Synopsis
Normal insect development requires a precisely timed, precipitous drop in hemolymph juvenile hormone (JH) titer. This drop occurs through a coordinated halt in JH biosynthesis and increase in JH metabolism. In many species, JH esterase (JHE) is critical for metabolism of the resonance-stabilized methyl ester of JH. JHE metabolizes JH with a high kcat/KM ratio that results primarily from an exceptionally low KM. Here we review the biochemistry and structure of authentic and recombinant JHEs from six insect orders, and present updated diagnostic criteria that help to distinguish JHEs from other carboxylesterases. The use of a JHE-encoding gene to improve the insecticidal efficacy of biopesticides is also discussed.
Keywords: juvenile hormone esterase (JHE), JH, inhibitor, carboxylesterase
Introduction
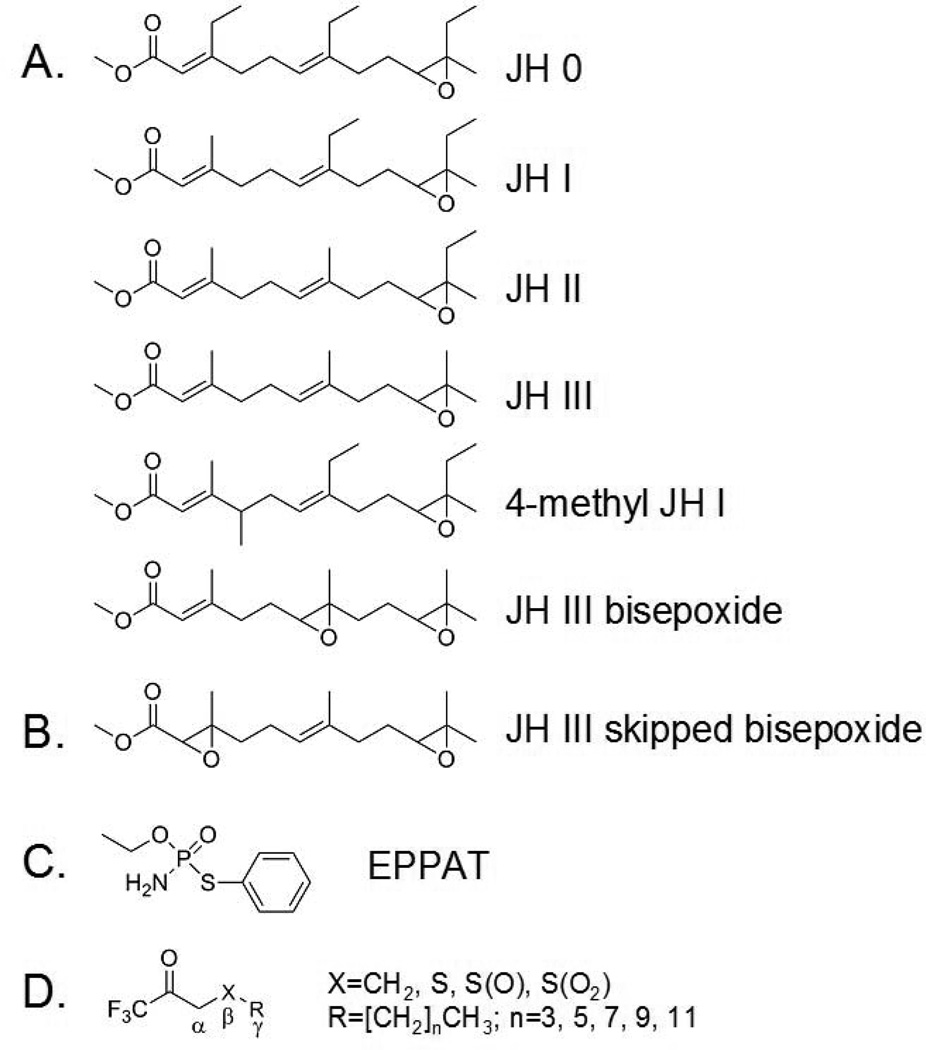
Juvenile hormones (JH) are a family of sesquiterpenes that regulate a wide spectrum of critical biological events in insects including development, metamorphosis, reproduction, polyphenisms, and alteration in behavior.1–6) Six major JH homologs (JH 0, JH I, JH II, JH III, 4-methyl JH I, and JH III bisepoxide; Fig. 1A) have been identified that possess an α,β-unsaturated methyl ester at one end of the molecule and an epoxide at the other.3) A novel JH (JH III skipped bisepoxide; Fig. 1B) with an epoxide moiety in place of the α,β-double bond has also been recently identified in the heteropteran insect Plautia stali.7) Because this epoxide is very difficult to form chemically, this heteropteran compound is likely generated by a specific cytochrome P450. In addition, hydroxylated JHs, JH metabolites, JH precursors (e.g., farnesoic acid, methyl farnesoate), and other molecules that are not typically thought to be hormones may also function as biologically active “JH”.2,3)
Fig.1.
Chemical structures of JH and inhibitors of JHE. Six major forms of JH have been identified that possess an α,β-unsaturated methyl ester at one end of the molecule and an epoxide at the other (A). A JH with an epoxide moiety in place of the α,β-double bond (B) has also been identified in a heteropteran insect. Phosphoramidothiolates such as O-ethyl-S-phenyl phosphoramidothiolate (EPPAT) (C) and trifluoromethyl ketone (TFK) containing compounds (D) are potent inhibitors of JHE.
Stringent regulation of JH and ecdysteroid titers is essential for normal insect development and metamorphosis. When hemolymph JH is present at low nanomolar levels, the status quo is maintained such that a larval-larval or nymphal-nymphal molt results (i.e., the juvenile stage of the insect is maintained). On the other hand, when hemolymph JH titer is dramatically reduced (by at least 50-fold to perhaps more than 1000-fold) from the status quo level and when there is a concurrent spike (of ca. 50 nM) in molting hormone, a developmentally more advanced larval-pupal or nymphal-pupal molt occurs. In the most simplistic terms, the reduction in JH titer occurs through a combination of a decrease in JH biosynthesis and increase in JH metabolism.3,8–10) The contribution of JH sequestration by JH binding proteins and/or the availability of JH receptor molecule(s), however, should not be overlooked when considering the availability and action of JH.3,6,11)
Two primary pathways for JH metabolism are known following the biosynthesis and release of JH from a pair of endocrine glands known as the corpora allata (Fig. 2). Both of these primary routes of metabolism involve enzymes in the α/β-hydrolase fold superfamily. The relative importance of these hydrolytic pathways varies with the species and developmental stage of the insect. The methyl ester of JH is hydrolyzed by a JH-specific esterase (JHE) and the epoxide by JH epoxide hydrolase (JHEH). JHE activity is largely found in the hemolymph whereas JHEH activity is membrane bound and found in tissues. Resonance stabilization of the α,β-unsaturated ester of JH (Fig. 1A) is hypothesized to make this ester more resistant (compared to a saturated form of JH) to chemical hydrolysis and to nucleophilic attack by non-JHE carboxylesterases that are found in the hemolymph.3,8) The ability of JHE to efficiently metabolize this resonance-stabilized ester suggests that JHE has uniquely evolved for this task. Although JH hydrolysis is generally considered a pathway of inactivation, the metabolite JH acid has been shown to possess hormonal activity12,13) suggesting that under certain situations JHE may function as a biosynthetic enzyme. Other common primary metabolic pathways including oxidation and conjugation by enzymes in the cytochrome P450 and glutathione S-transferase families, respectively, appear to be relatively unimportant for JH metabolism in the insect species that have been studied.8) In addition, a secondary metabolic pathway of JH involves JH diol kinase.3,9,14–17) This enzyme adds a phosphate group to the C10 of JH diol following the conversion of JH to JH diol by JHEH.
