Abstract
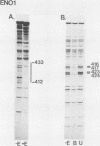
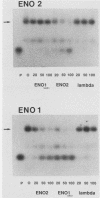
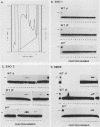
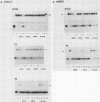
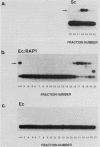
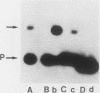
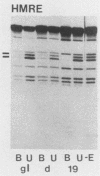
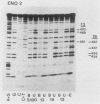
Binding sites for three distinct proteins were mapped within the upstream activation sites (UAS) of the yeast enolase genes ENO1 and ENO2. Sequences that overlapped the UAS1 elements of both enolase genes bound a protein which was identified as the product of the RAP1 regulatory gene. Sequences within the UAS2 element of the ENO2 gene bound a second protein which corresponded to the ABFI (autonomously replicating sequence-binding factor) protein. A protein designated EBF1 (enolase-binding factor) bound to sequences which overlapped the UAS2 element in ENO1. There was a good correlation among all of the factor-binding sites and the location of sequences required for UAS activity identified by deletion mapping analysis. This observation suggested that the three factors play a role in transcriptional activation of the enolase genes. UAS elements which bound the RAP1 protein or the ABFI protein modulated glucose-dependent induction of ENO1 and ENO2 expression. The ABFI-binding site in ENO2 overlapped sequences required for UAS2 activity in wild-type strains and for repression of ENO2 expression in strains carrying a null mutation in the positive regulatory gene GCR1. These latter results showed that the ABFI protein, like the RAP1 protein, bound sequences required for positive as well as negative regulation of gene expression. These observations strongly suggest that the biological functions of the RAP1 and ABFI proteins are similar.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker H. V. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986 Nov;6(11):3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucherie H. Protein synthesis during transition and stationary phases under glucose limitation in Saccharomyces cerevisiae. J Bacteriol. 1985 Jan;161(1):385–392. doi: 10.1128/jb.161.1.385-392.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol Cell Biol. 1987 Jan;7(1):403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Micklem G., Nasmyth K. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell. 1987 Dec 4;51(5):709–719. doi: 10.1016/0092-8674(87)90094-8. [DOI] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Lue N. F., Kornberg R. D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988 Dec;8(12):5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton D., Fraenkel D. G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981 Dec 25;256(24):13074–13078. [PubMed] [Google Scholar]
- Clifton D., Weinstock S. B., Fraenkel D. G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978 Jan;88(1):1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R., Holland J. P., Yokoi T., Holland M. J. Identification of a regulatory region that mediates glucose-dependent induction of the Saccharomyces cerevisiae enolase gene ENO2. Mol Cell Biol. 1986 Jul;6(7):2287–2297. doi: 10.1128/mcb.6.7.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R., Yokoi T., Holland J. P., Pepper A. E., Holland M. J. Transcription of the constitutively expressed yeast enolase gene ENO1 is mediated by positive and negative cis-acting regulatory sequences. Mol Cell Biol. 1987 Aug;7(8):2753–2761. doi: 10.1128/mcb.7.8.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Purification of a yeast protein that binds to origins of DNA replication and a transcriptional silencer. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2120–2124. doi: 10.1073/pnas.85.7.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Stillman B. Similarity between the transcriptional silencer binding proteins ABF1 and RAP1. Science. 1989 Nov 24;246(4933):1034–1038. doi: 10.1126/science.2511628. [DOI] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Identification of two factors which bind to the upstream sequences of a number of nuclear genes coding for mitochondrial proteins and to genetic elements important for cell division in yeast. Nucleic Acids Res. 1988 Aug 11;16(15):7287–7301. doi: 10.1093/nar/16.15.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S., Civalier C., Tye B. K. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc Natl Acad Sci U S A. 1988 Feb;85(3):743–746. doi: 10.1073/pnas.85.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter H., Müller U., Winnacker E. L., Gallwitz D. Isolation and DNA-binding characteristics of a protein involved in transcription activation of two divergently transcribed, essential yeast genes. EMBO J. 1989 Oct;8(10):3029–3037. doi: 10.1002/j.1460-2075.1989.tb08453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamil K. G., Nam H. G., Fried H. M. Constitutive transcription of yeast ribosomal protein gene TCM1 is promoted by uncommon cis- and trans-acting elements. Mol Cell Biol. 1988 Oct;8(10):4328–4341. doi: 10.1128/mcb.8.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruer M. H., Mager W. H., Woudt L. P., Nieuwint R. T., Wassenaar G. M., Groeneveld P., Planta R. J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987 Dec 23;15(24):10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann J. F., Laroche T., Brand A. H., Gasser S. M. RAP-1 factor is necessary for DNA loop formation in vitro at the silent mating type locus HML. Cell. 1989 Jun 2;57(5):725–737. doi: 10.1016/0092-8674(89)90788-5. [DOI] [PubMed] [Google Scholar]
