Background: Although AUG is the traditional eukaryotic initiation codon, near-cognate codons may serve this function.
Results: In N. crassa, some near-cognate codons are used as initiation codons at reasonable efficiency.
Conclusion: The hierarchy of near-cognate codon usage in N. crassa is similar to that in human cells.
Significance: Near-cognate initiation codons could provide additional coding capacity or have regulatory functions.
Keywords: Fungi, Molecular Genetics, mRNA, Neurospora, Translation, Near-cognate Codon, Translation Initiation
Abstract
In eukaryotic cells initiation may occur from near-cognate codons that differ from AUG by a single nucleotide. The stringency of start codon selection impacts the efficiency of initiation at near-cognate codons and the efficiency of initiation at AUG codons in different contexts. We used a codon-optimized firefly luciferase reporter initiated with AUG or each of the nine near-cognate codons in preferred context to examine the stringency of start codon selection in the model filamentous fungus Neurospora crassa. In vivo results indicated that the hierarchy of initiation at start codons in N. crassa (AUG ≫ CUG > GUG > ACG > AUA ≈ UUG > AUU > AUC) is similar to that in human cells. Similar results were obtained by translating mRNAs in a homologous N. crassa in vitro translation system or in rabbit reticulocyte lysate. We next examined the efficiency of initiation at AUG, CUG, and UUG codons in different contexts in vitro. The preferred context was more important for efficient initiation from near-cognate codons than from AUG. These studies demonstrated that near-cognate codons are used for initiation in N. crassa. Such events could provide additional coding capacity or have regulatory functions. Analyses of the 5′-leader regions in the N. crassa transcriptome revealed examples of highly conserved near-cognate codons in preferred contexts that could extend the N termini of the predicted polypeptides.
Introduction
In standard eukaryotic translation initiation, the preinitiation complex that is formed by the small ribosomal subunit, initiator tRNA, and multiple initiation factors binds at the mRNA 5′-cap and scans downstream for an initiation codon (1). The AUG codon that is closest to the mRNA 5′-cap and that is in a preferred context is typically selected as the initiation site (2). Start codon selection is aided through biases in the nucleotides surrounding the start codon and through the actions of initiation factors (3). In certain cases, initiation occurs at non-AUG codons, especially near-cognate codons that differ from AUG by a single nucleotide (4–8). In most cases, initiation from near-cognate codons uses Met-tRNAiMet (5). There are specific cases where viral internal ribosome entry sites use a different mechanism for non-AUG initiation (9), and recently it was established that leucyl-tRNA can be used for translation initiation at a CUG codon of a mammalian mRNA (10). Initiating translation from near-cognate codons may increase coding capacity of transcripts or contribute regulatory functions.
The initiation context surrounding the start codon, which is generally defined by nucleotides from −6 to +4 (the A of AUG is +1), has strong influence on initiation efficiency (11, 12). The Kozak consensus (GCC(A/G)CCXXXG) is optimal for initiation in mammalian cells (12, 13). A purine at position −3 and a G at position +4 are most important for efficient initiation (3, 12). Substitutions at positions −1 and −2 have less impact on translation efficiency, but the effects of changes at these positions are intensified when the −3 nucleotide is also suboptimal.
Codons other than AUG are generally less efficient initiation codons in vivo; however, codons differing from AUG in a single position, collectively referred to as near-cognate initiation codons, are known to support translation initiation in eukaryotes (5, 14). The presence of a good Kozak context is crucial for efficient use of near-cognate initiation codons in mammals, plants, and yeast (14–16). In these studies, the measured efficiencies of initiation from functional near-cognate codons vary, and values between ∼1 and 20% of initiation from AUG are reported. AAG and AGG do not serve as initiation codons: a purine at +2 evidently eliminates function as an initiation codon (4–6). A recent study in human cells reveals a hierarchy in initiation efficiencies at near-cognate codons (8). The most efficient near-cognate codons are CUG and GUG (19.5 and 9.2% of AUG-initiated translation). ACG (6.6%), AUA (3.3%), AUU (3.2%), UUG (1.9%), and AUC (1.7%) are used as initiation codons at lower efficiencies (8). A recent analysis in Saccharomyces cerevisiae demonstrated the hierarchy CUG > GUG ≈ UUG > ACG ≈ AUA > AUU > AUC (4), with efficiencies comparable with those observed in human cells. In plant protoplasts, CUG is the most active (30%) followed by GUG and ACG (each 15%), whereas AUA, AUU, UUG, and AUC are less active (2–5%) (7).
Initiation from near-cognate codons increases the capacity to generate protein isoforms with different regulatory functions (17). In some cases initiation from an upstream in-frame near-cognate codon produces an alternative N-terminal-extended isoform of protein in addition to that produced by initiation at the downstream AUG (2). Protein isoforms with different N-terminal sequences can serve different functions and/or be targeted to different locations (18–21). The synthesis of some proteins is thought to start exclusively at non-AUG codons. These include: mammalian eIF4G2, which uses GUG (22); Podospora anserina IDI-4, which uses CUG (23); yeast glycyl-tRNA synthetase, which uses UUG (24); Arabidopsis AGAMOUS, which uses ACG (25); Arabidopsis FCA, which uses CUG (26); some RpoT genes in Nicotiana spp., which use CUG (27, 28). In yeast, genome-wide analysis reveals that near-cognate codon-initiated upstream open reading frames (uORFs)3 can be translated at levels comparable with AUG-initiated uORFs (29). Data indicate that in response to amino acid starvation, levels of translation of these non-AUG yeast uORFs generally increase relative to levels of translation of the downstream coding sequences. Studies in mouse embryonic stem cells identified many unannotated initiation sites started by AUG or near-cognate codons that direct the translation of uORFs with regulatory potential (30).
The stringency of start codon selection can affect both the efficiency of initiation at AUG codons in different contexts and the efficiency of initiation at near-cognate codons. Stringency can be modulated by different physiological conditions, such as amino acid starvation (29). eIF1 and eIF5 play important roles in regulating the stringency of start codon selection (1, 8, 31, 32). The auto- and cross- regulatory functions of eIF1 and eIF5 that themselves utilize stringency of start codon selection demonstrates that varying the stringency of start codon selection can be used for gene regulation in eukaryotes.
The stringency of start codon selection can also be regulated by other parameters. Two structurally related molecules, isoquinoline-1-carboxylic and 7-amino-5-iodo-8-quinolinol, were identified in a high throughput screen to decrease the stringency of start codon selection in yeast (33). These compounds can also increase initiation at natural uORFs initiated by near-cognate codons. Their mechanism of action is currently not known. The concentration of Mg2+ can influence the fidelity of translation initiation; a high concentration of Mg2+ increases initiation at non-AUG codons in vitro (14).
Here, we used a firefly luciferase reporter that was codon-optimized for expression in Neurospora crassa to determine the efficiency of translation initiation at near-cognate codons. We stably integrated luciferase reporter genes that initiated with AUG or each of the nine near-cognate codons in preferred context at the N. crassa his-3 locus and determined luciferase levels in vivo. CUG and GUG were the most efficient near-cognate codons, with 11 and 7% of the efficiency of AUG. AUA, AUU, UUG, and AUC were less active (1–5%), whereas AAG and AGG did not function. The hierarchy of initiation efficiency at near-cognate codons was similar to that in human cells. Cell-free translation extracts of N. crassa were also used to analyze translation initiation at near-cognate codons. The hierarchy of utilization of near-cognate codons was similar to that observed in vivo, but overall efficiency was strongly dependent on the concentration of Mg2+. Our studies, which are one of a handful of analyses that systematically examine near-cognate codon initiation in eukaryotes and are the first for filamentous fungi, demonstrate that such initiation could substantially increase the coding capacity of mRNAs.
EXPERIMENTAL PROCEDURES
Logogram Generation
The frequencies for nucleotide occurrence at each position of the N. crassa initiation context, which were used to generate the logogram in Fig. 1, were obtained from the Transterm database (34).
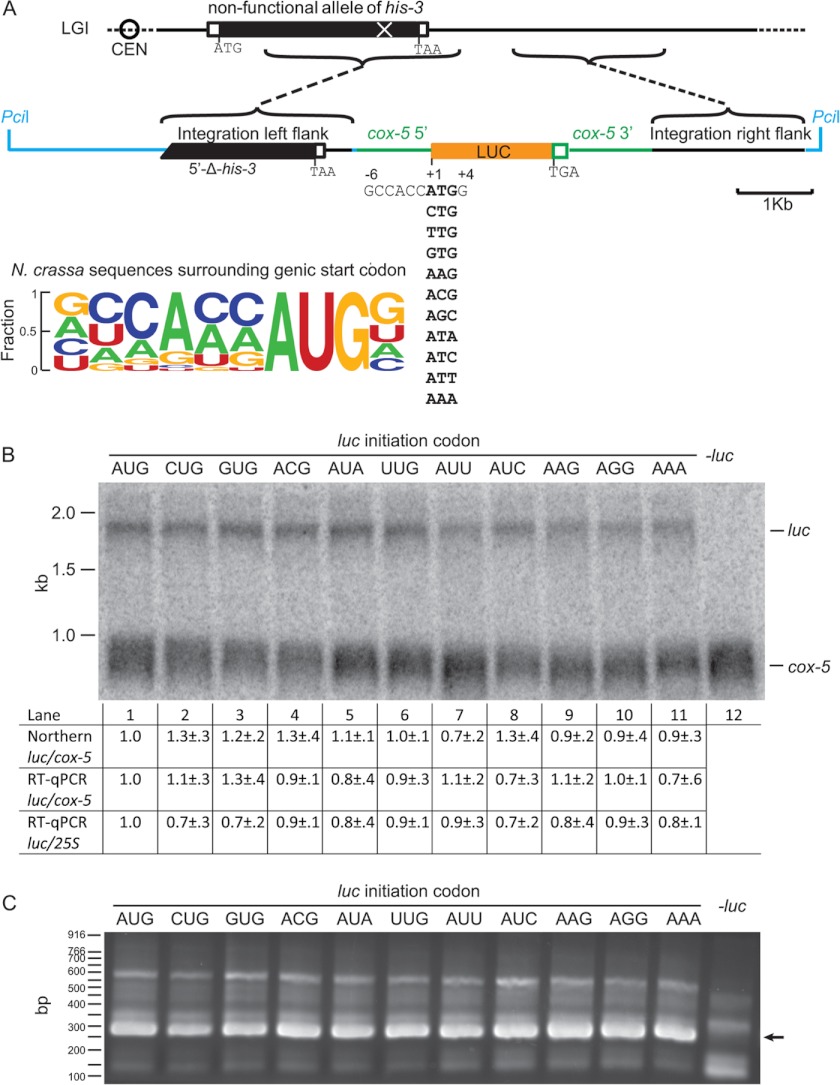
FIGURE 1.