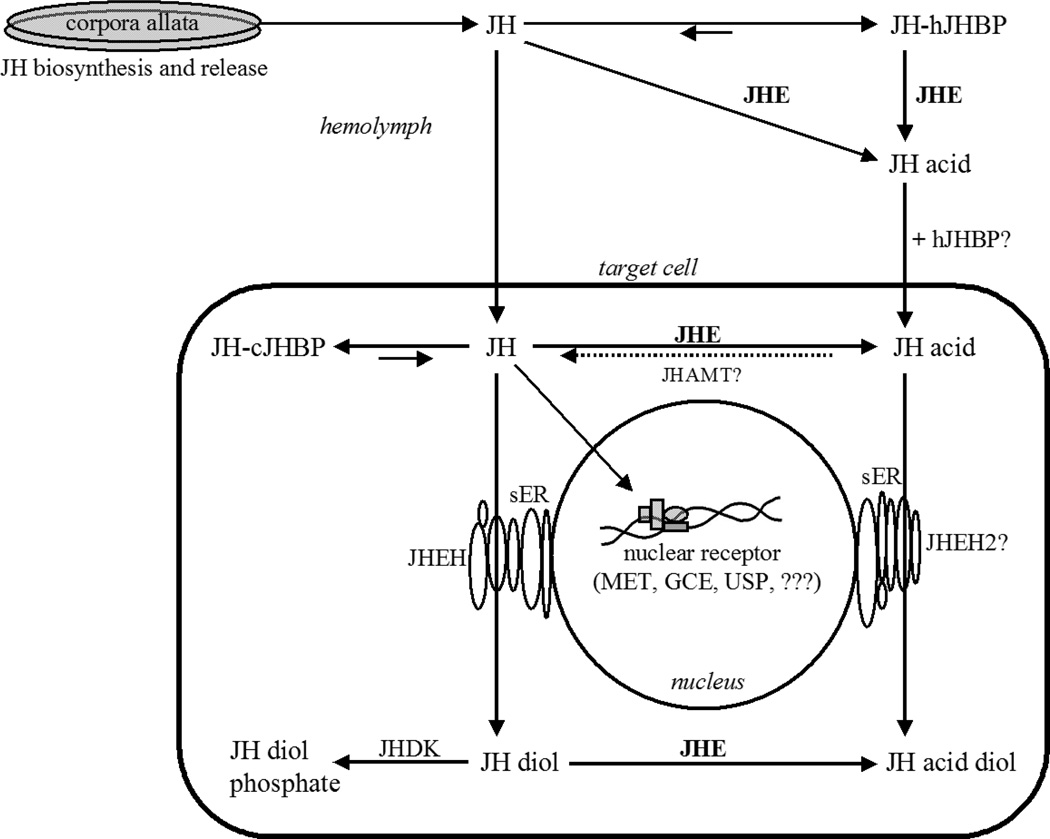
Fig.2.
The physiological fate of JH following its biosynthesis in the corpora allata glands and release into the hemolymph. The primary mode of JH metabolism in the hemolymph is through the action of JHE. JHE hydrolyzes JH in the presence or absence of hemolymph JH binding protein (hJHBP) forming JH acid. JHE is also be found inside the cell where JH or JH diol are potential substrates. Additional abbreviations: JHEH, JH epoxide hydrolase; JHAMT, JH acid methyl transferase; JHDK, JH diol kinase; cJHBP, cytosolic JHBP; sER, smooth endoplasmic reticulum; Met, methoprene tolerant protein; GCE, germ cell expressed protein; USP, ultraspiracle protein.
Twenty-five years ago, Hammock first proposed biological and biochemical criteria as a way to define a JH-specific esterase.8) Biologically, JHE was defined by two criteria as “a JH hydrolyzing esterase whose role is shown to be essential for the clearance of JH from the insect’s body; and whose titer correlates with suspected declines in JH titer”. Biochemically, JHE was defined by three criteria as “an esterase capable of rapidly hydrolyzing JH in the presence or absence of carrier protein; an enzyme which hydrolyzes JH with a high kcat/KM ratio; and an enzyme which hydrolyzes JH with low apparent KM”. In this review we revisit these criteria, which still remain valid, in light of the wealth of biological, biochemical, and structural data that are now available.
Catalytic mode of action of JHE and kinetic parameters
JHE, like other carboxylesterases, hydrolyze ester bonds in a two-step reaction.18) The first step of JH hydrolysis involves nucleophilic attack of the carbonyl carbon of JH by the catalytic serine residue that is found in the substrate-binding pocket, release of methyl alcohol, and formation of an acyl-enzyme intermediate. The second step involves release of JH acid and regeneration of JHE following nucleophilic attack of the carbonyl carbon of the acyl-enzyme by an activated water molecule. Tetrahedral transition state intermediates are formed in both steps. Decomposition or deacylation of the transition state intermediate that is formed during the second step is implicated as a rate-determining step in the hydrolysis of JH.8,19)
The kinetic parameters of JH hydrolysis by authentic and recombinant JHEs are well characterized from numerous species that are found in at least five insect orders (see Table 1).3,8) The apparent Michaelis constant (KM) of JHEs from lepidopteran and orthopteran insects are in the low nanomolar range whereas those of JHEs from coleopteran and hemipteran insects appear to be higher. Although the physiological substrate of JHE is clearly defined as JH, the hydrolysis of JH by JHE occurs at a relatively slow rate (kcat of generally less than 2 s−1). On the other hand, JHE turns over ester-containing substrates such as α-naphthyl acetate (a general carboxylesterase substrate) at a significantly higher rate. The authentic JHE of Drosophila melanogaster (DmJHE), for example, turns over α-naphthyl acetate with a kcat of 71 s−1 in comparison to 0.6 s−1 for JH III.20) Although the turnover of JH by JHE is relatively slow, the specificity constant (kcat/KM ratio) of JHE for JH is high. The driver for this high specificity constant is found in the exceptionally low KM. Thus, continuing with the example above, authentic DmJHE turns over JH significantly more slowly than α-naphthyl acetate but DmJHE also shows a higher specificity constant for JH because its KM for JH (89 nM) is dramatically lower than its KM for α-naphthyl acetate (120,000 nM).20) In general, JHEs show specificity constants of around 106 to 107 M−1s−1 (Table 1).
Table 1.
Kinetic parameters of recombinant and authentic JHEs from various insect orders using racemic JH III
| Order | Species | Sourcea |
Vmax (nmol min−1 mg−1) |
kcat (s−1) |
KM (nM) |
kcat/KM (M−1 s−1) |
Key reference |
|---|---|---|---|---|---|---|---|
| Lepidoptera | Galleria mellonella | Authentic | 590b | 0.6 | 21 | 2.8 × 107 | 73) |
| Heliothis virescens | AF037197 | 1,570 | 1.7 | 30 | 5.7 × 107 | 41) | |
| Manduca sexta | AF327882 | 1,400 | 1.4b | 52 | 2.8 × 107 | 74) | |
| Bombyx mori | AF287267 | 990b | 1.0 | 28 | 3.6 × 107 | 47) | |
| Coleoptera | Leptinotarsa decemlineata | Authentic | 950b | 0.9 | 1,400 | 6.4 × 105 | 75) |
| Tenebrio molitor | AF448479 | 1,100 | 1.2b | 315 | 3.7 × 107 | 76) | |
| Tribolium castaneum | XP_969709 | 160 | 0.164 | 657 | 2.5 × 105 | 50) | |
| Diptera | Drosophila melanogaster | Authentic | 590 | 0.6 | 89 | 6.8 × 106 | 20) |
| Drosophila melanogaster | AF304352 | 970b | 1.0 | 1,500 | 6.0 × 105 | 46) | |
| Orthoptera | Gryllus assimilis | Authentic | ~1,600 | 1.4 | 84 | 1.7 × 107 | 77) |
| Hemiptera | Nilaparvata lugens | EU380769 | 2,040b | 2.2 | 514 | 4.3 × 106 | 78) |
GenBank accession numbers are given for recombinant proteins; authentic proteins are indicated.
