Abstract
Genome sequencing data for Streptococcus equi subspecies equi and zooepidemicus were used to develop a novel diagnostic triplex quantitative PCR (qPCR) assay targeting two genes specific to S. equi (eqbE and SEQ2190) and a unique 100 base pair control DNA sequence (SZIC) inserted into the SZO07770 pseudogene of S. zooepidemicus strain H70. This triplex strangles qPCR assay can provide results within 2 h of sample receipt, has an overall sensitivity of 93.9% and specificity of 96.6% relative to the eqbE singlex assay and detects S. equi at levels below the threshold of the culture assay, even in the presence of contaminating bacteria.
Keywords: Equine, Strangles, Streptococcus equi, Triplex quantitative PCR
Introduction
Strangles, caused by Streptococcus equi subspecies equi, is the most frequently diagnosed infectious disease of horses worldwide and is responsible for significant welfare concerns and economic losses to the equine industry. The disease is characterised by abscessation of the lymph nodes of the head and neck. Abscesses formed in the retropharyngeal lymph nodes usually rupture into the guttural pouches, which drain via the nostrils, leading to the classical mucopurulent nasal discharge associated with strangles. Over time, the purulent material in the guttural pouches of some horses becomes inspissated, enabling S. equi to persist in horses that have recovered from the acute disease for up to several years in the absence of clinical signs (Newton et al., 1997).
S. equi is shed from persistently infected carrier horses periodically, allowing transmission to naïve individuals and resulting in new outbreaks of disease. The generation and persistence of carriers within equine populations is critical to the spread of S. equi infection. Efficient identification and treatment of carriers is important for prevention and eradication of this disease.
Diagnosis of S. equi infection has been based upon the culture of this β-haemolytic organism using selective media, followed by biochemical tests, which rely on the inability of S. equi to ferment trehalose, lactose or sorbitol (Bannister et al., 1985). Isolation of β-haemolytic colonies may be confounded by the presence of other bacteria, most notably Streptococcus equi subspecies zooepidemicus and Streptococcus dysgalactiae subspecies equisimilis, which may lead to the generation of false negative results. Isolation and identification of S. equi by routine methods is time consuming and requires a minimum of 48 h from receipt of clinical samples. This reporting delay often has consequences for the isolation of infected horses, providing S. equi with greater opportunity to transmit through naïve populations.
The first PCR-based test developed for S. equi targeted the 5′ region of the SeM gene. Using this target, up to three times more clinical samples were positive for S. equi than by culture and biochemical tests alone (Timoney and Artiushin, 1997; Newton et al., 2000). Historically, the SeM gene was thought to be non-variant, based upon its HindIII restriction pattern on Southern blotting (Galan and Timoney, 1988). However, it is now known that this region of the SeM gene is highly variable (Anzai et al., 2005; Kelly et al., 2006) and can be deleted in strains of S. equi isolated from persistently infected carriers (Chanter et al., 2000). The loss of diagnostic PCR targets leading to incorrect reporting can have deleterious consequences for the control of infectious disease. In Sweden, the occurrence of a 377 base pair (bp) deletion in CDS1 of the pSW2 plasmid of the human pathogen Chlamydia trachomatis resulted in the false negative diagnosis of many infected patients, and the rapid spread of this variant within the population (Seth-Smith et al., 2009).
Genes for the superantigens seeH, seeI, seeL and seem, encoded on the prophages φSeq4 and φSeq3, have potential as diagnostic candidates for S. equi subsp. equi (Baverud et al., 2007). However, the production of four superantigens confers a certain level of functional redundancy that could lead to loss of target sequences (Paillot et al., 2010). Furthermore, S. zooepidemicus multilocus sequence types ST-106, ST-118 and ST-120 were qPCR positive for seeL and seeM (Holden et al., 2009), suggesting that the use of these targets may lead to the identification of false positive results in some S. zooepidemicus strains. The gene encoding the factor H-binding protein Se18.9 has also been highlighted as a diagnostic target, but is present in ST-57 strains of S. zooepidemicus (Holden et al., 2009), suggesting that qPCRs targeting this sequence may also lead to false positive results.