Site-specific integration and expression of LUC constructs. A, shown is the strategy for placing LUC constructs at the N. crassa his-3 locus. The luc coding sequence is codon-optimized for N. crassa. It initiates with an ATG codon, one of the nine near-cognate codons, or an AAA codon. All codons tested were in the same surrounding consensus context as indicated. The LUC coding sequence is preceded by 5′ region of the N. crassa cox-5 gene that has promoter activity and followed by cox-5 3′ region, which provides the polyadenylation site to produce reporter transcripts with a cox-5 3′UTR. The plasmids used for integration contain a unique PciI site for linearization. The Integration left flank on the plasmid contains the distal region of the functional wild-type his-3 coding sequence and additional downstream genomic sequence; the Integration right flank contains additional sequence from the chromosomal region downstream of his-3. The positions of the corresponding segments on the N. crassa chromosome (Linkage Group I (LGI)) containing a non-functional his-3 allele are indicated. LUC coding sequence is orange; cox-5 sequences are green; Linkage Group I sequences are black; additional plasmid sequences are blue. Left lower panel, frequency logograms of the conservation of the initiation contexts, from −6 to +4, of all predicted ATG-initiated N. crassa genes. Letter heights are proportional to the frequency of occurrence of each nucleotide at each position. B, Northern analysis of luc and cox-5 mRNAs (450 ng of poly(A) mRNA/lane) shows luc mRNAs containing the indicated luc initiation codons are similarly expressed. The primary data for one set of three independent sets of transformants analyzed is shown. Lane 12 was loaded with mRNA from wild-type cells lacking luc. For quantification, signals representing luc mRNA was normalized to signals representing cox-5 mRNA for all three sets of transformants. Then this ratio for each initiation codon was normalized to the ratio obtained for the AUG initiation codon. For RT-qPCR quantification, cDNA prepared from total RNA from all transformants was analyzed; luc mRNA was normalized to cox-5 or 25 S rRNA. Then these ratios for each initiation codon were normalized to the ratios obtained for the AUG initiation codon. C, all luc-containing mRNAs were similarly polyadenylated. 3′-RACE analysis shows proper polyadenylation at the cox-5 poly(A) site for the luc mRNA. 3′-RACE was performed as described under “Experimental Procedures” with 50 ng poly(A) mRNA template. The major bands of all samples migrate at the position expected from proper polyadenylation (285 bp) (arrow); this was further confirmed by sequencing. The primary data is shown for one set of three independent sets of transformants analyzed.
Strains and Culture Conditions
Strains his-3 (Y234M723) mat A strain FGSC 6103 and the mat A wild-type (WT) reference sequenced strain FGSC 2489 (74-OR23–1V A) were obtained from the Fungal Genetics Stock Center (FGSC) (35).
Firefly luciferase reporters were targeted to the N. crassa his-3 locus using his-3 left flank and right flank integration sequences by transformation of Fungal Genetics Stock Center 6103 with PciI-linearized plasmid DNA (pJI301-pJI311). Each plasmid contained the luciferase (luc) reporter with an N. crassa cox-5 promoter and 3′-region (plasmid construction is described below and in supplemental Fig. S1). Transformants were obtained by electroporation (36) followed by selection for histidine prototrophy using plates containing Vogel's minimal medium (VM), 0.05% fructose, 0.05% glucose, 2% sorbose, 2% agar at 30 °C. Homokaryons were obtained by microconidiation essentially as described previously (37) except that microconidia obtained by filtration were pelleted in an Eppendorf 5415D centrifuge at 12,000 × g for 2 min and, after resuspension, germinated on VM, 0.05% fructose, 0.05% glucose, 2% sorbose, 2% agar at 30 °C.
Conidia were obtained from cultures in 125-ml flasks containing 25 ml of VM, 2% sucrose, and 2% agar (38). Cultures were grown at 25 °C with 12:12-h light:dark cycle for 7 days in a Percival Environmental Chamber (Model I36VL). Conidia were harvested by suspension in VM, 2% sucrose and filtration through two layers of cheesecloth. The concentration of conidia was determined using a hemacytometer.
For RNA isolation and preparation of cell extracts to measure luciferase (LUC) activity produced in vivo, conidia were inoculated into 25 ml of VM, 2% sucrose in a 125-ml flask at a concentration of 107 conidia/ml. Conidia were germinated under constant light at 30 °C for 6 h with 125 rpm shaking. Germlings were harvested by vacuum filtration onto Whatman 541 filter paper (42.5-mm circle); the pad of cells was washed with 4 °C sterile water, cut into ∼0.1-g pieces with a single-edged razor, transferred to 2-ml screw-cap Eppendorf tubes, quick-frozen in liquid nitrogen, and stored at −80 °C.
Measurements of LUC Activity in Vivo
For measurements of LUC activity in vivo using real-time detection of photon emission, conidia were inoculated into 0.15 ml VM, 2% sucrose containing 25 nm luciferin (NanoLight Technology, catalog #306) in a 96-well microtiter plate (PerkinElmer Life Sciences OptiPlate-96F) and incubated as stationary cultures at 25 °C in constant darkness. Light emission was measured with a microplate scintillation and luminescence counter (Topcount NXT, Packard Instrument Co.).
For measurements of LUC activity in soluble extracts, N. crassa extracts were prepared as described (39), quick-frozen in aliquots, and stored at −80 °C. Protein concentration was determined using the Coomassie Plus (Bradford) Assay Reagent (Thermo Scientific) with BSA as the standard (Albumin Standard, Thermo Scientific) using the microplate procedures (300 μl of the Coomassie Plus Reagent was added to 10 μl of sample). The absorbance at 595 nm was measured by a Victor 3 Multitask plate reader. The typical yield of this method was 10 μg of total protein/μl (approximately a yield of 0.1 mg of protein/mg of cells). 10 μl of cell extract (diluted to 0.5–1 μg of total protein/μl in breaking buffer (39)) were further diluted by adding 10 μl of 2× passive lysis buffer (Promega), and luciferase activity was measured using a Victor 3 Multitask plate reader (PerkinElmer Life Sciences). Firefly luciferase assay reagents were prepared as described (40).
Plasmids
A flowchart and full description for plasmid construction is given in supplemental Fig. S1. The N. crassa optimized firefly luc coding sequence in plasmid pRMP57 was a gift from Dr. Deborah Bell-Pedersen.4 Plasmids pJI301-pJI311 (described in supplemental Fig. S1 and Tables S1 and S3) were used in in vivo experiments. Plasmids pJI201-pJI211 and pJI601-pJI606 (described in supplemental Fig. S1 and Tables S1, S2, and S4) were used in in vitro assays.
RNA Isolation
Cells were kept at −80 °C until immediately before breakage; cells were not thawed on ice before breaking. Total RNA from cells was isolated by modification of the procedure previously described (39) using 1 g of Zirconia/Silica Beads (Biospec Products, baked at 180 °C overnight before use) and a Mini-BeadBeater 8 (Biospec Products) with ice-cold 0.5-ml freshly prepared extraction buffer (100 mm Tris-HCl, pH 7.5, 100 mm LiCl, 20 mm DTT in diethylpyrocarbonate-treated water), 0.36 ml of phenol, 0.36 ml of chloroform, and 0.072 ml of 10% SDS. Cells (∼0.1 g) were extracted by 1-min breakage using the bead-beater at full speed. Tubes were rotated end-over-end for 4 min (New Brunswick TC-6) and then centrifuged (Eppendorf 5415D) at 12,000 × g at 4 °C for 1 min to separate phases. The aqueous phase was removed and extracted once with 0.69 ml of phenol/chloroform and once with 0.69 ml of chloroform, precipitated with 0.875 ml of ethanol and 0.057 ml of 3 m sodium acetate, washed with 80% ethanol twice, and dissolved in sterile and filtered diethylpyrocarbonate-treated water. RNA concentration was determined using a Nanodrop spectrophotometer. The typical yield of total RNA was 500–1000 μg/0.1 g cells. DNA contamination was removed by DNase I treatment using the Turbo DNA-free kit (Ambion). Poly(A) RNAs were purified from 150 μg of total RNA using Poly(A) Purist MAG Kit (Ambion) and stored at −80 °C. The yield of poly(A) mRNA was determined using RiboGreen (Invitrogen).