Values were calculated on the basis of a deduced molecular weight (MW) or apparent MW of the protein lacking its putative signal peptide.
Only two carboxylesterases, JHE and acetylcholinesterase (AChE), have clearly defined physiologically substrates.3) Biochemical studies using authentic and recombinant JHEs indicate that JHEs prefer esters that are formed by a small alcohol (i.e., methyl or ethyl alcohol) and acid with a long alkyl chain of similar length to the JH backbone.21,22) Higher order esters (e.g., n-propyl or n-butyl esters) may be hydrolyzed by JHE but at significantly lower rates or may not be hydrolyzed. This preference for esters with small alcohols appears to result from steric limitations at the active site of JHE as will be discussed below. JHE activity levels and its regulation throughout development in hemolymph, tissues, and within cells have been reviewed previously.1,3,8–10,23) Interestingly, in some studies peak JHE activity is found after a drop in JH titer suggesting that JHE has an important role as a scavenger of JH in addition to its primary role in JH catabolism.3) Considering the physiological titer of JH in the hemolymph,24–27) the high specificity constant (kcat/KM ratio) and low KM of JHE is consistent with a role as a scavenger enzyme. The availability of jhe gene sequences from multiple insect species has allowed developmental expression profiles of jhe genes to be determined (Table 2). These expression profiles have shed additional light on the regulation of JHE activity.
Table 2.
Analysis of developmental expression of jhe in various insect orders
| Order | Species | Stage | Reference |
|---|---|---|---|
| Lepidoptera | Choristoneura fumiferana | Embryo, all larval stages, pupa | 79) |
| Bombyx mori | Embryo, all larval stages | 47,80) | |
| Coleoptera | Psacothea hilaris | 5th Instar | 81) |
| Tribolium castaneum | All life stages | 50) | |
| Diptera | Drosophila melanogaster | All life stages | 82) |
| Aedes aegypti | 4th Instar, pupa | 42) | |
| Orthoptera | Gryllus assimilis | Last instar nymph | 83) |
| Hemiptera | Nilaparvata lugens | 5th Instar nymph | 78) |
| Hymenoptera | Apis mellifera | 5th Instar larva, pupa | 43) |
Chemical inhibitors of JHE
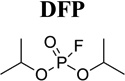
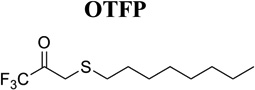
Potent and selective inhibitors are highly valuable tools for the in vitro and in vivo study of enzyme function. Phosphoramidothiolates such as O-ethyl-S-phenyl phosphoramidothiolate (EPPAT, Fig. 1C) and trifluoromethyl ketone (TFK) containing compounds (Fig. 1D) are the most selective and potent chemical inhibitors of JHE that have been identified to date. The development and use of these compounds are reviewed previously.3,8,28) Of the many variations of TFK containing compounds that have been developed, 3-octylthio-1,1,1-trifluoropropan-2-one (OTFP; Table 3)29) and 1-octyl[3,3,3-trifluoropropan-2,2-dihydroxy)] sulfone (OTPdOH-sulfone)30) are two of the most biologically potent. OTFP is a slow tight binding inhibitor that shows low nanomolar-level potency against lepidopteran, coleopteran, and dipteran JHEs in vitro (Table 3). In comparison, the general serine protease/esterase inhibitor diisopropyl fluorophosphate (DFP), even when pure, shows essentially no inhibition of lepidopteran and coleopteran JHEs, and relatively poor inhibition of dipteran JHEs (Table 3). EPPAT is one of the earliest compounds to show selective, potent, and long-lasting inhibition of JHE.31,32) Unlike TFK inhibitors, EPPAT binds irreversibly to JHE and is longer lived when applied to insects for biological studies. EPPAT and TFK containing inhibitors have been important for determining the physiological role of JHE. For example, larvae of Trichoplusia ni show an extended feeding state and delay in pupation following the application of EPPAT.31) Similar effects are found following the application of OTFP33) or OTFPdOH-sulfone.30) In adults of T. ni, topical application of OTFPdOH-sulfone also results in an increase in egg oviposition.30) The biology that is inferred in these studies is that JH metabolism by JHE is a critical requirement for normal development.
Table 3.
Inhibitory activity of DFP and OTFP on JH III hydrolysis by JHEs from various insect orders
| Order | Species | Sourcea | IC50 (nM) |
Reference | |
|---|---|---|---|---|---|
 |
 |
||||
| Lepidoptera | Manduca sexta | Authenticb | >100,000 | 84) | |
| Manduca sexta | AF327882b | 9.1 | 74) | ||
| Trichoplusia ni | Authenticc | >100,000 | 2.3 | 33,85) | |
| Bombyx mori | Authenticc | >100,000 | 2.7 | 86) | |
| Coleoptera | Tenebrio molitor | Authenticc | >500,000 | 87) | |
| Tenebrio molitor | AF448479b | 4.6 | 76) | ||
| Tribolium castaneum | XP_969709d | 2.0 | 50) | ||
| Diptera | Drosophila melanogaster | AF304352d | 2,000 | < 1 | 49) |
| Culex quinquefasciatus | Recombinantb | 600 | < 1 | 88) | |
| Orthoptera | Gryllus rubens | Authenticc | ~40,000 | ~30 | 89) |
| Gryllus assimilis | Authenticb | ~100 | ~100 | 77) | |
GenBank accession numbers are given for recombinant proteins.
Purified protein.
Diluted hemolymph or diluted hemolymph plasma.
Diluted cell culture supernatant.
TFK containing inhibitors function as transition state analogs of the JH substrate34,35) in a manner that is analogous to the first step of the JH hydrolysis reaction. Specifically, the catalytic serine of JHE attacks the carbonyl carbon of the TFK containing inhibitor and forms a tetrahedral acyl-enzyme complex. However, because the trifluoromethyl group is a very poor leaving group, the second step in the reaction involving nucleophilic attack of the acyl-enzyme complex does not occur and the enzyme is inhibited. The physical properties of TFK inhibitors such as mimicry of the carbon backbone of JH, role of sulfur or sulfone at the β position, effect of ketone hydration, and lipophilicity are discussed previously.3,33,36,37) The most potent TFK inhibitors have long aliphatic tails that mimic the length of the JH backbone and a sulfur29) or sulfone30) at the β position of the inhibitor (Fig. 1D). TFK inhibitors have been attached to Sepharose and used as highly selective ligands for the affinity purification of authentic JHE from hemolymph.38) In some cases, this strategy is made more efficient by pretreating the hemolymph with DFP in order to remove serine protease/esterase activity prior to affinity purification.39)
Primary sequence and phylogeny of JHE
The first JHE-encoding gene was cloned from the tobacco budworm Heliothis virescens.40) To date, at least 16 JHE-encoding or putative JHE-encoding genes have been cloned and characterized from at least 15 species found in six insect orders (Table 4). In addition, a large number other putative JHE-encoding sequences that are not yet well characterized can be found in National Center for Biotechnology Information (NCBI) databases. Ward et al. analyzed the catalytic mechanism of the JHE of H. virescens (HvJHE) by multiple sequence alignment and site-directed mutagenesis of conserved motifs.41) They identified five motifs in HvJHE (R47F, D173Q; GxS201xG; E332; and H446GxD/E) that are highly conserved in carboxylesterase/lipase sequences. (Note: The numbers refer to the location the amino acid residues in an HvJHE protein that lacks its 19 amino acid residue-long signal peptide.) The Ser-201, His-446, and Glu-332 residues were predicted to form the catalytic triad whereas the Arg-47 and Asp-173 residues were found to be required for efficient functioning of HvJHE. The multiple sequence alignments of Ward et al. were performed with a wide diversity of carboxylesterase/lipase sequences but with only one JHE sequence.41) With the availability of additional JHE sequences (Table 4), we are now able to revisit the multiple sequence alignment analysis of Ward et al. using only JHE and putative JHE sequences.