To ensure the robust and sensitive identification of horses infected with S. equi, we have developed a triplex qPCR test that targets two S. equi-specific genes and employs an internal control strain of S. zooepidemicus, which serves as a DNA extraction and within-assay PCR control for every sample to reduce the risk of false negative reporting.
Materials and methods
Bacterial strains, media and growth conditions
S. equi strain 4047 (Se4047) was isolated from a submandibular lymph node abscess of a New Forest pony in 1990 and S. zooepidemicus strain H70 (SzH70) was isolated from a Thoroughbred horse in 2000. Strains were cultured in Todd Hewett broth (Oxoid) at 37 °C with 5% CO2.
Target selection
S. equi-specific targets were selected by use of comparative genome analysis of the published Se4047 and SzH70 genomes (Heather et al., 2008; Holden et al., 2009). eqbE and SEQ2190 were amplified by PCR using DNA samples previously extracted from 26 S. equi strains (Holden et al., 2009) and the primers ZM435/ZM436 and 2190A/2190B, respectively (Table 1). Products were sequenced on an ABI3100 DNA sequencer with BigDye fluorescent terminators using the same primers, with the addition of ZM437 for sequencing eqbE.
Table 1.
Primer and probe sequences.
| Primer/probe name | Sequence (5′–3′)a | Label/restriction site |
|---|---|---|
| ZM435 | CCGAATTTGTCCAAGTGGTATG | |
| ZM436 | GCACTCCGTTATACTCACTG | |
| ZM437 | TTTGCTAGTGCTACTCCTGC | |
| 2190A | ATGGGAACAGGACTACTTG | |
| 2190B | GTCTTAGCTTCCTCTTTCGC | |
| Ec07770Fwd1 | GACGACGAATTCTGAGAGGCAAGTGACGAGTC | EcoRI |
| Ec07770Rev1a | GACGACCGATCGGACGACACCGGTTTGCCAAACTCCCTTCCAAG | PvuI, AgeI |
| Ec07770Fwd2 | GACGACCGATCGCACTTGCTTGTTCTAGCTGAG | PvuI |
| Ec07770Rev2 | GACGACGTCGACGGAACGAACCTCTTACCACA | SalI |
| 5′pGhost9 | TTGGAAAGTTACACGTTACTAAAG | |
| 3′pGhost9 | GGGCGAATTGGGGTACCGGGC | |
| eqbE2 forward | TGGGATTCTGTGCCGATTTT | |
| eqbE2 reverse | CCCTGAAAGCATCACAATTCTAAA | |
| 2190 forward | CAACGCGTAGAAGAACGATCTAAA | |
| 2190 reverse | CCTCCAATTGAGCTTTTTGGTT | |
| SZIC forward | CGCATGCGGGTAGATTATGTAG | |
| SZIC reverse | TCCCACGAGAAGGTCGAGAA | |
| eqbE2 probe | ATTGTTACTATGGCTGAAGGT | FAM |
| 2190 probe | AAGCCAAGGAAGCCACT | VIC |
| SZIC probe | AGAGACATCCAGGTCAA | NED |
| Artificial 82 bp DNA sequence | CCGTGTATTACGCATGCGGGTAGATTATGTAGGTAGAGACATCCAGGTCAAGTTCTCGACCTTCTCGTGGGAGGTGAACCAG |
Restriction sites are underlined.
Design and generation of the Sz07770c internal control strain
Two 500 bp fragments of the SZO07770 pseudogene were amplified by PCR from SzH70 using primers Ec07770Fwd1/Ec07770Rev1a and Ec07770Fwd2/Ec07770Rev2 (Table 1). PCR products were digested with EcoR1/Pvu1 and Pvu1/Sal1, respectively, and ligated into the multiple cloning site of the pGHost9 plasmid to generate pGHost9ΔSZO07770, which was sequenced using primers 5′pGhost9 and 3′pGhost9 (Table 1).
An artificial 82 bp DNA sequence with no significant homology with any nucleotide sequences on the NCBI database was designed (Table 1). Restriction sites for PvuI and AgeI were engineered to the 5′ and 3′ end of this DNA fragment, respectively, and the resultant DNA sequence, SZIC, was produced using the Gene Oracle gene synthesis service. The synthesised SZIC DNA was digested with PvuI and AgeI restriction enzymes and sub-cloned into pGHost9ΔSZO07770 digested with the same restriction enzymes, such that the resultant plasmid, pGHost9SZIC, contained SZIC flanked by 500 bp SZO07770 gene fragments.
