on these findings, we propose that EXTL2 functions to suppress the GAG synthesis that is enhanced by FAM20B and that this EXTL2-mediated suppression regulates GAG synthesis.
FIGURE 6.
GAG biosynthesis and the EXTL2-dependent mechanism for termination of GAG chains. Synthesis of the linkage region is initiated by the addition of a Xyl residue to a specific serine residue on a core protein, followed by the sequential transfer of two Gal residues, and completed by transfer of a GlcUA residue. During synthesis of the linkage region, a transient FAM20B-catalyzed phosphorylation of the Xyl residue occurs and enhances linkage region synthesis. Before subsequent polymerization, the Xyl residue is dephosphorylated by an unknown phosphatase, and N-acetylhexosamine and GlcUA residues are then alternatively added. HS chain synthesis occurs when a GlcNAc is transferred to the linkage tetrasaccharide, but CS chain synthesis occurs when a GalNAc is the first residue added to the linker. If the formation of linkage region is excessively accelerated by FAM20B, biosynthetic intermediates (phosphorylated linkage tetrasaccharide) could accumulate; under this condition, EXTL2 may transfer a GlcNAc to the phosphorylated linkage tetrasaccharide and, thereby, induce chain termination.
DISCUSSION
Since we cloned an EXTL2 gene in 1999 (16), we have sought to understand the role of EXTL2 in GAG biosynthesis. We demonstrated that EXTL2 has dual glycosyltransferase activities based on in vitro studies (16), including an α-N-acetylgalactosaminyltransferase (α-GalNAcT) activity that transfers the first GalNAc residue to the GAG-protein linkage tetrasaccharide, GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser, via an α1,4-linkage. However, a reaction product of the α-GalNAcT, GalNAcα1–4GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser, is not used as an acceptor for the glucuronyltransferase involved in the CS biosynthesis (31), suggesting that the addition of an α-GalNAc residue may serve as a stop signal that precludes further chain elongation. EXTL2 also has a GlcNAcT-I activity that transfers the first GlcNAc residue to the GAG-protein linkage tetrasaccharide, GlcUAβ1–3Galβ1–3Galβ1–4Xylβ1-O-Ser, via an α1,4-linkage. Thus, to date, EXTL2 has been thought to function as a GlcNAcT-I in initiation of the biosynthesis of HS. Here, we generated EXTL2-deficient mice and investigated what effect loss of EXTL2 had on GAG biosynthesis. Interestingly, both HS and CS levels were higher in EXTL2-deficient mice than in wild-type mice (Fig. 2F). These results indicated that EXTL2 might not be involved in HS biosynthesis as a HS biosynthetic enzyme in vivo. Therefore, we explored a naturally occurring truncated GAG structure on the assumption that EXTL2 could induce chain termination by its α-GalNAcT or GlcNAcT-I activity. Based on structural analysis by a combination of glycosidase digestion and 500-MHz 1H NMR spectroscopy, an oligosaccharide isolated from the mouse liver was found to be GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1-4Xyl(2-O-phosphate), which was considered to be a biosynthetic intermediate of an immature GAG chain. This result indicated that EXTL2 transferred a GlcNAc residue to a phosphorylated linkage tetrasaccharide to stop chain polymerization (Figs. 4 and 6). Remarkably, the phosphorylated linkage pentasaccharide, GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1-4Xyl(2-O-phosphate), was not used as an acceptor for HS polymerases (Fig. 4) or for CS polymerases. Furthermore, our previous study raised the possibility that dephosphorylated linkage pentasaccharide, GlcNAcα1–4GlcUAβ1–3Galβ1–3Galβ1-4Xyl, is utilized as a primer for HS biosynthesis (21). These results suggest that persistent xylose phosphorylation of the pentasaccharide may prevent further polymerization of HS. On the other hand, our preliminary results have indicated that phosphorylated linkage tetrasaccharide is a good acceptor for chondroitin β1,4-N-acetylgalactosaminyltransferase-1 (33), and the resulting phosphorylated linkage pentasaccharide, GalNAcβ1–4GlcUAβ1–3Galβ1–3Galβ1–4Xyl(2-O-phosphate), can be utilized as a primer for subsequent polymerization of CS chains (data not shown), suggesting that phosphorylation of Xyl alone does not necessarily function as a stop signal. Based on these findings, we propose that both steps, the phosphorylation of a Xyl residue and EXTL2-mediated transfer of a GlcNAc residue, are involved in chain termination.