Northern Blots
Northern blots were performed as described (41) except that dextran sulfate was omitted from the hybridization buffer. 32P-Labeled probes were obtained from the cox-5 and luc templates by random priming and then purified with Mini Quick Spin Columns (Roche Applied Science).
cDNA Synthesis
1.5 μg of DNA-free total RNA was used as the template to synthesize first-strand cDNA using SuperScript III reverse transcriptase (Invitrogen). First, 1.5 μg of total RNA, 5 nmol of dNTP mix, 125 ng of oligo(dT)18, and 50 ng of random hexamer were mixed together in water to a total volume of 6.3 μl. These components were incubated at 65 °C for 5 min and transferred to ice for 2 min. Then the reaction mixture was adjusted to a final volume of 10 μl containing 1× First Strand buffer (Invitrogen), 5 mm DTT, and 40 units of SuperScript III reverse transcriptase and then incubated at 25 °C for 5 min and then 50 °C for 50 min. The reverse transcriptase was inactivated at 70 °C for 15 min, and the cDNA was stored at −80 °C.
Quantitative PCR (qPCR)
Aliquots of cDNA representing 8 and 16 ng of total RNA were used as qPCR templates in triplicate 10-μl reactions containing 1× Platinum TaqPCR buffer (200 mm Tris-HCl, pH 8.4, 500 mm KCl), 2.5 mm MgCl2, 0.2 mm dNTPs, 1× ROX Reference Dye (Invitrogen), 1 unit of Platinum TaqDNA Polymerase (Invitrogen), 1× SYBR Green I (Invitrogen), and 500 nm concentrations of each primer (chosen using Primer Express software). Thermal cycling was performed using an ABI 7300 real-time PCR machine (Applied Biosystems) as follows: 50 °C for 2 min and 95 °C for 10 min followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min.
3′-Rapid Amplification of cDNA Ends (3′-RACE)
First stand cDNA was first synthesized from poly(A) RNA using the Clontech SMART RACE kit protocol. A 5-μl mixture containing 50 ng of poly(A) RNA and 1.2 μm 3′-RACE CDS Primer A (oYZ291, 5′-AAGCAGTGGTATCAACGCAGAGTAC(T)30VN-3′, N = A, C, G, or T; V = A, G, or C) in water was denatured at 65 °C for 5 min. The mixture was cooled on ice for 2 min and combined with 2 μl of 5× First Strand buffer (250 mm Tris-HCl pH 8.3, 375 mm KCl, 30 mm MgCl2), 0.5 μl 0.1 m DTT, 1 μl 10 mm dNTPs, and 0.5 μl of 200 units/μl SuperScript III reverse transcriptase (Invitrogen) in a final volume of 10 μl. First-strand cDNA was synthesized at 55 °C for 1 h. The reverse transcriptase was inactivated at 70 °C for 15 min, and the reaction mixture was diluted with 90 μl of 10 mm Tris-HCl, pH 7.5. RACE was performed with 1 μl of first strand cDNA as template in a 20-μl reaction containing 1× TaKaRa PCR buffer (10 mm Tris-HCl, pH 8.3, 50 mm KCl, 1.5 mm MgCl2), 0.2 mm dNTP mix, 0.25 units of rTaq DNA polymerase (TaKaRa), 0.5 μm luc mRNA-specific primer_1 (MSP_1, oYZ365 5′-CGTCTTCGTCGACGAGGTCC-3′), and 0.5 μm nested universal primer (NUP, oYZ294 5′-AAGCAGTGGTATCAACGCAGAGT-3′). First-round PCR was accomplished by (i) denaturing at 94 °C for 30 s then (ii) 25 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s and (iii) a final extension step at 72 °C for 10 min. Then 1 μl of this reaction mixture containing the PCR products was mixed with 99 μl of water, and 1 μl of this 100-fold diluted first round PCR product was used as the template for a second round of PCR with different primers using the same reaction components and PCR conditions as the first round. The primers used in the second round were oYZ294 and luc mRNA-specific primer_2 (MSP_2, oYZ287 5′-GGCCAAGAAGGGCGGCAAGATCGCCGTC-3′), which is complementary to the luc mRNA 3′ distal to the region complementary to oYZ365. After the second round of PCR, 5 μl of the reaction mixture was examined by electrophoresis in a 2% TAE (40 mm Tris-acetate pH 8, 1 mm EDTA) agarose gel. For sequencing, bands of interest were excised from a 2% TAE-agarose gel containing 1 mm guanosine to protect DNA from UV damage (42). The PCR product representing the equivalent of 30 μl of the reaction mixture was purified from the gel using the QIAquick Gel Extraction kit (Qiagen), the concentration of recovered DNA determined by A260/A280 measurement (Nanodrop), and sequenced with primer 0YZ287 to determine the mRNA 3′ sequence including the poly(A) site.
Cell-free Translation Analysis
Capped and polyadenylated RNAs were transcribed in vitro by T7 RNA polymerase from plasmid DNA templates (pJI201-pJI211) that were linearized with HindIII (43). The relative amount of RNA was determined as described (44). Equal amounts (60 ng) of each luc mRNA were used to program N. crassa extracts as described (44) except that Mg2+ and K+ concentrations were varied as specified under “Results.”
For in vitro translation using rabbit reticulocyte lysate (Invitrogen), translation reaction mixtures (10 μl) were incubated at 30 °C for 30 min, and translation was halted by adding 50 μl of 1.2× passive lysis buffer (Promega). Equal amounts of each luc mRNA (6 ng) were used to program extracts.
For cell-free translation in N. crassa and in rabbit reticulocyte lysate, 15-μl samples containing 2.5 μl of translation reaction and 12.5 μl of 1.2× passive lysis buffer were used to measure luciferase activity with a Victor 3 Multitask plate reader. Firefly luciferase assay reagents were prepared as described (40).
Primer extension inhibition (toeprint) assays to map ribosomes at initiation codons were accomplished using 32P-labeled primer oJI105 as described (45, 46), except that 0.5 mg/ml cycloheximide or 2 μg/ml harringtonine (Santa Cruz Biotechnology) were added to the reactions as indicated under “Results.”
Bioinformatics Analysis of Near-cognate Initiated N-terminal Extensions in N. crassa
The starting point of the analysis was a FASTA file of the N. crassa mRNA transcriptome.5 Genes represented by multiple transcripts were eliminated from the analysis because a pilot indicated they generating a large number of false positives. As a result, only the 6,804 genes represented by unique transcripts were subjected to systematic analysis. In the first step the sequence starting with the annotated AUG start codon of each mRNA and extending 5′ to the nearest in-frame stop codon (UAA, UAG, or UGA) was extracted. In the next step the coordinates of all in-frame functional near-cognate start codons (i.e. CUG, GUG, UUG, ACG, AUA, AUC, and AUU) in these sequences were determined. Finally, the coordinates of those in-frame near-cognate sequences that have A at position −3 or G at position −3 in combination with G at position +4 were extracted. For identifying conserved N-terminal near-cognate initiated extensions to existing N. crassa ORFs, an approach similar to one previously used to identify such sequences in humans (47) was employed. Briefly, all 5′-UTRs, which in the previous step were shown to have an in-frame near-cognate start codon in good context at least 25 codons 5′ of the annotated AUG start, were extracted (representing a total of 1,185 mRNAs). These 5′-UTRs were subjected to conceptual translation starting with the annotated AUG start site and extending 5′ to the nearest in-frame stop codon. The conceptually translated peptides were then used as query in a BLAST search (48) against the genomic sequence of the filamentous fungus Chaetomium globosum. It was empirically determined that C. globosum was suitably close evolutionarily to allow sufficient sensitivity and at the same time was adequately distant to allow desired selectivity. BLAST hits with expected values of 0.001 or lower were then subjected to manual inspection. In this step all available orthologous sequences from other filamentous fungi (i.e. Pezizomycotina) were obtained and analyzed both for the presence of a homologous N-terminal extension and also for the conservation of the putative near-cognate start codon in good context. For alignment of conserved extensions from Sordariomycetidae species, the N. crassa sequence (the extension plus 50–100 amino acids of the main ORF) were used as queries in a BLAST (tblastn) search in the “whole-genome shotgun contigs” (wgs) data bank restricted to Sordariomycetidae. The nucleotide sequences of the positive hits were extracted and then conceptually translated into amino acid sequences. These back-translated sequences were then aligned with ClustalX2 and subjected to minor manual realignment.
RESULTS
In Vivo luc Reporters Containing AUG or Near-cognate Initiation Codons Produce Similar Levels of mRNA
To examine the stringency of start codon selection in N. crassa in vivo, plasmids containing N. crassa codon-optimized luciferase (luc) coding sequences were constructed to enable site-specific integration of luc reporter genes at the N. crassa his-3 locus (Fig. 1A). In these constructs, either an AUG codon, one of nine near-cognate codons, or an AAA codon was placed at the start of the luc coding sequence (Fig. 1A). The AAA codon serves as a negative control, as it differs by two nucleotides from AUG and is not normally used as an initiation codon. All 11 codons were placed in the most-preferred consensus sequence GCCACCXXXG determined from census of N. crassa sequences surrounding the genic start codon (Fig. 1A); this preferred sequence is nearly identical to the most-preferred consensus sequence for human genes (13, 49). All in vivo data reported here represent the results of analyses from three independent transformants of each construct.