The pGHost9SZIC plasmid was transformed into electro-competent SzH70. Integration of the construct, inserting the SZIC sequence into the chromosomal copy of the SZO07770 gene and excision of the pGHost9 plasmid, was achieved by allelic replacement mutagenesis, as described for a ΔprtM mutant (Hamilton et al., 2006), creating the strain Sz07770c. The fidelity of SZIC was confirmed by PCR and sequencing across the SZO07770 insertion site.
The Sz07770c strain was grown to an optical density of 0.3 at 600 nm in Todd Hewitt Broth and inactivated by heating at 95 °C for 30 min. No colonies of Sz07770c were obtained when 100 μL of this culture was incubated overnight at 37 °C in 5% CO2 on colistin-oxolinic acid blood agar (COBA) streptococcal selective agar (Oxoid). The killed culture was diluted to obtain a bacterial density equivalent to 80,000 colony-forming units (cfu)/mL in phosphate buffered saline and stored at −20 °C prior to use.
DNA extraction for assay validation
One millilitre of excess clinical sample was spiked with 25 μL killed diluted Sz07770c, containing bacteria equivalent to 2000 cfu in the original live culture. DNA was extracted using the GenElute kit (Sigma) and eluted in 200 μL double distilled H2O.
Development of the triplex qPCR assay
Compatible primers and minor groove binder (MGB) probes were designed against eqbE, SEQ2190 and SZIC (Table 1). Reaction conditions were optimised for a fast cycling assay using an ABI StepOne Plus instrument using KAPA Fast probe mix (Kapa Biosystems) in a ratio of 50:50 master mix:DNA sample. Each reaction contained 10 μL KAPA Fast probe mix, 450 nM eqbE2 forward, 450 nM eqbE2 reverse, 450 nM 2190 forward, 450 nM 2190 reverse, 200 nM SZIC forward, 200 nM SZIC reverse, 150 nM eqbE2 probe, 2190 probe and SZIC probe and 10 μL DNA extracted from the clinical sample. Thermal cycling conditions were 3 min at 95 °C, followed by 40 cycles of 95 °C for 3 s and 60 °C for 10 s. All qPCR experiments were performed in triplicate.
Validation of the triplex qPCR assay and comparison with the eqbE singlex qPCR assay
Clinical samples (n = 213; >2 mL) received by the Animal Health Trust (AHT) diagnostic laboratories for detection of S. equi via the eqbE singlex qPCR assay were used in this study (see Appendix A: Supplementary Table 1). Two 1 mL aliquots of the original clinical sample were removed and blinded. Heat-killed Sz07770c (25 μL) was added to one aliquot and both aliquots were then centrifuged at 16,100 g. DNA was isolated from the pellets using the GenElute kit (Sigma). Samples containing spiked Sz07770c were analysed using the triplex qPCR assay and the number of copies of eqbE in the DNA isolated from the other aliquot was quantified by qPCR using the eqbE singlex assay by the AHT diagnostic laboratory.
Streptococcus equi culture test
Clinical samples (n = 194) submitted to the AHT diagnostic laboratory were tested for S. equi by routine culture and identification.
Statistical analysis
Receiver operator characteristic (ROC) curves, sensitivity and specificity of the qPCR and culture assays were calculated using STATA (StataCorp LP). Two-tailed Spearman’s Coefficients were calculated using PASW (version 18, SPSS). A two-tailed unpaired student’s t test was used to determine the significance of continuous data.
Results
Conservation of eqbE and SEQ2190 targets
PCR and sequencing of a 1063 bp region of eqbE (positions 1,229,518 to 1,228,456 of the Se4047 genome) and a 449 bp region of SEQ2190 (positions 2,203,349 to 2,203,797 of the Se4047 genome) demonstrated that these sequences were identical across all 26 strains of S. equi examined.