Whether the phosphorylated linkage pentasaccharide produced by EXTL2 can be recycled for the HS biosynthesis or be degraded remains unclear. Our previous studies suggest that the phosphorylated linkage pentasaccharide might be metabolized. In EXT1-deficient cells, a large number of phosphorylated linkage pentasaccharides may be generated, because the hetero-oligomeric EXT1·EXT2 complex is the biologically relevant form of HS polymerase. Notably, we have shown that EXT2 alone can achieve HS polymerization with the aid of GlcNAcT-I activity of EXTL2, even in the absence of EXT1 (21). These results indicate that a fraction of the intermediates that accumulate in EXT1-deficient cells could serve as a primer for the elongation of HS chains after dephosphorylation of each chain by an unknown phosphatase. Therefore, the dephosphorylation may be a way of quickly increasing HS synthesis upon physiological stimuli that require increased HS. Furthermore, we believe that the linkage pentasaccharide marks a regulatory branch point between synthesis and degradation of nascent HS chains, although we have not demonstrated that this pentasaccharide is degraded (Fig. 6).
We next considered the biological significance of the EXTL2-dependent mechanism that regulates GAG synthesis. If chain polymerization takes place at a slow rate despite accelerated formation of the linkage region, the synthesis of a linkage region will be uncoupled from subsequent polymerization. In this situation, it is thought that EXTL2 functions as a brake to control GAG biosynthesis. Unless the EXTL2-dependent regulation system operates, an increase in supply of tetrasaccharide linkage (primers for chain polymerization) would cause a shortage of GAG-modifying enzymes, and consequently abnormally modified PGs might be produced. In fact, sulfation patterns of GAG chains produced in EXTL2−/− MEFs were different from those in EXTL2+/+ MEFs (Tables 1 and 2). These poor quality PGs may be associated with some pathological conditions. Thus, we are now studying the EXTL2-dependent mechanism that regulates GAG biosynthesis from the perspective that it is a “quality control system” for PGs.
Recently,4 we found that abnormal biosynthesis of GAGs due to lack of EXTL2 affected the liver injury and regeneration processes. Proliferation of EXTL2−/− hepatocytes after carbon tetrachloride (CCl4) administration was suppressed, and EXTL2−/− hepatocytes exhibit higher sensitivity to CCl4 than did EXTL2+/+ hepatocytes. In fact, EXTL2−/− mice experienced less hepatocyte growth factor-mediated signaling than did wild-type mice specifically because GAG synthesis was altered in these mutant mice. Therefore, EXTL2-mediated regulation of GAG synthesis is important to the tissue regeneration processes that follow liver injury.
Here we report on the functional importance of EXTL2 in the GAG biosynthesis. EXTL2 orthologues have not been identified in invertebrates, such as Drosophilla, and they have only been found in vertebrates. Thus, it might be suggested that EXTL2 is involved in higher functions (e.g. functions in the nervous system). In addition, we found that EXTL2 could control GAG biosynthesis in a FAM20B-dependent manner. FAM20B is reportedly a Xyl kinase that regulates GAG biosynthesis (5) and controls the timing of endochondral ossification by inhibiting chondrocyte maturation (32). In addition, the expression level of EXTL2 reportedly increases during chondrogenesis (34). Based on these findings, we propose that EXTL2 plays an important role in the regulation of GAG synthesis in concert with FAM20B and that an imbalance between EXTL2 and FAM20B functions may lead to abnormal endochondral ossification. Moreover, the EXTL2 gene was localized to chromosome 1q11-12, a position adjacent to the chromosomal location of the CSF-1 gene, which is mutated in the op/op mouse model for osteopetrosis (35). EXTL2 might be also associated with pathological conditions such as osteopetrosis.