Table 4.
JHE, putative JHE, and JHE-related sequences from various insect orders
| Order | Species | GenBank accession no. |
Reference |
|---|---|---|---|
| Lepidoptera | Heliothis virescens | AF037197 | 40) |
| Trichoplusia ni | not given | 90) | |
| Choristoneura fumiferana | AF153367 | 79) | |
| Manduca sexta | AF327882 | 39) | |
| Bombyx mori | AF287267 | 80) | |
| Helicoverpa armigera | FJ997319 | 91) | |
| Coleoptera | Leptinotarsa decemlineata | AF035423 | 92) |
| AF039135 | 93) | ||
| Tenebrio molitor | AF448479 | 76) | |
| Psacothea hilaris | AB259898 | 81) | |
| Tribolium castaneum | XP_969709 | 50) | |
| Diptera | Drosophila melanogaster | AF304352 | 51) |
| Aedes aegypti | EAT43357 | 42) | |
| Orthoptera | Gryllus assimilis | EF558769 | 46) |
| Hemiptera | Nilaparvata lugens | EU380769 | 78) |
| Hymenoptera | Apis mellifera | AY647436 | 43,94) |
Multiple sequence alignment analyses with currently available JHE and putative JHE sequences show that the five motifs (RF, DQ, GQSAG, E, and GxxHxxD/E) identified by Ward et al.41) and others (see references in Table 4) are highly conserved (Table 5). The serine and histidine residues that are part of the GQSAG and GxxHxxD/E motifs, respectively, form part of the catalytic triad and are completely conserved. The third member of the carboxylesterase catalytic triad, an acidic amino acid residue, is also completely conserved. Specifically, a glutamic acid (i.e., the E motif) is found in all of the identified JHE and putative JHE sequences (Table 5) except in the case of the JHE of Aedes aegypti (AaJHE; EAT43357)42) where an aspartic acid residue is found. Interestingly, another candidate jhe gene in A. aegypti (EAT43353) has been identified by Bai et al. with conserved E and GxxHxxD/E motifs as well as developmental expression profiles consistent with the physiological function of JHE.42) Unfortunately, however, the JH hydrolytic activity of this putative JHE is unknown. The GQSAG motif (a variant of the carboxyl/cholinesterase nucleophilic elbow GxSxG motif) is highly uncommon in general esterases and has been generally considered as invariant among JHEs.18) The GQSAG motif, however, was not identified in an analysis of the 21 putative carboxylesterases present in the honey bee genome suggesting that the honey bee encodes instead a JHE with a GLSAG motif.43) Taken together, the multiple sequence alignments suggest that the RF, DQ, GQSAG, E, and GxxHxxD/E are highly diagnostic for JHE but outside of the three catalytic residues there is some latitude in the amino acid residues. In addition, Thomas et al. identified a second serine in the active site of HvJHE that putatively helps to orient the water molecule that attacks the acyl-enzyme.44) This serine is completely conserved in the currently available JHE and putative JHE sequences. The crystal structure of MsJHE,45) however, indicates that this serine is not one of the 30 amino acid residues that directly form the active site. On the other hand this serine appears to be highly conserved among non-JHE esterases suggesting that it is important for maintaining the structure of the catalytic site or in some other aspect of catalysis.
Table 5.
Conserved sequence motifs in JHE and putative JHE sequences from various insect orders
| Order | Enzyme | Signal peptidea |
Motifb |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RF | DQ | GQSAG | E | GxxHxxD | R/Kx(6)R/KxxxR | T | ||||
| Lepidoptera | HvJHE | 17 | + | + | + | + | + | Rx(6)RxxxR | + | 40) |
| CfJHE | 21 | + | DM | + | + | + | Rx(6)RxxxR | + | 79) | |
| MsJHE | 22 | + | DM | + | + | + | Rx(6)KxxxR | + | 39) | |
| BmJHE | 19 | + | DM | + | + | + | Rx(6)RxxxR | + | 80) | |
| Coleoptera | TmJHE | 22 | + | + | + | + | + | Kx(6)KxxxE | + | 76) |
| PhJHE | 21 | + | + | + | + | + | Kx(6)KxxxS | N | 81) | |
| TcJHE | 23 | + | + | + | + | + | Kx(6)KxxxE | + | 50) | |
| Diptera | DmJHE | 20 | + | + | + | + | + | Kx(6)RxxxR | + | 51) |
| AaJHE | 23 | + | + | + | D | GxxHxxE | Kx(6)KxxxR | + | 42) | |
| Orthoptera | GaJHE | n.a.b | + | + | + | + | + | Kx(6)RxxxR | + | 46) |
| Hemiptera | NlJHE | 20 | RY | + | + | + | + | Lx(6)KxxxR | H | 78) |
| Hymenoptera | AmJHE | 18 | + | + | GLSAG | + | + | Kx(6)RxxxE | + | 43) |
Length of predicted secretion signal peptide. Some secretion signal peptide sequences were predicted using the Signal IP 3.0 server.95)
The complete N-terminal sequence is not available (n.a.).
The “+” indicates that the consensus sequence is maintained.
Another diagnostic indicator of JHE is the presence of a signal peptide sequence of roughly 17–23 amino acid residues in length at the N-terminal (Table 5). Since JHE activity is primarily found as a soluble hemolymph protein (at least in insects that have been well studied), the presence of this signal peptide for secretion is highly expected in the primary sequence.
Phylogenetic analyses indicate that the lepidopteran JHEs form a distinct clade from the non-lepidopteran JHEs.18,46,47) Of the non-lepidopteran JHEs, the dipteran/orthopteran, coleopteran, and hemipteran JHEs may form their own clades (Fig. 3). A putative hymenopteran JHE (GB18660) may belong to the lepidopteran clade48) whereas another putative hymenopteran JHE (AY647436)43) may belong to its own clade (Fig. 3). Biological activity, however, has not been associated with these putative hymenopteran JHEs. Interestingly, although some insect species encode multiple JHE-like esterases that are capable of hydrolyzing JH to some extent, it appears that there is only one biologically active and physiologically relevant jhe gene in a single species at least in the case of the dipteran, lepidopteran, and coleopteran insects that have been studied.47,49,50)
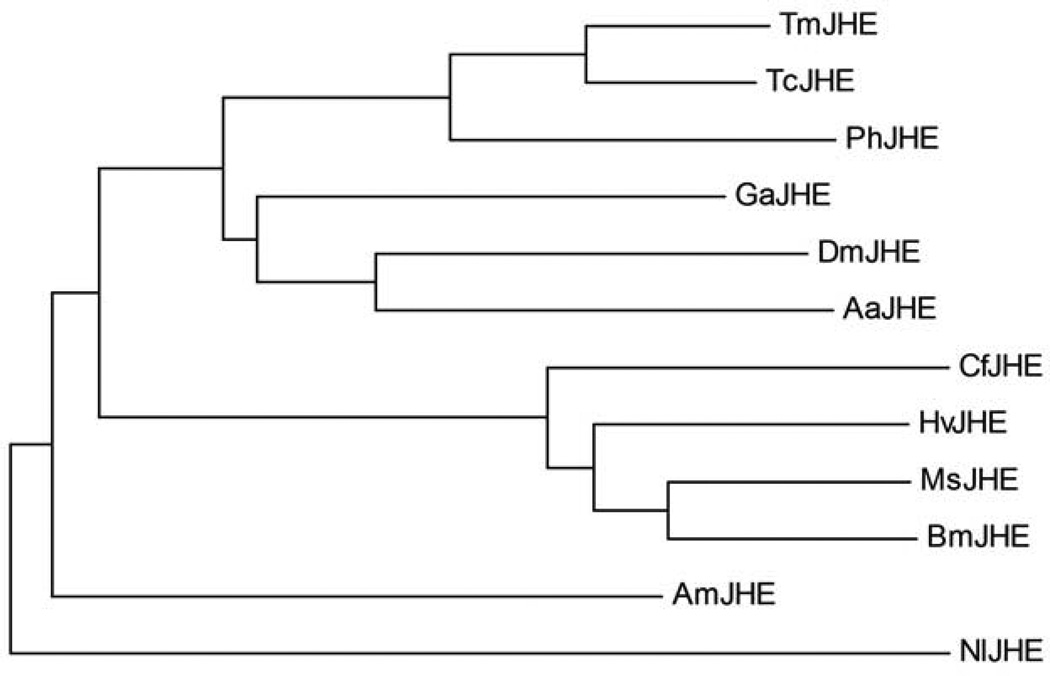
Fig.3.
