Abstract
Objective
Despite the established role of MRI in the diagnosis of brain tumours, histopathological assessment remains the clinically used technique, especially for the glioma group. Relative cerebral blood volume (rCBV) is a dynamic susceptibility-weighted contrast-enhanced perfusion MRI parameter that has been shown to correlate to tumour grade, but assessment requires a specialist and is time consuming. We developed analysis software to determine glioma gradings from perfusion rCBV scans in a manner that is quick, easy and does not require a specialist operator.
Methods
MRI perfusion data from 47 patients with different histopathological grades of glioma were analysed with custom-designed software. Semi-automated analysis was performed with a specialist and non-specialist operator separately determining the maximum rCBV value corresponding to the tumour. Automated histogram analysis was performed by calculating the mean, standard deviation, median, mode, skewness and kurtosis of rCBV values. All values were compared with the histopathologically assessed tumour grade.
Results
A strong correlation between specialist and non-specialist observer measurements was found. Significantly different values were obtained between tumour grades using both semi-automated and automated techniques, consistent with previous results. The raw (unnormalised) data single-pixel maximum rCBV semi-automated analysis value had the strongest correlation with glioma grade. Standard deviation of the raw data had the strongest correlation of the automated analysis.
Conclusion
Semi-automated calculation of raw maximum rCBV value was the best indicator of tumour grade and does not require a specialist operator.
Advances in knowledge
Both semi-automated and automated MRI perfusion techniques provide viable non-invasive alternatives to biopsy for glioma tumour grading.
MRI is an important tool for the diagnosis of brain tumours; however, despite its established role, histopathological assessment forms the basis of prognosis calculation and treatment planning in most cases, especially for the glioma group [1-6]. Recently developed radiology-based techniques have been utilised to accurately grade intra-axial tumours, avoiding the need for time-consuming and invasive histopathological examination. These techniques include dynamic susceptibility-weighted contrast-enhanced perfusion MRI parameters such as cerebral blood flow [7-10], and metabolic measurements such as the choline–creatine ratio [11], as well as relative cerebral blood volume (rCBV) [12-17].
One of the most promising and least intrusive of these new techniques uses rCBV measurements following intravenous contrast injection. rCBV is expected to correlate with tumour grade as it has been correlated with vascular endothelial growth factor expression and thus probable angiogenic ability [18]. Two main techniques have been used to measure rCBV in tumours: region of interest (ROI) analysis and histogram analysis. In ROI analyses, small ROIs are chosen by experienced radiologists in areas representing the maximum perfusion of the tumour, while avoiding artefacts such as large vessels. Good correlation with tumour grade has been observed [12,13], and this technique can also help differentiate oligodendroglioma from astrocytoma [14,15]. Histogram analyses similarly utilise experienced radiologists to draw a large analytical region around the tumour margins. All pixels contained within it are analysed to calculate various statistical values, which have also been shown to correlate with tumour grade [16,17,19,20].
The primary limitation of a ROI analysis is the labour-intensive nature of finding the maximum rCBV value (rCBVmax) in the ROI and differentiating this from a possible vessel-perfusion effect, which requires the skills of a specialist. For a histogram analysis, vessel perfusion skews results, as has been shown in a recent study in which vessels were either included or excluded during ROI placement [21].
We have developed easy-to-use analysis software capable of both semi-automated and automated glioma grading using ROI and histogram analyses based on perfusion scans. Our program is designed to enable non-specialists to perform the analysis, removing the need for a neuroradiologist operator, and to avoid the skewing effect of tumour vessels. The correlation of tumour parameters, calculated with the software, to glioma grade is determined using patient data previously classified from histology.
Methods
All images were acquired at the Academic Unit of Radiology, Hallamshire Hospital, Sheffield, UK, on a 3 T MR scanner (Achieva, Philips Medical Systems, Best, Netherlands). Written consent was acquired from all participants and approval was obtained from the Sheffield Regional Ethics Committee.
Patient group
A total of 47 patients with newly identified gliomas were included in the study, all of whom underwent histological diagnosis to determine the tumour grade: II, III or IV. General details and histological diagnoses are given in Table 1.
Table 1. The sex, age and histological glioblastoma multiforme tumour grading of 47 patients.
| Sex |
Histological grade |
||||
| Male | Female | Age (years) | II | III | IV |
| 30 | 17 | 53±13 | 8 | 4 | 35 |
Data are given as number or mean ± standard deviation.
Imaging protocols
Images were acquired with a 3 T MRI system (Intera 3.0 T; Philips Medical Systems).
Anatomical imaging
Standard axial and coronal images of the brain were acquired using the proton-density and T2 weighted imaging dual-echo turbo spin echo technique. Subsequently, two pre-contrast spin echo T1 weighted images were acquired in the axial and sagittal planes. Post-contrast T1 weighted images were acquired after the administration of the contrast agent (Gadovist®; Schering AG, Berlin, Germany).