Acknowledgments
We thank Hiromi Shimakawa and Noriyuki Egusa for technical support.
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT)-supported Program for the Strategic Research Foundation at Private Universities, 2012–2017 (to H. K.) and was supported in part by a Grant-in-Aid for Scientific Research in Innovative Areas 23110002 (Deciphering Sugar Chain-based Signals Regulating Integrative Neuronal Functions (to H. K.)) from MEXT, Japan.

This article contains supplemental Table 1 and Figs. 1 and 2.
S. Nadanaka, S. Kagiyama, and H. Kitagawa, submitted for publication.
- GAG
- glycosaminoglycan
- 2AB
- 2-aminobenzamide
- α-GalNAcT
- α1,4-N-acetylgalactosaminidase
- α-TM
- α-thrombomodulin
- CS
- chondroitin sulfate
- EXT
- exostosin
- GlcNAcT-I
- N-acetylglucosaminyltransferase-I
- HS
- heparan sulfate
- MEF
- mouse embryonic fibroblasts
- PG
- proteoglycan.
REFERENCES
- 1. Sugahara K., Kitagawa H. (2000) Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 10, 518–527 [DOI] [PubMed] [Google Scholar]
- 2. Bernfield M., Götte M., Park P. W., Reizes O., Fitzgerald M. L., Lincecum J., Zako M. (1999) Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68, 729–777 [DOI] [PubMed] [Google Scholar]
- 3. Salmivirta M., Lidholt K., Lindahl U. (1996) Heparan sulfate. A piece of information. FASEB J. 10, 1270–1279 [DOI] [PubMed] [Google Scholar]
- 4. Fransson L. A., Belting M., Jönsson M., Mani K., Moses J., Oldberg A. (2000) Biosynthesis of decorin and glypican. Matrix Biol. 19, 367–376 [DOI] [PubMed] [Google Scholar]
- 5. Koike T., Izumikawa T., Tamura J., Kitagawa H. (2009) FAM20B is a kinase that phosphorylates xylose in the glycosaminoglycan-protein linkage region. Biochem. J. 421, 157–162 [DOI] [PubMed] [Google Scholar]
- 6. Wise C. A., Clines G. A., Massa H., Trask B. J., Lovett M. (1997) Identification and localization of the gene for EXTL, a third member of the multiple exostoses gene family. Genome Res. 7, 10–16 [DOI] [PubMed] [Google Scholar]
- 7. Wuyts W., Van Hul W., Hendrickx J., Speleman F., Wauters J., De Boulle K., Van Roy N., Van Agtmael T., Bossuyt P., Willems P. J. (1997) Identification and characterization of a novel member of the EXT gene family, EXTL2. Eur. J. Hum. Genet. 5, 382–389 [PubMed] [Google Scholar]
- 8. Saito T., Seki N., Yamauchi M., Tsuji S., Hayashi A., Kozuma S., Hori T. (1998) Structure, chromosomal location, and expression profile of EXTR1 and EXTR2, new members of the multiple exostoses gene family. Biochem. Biophys. Res. Commun. 243, 61–66 [DOI] [PubMed] [Google Scholar]
- 9. Senay C., Lind T., Muguruma K., Tone Y., Kitagawa H., Sugahara K., Lidholt K., Lindahl U., Kusche-Gullberg M. (2000) The EXT1/EXT2 tumor suppressors. Catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 1, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCormick C., Duncan G., Goutsos K. T., Tufaro F. (2000) The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc. Natl. Acad. Sci. U.S.A. 97, 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hecht J. T., Hogue D., Strong L. C., Hansen M. F., Blanton S. H., Wagner M. (1995) Hereditary multiple exostosis and chondrosarcoma. Linkage to chromosome II and loss of heterozygosity for EXT-linked markers on chromosomes II and 8. Am. J. Hum. Genet. 56, 1125–1131 [PMC free article] [PubMed] [Google Scholar]