Evolutionary relationship of JHE and putative JHE sequences. The phylogenetic analysis was conducted using MEGA4.72) The tree was generated by the Neighbor-Joining method using a ClustalW generated alignment. A bootstrap analysis of 500 replicates was performed to evaluate the branch strength of the model. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The insect order and GenBank accession number of the sequences are as follows. Coleoptera: TmJHE (AF448479), PhJHE (AB259898), TcJHE (XP_969709); Orthoptera: GaJHE (EF558769); Diptera: DmJHE (AF304352), AaJHE (EAT43357); Hymenoptera: AmJHE (AY647436); Lepidoptera: CfJHE (AF153367), HvJHE (AF037197), MsJHE (AF327882), BmJHE (AF287267); and Hemiptera: NlJHE (EU380769). Key references for each of the sequences are given in Table 5.
JHE structure
Thomas et al. were the first to generate a three dimensional homology-based model of JHE.44) The HvJHE homology models were constructed using available crystal structures of an acetylcholinesterase (AChE) and lipase as templates. The homology models led to several hypotheses regarding interactions of JH within the substrate-binding pocket and the identification of an alpha helix on the surface of HvJHE with unique amphipathic characteristics. Although a homologous alpha helix is found on the surface of AChE, lipase and other carboxylesterases, the amphipathic characteristic appears to be unique to JHEs.18,44,51) Thomas et al. found that the basic amino acid residues of the amphipathic helix form a concentrated region of positive charge on the surface of HvJHE and hypothesized that this positively charged patch is important for recognition and binding of JHE to the insect’s pericardial cells.44) The pericardial cell complex is involved in the receptor-mediated endocytotic uptake of JHE from the hemolymph and its subsequent degradation.52–54) A comparison of currently available JHE and putative JHE sequences from six insect orders indicates that a line of three basic amino acid residues (arginine or lysine) is highly conserved on the hemolymph side of this amphipathic alpha helix (Table 5). We propose that this amphipathic alpha helix motif (R/Kx(6)R/KxxxR) can be used as a diagnostic indicator of JHE.
Wogulis et al. were the first to generate a crystal structure of JHE; a 2.7 Å resolution structure of the JHE of Manduca sexta (MsJHE) in complex with the TFK inhibitor OTFP.45) This structure has provided valuable insight into the overall structure, substrate-binding pocket, and catalytic mechanism of JHEs. The substrate-binding pocket of MsJHE is a deep and narrow pit that is lined with hydrophobic amino acid residues. The catalytic triad is found at the bottom of this pit. In contrast to the structure of the catalytic triad of MsJHE, the catalytic triad of AChE of the Pacific electric ray is highly accessible to water.55) This difference in binding pocket structure helps to explain some of the biochemical characteristics of JHEs (e.g., slow substrate turnover and exceptionally low KM) that are in contrast to those of AChE (e.g., high kcat and high KM).19)
Modeling of JH within the substrate-binding pocket of MsJHE predicts that the fit of JH within the binding pocket is very tight (Fig. 4). Interestingly, this tight fit places two non-catalytic amino acid residues, Phe-259 and Thr-314 of MsJHE, within the appropriate distance and geometry to interact the α,β-unsaturated ester (π-stacking interactions) and epoxide (hydrogen bonding interactions), respectively, of JH III.19,45) Analyses of the roles of these non-catalytic amino acid residues in the catalytic cycle of JHE by site-directed mutagenesis indicate that both the phenylalanine and threonine contribute to the low nanomolar KM of MsJHE for JH. This low KM, however, comes at the cost of reduced substrate turnover (i.e., a lower kcat) and lower Vmax. Multiple sequence alignments of biologically active JHEs show that an aromatic amino acid residue (phenylalanine or tryptophan) is absolutely conserved at a position corresponding to Phe-259 of MsJHE; whereas a threonine residue corresponding to Thr-314 of MsJHE is highly conserved (Table 5). A survey of carboxyl/cholinesterases of insect and mammalian origin indicates that the threonine residue appears to be conserved only in JHEs.19) Thus, we propose that this threonine residue (i.e., the T motif) can be used as another diagnostic indicator of JHE.
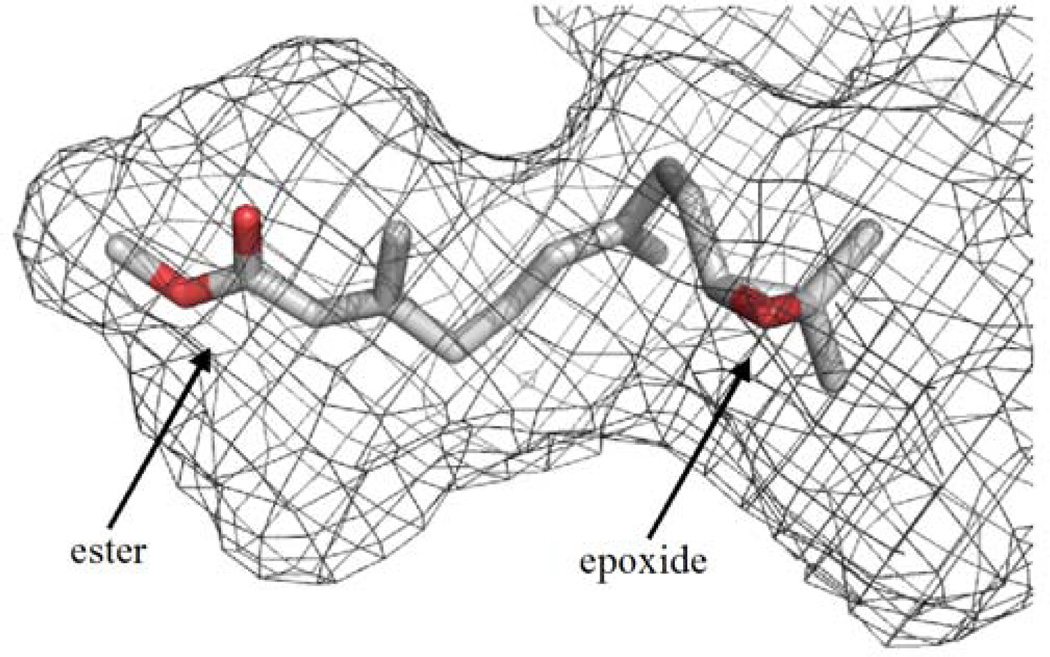
Fig.4.
Model of JH III within a surface representation of the substrate-binding pocket of MsJHE. The substrate-binding pocket of MsJHE is a deep and narrow pit that is approximately 20 Å deep and primarily lined by hydrophobic amino acid residues. JH III is modeled into the substrate-binding pocket so that the ester is nearest the catalytic triad, which is found at the end of the pit.