Generation of the Sz07770c strain
PCR and sequencing across the insertion site confirmed that the SZIC sequence had been incorporated into the SZO07770 pseudogene in the Sz07770c strain and that all plasmid DNA had been excised.
Limit of detection of the triplex qPCR assay
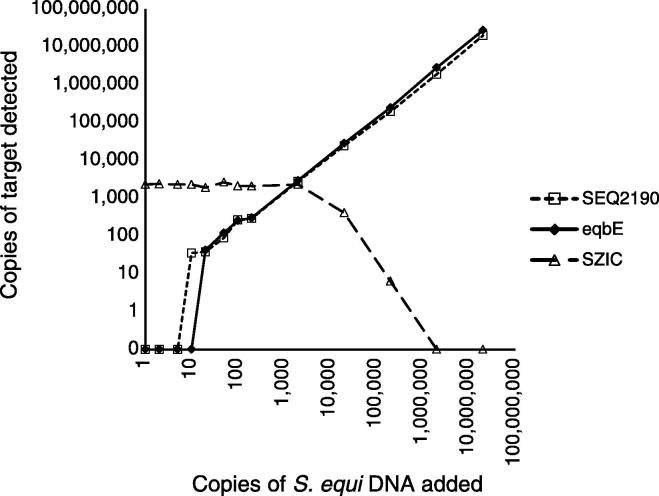
Quantification of eqbE, SEQ2190 and SZIC in samples containing a dilution series of Se4047 DNA and approximately 2000 copies of DNA from Sz07770c determined that the triplex qPCR assays had a limit of detection of 20 copies (eqbE assay) and 10 copies (SEQ2190 assay) of S. equi DNA, respectively (Fig. 1). Increasing the quantities of Se4047 DNA above 10,000 copies led to competition and inhibition of the SZIC assay (Fig. 1).
Fig. 1.
Effect of increasing concentration of S. equi DNA in the triplex qPCR reaction. Dilution series of Se4047 genomic DNA in the presence of 2000 copies of SZIC internal control target.
Validation of the triplex qPCR assay
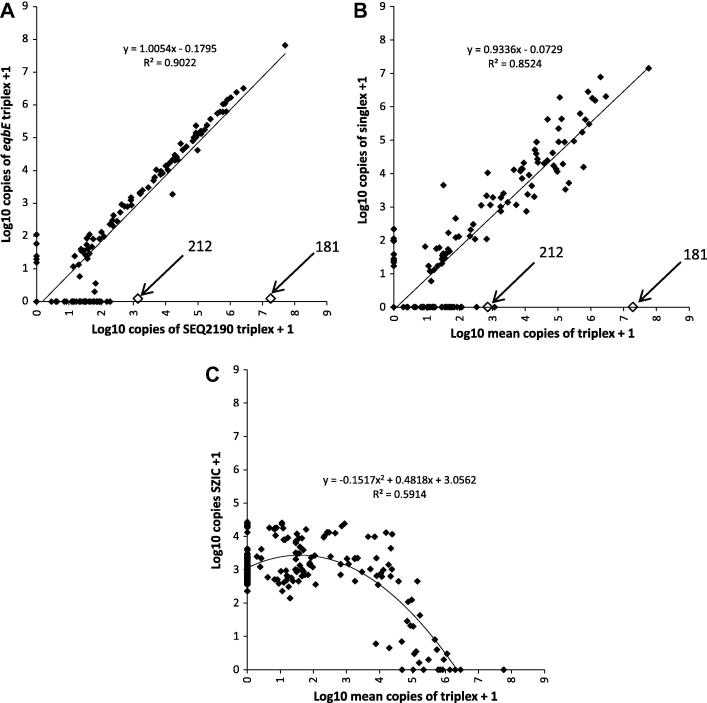
Sixty-six of 213 clinical samples tested positive for S. equi via the eqbE singlex qPCR assay (eqbE copy number ⩾100) and 147 tested negative. Blinded details of all clinical samples tested, including culture, singlex and triplex assay results, are presented in Supplementary Table 1 (see Appendix A). Copy numbers for eqbE and SEQ2190 quantified within the triplex assay correlated well with each other (Fig. 2A) and generated a Spearman’s Coefficient of 0.84 (P < 0.001). Mean triplex assay copy numbers also correlated well with the singlex eqbE results (Spearman’s Coefficient of 0.80, P < 0.001) (Fig. 2B).