The luc transcripts from N. crassa strains containing luc genes initiated with different codons were examined by Northern blot and RT-qPCR (Fig. 1B). Northern analysis using cox-5 mRNA as internal control indicated that luc mRNA was of the predicted size (1850 nucleotides) and the ratios of luc mRNA to cox-5 mRNA were similar in all 11 strains. The WT strain, which does not contain a luc gene, expressed cox-5 mRNA but not luc mRNA as expected (Fig. 1B, lane 12). The similarities in relative luc mRNA levels among strains expressing different genes were confirmed by RT-qPCR using either cox-5 mRNA or 25 S rRNA for normalizing RNA levels.
To determine that all luc transcripts were similarly processed at their 3′ ends, poly(A) mRNA was isolated, and 3′ RACE was performed for all 11 strains containing luc mRNAs and for WT. The major 3′-RACE product for all luc-containing strains, but not the WT strain lacking luc, migrated with the size expected (285 bp) for proper polyadenylation in the reporter gene cox-5 3′-UTR; this was confirmed directly by sequencing selected 3′-RACE products.
The Stringency of Selection of Near-cognate Codons Determined by Real-time Measurements of LUC Enzyme Activity in N. crassa Cultures
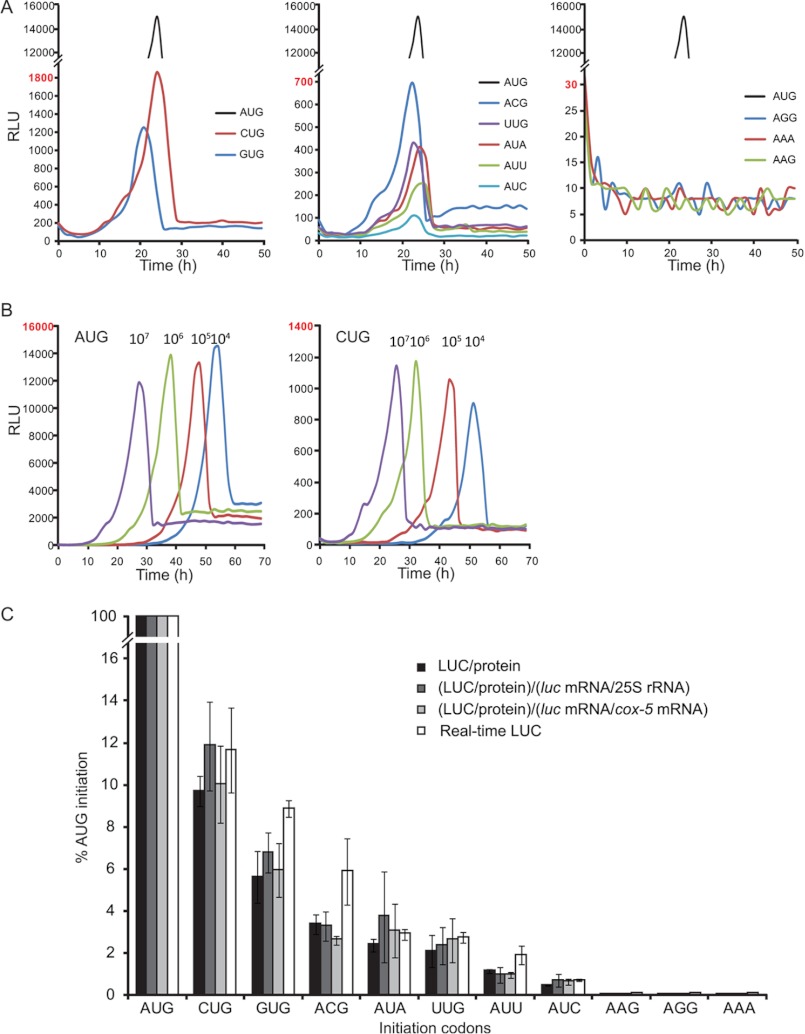
LUC production in vivo was measured using real-time detection of photon emission by growing cells in microtiter plates with luciferin included in the growth medium and imaging with a CCD camera. Different concentrations of conidia (107 conidia/ml in Fig. 2A; 104–107 conidia/ml in Fig. 2B) from strains containing AUG-initiated or near-cognate codon-initiated luc were inoculated into wells containing 0.15 ml of medium. During growth at 25 °C in constant darkness, LUC activity accumulated, reached a peak value, and then fell-off (Fig. 2, A and B). The magnitude of the peak values depended on the initiation codon of luc coding sequence, with an ∼10-fold higher value for AUG than for CUG. The LUC activity value peaked earlier when more conidia were used for inoculation, but the maximum value for each construct did not differ markedly with different amounts of conidia inoculated (Fig. 2B). We presume that the falloff in LUC activity arises from depletion of luciferin reagent as a consequence of cell growth, but this was not tested.
FIGURE 2.
Translation initiation efficiency in vivo at different initiation codons. A and B, measurements of LUC levels in vivo using real-time detection of photon emission is shown. The lines represent triplicate measurements of one set of three independent sets of transformants. A, conidia (107 conidia/ml) from strains expressing luc with initiation codons as indicated were incubated into 0.15 ml of luciferin-containing media in microtiter dish wells and incubated as stationary cultures at 25 °C in constant darkness. Light emission was measured with a microplate scintillation and luminescence counter. Relative light units (RLU) are plotted versus hours of incubation time. Left panel, CUG and GUG compared with AUG. Middle panel, ACG, UUG, AUA, AUU, and AUC compared with AUG. Right panel, AGG, AAA, and AAG compared with AUG. Cultures representing all codons were grown in parallel. B, different amounts of conidia (104–107 conidia/ml) from strains expressing LUC were inoculated. Left panel, cultures expressing AUG-initiated luc. Right panel, cultures expressing CUG-initiated luc. C, translation initiation efficiencies of non-AUG codons are calculated relative to the efficiency of the AUG codon using N. crassa strains with luc reporter genes containing AUG or the indicated codons at the initiation site. White bars, relative real time LUC activities measured in vivo using CCD imaging. The mean peak value from triplicate cultures of a given strain inoculated at 107 conidia/ml was used for calculation. Black, dark gray, and light gray bars, LUC activities were measured from cell extracts normalized in different ways: Black bar, normalized to total extracted protein (determined by Bradford assay); dark gray bar, normalized first to total protein and then calculated relative to the luc mRNA/25 S rRNA level (determined by qPCR, Fig. 1B); light gray bar, normalized first to total protein and then calculated relative to luc mRNA/cox-5 mRNA level (determined by qPCR, Fig. 1B). Mean values and S.D. for all measurements are derived from three independent experiments, each using one set of independent transformants.
The real-time assay was used to compare all 11 luc-containing strains in parallel inoculated at 107 conidia/ml (Fig. 2A). Eight strains showed the accumulation of LUC enzyme activity and reached peak values between 20 and 24 h of incubation (Fig. 2A). The three strains whose luc coding sequences initiated with AAG, AGG, and AAA did not show detectable LUC production, indicating that these three codons were not used as initiation codons to translate luc. Peak values from the eight strains expressing LUC were used to calculate the efficiency of near-cognate codon initiation (Fig. 2C, white bars) by normalizing levels of LUC produced from near-cognate codons to that from the AUG codon. Mean values and S.D. were derived from three independent experiments, each using one set of independent transformants and performed in triplicate. The real-time detection of LUC activity revealed the following hierarchy: AUG ≫ CUG (11.6%) > GUG (8.9%) > ACG (5.9%) > AUA (2.9%) ≈ UUG (2.7%) > AUU (1.9%) > AUC (0.7%).
The Stringency of Selection of Near-cognate Codons in Vivo Was Determined by Measurements of LUC Enzyme Activity in N. crassa Extracts
The stringency of start codon selection in N. crassa was also analyzed by a measurement of LUC activity in soluble cell extracts prepared from all 11 luc-containing strains (Fig. 2C). N. crassa cells were harvested after 6 h of germination, and a portion of cells was used to make extracts for measuring LUC activity and protein concentration, and a portion of cells was used for RNA isolation. LUC enzyme activity was normalized to protein concentration for each extract (LUC/mg of protein). The expression of LUC produced from genes containing non-AUG codons was then calculated relative to that from the AUG codon (Fig. 2C, black bars). To account for (the rather small) differences in luc mRNA levels found in these cells (Fig. 1B), we also calculated differences in the synthesis of luciferase after normalization of the values of LUC/mg of protein to relative cellular luc mRNA levels (using either 25 S rRNA or cox-5 mRNA as internal RNA controls) (Fig. 2C, dark gray and light gray bars). All of these measurements of relative expression of LUC using soluble extracts (Fig. 2C, black, dark gray, and light gray bars) corresponded closely to those obtained by direct measurement of LUC activity in living cells (Fig. 2C, white bars).