The structure of the substrate-binding pocket of MsJHE (i.e., a catalytic triad located at the bottom of a long and narrow pit) helps to explain the results of the biochemical studies discussed above that show JHE’s preference for JH as well as surrogate substrates21) and inhibitors28,34,56) that are formed with a small alcohol or show similar physical characteristics as JH. The MsJHE structure indicates that there simply is insufficient space for a large alcohol or other large constituent within the substrate-binding pocket of JHE. Biochemical studies,19,57) however, provide evidence that there is some flexibility in the substrate-binding pocket that is not immediately evident in the structure. Biochemical, primary sequence, and structural information can be used to help define an esterase as JH specific. However, the true definition of a JHE (or JH epoxide hydrolase) really depends upon demonstrating its biological role by correlating the enzymes presence with an endocrine change attributable to JH and ideally seeing the predicted biological effects when enzyme titer is increased or decreased by experimental manipulation.
Use of the jhe gene for pest insect control
The insect nervous system is the target of the majority of synthetic chemical insecticides that are in agricultural use. Insecticides that target the insect nervous system are generally highly selective, however, this selectivity often results from differences in the ability of the targeted insect and non-targeted organisms (e.g., humans) to metabolize or excrete the insecticide before it reaches its target site (sodium channel, γ-aminobutyric acid receptor, nicotinic acetylcholine receptor, etc.). In fact, the selectivity of many synthetic chemical insecticides at the target site may be limited. In contrast, targets within the insect gut or endocrine system are generally different from those found in non-targeted organisms. JH analog insecticides,58,59) for example, do not target the endocrine system of mammals and can show selectivity within an insect order or even family, a level of selectivity that is not obtained with classical insecticides. Another approach that targets the insect endocrine system involves the expression of JHE by a recombinant baculovirus.
Baculoviruses are arthropod (primarily insect)-specific pathogens that are used in developing regions as highly selective, economical, and effective biopesticides for the protection of agricultural crops such as soybean and cotton, and developed regions for the protection of forest lands.60–62) The natural insecticidal efficacy of the baculovirus can be improved by genetic modification. One such genetic modification involves the insertion of transgenes into the baculovirus genome that express a peptide hormone or enzyme such as JHE that can alter normal host physiology.61) Hammock et al. were the first to express a JHE in a recombinant baculovirus and test its efficacy as a biopesticide.63) In comparison to caterpillars infected with wildtype baculovirus, caterpillars infected with a first generation recombinant baculovirus construct, a recombinant Autographa californica nucleopolyhedrovirus construct carrying the jhe gene of H. virescens (AcRP23.JHE) showed profoundly reduced feeding and growth.
Two approaches to improve the insecticidal efficacy of AcRP23.JHE and similar constructs have been investigated. The first involves the use of various promoters to increase or alter the timing of jhe gene expression,64,65) whereas the second involves attempts decrease the degradation of the baculovirus expressed JHE protein.61) As discussed above, when recombinant HvJHE is injected into the hemolymph of lepidopteran insects it is rapidly removed from the hemolymph by a receptor-mediated, endocytotic process into pericardial cells.52–54,66) As discussed above, the amphipathic alpha helix motif of JHE is hypothesized to be involved in the recognition of JHE by pericardial cells. We have generated a recombinant baculovirus expressing a JHE from Manduca sexta (MsJHE) with mutations in its amphipathic alpha helix (AcMsJHE-HH, El-Sheikh and Hammock, unpublished). Caterpillars infected with AcMsJHE-HH show roughly 40% lower weight gain in comparison to control caterpillars infected with a recombinant baculovirus expressing the wild type MsJHE suggesting that a modest improvement in JHE stability can have significant effects in terms of insecticidal action.
Bonning et al. have identified and tested two lysine residues (Lys-29 and Lys-524) that are predicted to be located on the surface of HvJHE and involved JHE degradation within cells.67,68) Specifically, Lys-29 was predicted to be a ubiquitin conjugation site, and Lys-524 was predicted to be part of a KFERQ-like lysosome targeting motif. A recombinant baculovirus expressing a mutant HvJHE in which the lysine residues were mutated (AcJHE-KK) showed an improvement in speed of kill of between 0 and 27% (depending upon the insect species and developmental stage) in comparison to a recombinant virus expressing the wildtype HvJHE.68,69) Feeding damage by the AcJHE-KK-infected insects (in comparison to larvae infected with wildtype virus) was reduced by up to 50%. Interestingly, a similar reduction in feeding damage was obtained with a recombinant baculovirus expressing a catalytically inactive HvJHE mutant in which both lysine residues (Lys-29 and Lys-524) and the catalytic serine (Ser-203) were mutated70) suggesting that multiple mechanisms of insecticidal action may be at work. In an alternative approach, a binary GAL4/UAS system was used to over express JHE (8- to 10-fold higher hemolymph JHE levels in first to third stadia larvae) in the silkworm.71) Over expression of JHE starting from the embryonic stage using this system resulted in precocious larval-pupal metamorphosis following the third stadium. These findings further confirm the utility of the jhe gene as a viable transgene for insecticidal activity.
Summary
JHE is one of two carboxylesterases that has a clearly defined physiological substrate. JHE hydrolyzes JH in the presence or absence of JH binding protein with an apparent KM in the nanomolar range. JHE turns over JH at a relatively slow rate (ca. 2 s−1 or lower). The specificity constant (kcat/KM ratio) of JHE for JH, however, is high because of the low KM. Highly potent and selective inhibitors of JHE are available for study of the physiological role of JHE and its endogenous substrate JH. Seven amino acid sequence motifs (RF, DQ, GQSAG, E, GxxHxxD/E, R/Kx(6)R/KxxxR, and T) are highly conserved in JHE sequences. JHE encodes a signal peptide for secretion at the N-terminal of the protein. The jhe gene has potential as a highly insecticidal transgene given an appropriate vector for expression.
Acknowledgments
This work was supported in part by a grant from the USDA (2007-35607-17830) and NIEHS (R01 ES0002710). We thank Aman I. Samra and Mark D. Wogulis for help with phylogenetic and molecular model analyses, respectively.