Fig. 2.
(A) Relationship of log10 qPCR copy numbers for eqbE and SEQ2190 within the triplex assay. The location of the results for samples 181 and 212 are highlighted. (B) Log10 mean quantity of S. equi DNA detected in the triplex assay compared with eqbE quantified by the singlex assay. The location of the results for samples 181 and 212 are highlighted. (C) Effect of increasing quantities of S. equi in clinical samples on log10SZIC copy number. A copy number of one was added to all data, which was then transformed into a logarithmic format for presentation purposes. The line of best fit and its equation is shown on each graph.
Sample 181, recovered from the guttural pouches of a horse in Dorset, contained SEQ2190 copy numbers of 14,030,667, but did not amplify eqbE in the triplex or singlex assays. Sample 212, taken from the guttural pouches of another horse from the same yard in Dorset, similarly tested negative for eqbE using both assays, but was positive for SEQ2190 (Fig. 2A and B). These samples were removed from the data set for the purposes of plotting the associated lines of best fit.
Five clinical samples (2.3%) originally tested negative for the SZIC control, highlighting a potential failure in the DNA extraction method. These samples were re-extracted, retested and the eqbE and SEQ2190 triplex results for two discrepant samples, 194 and 211, then were found to be in agreement with singlex assay results.
The SZIC assay detected a mean of 3994 copies in all reactions where the amount of S. equi DNA was ⩽10,000 copies (n = 175) (Fig. 2C). However, the SEQ2190 and eqbE assays significantly interfered with the SZIC assay when the amount of S. equi DNA exceeded 10,000 copies per reaction (P = 0.0038), leading to a mean of 940 copies of SZIC in these clinical samples (n = 38) and a non-linear line of best fit (Fig. 2C).
To identify the optimal assay breakpoints and so ensure that the most robust diagnosis was obtained, triplex data were compared with the singlex qPCR assay as the gold-standard using STATA. Optimal sensitivity (90.9%) and specificity (96.6%) for the eqbE component of the triplex assay were achieved with a breakpoint of 110 copies, generating an area under the ROC curve of 0.95, where a value of 1.0 indicates complete concordance of data. Optimal sensitivity for the SEQ2190 component of the triplex assay (sensitivity 92.4%, specificity 97.3%) was achieved with a breakpoint of 150 copies, generating an area under the ROC curve of 0.9727.
Using these breakpoints and combining the results of the eqbE and SEQ2190 assays, so that a sample was reported as positive if either assay reached its cut-off, 67 positive samples were identified, of which five were negative by the singlex assay, giving an overall sensitivity and specificity for the triplex assay of 93.9% and 96.6%, respectively.
Comparison of qPCR with the culture assay
The culture test identified no additional positive clinical samples and had an overall sensitivity of 60.3% and specificity of 100% relative to the combined qPCR results. Other β-haemolytic streptococci were present in 15/27 (56%) culture negative, qPCR positive samples (10 contained S. zooepidemicus and five contained S. equisimilis; see Appendix A: Supplementary Table 1). The mean qPCR copy number of culture positive/qPCR positive samples was 1,388,580 (n = 41). Culture negative, qPCR positive samples that did not contain other β-haemolytic streptococci had a mean copy number of 415 (n = 12) and culture negative/qPCR samples that contained other β-haemolytic streptococci had a mean S. equi DNA copy number of 170,312 (n = 15).
Discussion
Emerging genome sequencing data and an extensive collection of S. equi and S. zooepidemicus isolates were exploited to select two conserved S. equi-specific targets that are located in separate regions of the Se4047 genome. The eqbE gene forms part of the equibactin locus, which encodes a non-ribosomal peptide synthesis system that enhances the ability of S. equi to acquire iron (Heather et al., 2008). SEQ2190 encodes a putative sortase-processed protein, which was conserved in all strains of S. equi examined. Targeting both eqbE and SEQ2190 is predicted to significantly reduce the likelihood that both diagnostic targets are lost in a strain, resulting in the failure of the test and subsequent clonal expansion of the modified strain.