The Stringency of Selection of Near-cognate Codons in N. crassa Cell-free Translation Extracts
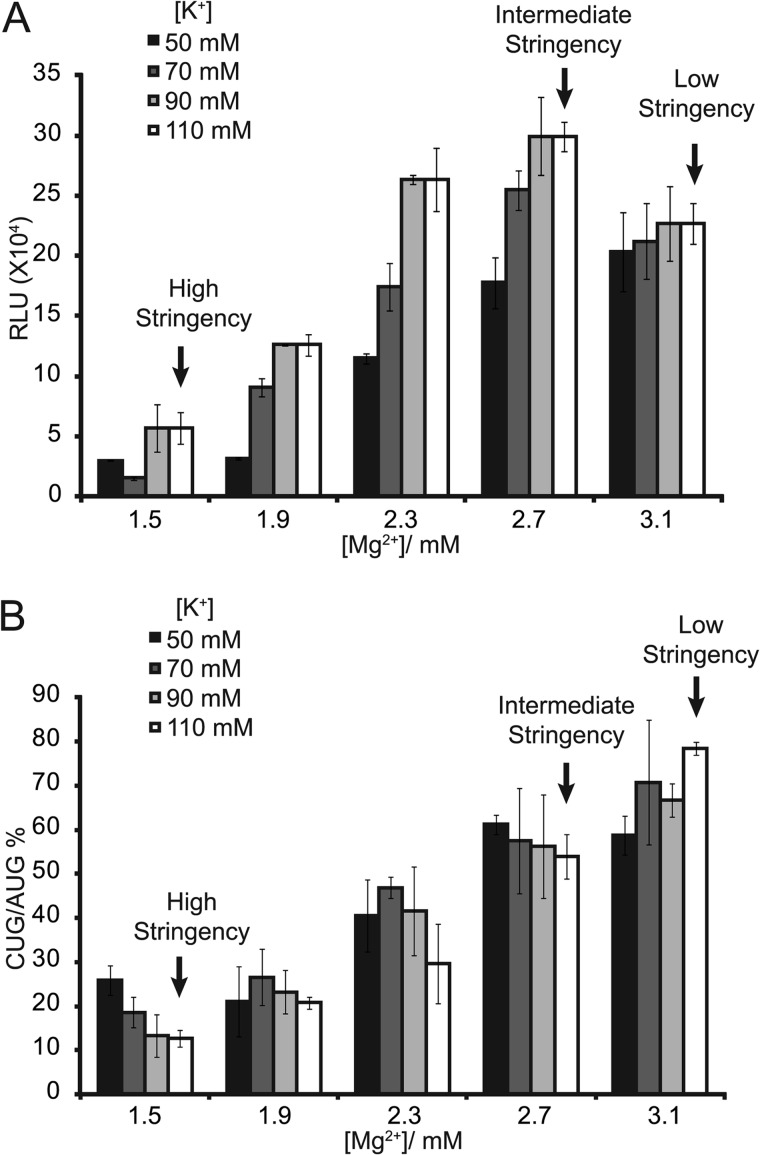
Plasmids were constructed to produce mRNA to test initiation from near-cognate codons in N. crassa cell-free translation extracts. The 11 plasmid templates used for in vitro studies contain the same luc coding and initiation contexts used for in vivo assays. Synthetic capped and polyadenylated mRNAs produced from linearized templates were used to program translation extracts. Mg2+ and K+ concentrations can potentially affect the efficiency of translation and/or the stringency of start codon selection (14, 50). All combinations of five different Mg2+ concentrations and four different K+ concentrations were tested in parallel in triplicate in the N. crassa cell-free translation system using AUG-initiated luc mRNA (Fig. 3A). The overall production of LUC was highest when 110 mm K+ and 2.7 mm Mg2+ were added. Next, the same amounts of CUG-initiated luc mRNA and AUG-initiated luc mRNA were directly compared with the same combinations of Mg2+ and K+ concentrations to determine the effects of these cations on the stringency of start codon selection (Fig. 3B). The CUG/AUG ratio was plotted for each condition used. When reaction mixtures contained 110 mm K+ and 1.5 mm Mg2+, the efficiency of CUG-initiated translation was 12% that of AUG-initiated translation. However, when the concentration of Mg2+ increased to 3.1 mm and K+ was not changed, the efficiency of CUG-initiated translation increased to 80% that of AUG-initiated translation. Three different Mg2+ concentrations and a fixed K+ concentration (110 mm) were chosen to represent high stringency (1.5 mm Mg2+), intermediate stringency (2.7 mm Mg2+), and low stringency (3.1 mm Mg2+) conditions for subsequent analyses of start codon selection in vitro.
FIGURE 3.
Analyses of initiation efficiency using cell-free translation systems. Mean values and S.D. from three independent experiments, each performed in triplicate, are given. A, effects of Mg2+ and K+ concentrations on LUC synthesis from the AUG codon are shown. Capped and polyadenylated luc mRNA (60 ng) was used to program N. crassa translation reactions (10 μl) with all combinations of five different Mg2+ and four different K+ concentrations. Mg2+ and K+ concentrations representing high, intermediate, and low stringency start-codon selection conditions in subsequent experiments are indicated. LUC activity obtained after 30 min of incubation at 26 °C is given in relative light units (RLU). Low stringency conditions are similar to previously established standard conditions (43, 73). B, effects of Mg2+ and K+ concentrations on the stringency of start codon selection are shown. Translation reactions were programmed and incubated as described in A using either CUG or AUG as the LUC initiation codon. The CUG/AUG ratio was plotted as a function of Mg2+ and K+ concentrations.
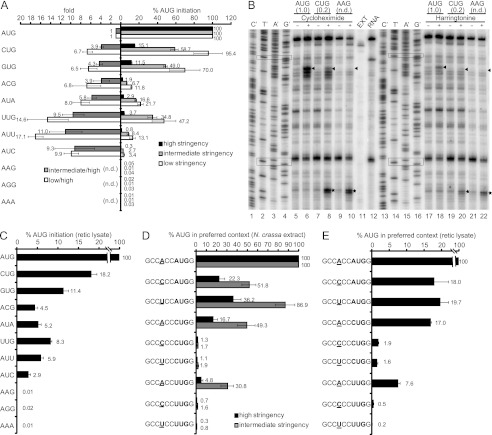
Equal amounts of all 11 mRNAs were tested in parallel in N. crassa translation extracts at salt concentrations conferring high, intermediate, and low stringency conditions for start codon selection (Fig. 4A). Similar to the results obtained in vivo, the use of AAG, AGG, and AAA as start codons was not detected under any condition. Although changing Mg2+ concentration changed the extent of translation from functional near-cognate codons relative to AUG, the relative hierarchy of start codon utilization was not changed. The most efficient near-cognate codon was CUG followed by GUG; codons ACG, AUA, UUG, AUU, and AUC were less efficient as start codons. Under high stringency conditions, the efficiency of utilization of near-cognate codons was generally comparable with those observed in vivo.
FIGURE 4.
Analyses of initiation efficiency and stringency of start codon selection using cell-free translation systems. A, shown is relative initiation efficiency of non-AUG start codons at different Mg2+ concentrations (high, intermediate, and low stringency conditions). On the right, the black, gray, and white bars represent relative initiation at high, intermediate, and low stringency based on LUC activity assays. LUC synthesis from non-AUG codons was calculated relative to synthesis from the AUG codon. On the left, gray and white bars represent the ratios of LUC synthesis between intermediate and high stringency conditions and between low and high stringency conditions, respectively. The ratios for AAG, AGG, and AAA are not given because these codons at the luc initiation site did not yield detectable LUC. B, shown is toeprint analysis to assess initiation at AUG, CUG, and AAG. N. crassa extract was programmed with equal amounts (60 ng) of the indicated luc mRNAs. Cycloheximide or harringtonine were omitted (−) or added (+) before incubation of translation reactions for 5 min at 26 °C. Radiolabeled primer oJI105 was used for primer extension analysis and for sequencing the AUG template (lanes 1–4 and 13–16). The nucleotide complementary to the dideoxynucleotide added to each sequencing reaction is indicated above the corresponding lane. Arrowheads, toeprint products corresponding to ribosomes at the luc initiation codon. Signals from CUG were normalized to signals from AUG, and the results are shown in parentheses. Values were calculated from two independent experiments (cycloheximide 0.24 ± 0.06 and harringtonine 0.24 ± 0.02). Asterisks, toeprint products corresponding to ribosomes at the first downstream AUG codon within the luc coding region. Boxes (top to bottom), luc initiation codon and the first downstream AUG codon. EXT, extract alone; RNA, RNA alone (n.d., not determined). C, translation initiation efficiency in rabbit reticulocyte lysate is shown. Equal amounts of each luc mRNA (6 ng) were translated in rabbit reticulocyte lysate (10 μl translation reaction incubated for 30 min at 30 °C). LUC synthesis from non-AUG codons was calculated relative to synthesis from the AUG codon. D, relative initiation efficiency of AUG, CUG, and UUG in preferred context versus poor contexts in N. crassa under high (black bars) and intermediate (gray bars) stringency conditions is shown. E, relative initiation efficiency of AUG, CUG, and UUG in preferred context versus poor contexts in rabbit reticulocyte lysate is shown. LUC synthesis from non-AUG codons and AUG codon in poor contexts was calculated relative to synthesis from the AUG codon in the preferred context. For A–E, mean values and S.D. from three independent experiments, each performed in triplicate, are given.
When conditions changed from high to intermediate stringency and from high to low stringency, translation from the AUU codon increased 11- and 17.1-fold, respectively; in comparison, ACG initiated luc increased 3.8- and 6.8-fold, respectively (Fig. 4A). This result suggested that different near-cognate codons responded differently to conditions that altered stringency, a phenomenon also observed in human cells (8). The biological bases for this differential response are unknown.
We next directly examined the capacity of AUG, CUG, and AAG to initiate translation in N. crassa extracts under high stringency conditions using the toeprinting assay, which shows the positions of ribosomes engaged in the translation of mRNA (Fig. 4B). Cycloheximide (CYH), which blocks translation elongation, was added before the translation reaction started to increase the signals from ribosomes at translation initiation sites (51). When CYH was added, mRNA encoding AUG-initiated luc showed a strong toeprinting signal corresponding to that start codon (Fig. 4B, lane 6). A weaker signal (24% of the signal from AUG) was observed for the CUG codon at the corresponding position (Fig. 4B, lane 8). For AAG-initiated luc, a signal corresponding to ribosomes at the corresponding position was not detected (Fig. 4B, lane 10), consistent with AAG not serving as an initiation codon. For CUG- and AAG-initiated luc, another signal corresponding to ribosomes at the first downstream AUG codon (which is in the 0 frame) within the luc coding region was observed (Fig. 4B, lanes 8 and 10), consistent with ribosomes scanning pass the CUG and AAG codons and initiating at the first downstream AUG codon. As expected, none of these signals was observed with extract alone (Fig. 4B, lane 11) or RNA alone (Fig. 4B, lane 12). Thus, with respect to the stringency of start codon selection, toeprinting data were similar to the luciferase reporter data both in vivo and in vitro (Figs. 2C and 4A).