References
- 1.Gilbert LI, Granger NA, Roe RM. Insect Biochem. Mol. Biol. 2000;30:617–644. doi: 10.1016/s0965-1748(00)00034-5. [DOI] [PubMed] [Google Scholar]
- 2.Kamimura M. Jpn. J. Appl. Entomol. Zool. 2004;48:1–12. [Google Scholar]
- 3.Goodman WG, Granger NA. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol.3. Oxford: Elsevier; 2005. pp. 319–408. [Google Scholar]
- 4.Minakuchi C, Riddiford LM. J. Pestic. Sci. 2006;31:77–84. [Google Scholar]
- 5.Truman JW, Riddiford LM. Insect Biochem. Mol. Biol. 2007;37:761–770. doi: 10.1016/j.ibmb.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Riddiford LM. J. Insect Physiol. 2008;54:895–901. doi: 10.1016/j.jinsphys.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Kotaki T, Shinada T, Kaihara K, Ohfune Y, Numata H. Org. Lett. 2009;11:5234–5237. doi: 10.1021/ol902161x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammock BD. In: Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Kerkut GA, Gilbert LI, editors. New York: Pergamon Press; 1985. pp. 431–472. [Google Scholar]
- 9.Roe RM, Venkatesh K. In: Morphogenetic Hormones of Arthropods. Gupta AP, editor. Vol. I. New Brunswick: Rutgers University Press; 1990. pp. 126–179. [Google Scholar]
- 10.de Kort CAD, Granger NA. Arch. Insect Biochem. Physiol. 1996;33:1–26. [Google Scholar]
- 11.Nijhout HF, Reed MC. J. Insect Physiol. 2008;54:255–264. doi: 10.1016/j.jinsphys.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 12.Ismail SM, Satyanarayana K, Bradfield JY, Dahm KH, Bhaskaran G. Arch. Insect Biochem. Physiol. 1998;37:305–314. doi: 10.1002/(SICI)1520-6327(1998)37:4<305::AID-ARCH6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 13.Ismail SM, Goin C, Muthumani K, Kim M, Dahm KH, Bhaskaran G. J. Insect Physiol. 2000;46:59–68. doi: 10.1016/s0022-1910(99)00102-x. [DOI] [PubMed] [Google Scholar]
- 14.Halarnkar PP, Jackson GP, Straub KM, Schooley DA. Experientia. 1993;49:988–994. [Google Scholar]
- 15.Grieneisen ML, Kieckbusch TD, Mok A, Dorman G, Latli B, Prestwich GD, Schooley DA. Arch. Insect Biochem. Physiol. 1995;30:255–270. [Google Scholar]
- 16.Maxwell RA, Welch WH, Schooley DA. J. Biol. Chem. 2002;277:21874–21881. doi: 10.1074/jbc.M201510200. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell RA, Welch WH, Horodyski FM, Schegg KM, Schooley DA. J. Biol. Chem. 2002;277:21882–21890. doi: 10.1074/jbc.M201509200. [DOI] [PubMed] [Google Scholar]
- 18.Oakeshott JG, Claudianos C, Campbell PM, Newcomb RD, Russell RJ. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 5. Oxford: Elsevier; 2005. pp. 309–381. [Google Scholar]
- 19.Kamita SG, Wogulis MD, Law CS, Morisseau C, Tanaka H, Huang H, Wilson DK, Hammock BD. Biochemistry. 2010;49:3733–3742. doi: 10.1021/bi901641x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell PM, Oakeshott JG, Healy MJ. Insect Biochem. Mol. Biol. 1998;28:501–515. doi: 10.1016/s0965-1748(98)00037-x. [DOI] [PubMed] [Google Scholar]
- 21.McCutchen BF, Uematsu T, Székács A, Huang TL, Shiotsuki T, Lucas A, Hammock BD. Arch. Biochem. Biophys. 1993;307:231–241. doi: 10.1006/abbi.1993.1584. [DOI] [PubMed] [Google Scholar]
- 22.Grieneisen M, Mok A, Kieckbusch T, Schooley DA. Insect Biochem. Mol. Biol. 1997;27:365–376. [Google Scholar]
- 23.Roe RM, Jesudason P, Venkatesh K, Kallapur VL, Anspaugh DD, Majumder C. Am. Zool. 1993;33:375–383. [Google Scholar]
- 24.Fain MJ, Riddiford LM. Biol. Bull. 1975;149:506–521. doi: 10.2307/1540383. [DOI] [PubMed] [Google Scholar]
- 25.Baker FC, Tsai LW, Reuter CC, Schooley DA. Insect Biochem. 1987;17:989–996. [Google Scholar]
- 26.Jones G, Hanzlik T, Hammock BD, Schooley DA, Miller CA, Tsai LW, Baker FC. J. Insect Physiol. 1990;36:77–83. [Google Scholar]
- 27.Hidayat P, Goodman WG. Insect Biochem. Mol. Biol. 1994;24:709–715. [Google Scholar]
- 28.Roe RM, Linderman RJ, Lonikar M, Venkatesh K, Abdelaal YAI, Leazer J, Upchurch L. J. Agr. Food Chem. 1990;38:1274–1278. [Google Scholar]
- 29.Abdel-Aal YAI, Hammock BD. Insect Biochem. 1985;15:111–122. [Google Scholar]
- 30.Roe RM, Anspaugh DD, Venkatesh K, Linderman RJ, Graves DM. Arch. Insect Biochem. Physiol. 1997;36:165–179. [Google Scholar]
- 31.Sparks TC, Hammock BD. Pestic. Biochem. Physiol. 1980;14:290–302. [Google Scholar]
- 32.Abdel-Aal YAI, Roe RM, Hammock BD. Pest. Biochem. Physiol. 1984;21:232–241. [Google Scholar]
- 33.Hammock BD, Abdel-Aal YAI, Mullin CA, Hanzlik TN, Roe RM. Pestic. Biochem. Physiol. 1984;22:209–223. [Google Scholar]
- 34.Hammock BD, Wing KD, McLaughlin J, Lovell VM, Sparks TC. Pestic. Biochem. Physiol. 1982;17:76–88. [Google Scholar]
- 35.Szekacs A, Halarnkar PP, Olmstead MM, Prag KA, Hammock BD. Chem. Res. Toxicol. 1990;3:325–332. doi: 10.1021/tx00016a009. [DOI] [PubMed] [Google Scholar]
- 36.Wheelock CE, Colvin ME, Uemura I, Olmstead MM, Sanborn JR, Nakagawa Y, Jones AD, Hammock BD. J. Med. Chem. 2002;45:5576–5593. doi: 10.1021/jm020072w. [DOI] [PubMed] [Google Scholar]
- 37.Wheelock CE, Nishi K, Ying A, Jones PD, Colvin ME, Olmstead MM, Hammock BD. Bioorgan. Med. Chem. 2008;16:2114–2130. doi: 10.1016/j.bmc.2007.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdel-Aal YAI, Hammock BD. Science. 1986;233:1073–1076. doi: 10.1126/science.3738525. [DOI] [PubMed] [Google Scholar]
- 39.Hinton AC, Hammock BD. Insect Biochem. Mol. Biol. 2001;32:57–66. doi: 10.1016/s0965-1748(01)00079-0. [DOI] [PubMed] [Google Scholar]
- 40.Hanzlik TN, Abdel-Aal YAI, Harshman LG, Hammock BD. J. Biol. Chem. 1989;264:12419–12425. [PubMed] [Google Scholar]
- 41.Ward VK, Bonning BC, Huang T, Shiotsuki T, Griffeth VN, Hammock BD. Int J Biochem. 1992;24:1933–1941. doi: 10.1016/0020-711x(92)90289-d. [DOI] [PubMed] [Google Scholar]
- 42.Bai H, Ramaseshadri P, Palli SR. Insect Biochem. Mol. Biol. 2007;37:829–837. doi: 10.1016/j.ibmb.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackert A, do Nascimento AM, Bitondi MMG, Hartfelder K, Simoes ZLP. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 2008;150:33–44. doi: 10.1016/j.cbpb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Thomas BA, Church WB, Lane TR, Hammock BD. Proteins: Struct., Funct, Genet. 1999;34:184–196. [PubMed] [Google Scholar]
- 45.Wogulis M, Wheelock CE, Kamita SG, Hinton AC, Whetstone PA, Hammock BD, Wilson DK. Biochemistry. 2006;45:4045–4057. doi: 10.1021/bi0521644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crone EJ, Zera AJ, Anand A, Oakeshott JG, Sutherland TD, Russell RJ, Harshman LG, Hoffmann FG, Claudianos C. Insect Biochem. Mol. Biol. 2007;37:1359–1365. doi: 10.1016/j.ibmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 47.Tsubota T, Shimomura M, Ogura T, Seino A, Nakakura T, Mita K, Shinoda T, Shiotsuki T. Insect Biochem. Mol. Biol. 2010;40:100–112. doi: 10.1016/j.ibmb.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Claudianos C, Ranson H, Johnson RM, Biswas S, Schuler MA, Berenbaum MR, Feyereisen R, Oakeshott JG. Insect Mol. Biol. 2006;15:615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crone EJ, Sutherland TD, Campbell PM, Coppin CW, Russell RJ, Oakeshott JG. Insect Biochem. Mol. Biol. 2007;37:540–549. doi: 10.1016/j.ibmb.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 50.Tsubota T, Minakuchi C, Nakakura T, Shinada T, Shiotsuki T. Insect Mol. Biol. 2010 doi: 10.1111/j.1365-2583.2010.01019.x. in press. [DOI] [PubMed] [Google Scholar]