- 12. Raskind W. H., Conrad E. U., Chansky H., Matsushita M. (1995) Loss of heterozygosity in chondrosarcomas for markers linked to hereditary multiple exostoses loci on chromosomes 8 and 11. Am. J. Hum. Genet. 56, 1132–1139 [PMC free article] [PubMed] [Google Scholar]
- 13. Nadanaka S., Kitagawa H. (2008) Heparan sulphate biosynthesis and disease. J. Biochem. 144, 7–14 [DOI] [PubMed] [Google Scholar]
- 14. Wuyts W., Spieker N., Van Roy N., De Boulle K., De Paepe A., Willems P. J., Van Hul W., Versteeg R., Speleman F. (1999) Refined physical mapping and genomic structure of the EXTL1 gene. Cytogenet. Cell Genet. 86, 267–270 [DOI] [PubMed] [Google Scholar]
- 15. Arai T., Akiyama Y., Nagasaki H., Murase N., Okabe S., Ikeuchi T., Saito K., Iwai T., Yuasa Y. (1999) EXTL3/EXTR1 alterations in colorectal cancer cell lines. Int. J. Oncol. 15, 915–919 [DOI] [PubMed] [Google Scholar]
- 16. Kitagawa H., Shimakawa H., Sugahara K. (1999) The tumor suppressor EXT-like gene EXTL2 encodes an α1,4-N-acetylhexosaminyltransferase that transfers N-acetylgalactosamine and N-acetylglucosamine to the common glycosaminoglycan-protein linkage region. The key enzyme for the chain initiation of heparan sulfate. J. Biol. Chem. 274, 13933–13937 [DOI] [PubMed] [Google Scholar]
- 17. Kitagawa H., Taoka M., Tone Y., Sugahara K. (2001) Human glycosaminoglycan glucuronyltransferase I gene and a related processed pseudogene. Genomic structure, chromosomal mapping and characterization. Biochem. J. 358, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kühn R., Rajewsky K., Müller W. (1991) Generation and analysis of interleukin-4 deficient mice. Science 254, 707–710 [DOI] [PubMed] [Google Scholar]
- 19. Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nadanaka S., Ishida M., Ikegami M., Kitagawa H. (2008) Chondroitin 4-O-sulfotransferase-1 modulates Wnt-3a signaling through control of E disaccharide expression of chondroitin sulfate. J. Biol. Chem. 283, 27333–27343 [DOI] [PubMed] [Google Scholar]
- 21. Okada M., Nadanaka S., Shoji N., Tamura J., Kitagawa H. (2010) Biosynthesis of heparan sulfate in EXT1-deficient cells. Biochem. J. 428, 463–471 [DOI] [PubMed] [Google Scholar]
- 22. Heinegård D. (1972) Hyaluronidase digestion and alkaline treatment of bovine tracheal cartilage proteoglycans. Isolation and characterisation of different keratan sulfate proteins. Biochim. Biophys. Acta 285, 193–207 [DOI] [PubMed] [Google Scholar]
- 23. Nadanaka S., Kitagawa H., Sugahara K. (1998) Demonstration of the immature glycosaminoglycan tetrasaccharide sequence GlcAβ1–3Galβ1-3Galβ1–4Xyl on recombinant soluble human α-thrombomodulin. An oligosaccharide structure on a “part-time” proteoglycan. J. Biol. Chem. 273, 33728–33734 [DOI] [PubMed] [Google Scholar]
- 24. Nadanaka S., Kinouchi H., Taniguchi-Morita K., Tamura J., Kitagawa H. (2011) Down-regulation of chondroitin 4-O-sulfotransferase-1 by Wnt signaling triggers diffusion of Wnt-3a. J. Biol. Chem. 286, 4199–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitagawa H., Tanaka Y., Tsuchida K., Goto F., Ogawa T., Lidholt K., Lindahl U., Sugahara K. (1995) N-Acetylgalactosamine (GalNAc) transfer to the common carbohydrate-protein linkage region of sulfated glycosaminoglycans. Identification of UDP-GalNAc:chondro-oligosaccharide α-N-acetylgalactosaminyltransferase in fetal bovine serum. J. Biol. Chem. 270, 22190–22195 [DOI] [PubMed] [Google Scholar]