Harringtonine blocks initiation by inhibiting elongation during the first rounds of peptide bond formation after subunit joining, causing ribosomes to accumulate at sites of translation initiation (52, 53). We examined AUG, CUG, and AAG codons as initiators using harringtonine in the toeprinting assay in parallel with CYH (Fig. 4B, lanes 17–22). The results were similar to the results obtained for CYH, except that the signals corresponding to ribosomes at initiation codons were weaker than when CYH was used.
The Stringency of Selection of Near-cognate Codons in Reticulocyte Lysate
To examine the stringency of selection of near-cognate codons in a mammalian system, we used rabbit reticulocyte lysate (Invitrogen), which the manufacturer reports to be a high fidelity system with respect to initiation. Similar to the results obtained with N. crassa, CUG was the most efficient near-cognate codon followed by GUG. ACG, AUA, UUG, AUU, and AUC conferred intermediate translation efficiency, and AAG, AGG, and AAA yielded no detectable LUC activity (Fig. 4C).
The Effect of Altering the Initiation Context on Stringency of Start Codon Selection
To examine the effect of poor initiation context on selection of AUG, CUG, and UUG start codons, the nucleotide A at the −3 position, which is the most important upstream position for a preferred context, was mutated to C or U. These poor initiation contexts were compared in parallel to the preferred initiation context in N. crassa translation extract under high stringency conditions (Fig. 4D). As expected, mutating the −3 A to C or U decreased the efficiency of translation initiation from AUG, CUG, and UUG codons. The efficiency of LUC synthesis from CUG and UUG decreased more compared with AUG in these poor contexts. Under intermediate stringency conditions, the efficiency of translation initiation increased for AUG in poor contexts and for CUG and UUG in the preferred context. However, translation from CUG and UUG in poor contexts barely improved. In rabbit reticulocyte lysate, similar results were observed (Fig. 4E). Mutating the −3 A to C or U decreased the efficiency of translation initiation for CUG and UUG more than for AUG. These results indicated that a preferred context is crucial for efficient translation initiation from near-cognate codons.
Prevalence and Relevance of Potential Initiation at Near-cognate Codons in N. crassa
Experimentally verified or bioinformatically predicted cases of translation initiation at near-cognate codons have been described in animals, plants, and fungi, including filamentous fungi (23, 54, 55). However, the prevalence of initiation at near-cognate codons in N. crassa has not been previously investigated. This is important to consider given the results presented here. When examined systematically, for example by taking advantage of ribosome profiling to identify initiation codons (30, 56, 57), initiation at near-cognate codons is seen mostly 5′ of the previously annotated AUG start codons. These near-cognate codons either initiate a uORF or an N-terminal extension to the main ORF. Individual examples of conserved non-AUG-initiated uORFs are known (58); however, a systematic bioinformatics search for such uORFs is complex. Identifying N-terminal-extended conserved ORFs initiated with near-cognate codons is a more tractable problem (47).
RNA-Seq has been used to determine the N. crassa mRNA transcriptome.5 This data set includes sequences of 10,785 different N. crassa mRNA transcripts. Genes represented by more than one transcript were removed from further analysis to avoid redundancy. This yielded a total of 6804 genes represented by unique transcripts. Using these data, we investigated bioinformatically the prevalence of potential translation initiation at near-cognate codons in N. crassa. We defined “optimal” context as having A at position −3 regardless of the identity of position +4 or having G at position −3 in combination with G at position +4. All other contexts were considered suboptimal. AAG and AGG codons were considered incompatible with initiation. Using these criteria we identified 5688 near-cognate codons 5′ of the annotated starting AUG and in-frame with it in optimal context and without intervening in-frame stop codons. Because in some instances more than one of these near-cognate codons is present in an mRNA, there are a total of 3030 (45% of all examined) ORFs with potential N-terminal extensions. Of these, 2172 in-frame near-cognate codons from 1185 genes could yield extensions of at least 25 amino acids, with 163 in-frame near-codons from 73 genes able to initiate extensions of at least 100 amino acids (data not shown).
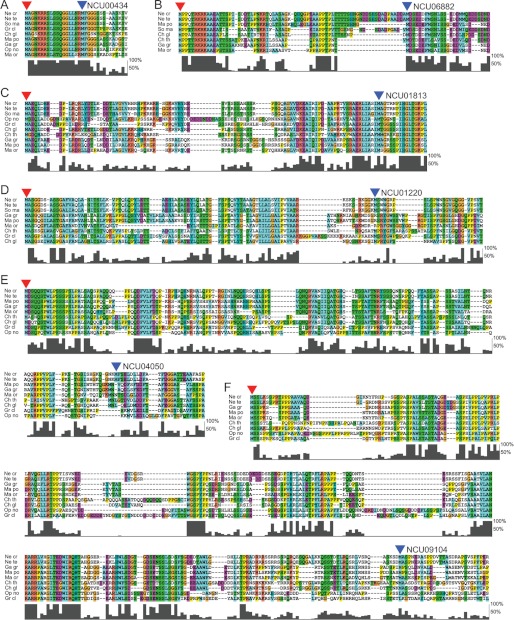
We next asked whether cases of physiologically relevant near-cognate-initiated N-terminal extensions might be identifiable in this set. We reasoned that many physiologically relevant near-cognate-initiated extensions will be conserved even in more distant relatives of N. crassa. We applied a comparative genomics approach similar to one that allowed the identification of conserved near-cognate-initiated N-terminal extensions in mammals to search for such extensions in N. crassa (Ref. 47; also see “Experimental Procedures”). We limited the search to extension queries that are at least 25 amino acids long. Using a conservative approach, we identified six N. crassa genes with conserved near-cognate-initiated N-terminal extensions. The genes (and annotations in N. crassa genome release NC10 at Broad Institute) are: NCU00434 (protein phosphatase 2C isoform β); NCU01220 (BAG domain-containing protein); NCU01813 (high affinity glucose transporter); NCU04050 (cross-pathway control protein 1, cpc-1); NCU06882 (RING-5); NCU09104 (hypothetical protein). The predicted extensions add 18, 88, 91, 153, 71, and 262 N-terminal amino acids, respectively. The identified near-cognate codons are CUG, GUG, AUC, ACG, AUU, and ACG, respectively, and each is highly conserved in the orthologs whose sequences are available. An interesting exception is the N-terminal extension of NCU01813, which is mostly initiated by AUU in homologs, but in a minority of filamentous fungi, for example Cryphonectria parasitica, is initiated by a conventional AUG. In all cases the putative N-terminal extensions show phylogenetic conservation in almost all or all Pezizomycotina orthologs for which sequences are available. The conservation of the identified extensions from subclass Sordariomycetidae is shown in Fig. 5 and their respective annotated N. crassa nucleotide sequences are shown in Supplemental Fig. S2. In all but one case, NCU04050, no out-of-frame AUG codon is located between the conserved in-frame near-cognate and conventional initiation codon of the main ORF. This indicates that both the near-cognate and the conventional initiation codons could potentially be used for initiation, resulting in distinct long and short isoforms of the given protein.
FIGURE 5.
Amino acid conservation of the newly predicted non-AUG initiated N-terminal extensions from Pezizomycotina, subclass Sordariomycetidae. The alignments were generated with ClustalX2 using available sequences from Sordariomycetidae. In each case the N. crassa sequence is presented on the top row. Amino acids with similar chemical properties are highlighted with the same color. The alignment clusters are as follows: A, NCU00434; B, NCU06882; C, NCU01813; D, NCU01220; E, NCU04050; F, NCU09104. The level of conservation for the amino acids in each column, expressed as a percentage, is indicated on the right side of each alignment. In each case it is assumed that the near cognate start codon is initiated with methionine. This amino acid is indicated by a red arrow. The position of the predominant methionine corresponding to the first in-frame AUG of the main open reading frame is indicated by blue arrow above it. In each case only alignment for the first 15–35 amino acids on the main open reading frame is shown. Species name abbreviations (shown on the left of each alignment) are as follows: Ne cr, N. crassa; Ne te, Neurospora tetrasperma; Ma or, Magnaporthe oryzae; Ma po, Magnaporthe poae; Ch gl, C. globosum; Ch th, Chaetomium thermophilum; Gr cl, Grosmannia clavigera; So ma, Sordaria macrospora; Op no, Ophiostoma novo-ulmi; Ga gr, Gaeumannomyces graminis.
DISCUSSION
We examined the stringency of start codon selection in N. crassa in vivo and in vitro using a codon-optimized firefly luciferase reporter gene. Translation initiation from the nine near-cognate codons in the preferred initiation context was compared in parallel with AUG. CUG and GUG are the most efficient near-cognate codons followed by ACG, AUA, UUG, AUU and AUC; AAG and AGG are not used for initiation. The efficiency of near-cognate start-codon selection in vitro was affected by Mg2+ concentration; under the most stringent conditions examined, translation from CUG in a preferred context was ∼12–15% compared with AUG, similar to the level observed in vivo. Additional analyses in vitro showed that the preferred −3 nucleotide was important for maintaining translation initiation efficiency, particularly for near-cognate start codons.