This risk and the usefulness of the triplex assay are highlighted in samples 181 and 212, which tested negative for eqbE, but positive for the SEQ2190 gene. Decay of the SeM gene in isolates recovered from the guttural pouch has been reported previously (Chanter et al., 2000). Recently, we identified significant decay of the S. equi genome on sequencing >230 isolates using the Illumina HiSeq platform (data not shown), which suggests that reliance on only one diagnostic target is likely to lead to the reporting of false negative results.
The novel internal control strain, Sz07770c, which serves as a DNA extraction and assay control, ensures that all clinical samples should generate a qPCR result. High levels of S. equi DNA (>10,000 copies) out-compete the SZIC assay and generate eqbE positive/SEQ2190 positive/SZIC negative results. Similarly, high levels of Bacillus anthracis, Francisella tularensis and Yersinia pestis inhibited the detection of a Bacillus thuringensis internal control in a multiplex qPCR assay (Janse et al., 2010). The triplex qPCR assay was optimised to ensure that amplification of low concentrations of S. equi DNA (<1000 copies) was not inhibited by the presence of SZIC. The close agreement between the singlex and triplex assays in this study (95.8%) and low limit of detection (10–20 copies of S. equi DNA) suggests that no compromise on sensitivity was made through inclusion of the SZIC control.
The inclusion of the SZIC assay permits robust quality control of all assay stages for each clinical sample. Five clinical samples analysed using the triplex assay failed these criteria and were re-tested, two of which were positive. The five discrepant clinical samples that tested negative on singlex, but positive on triplex, similarly may be due to assay failures that would have gone unnoticed using the singlex assay system. Therefore, the triplex assay will reduce the number of false negative results and so enable the detection, isolation and treatment of infected horses before they can transmit S. equi to others. Since the triplex assay requires only the addition of primers and probes specific to SEQ2190 and SZIC, the extra cost per sample is £0.16,2 which enables the implementation of the triplex assay without additional cost to clients.
Comparison of results from the singlex and triplex qPCR assays with the culture assay highlighted a lack of sensitivity of the culture assay (60.3%). The lowest mean quantity of S. equi DNA in a culture positive sample was 960 copies. The presence of other β-haemolytic streptococci accounted for more than half (15/27) of culture negative, qPCR positive samples, emphasising the problem of isolating S. equi from mixed cultures. Our data demonstrate that the S. equi culture test can no longer be considered the to be gold-standard test for this organism.
Conclusions
The use of sensitive qPCR assays will improve the identification and treatment of horses infected with S. equi, particularly carriers where intermittent low level shedding of this bacterium is common and can be missed by traditional culture and PCR assays. The strangles triplex assay provides a rapid, sensitive and robust method for the detection of S. equi infection for minimal additional cost.
Conflict of interest statement
The use of eqbE, SEQ2190 and the SZIC control for the detection of S. equi has been patented (US 2011/0201007, US 2007/0243195 and GB1122121.5).
Acknowledgement
The authors wish to acknowledge the Wellcome Trust for funding this work.
Footnotes
UK£1.00 = US$1.55 = €1.24 (13 June 2012).
Supplementary data associated with this article, which provides blinded details of all clinical samples used in this study including culture, singlex and triplex assay results can be found, in the online version, at http://dx.doi.org/10.1016/j.tvjl.2012.07.007.