- 51.Campbell PM, Harcourt RL, Crone EJ, Claudianos C, Hammock BD, Russell RJ, Oakeshott JG. Insect Biochem. Mol. Biol. 2001;31:513–520. doi: 10.1016/s0965-1748(01)00035-2. [DOI] [PubMed] [Google Scholar]
- 52.Ichinose R, Kamita SG, Maeda S, Hammock BD. Pestic. Biochem. Physiol. 1992;42:13–23. [Google Scholar]
- 53.Ichinose R, Nakamura A, Yamoto T, Booth TF, Maeda S, Hammock BD. Insect Biochem. Mol. Biol. 1992;22:893–904. [Google Scholar]
- 54.Booth TF, Bonning BC, Hammock BD. Tissue Cell. 1992;24:267–282. doi: 10.1016/0040-8166(92)90100-l. [DOI] [PubMed] [Google Scholar]
- 55.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 56.Prestwich GD, Eng W-S, Roe RM, Hammock BD. Arch. Biochem. Biophys. 1984;228:639–645. doi: 10.1016/0003-9861(84)90033-x. [DOI] [PubMed] [Google Scholar]
- 57.Kamita SG, Hinton AC, Wheelock CE, Wogulis MD, Wilson DK, Wolf NM, Stok JE, Hock B, Hammock BD. Insect Biochem. Mol. Biol. 2003;33:1261–1273. doi: 10.1016/j.ibmb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Wilson TG. J. Insect Physiol. 2004;50:111–121. doi: 10.1016/j.jinsphys.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Henrick CA. J. Am. Mosq. Control Assoc. 2007;23:225–239. doi: 10.2987/8756-971X(2007)23[225:M]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 60.Moscardi F. Annu. Rev. Entomol. 1999;44:257–289. doi: 10.1146/annurev.ento.44.1.257. [DOI] [PubMed] [Google Scholar]
- 61.Kamita SG, Kang K-D, Inceoglu AB, Hammock BD. In: Comprehensive Molecular Insect Science. Gilbert LI, Iatrou K, Gill SS, editors. Vol. 6. Oxford: Elsevier; 2005. pp. 271–322. [Google Scholar]
- 62.Inceoglu AB, Kamita SG, Hammock BD. Adv. Virus Res. 2006;68:323–360. doi: 10.1016/S0065-3527(06)68009-3. [DOI] [PubMed] [Google Scholar]
- 63.Hammock BD, Bonning BC, Possee RD, Hanzlik TN, Maeda S. Nature. 1990;344:458–461. [Google Scholar]
- 64.Bonning BC, Hirst M, Possee RD, Hammock BD. Insect Biochem. Mol. Biol. 1992;22:453–458. [Google Scholar]
- 65.Jarvis DL, Reilly LM, Hoover K, Schultz CM, Hammock BD, Guarino LA. Biol. Control. 1996;7:228–235. [Google Scholar]
- 66.Bonning BC, Booth TF, Hammock BD. Arch. Insect Biochem. Physiol. 1997;34:275–286. doi: 10.1002/(SICI)1520-6327(1997)34:3<275::AID-ARCH3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 67.Bonning BC, Ward VK, VanMeer MMM, Booth TF, Hammock BD. Proc. Natl. Acad. Sci. U. S. A. 1997;94:6007–6012. doi: 10.1073/pnas.94.12.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bonning BC, Possee RD, Hammock BD. J. Invertebr. Pathol. 1999;73:234–236. doi: 10.1006/jipa.1998.4839. [DOI] [PubMed] [Google Scholar]
- 69.Kunimi Y, Fuxa JR, Richter AR. Biol. Control. 1997;9:129–135. [Google Scholar]
- 70.van Meer MMM, Bonning BC, Ward VK, Vlak JM, Hammock BD. Biol. Control. 2000;19:191–199. [Google Scholar]
- 71.Tan A, Tanaka H, Tamura T, Shiotsuki T. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11751–11756. doi: 10.1073/pnas.0500954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, Dudley J, Nei M, Kumar S. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 73.Rudnicka M, Kochman M. Insect Biochem. 1984;14:189–198. [Google Scholar]
- 74.Hinton AC, Hammock BD. Insect Biochem. Mol. Biol. 2003;33:317–329. doi: 10.1016/s0965-1748(02)00246-1. [DOI] [PubMed] [Google Scholar]
- 75.Vermunt AMW, Vermeesch AMG, de Kort CAD. Arch. Insect Biochem. Physiol. 1997;35:261–277. doi: 10.1002/(SICI)1520-6327(199705)35:3<261::AID-ARCH2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 76.Hinton AC, Hammock BD. Insect Biochem. Mol. Biol. 2003;33:477–487. doi: 10.1016/s0965-1748(03)00010-9. [DOI] [PubMed] [Google Scholar]
- 77.Zera AJ, Sanger T, Hanes J, Harshman L. Arch. Insect Biochem. Physiol. 2002;49:41–55. doi: 10.1002/arch.10004. [DOI] [PubMed] [Google Scholar]
- 78.Liu SH, Yangb BJ, Go JH, Yao XM, Zhang YX, Song F, Liu ZW. J. Insect Physiol. 2008;54:1495–1502. doi: 10.1016/j.jinsphys.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 79.Feng QL, Ladd TR, Tomkins BL, Sundaram M, Sohi SS, Retnakaran A, Davey KG, Palli SR. Mol. Cell. Endocrinol. 1999;148:95–108. doi: 10.1016/s0303-7207(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 80.Hirai M, Kamimura M, Kikuchi K, Yasukochi Y, Kiuchi M, Shinoda T, Shiotsuki T. Insect Biochem. Mol. Biol. 2002;32:627–635. doi: 10.1016/s0965-1748(01)00141-2. [DOI] [PubMed] [Google Scholar]
- 81.Munyiri FN, Ishikawa Y. Insect Biochem. Mol. Biol. 2007;37:497–505. doi: 10.1016/j.ibmb.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 82.Kethidi DR, Xi ZY, Palli SR. J. Insect Physiol. 2005;51:393–400. doi: 10.1016/j.jinsphys.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 83.Anand A, Crone EJ, Zera AJ. J. Insect Physiol. 2008;54:1323–1331. doi: 10.1016/j.jinsphys.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 84.Sanburg LL, Kramer KJ, Kezdy FJ, Law JH. J. Insect Physiol. 1975;21:873–887. doi: 10.1016/0022-1910(75)90015-3. [DOI] [PubMed] [Google Scholar]
- 85.Wing KD, Sparks TC, Lovell VM, Levinson SO, Hammock BD. Insect Biochem. 1981;11:473–485. [Google Scholar]
- 86.Shiotsuki T, Bonning BC, Hirai M, Kikuchi K, Hammock BD. Biosci. Biotechnol. Biochem. 2000;64:1681–1687. doi: 10.1271/bbb.64.1681. [DOI] [PubMed] [Google Scholar]
- 87.McCaleb DC, Reddy G, Kumaran AK. Insect Biochem. 1980;10:273–277. [Google Scholar]
- 88.Kamita SG, Hammock BD. (unpublished). [Google Scholar]
- 89.Zera AJ, Gu X, Zeisset M. Insect Biochem. Mol. Biol. 1992;22:829–839. [Google Scholar]
- 90.Jones G, Venkataraman V, Ridley B, Omahony P, Turner H. Biochem. J. 1994;302:827–835. doi: 10.1042/bj3020827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Teese MG, Campbell PM, Scott C, Gordon KHJ, Southon A, Hovan D, Robin C, Russell RJ, Oakeshott JG. Insect Biochem. Mol. Biol. 2010;40:1–16. doi: 10.1016/j.ibmb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 92.Vermunt AMW, Koopmanschap AB, Vlak JM, De Kort CAD. Insect Biochem. Mol. Biol. 1997;27:919–928. doi: 10.1016/s0965-1748(97)00073-8. [DOI] [PubMed] [Google Scholar]
- 93.Vermunt AMW, Koopmanschap AB, Vlak JM, de Kort CAD. Insect Mol. Biol. 1998;7:327–336. doi: 10.1046/j.1365-2583.1998.740327.x. [DOI] [PubMed] [Google Scholar]
- 94.Kamikouchi A, Morioka M, Kubo T. Zool. Sci. 2004;21:53–62. doi: 10.2108/0289-0003(2004)21[53:IOHAGE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 95.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. J Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]