There is a good correlation between the efficiency of initiation from near-cognate codons in N. crassa and human cells (compare Fig. 2C with Fig. 3B in Ref. 8). In each case the efficiency of the near-cognate codons can be grouped in three categories: high, CUG and GUG; intermediate, ACG, AUA, UUG, AUU, and AUC; inactive, AAG and AGG. The difference in initiation efficiency between the most active (CUG) and least active (AUC) near-cognate codon in N. crassa and human cells is ∼10-fold. In each case the context for comparing near-cognate codon initiation was GCCACCXXXG. In contrast, one recent analysis in S. cerevisiae shows that the efficiencies of the different near-cognate codons do not differ as greatly, with only the least active (also AUC) showing a substantial difference from the others (4). The reasons why N. crassa more closely resembles mammals than does S. cerevisiae in this regard remains to be determined. For at least some initiation factors, such as eIF3, N. crassa more closely resembles mammals than does S. cerevisiae (59); this might be important for determining initiation efficiency at different start codons. Possibly, additional considerations can impact choice of near-cognate codons as initiation codons. The differences in initiation among near-cognate codons can depend on the specific initiation context (60). Earlier studies in S. cerevisiae yielded different results with respect to the relative efficiencies of different near-cognate codons (6, 61). Thus, some differences may arise because different initiation contexts were used. For example, the respective contexts used in S. cerevisiae studies were CUCUCUXXXC (4), GACAAGXXXA (60), GAAAAAXXXU (24, 60), UGAAUAXXXG (61), and CAAAACXXXG (6).
Firefly luciferase has been used in N. crassa for circadian studies but has not been used quantitatively. Few reporters have been used quantitatively in this organism. Here we showed that independent transformants containing this reporter integrated as a fusion with cox-5 5′ promoter and 3′ regions showed similar relative levels of expression by both real-time luciferase measurements and assays using cell-free extracts. The luc reporter we used can be exchanged between vectors for in vivo and in vitro expression. The in vivo reporter construct is designed to enable exchange of promoter, 5′-UTR, and 3′ sequences. This system should thus be adaptable for analyses of other elements that control translation as well as other processes affecting gene expression.
When N. crassa extracts are optimized by titrating Mg2+ for overall translational yield from the AUG-initiated luciferase reporter mRNA (Fig. 3A), higher concentrations of Mg2+ both increase yield and reduce the stringency of start codon selection (Fig. 3B). It is known that Mg2+ levels in cell-free extracts affect stringency (14). Although the overall hierarchy of near-cognate codon utilization did not change, UUG and AUU were used relatively more efficiently under reduced stringency conditions (Fig. 4A). In cultured human cells, UUG responded similarly to reduced stringency, which was achieved by eIF1 overexpression (8). In vitro experiments demonstrate that eIF1 is crucial for the stringency of start codon selection, impacting both discrimination between AUG and near-cognate codons and bias between good and poor initiation contexts (62). Genetic, biochemical, and molecular studies suggest that increased levels of eIF5 can cause eIF1 to be dissociated from the preinitiation complex, decreasing the stringency of start codon selection (32, 63–66). We speculate that higher Mg2+ concentrations could increase release of eIF1 from the preinitiation complex N. crassa extracts, resulting in increased initiation from near-cognate codons. Increased intracellular Mg2+ in S. cerevisiae is known to affect the fidelity of translation termination (67); effects of Mg2+ levels in vivo on stringency of initiation have yet to be determined. It is possible that the changes of Mg2+ concentration in response to different physiological conditions (68) may provide regulatory function by impacting the stringency of start codon selection and, therefore, gene expression.
In the preferred consensus (GCC(A/G)CCXXXG) that is optimal for initiation (12, 13), a purine at position −3 and a G at position +4 is most important (12). Mutations that depart from the consensus at position −3 can reduce initiation by more than an order of magnitude in mammalian cells (12). The comparison of initiation from AUG, CUG, and UUG in the preferred versus poor contexts using N. crassa extract indicated that mutation at position −3 reduced translation initiation from each codon. The reduction of initiation from CUG and UUG was greater than from AUG, indicating that a preferred context is crucial for initiation at non-AUG codons (Fig. 4D and Refs. 14, 69, and 70). When the stringency of initiation is relaxed by increasing Mg2+ concentration, the efficiency of initiation from these near-cognate codons in poor contexts did not improve, although efficiency improved when these codons were in a good context. A possible explanation for this is that near-cognate codons in poor contexts are below a threshold for conversion of the open preinitiation complex (PIC) to a closed PIC independent of Mg2+. For example, the scanning PIC might fail to recognize near-cognate codons in poor contexts, but when near-cognate codons in a preferred context are recognized, Mg2+ levels would increase the likelihood of formation of a closed PIC.
The ramifications of initiation from near-cognate start codons for the biology of eukaryote organisms are beginning to be widely appreciated. Seven of the nine near-cognate codons in a preferred context demonstrably initiated translation in N. crassa in vivo, showing that in this organism non-AUG codons initiate translation. Near-cognate codons can initiate translation of uORFs (71) and synthesis of alternative N-terminally extended protein isoforms (2). Some proteins are synthesized exclusively from near-cognate codons, including mammalian eIF4G2 (22), P. anserina IDI-4 (23), and S. cerevisiae glycyl-tRNA synthetase (24). Ribosome profiling has revealed a multitude of eukaryotic near-cognate initiation events (29, 30, 56). In S. cerevisiae, there is evidence for widespread regulated initiation at non-AUG codons (29). Studies using mouse embryonic stem cells provide evidence that near-cognate codons initiate translation of longer and shorter forms of proteins as well as uORFs and that initiation at these codons changes during differentiation (30). Our studies, which demonstrate that near-cognate codons can substitute for AUG to initiate synthesis of a luciferase reporter at substantial levels, provide additional direct experimental support that the traditional view that AUG is the translation initiation codon must be expanded. Evolutionarily speaking, near-cognate codon initiation, although generally less efficient than AUG initiation, could serve important roles. Consistent with the idea that high efficiency is not always evolutionarily preferred and low efficiency can be advantageous, recently it was demonstrated that non-optimal codon preference in the elongation phase is crucial for the synthesis of a functional eukaryotic protein central to establishing a circadian rhythm (72). The potential functions of 5′ proximal near-cognate codons need to be considered in evaluating the coding capacity of mRNA.
Acknowledgments
We thank Renato de Paula and Deborah Bell-Pedersen for providing the codon-optimized luc construct, Virag Sharma, Anmol Kiran, and Matthew Peterson for assistance with the bioinformatic analysis of near-cognate codons in the N. crassa transcriptome, and Cheng Wu for help with the toeprint analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants GM47498 and GM068087 (to M. S. S.).

This article contains supplemental Tables S1–S4 and Figs. S1 and S2.
R. de Paula and D. Bell-Pedersen, unpublished information.
M. Sachs, unpublished information.
- uORF
- upstream open reading frame
- LUC
- luciferase
- VM
- Vogel's minimal medium
- qPCR
- quantitative PCR
- CYH
- cycloheximide
- PIC
- preinitiation complex
- RACE
- rapid amplification of cDNA ends
- contig
- group of overlapping clones.