Appendix A. Supplementary material
Supplementary table
References
- Anzai T., Kuwamoto Y., Wada R., Sugita S., Kakuda T., Takai S., Higuchi T., Timoney J.F. Variation in the N-terminal region of an M-like protein of Streptococcus equi and evaluation of its potential as a tool in epidemiologic studies. American Journal of Veterinary Research. 2005;66:2167–2171. doi: 10.2460/ajvr.2005.66.2167. [DOI] [PubMed] [Google Scholar]
- Bannister M.F., Benson C.E., Sweeney C.R. Rapid species identification of group C streptococci isolated from horses. Journal of Clinical Microbiology. 1985;21:524–526. doi: 10.1128/jcm.21.4.524-526.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baverud V., Johansson S.K., Aspan A. Real-time PCR for detection and differentiation of Streptococcus equi subsp. equi and Streptococcus equi subsp. zooepidemicus. Veterinary Microbiology. 2007;124:219–229. doi: 10.1016/j.vetmic.2007.04.020. [DOI] [PubMed] [Google Scholar]
- Chanter N., Talbot N.C., Newton J.R., Hewson D., Verheyen K. Streptococcus equi with truncated M-proteins isolated from outwardly healthy horses. Microbiology. 2000;146:1361–1369. doi: 10.1099/00221287-146-6-1361. [DOI] [PubMed] [Google Scholar]
- Galan J.E., Timoney J.F. Immunologic and genetic comparison of Streptococcus equi isolates from the United States and Europe. Journal of Clinical Microbiology. 1988;26:1142–1146. doi: 10.1128/jcm.26.6.1142-1146.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A., Robinson C., Sutcliffe I.C., Slater J., Maskell D.J., Davis-Poynter N., Smith K., Waller A., Harrington D.J. Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infection and Immunity. 2006;74:6907–6919. doi: 10.1128/IAI.01116-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather Z., Holden M.T., Steward K.F., Parkhill J., Song L., Challis G.L., Robinson C., Davis-Poynter N., Waller A.S. A novel streptococcal integrative conjugative element involved in iron acquisition. Molecular Microbiology. 2008;70:1274–1292. doi: 10.1111/j.1365-2958.2008.06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M.T., Heather Z., Paillot R., Steward K.F., Webb K., Ainslie F., Jourdan T., Bason N.C., Holroyd N.E., Mungall K., Quail M.A., Sanders M., Simmonds M., Willey D., Brooks K., Aanensen D.M., Spratt B.G., Jolley K.A., Maiden M.C., Kehoe M., Chanter N., Bentley S.D., Robinson C., Maskell D.J., Parkhill J., Waller A.S. Genomic evidence for the evolution of Streptococcus equi: Host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathogens. 2009;5:e1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse I., Hamidjaja R.A., Bok J.M., van Rotterdam B.J. Reliable detection of Bacillus anthracis, Francisella tularensis and Yersinia pestis by using multiplex qPCR including internal controls for nucleic acid extraction and amplification. BMC Microbiology. 2010;10:314. doi: 10.1186/1471-2180-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C., Bugg M., Robinson C., Mitchell Z., Davis-Poynter N., Newton J.R., Jolley K.A., Maiden M.C., Waller A.S. Sequence variation of the SeM gene of Streptococcus equi allows discrimination of the source of strangles outbreaks. Journal of Clinical Microbiology. 2006;44:480–486. doi: 10.1128/JCM.44.2.480-486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton J.R., Verheyen K., Talbot N.C., Timoney J.F., Wood J.L., Lakhani K.H., Chanter N. Control of strangles outbreaks by isolation of guttural pouch carriers identified using PCR and culture of Streptococcus equi. Equine Veterinary Journal. 2000;32:515–526. doi: 10.2746/042516400777584721. [DOI] [PubMed] [Google Scholar]
- Newton J.R., Wood J.L., Dunn K.A., DeBrauwere M.N., Chanter N. Naturally occurring persistent and asymptomatic infection of the guttural pouches of horses with Streptococcus equi. Veterinary Record. 1997;140:84–90. doi: 10.1136/vr.140.4.84. [DOI] [PubMed] [Google Scholar]
- Paillot R., Robinson C., Steward K., Wright N., Jourdan T., Butcher N., Heather Z., Waller A.S. Contribution of each of four superantigens to Streptococcus equi-induced mitogenicity, gamma interferon synthesis, and immunity. Infection and Immunity. 2010;78:1728–1739. doi: 10.1128/IAI.01079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith H.M., Harris S.R., Persson K., Marsh P., Barron A., Bignell A., Bjartling C., Clark L., Cutcliffe L.T., Lambden P.R., Lennard N., Lockey S.J., Quail M.A., Salim O., Skilton R.J., Wang Y., Holland M.J., Parkhill J., Thomson N.R., Clarke I.N. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoney J.F., Artiushin S.C. Detection of Streptococcus equi in equine nasal swabs and washes by DNA amplification. Veterinary Record. 1997;141:446–447. doi: 10.1136/vr.141.17.446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table