- 26. Vliegenthart J. F., Dorland L., van Halbeek H. (1983) High-resolution 1H-nuclear magnetic resonance spectroscopy as a tool in the structural analysis of carbohydrates related to glycoproteins. Adv. Carbohydr. Chem. Biochem. 41, 209–374 [Google Scholar]
- 27. Kim B. T., Kitagawa H., Tanaka J., Tamura J., Sugahara K. (2003) In vitro heparan sulfate polymerization. Crucial roles of core protein moieties of primer substrates in addition to the EXT1-EXT2 interaction. J. Biol. Chem. 278, 41618–41623 [DOI] [PubMed] [Google Scholar]
- 28. Kitagawa H., Tsuchida K., Ujikawa M., Sugahara K. (1995) Detection and characterization of UDP-GalNAc. Chondroitin N-acetylgalactosaminyltransferase in bovine serum using a simple assay method. J. Biochem. 117, 1083–1087 [DOI] [PubMed] [Google Scholar]
- 29. Kitagawa H., Egusa N., Tamura J. I., Kusche-Gullberg M., Lindahl U., Sugahara K. (2001) rib-2, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes, encodes a novel α1,4-N-acetylglucosaminyltransferase involved in the biosynthetic initiation and elongation of heparan sulfate. J. Biol. Chem. 276, 4834–4838 [DOI] [PubMed] [Google Scholar]
- 30. Kitagawa H., Tsutsumi K., Ujikawa M., Goto F., Tamura J., Neumann K. W., Ogawa T., Sugahara K. (1997) Regulation of chondroitin sulfate biosynthesis by specific sulfation. Acceptor specificity of serum β-GalNAc transferase revealed by structurally defined oligosaccharides. Glycobiology 7, 531–537 [DOI] [PubMed] [Google Scholar]
- 31. Kitagawa H., Kano Y., Shimakawa H., Goto F., Ogawa T., Okabe H., Sugahara K. (1999) Identification and characterization of a novel UDP-GalNAc:GlcAβ-R α1,4-N-acetylgalactosaminyltransferase from a human sarcoma cell line. Glycobiology 9, 697–703 [DOI] [PubMed] [Google Scholar]
- 32. Eames B. F., Yan Y. L., Swartz M. E., Levic D. S., Knapik E. W., Postlethwait J. H., Kimmel C. B. (2011) Mutations in fam20b and xylt1 reveal that cartilage matrix controls timing of endochondral ossification by inhibiting chondrocyte maturation. PLoS Genet. 7, e1002246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uyama T., Kitagawa H., Tanaka J., Tamura J., Ogawa T., Sugahara K. (2003) Molecular cloning and expression of a second chondroitin N-acetylgalactosaminyltransferase involved in the initiation and elongation of chondroitin/dermatan sulfate. J. Biol. Chem. 278, 3072–3078 [DOI] [PubMed] [Google Scholar]
- 34. Prante C., Bieback K., Funke C., Schön S., Kern S., Kuhn J., Gastens M., Kleesiek K., Götting C. (2006) The formation of extracellular matrix during chondrogenic differentiation of mesenchymal stem cells correlates with increased levels of xylosyltransferase I. Stem Cells 24, 2252–2261 [DOI] [PubMed] [Google Scholar]
- 35. Van Hul W., Bollerslev J., Gram J., Van Hul E., Wuyts W., Benichou O., Vanhoenacker F., Willems P. J. (1997) Localization of a gene for autosomal dominant osteopetrosis (Albers-Schönberg disease) to chromosome 1p21. Am. J. Hum. Genet. 61, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Waard P., Vliegenthart J. F., Harada T., Sugahara K. (1992) Structural studies on sulfated oligosaccharides derived from the carbohydrate-protein linkage region of chondroitin 6-sulfate proteoglycans of shark cartilage. II. Seven compounds containing 2 or 3 sulfate residues J. Biol. Chem. 267, 6036–6043 [PubMed] [Google Scholar]