REFERENCES
- 1. Aitken C. E., Lorsch J. R. (2012) A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576 [DOI] [PubMed] [Google Scholar]
- 2. Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kolitz S. E., Takacs J. E., Lorsch J. R. (2009) Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA 15, 138–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peabody D. S. (1989) Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264, 5031–5035 [PubMed] [Google Scholar]
- 6. Clements J. M., Laz T. M., Sherman F. (1988) Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 4533–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gordon K., Fütterer J., Hohn T. (1992) Efficient initiation of translation at non-AUG triplets in plant cells. Plant J. 2, 809–813 [PubMed] [Google Scholar]
- 8. Ivanov I. P., Loughran G., Sachs M. S., Atkins J. F. (2010) Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 107, 18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kieft J. S. (2008) Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33, 274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Starck S. R., Jiang V., Pavon-Eternod M., Prasad S., McCarthy B., Pan T., Shastri N. (2012) Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336, 1719–1723 [DOI] [PubMed] [Google Scholar]
- 11. Kozak M. (1984) Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 12, 857–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44, 283–292 [DOI] [PubMed] [Google Scholar]
- 13. Kozak M. (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196, 947–950 [DOI] [PubMed] [Google Scholar]
- 14. Kozak M. (1989) Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 9, 5073–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vaughn J. N., Ellingson S. R., Mignone F., Arnim A. (2012) Known and novel post-transcriptional regulatory sequences are conserved across plant families. RNA 18, 368–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang F., Hinnebusch A. G. (2011) An upstream ORF with non-AUG start codon is translated in vivo but dispensable for translational control of GCN4 mRNA. Nucleic Acids Res. 39, 3128–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003) Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol. Cell 95, 169–178 [DOI] [PubMed] [Google Scholar]
- 18. Park E. H., Walker S. E., Lee J. M., Rothenburg S., Lorsch J. R., Hinnebusch A. G. (2011) Multiple elements in the eIF4G1 N terminus promote assembly of eIF4G1·PABP mRNPs in vivo. EMBO J. 30, 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilke C. O., Drummond D. A. (2010) Signatures of protein biophysics in coding sequence evolution. Curr. Opin. Struct. Biol. 20, 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang H. L., Yeh L. S., Chen N. K., Ripmaster T., Schimmel P., Wang C. C. (2004) Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J. Biol. Chem. 279, 49656–49663 [DOI] [PubMed] [Google Scholar]
- 21. Christensen A. C., Lyznik A., Mohammed S., Elowsky C. G., Elo A., Yule R., Mackenzie S. A. (2005) Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17, 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi K., Maruyama M., Tokuzawa Y., Murakami M., Oda Y., Yoshikane N., Makabe K. W., Ichisaka T., Yamanaka S. (2005) Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85, 360–371 [DOI] [PubMed] [Google Scholar]
- 23. Dementhon K., Saupe S. J., Clavé C. (2004) Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Mol. Microbiol. 53, 1625–1640 [DOI] [PubMed] [Google Scholar]
- 24. Chang K. J., Wang C. C. (2004) Translation initiation from a naturally occurring non-AUG codon in Saccharomyces cerevisiae. J. Biol. Chem. 279, 13778–13785 [DOI] [PubMed] [Google Scholar]
- 25. Riechmann J. L., Ito T., Meyerowitz E. M. (1999) Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol. Cell. Biol. 19, 8505–8512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simpson G. G., Laurie R. E., Dijkwel P. P., Quesada V., Stockwell P. A., Dean C., Macknight R. C. (2010) Noncanonical translation initiation of the Arabidopsis flowering time and alternative polyadenylation regulator FCA. Plant Cell 22, 3764–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hedtke B., Legen J., Weihe A., Herrmann R. G., Börner T. (2002) Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J. 30, 625–637 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi Y., Dokiya Y., Kumazawa Y., Sugita M. (2002) Non-AUG translation initiation of mRNA encoding plastid-targeted phage-type RNA polymerase in Nicotiana sylvestris. Biochem. Biophys. Res. Commun. 299, 57–61 [DOI] [PubMed] [Google Scholar]
- 29. Ingolia N. T., Ghaemmaghami S., Newman J. R., Weissman J. S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ingolia N. T., Lareau L. F., Weissman J. S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martin-Marcos P., Cheung Y. N., Hinnebusch A. G. (2011) Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 31, 4814–4831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loughran G., Sachs M. S., Atkins J. F., Ivanov I. P. (2012) Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res. 40, 2898–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xie W., Ling T., Zhou Y., Feng W., Zhu Q., Stunnenberg H. G., Grummt I., Tao W. (2012) The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc. Natl. Acad. Sci. U.S.A. 109, 8161–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jacobs G. H., Chen A., Stevens S. G., Stockwell P. A., Black M. A., Tate W. P., Brown C. M. (2009) Transterm. A database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 37, D72–D76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McCluskey K., Wiest A., Plamann M. (2010) The Fungal Genetics Stock Center. A repository for 50 years of fungal genetics research. J. Biosci. 35, 119–126 [DOI] [PubMed] [Google Scholar]
- 36. Margolin B. S., Freitag M., Selker E. U. (1997) Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newsl. 44, 34–36 [Google Scholar]
- 37. Ebbole D., Sachs M. S. (1990) A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newsl. 37, 17–18 [Google Scholar]
- 38. Vogel H. J. (1956) A convenient growth medium for Neurospora (Medium N). Microbiol. Genet. Bull. 13, 42–43 [Google Scholar]
- 39. Luo Z., Freitag M., Sachs M. S. (1995) Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol. Cell. Biol. 15, 5235–5245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dyer B. W., Ferrer F. A., Klinedinst D. K., Rodriguez R. (2000) A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 282, 158–161 [DOI] [PubMed] [Google Scholar]
- 41. Sachs M. S., Yanofsky C. (1991) Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev. Biol. 148, 117–128 [DOI] [PubMed] [Google Scholar]
- 42. Gründemann D., Schömig E. (1996) Protection of DNA during preparative agarose gel electrophoresis against damage induced by ultraviolet light. BioTechniques 21, 898–903 [DOI] [PubMed] [Google Scholar]
- 43. Fang P., Wang Z., Sachs M. S. (2000) Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem. 275, 26710–26719 [DOI] [PubMed] [Google Scholar]
- 44. Wei J., Wu C., Sachs M. S. (2012) The arginine attenuator peptide interferes with the ribosome peptidyl transferase center. Mol. Cell. Biol. 32, 2396–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Z., Sachs M. S. (1997) Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 17, 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sachs M. S., Wang Z., Gaba A., Fang P., Belk J., Ganesan R., Amrani N., Jacobson A. (2002) Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 26, 105–114 [DOI] [PubMed] [Google Scholar]
- 47. Ivanov I. P., Firth A. E., Michel A. M., Atkins J. F., Baranov P. V. (2011) Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic Acids Res. 39, 4220–4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 49. Nakagawa S., Niimura Y., Gojobori T., Tanaka H., Miura K. (2008) Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 36, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu C., Amrani N., Jacobson A., Sachs M. S. (2007) in Translation Initiation: Extract Systems and Molecular Genetics (Lorsch J., ed.) pp. 203–225, Elsevier, San Diego, CA [Google Scholar]
- 51. Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J. 20, 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fresno M., Jiménez A., Vázquez D. (1977) Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72, 323–330 [DOI] [PubMed] [Google Scholar]
- 53. Robert F., Carrier M., Rawe S., Chen S., Lowe S., Pelletier J. (2009) Altering chemosensitivity by modulating translation elongation. PLoS One 4, e5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Conlon H., Zadra I., Haas H., Arst H. N., Jr., Jones M. G., Caddick M. X. (2001) The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol. Microbiol. 40, 361–375 [DOI] [PubMed] [Google Scholar]
- 55. Gutiérrez S., Díez B., Montenegro E., Martín J. F. (1991) Characterization of the Cephalosporium acremonium pcbAB gene encoding α-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase. Linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J. Bacteriol. 173, 2354–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee S., Liu B., Lee S., Huang S. X., Shen B., Qian S. B. (2012) Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 109, E2424–E2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fritsch C., Herrmann A., Nothnagel M., Szafranski K., Huse K., Schumann F., Schreiber S., Platzer M., Krawczak M., Hampe J., Brosch M. (2012) Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 22, 2208–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ivanov I. P., Loughran G., Atkins J. F. (2008) uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. U.S.A. 105, 10079–10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Borkovich K. A., Alex L. A., Yarden O., Freitag M., Turner G. E., Read N. D., Seiler S., Bell-Pedersen D., Paietta J., Plesofsky N., Plamann M., Goodrich-Tanrikulu M., Schulte U., Mannhaupt G., Nargang F. E., Radford A., Selitrennikoff C., Galagan J. E., Dunlap J. C., Loros J. J., Catcheside D., Inoue H., Aramayo R., Polymenis M., Selker E. U., Sachs M. S., Marzluf G. A., Paulsen I., Davis R., Ebbole D. J., Zelter A., Kalkman E. R., O'Rourke R., Bowring F., Yeadon J., Ishii C., Suzuki K., Sakai W., Pratt R. (2004) Lessons from the genome sequence of Neurospora crassa. Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68, 1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang C. P., Chen S. J., Lin C. H., Wang T. L., Wang C. C. (2010) A single sequence context cannot satisfy all non-AUG initiator codons in yeast. BMC Microbiol. 10, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Donahue T. F., Cigan A. M. (1988) Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol. Cell. Biol. 8, 2955–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pestova T. V., Kolupaeva V. G. (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 16, 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Valásek L., Phan L., Schoenfeld L. W., Valásková V., Hinnebusch A. G. (2001) Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 20, 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nanda J. S., Cheung Y. N., Takacs J. E., Martin-Marcos P., Saini A. K., Hinnebusch A. G., Lorsch J. R. (2009) eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 394, 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luna R. E., Arthanari H., Hiraishi H., Nanda J., Martin-Marcos P., Markus M. A., Akabayov B., Milbradt A. G., Luna L. E., Seo H. C., Hyberts S. G., Fahmy A., Reibarkh M., Miles D., Hagner P. R., O'Day E. M., Yi T., Marintchev A., Hinnebusch A. G., Lorsch J. R., Asano K., Wagner G. (2012) The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep. 1, 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Singh C. R., Watanabe R., Chowdhury W., Hiraishi H., Murai M. J., Yamamoto Y., Miles D., Ikeda Y., Asano M., Asano K. (2012) Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48 S preinitiation complex and promotes its shift to the initiation mode. Mol. Cell. Biol. 32, 3978–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johansson M. J., Jacobson A. (2010) Nonsense-mediated mRNA decay maintains translational fidelity by limiting magnesium uptake. Genes Dev. 24, 1491–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang A., Cheng T. P., Wu X. Y., Altura B. T., Altura B. M. (1997) Extracellular Mg2+ regulates intracellular Mg2+ and its subcellular compartmentation in fission yeast, Schizosaccharomyces pombe. Cell. Mol. Life Sci. 53, 69–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen S. J., Lin G., Chang K. J., Yeh L. S., Wang C. C. (2008) Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J. Biol. Chem. 283, 3173–3180 [DOI] [PubMed] [Google Scholar]
- 70. Portis J. L., Spangrude G. J., McAtee F. J. (1994) Identification of a sequence in the unique 5' open reading frame of the gene encoding glycosylated Gag, which influences the incubation period of neurodegenerative disease induced by a murine retrovirus. J. Virol. 68, 3879–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kochetov A. V., Prayaga P. D., Volkova O. A., Sankararamakrishnan R. (2013) Hidden coding potential of eukaryotic genomes. NonAUG started ORFs. J. Biomol. Struct. Dyn. 1, 103–114 [DOI] [PubMed] [Google Scholar]
- 72. Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J. M., Sachs M. S., Liu Y. (2013) Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang Z., Fang P., Sachs M. S. (1998) The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it. Mol. Cell. Biol. 18, 7528–7536 [DOI] [PMC free article] [PubMed] [Google Scholar]