- Holland J. P., Brindle P. K., Holland M. J. Sequences within an upstream activation site in the yeast enolase gene ENO2 modulate repression of ENO2 expression in strains carrying a null mutation in the positive regulatory gene GCR1. Mol Cell Biol. 1990 Sep;10(9):4863–4871. doi: 10.1128/mcb.10.9.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. J., Yokoi T., Holland J. P., Myambo K., Innis M. A. The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol. 1987 Feb;7(2):813–820. doi: 10.1128/mcb.7.2.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Sentenac A. TUF, the yeast DNA-binding factor specific for UASrpg upstream activating sequences: identification of the protein and its DNA-binding domain. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3648–3652. doi: 10.1073/pnas.84.11.3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jigami Y., Toshimitsu N., Fujisawa H., Uemura H., Tanaka H., Nakasato S. Analysis of expression of yeast enolase 1 gene containing a longer pyrimidine-rich region located between the TATA box and transcription start site. J Biochem. 1986 Apr;99(4):1111–1125. doi: 10.1093/oxfordjournals.jbchem.a135575. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Machida M., Uemura H., Jigami Y., Tanaka H. The protein factor which binds to the upstream activating sequence of Saccharomyces cerevisiae ENO1 gene. Nucleic Acids Res. 1988 Feb 25;16(4):1407–1422. doi: 10.1093/nar/16.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra P. K., Lobo Z. A kinetic study of glycolytic enzyme synthesis in yeast. J Biol Chem. 1971 Jan 25;246(2):475–488. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister L., Holland M. J. Targeted deletion of a yeast enolase structural gene. Identification and isolation of yeast enolase isozymes. J Biol Chem. 1982 Jun 25;257(12):7181–7188. [PubMed] [Google Scholar]
- Nishizawa M., Araki R., Teranishi Y. Identification of an upstream activating sequence and an upstream repressible sequence of the pyruvate kinase gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):442–451. doi: 10.1128/mcb.9.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden J. E., Stanway C., Kim S., Mellor J., Kingsman A. J., Kingsman S. M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986 Dec;6(12):4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlović B., Hörz W. The chromatin structure at the promoter of a glyceraldehyde phosphate dehydrogenase gene from Saccharomyces cerevisiae reflects its functional state. Mol Cell Biol. 1988 Dec;8(12):5513–5520. doi: 10.1128/mcb.8.12.5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper P. W., Curran B., Davies M. W., Hirst K., Lockheart A., Ogden J. E., Stanway C. A., Kingsman A. J., Kingsman S. M. A heat shock element in the phosphoglycerate kinase gene promoter of yeast. Nucleic Acids Res. 1988 Feb 25;16(4):1333–1348. doi: 10.1093/nar/16.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanway C., Mellor J., Ogden J. E., Kingsman A. J., Kingsman S. M. The UAS of the yeast PGK gene contains functionally distinct domains. Nucleic Acids Res. 1987 Sep 11;15(17):6855–6873. doi: 10.1093/nar/15.17.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. E., Holland M. J. RNA polymerase I-dependent selective transcription of yeast ribosomal DNA. Identification of a new cellular ribosomal RNA precursor. J Biol Chem. 1983 Mar 10;258(5):3242–3250. [PubMed] [Google Scholar]
- Sweder K. S., Rhode P. R., Campbell J. L. Purification and characterization of proteins that bind to yeast ARSs. J Biol Chem. 1988 Nov 25;263(33):17270–17277. [PubMed] [Google Scholar]
- Uemura H., Jigami Y., Tanaka H., Toshimitsu N., Paterson M., Nakasato S. Nucleotide sequence of the 5' flanking region responsible for the enhancement of the expression of yeast enolase 1 gene. J Biochem. 1985 Sep;98(3):859–862. doi: 10.1093/oxfordjournals.jbchem.a135345. [DOI] [PubMed] [Google Scholar]
- Uemura H., Shiba T., Machida M., Matsui I., Jigami Y., Tanaka H. A positive regulatory sequence of the Saccharomyces cerevisiae ENO1 gene. J Biochem. 1987 Jul;102(1):181–189. doi: 10.1093/oxfordjournals.jbchem.a122031. [DOI] [PubMed] [Google Scholar]
- Uemura H., Shiba T., Paterson M., Jigami Y., Tanaka H. Identification of a sequence containing the positive regulatory region of Saccharomyces cerevisiae gene ENO1. Gene. 1986;45(1):67–75. doi: 10.1016/0378-1119(86)90133-2. [DOI] [PubMed] [Google Scholar]
- Walker S. S., Francesconi S. C., Tye B. K., Eisenberg S. The OBF1 protein and its DNA-binding site are important for the function of an autonomously replicating sequence in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Jul;9(7):2914–2921. doi: 10.1128/mcb.9.7.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]