Perfusion imaging
Perfusion imaging was performed using a gradient-echo fat-suppressed T2* weighted echoplanar imaging sequence, which was acquired during the first pass of a standard dose of the contrast agent (Gadovist). One dose (0.05 mmol kg−1) of contrast agent was used. The following sequence parameters were used: repetition time (TR), 957 ms; echo time (TE), 30 ms; section thickness, 4 mm; field of view (FOV), 230×158 mm; data matrix, 256×256 pixels; flip angle, 40°, and no intersection gap. 20 slices were acquired to cover the entire volume. A series of 45 multisection dynamic acquisitions was acquired at 1.8-s intervals. The first 10 dynamic sequences were performed without contrast administration to obtain the baseline signal. At the 10th acquisition, 0.05 mmol kg−1 of contrast agent was injected at a rate of 5 ml s−1 using a power injector. Contrast administration was followed by a 20-ml continuous saline flush at the same rate.
Perfusion images were transferred to a workstation (View Forum; Philips Medical Systems) for processing. rCBV was calculated on a pixel-by-pixel basis and parametric colour maps were generated by the proprietary software. These images were converted to greyscale with upper rCBV boundaries set to the maximum pixel value and transferred to our custom-made analysis software.
Analysis software details
The custom analysis software is available under a standard free open source licence and written for the Matlab (The MathWorks Inc., Natick, MA) environment. The graphical user interface (GUI) of the program enables the user to simultaneously view rCBV data sets and corresponding high-resolution anatomical scans, and perform a histogram analysis of tumour regions in the perfusion data.
Slices containing the tumour are manually selected for analysis, and co-registration between perfusion and anatomical data is achieved by the user based on knowledge and recognition of anatomical landmarks. When the analysis is initiated, the user is prompted for a rectangular tumour ROI to be selected within the GUI. Tools are available to show which pixels within the ROI correspond to selected ranges of rCBV values, enabling the user to isolate pixel values corresponding to the tumour from those of surrounding normal brain tissue as well as blood vessels. The user can graphically manipulate the range of histogram values corresponding to the tumour, and a statistical analysis is performed on the values and made available to the user as part of the histogram analysis. Calculated values include the mean, standard deviation, median, skewness and kurtosis of all data within the perfusion slice and tumour ROI, as well as the mean and standard deviation calculated from only the top 10%, 25% and 50% rCBV values within each. The user may also choose to normalise tumour rCBV values by a mean rCBV value calculated from a normal ROI of the brain within the slice before performing a histogram analysis.
Relative cerebral blood volume analyses
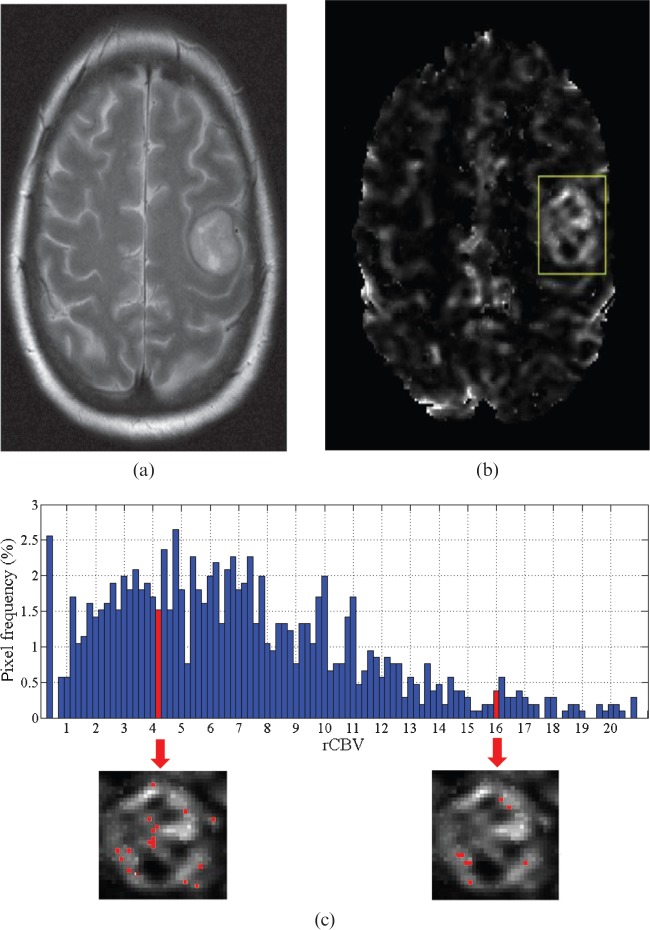
Tumour perfusion images were selected for each patient corresponding to the section with the largest mass defect. Manual correlation of each tumour mass edge in the perfusion image and corresponding conventional T1 image was performed. A rectangular ROI was drawn around the tumour and all pixels within the rectangle were analysed. A histogram showing rCBV frequency using a bin width of 0.2 was produced. This was either analysed in its present form, which we will call the raw data, or normalised by the averaged rCBV in a region of contralateral white matter, which we will call the normalised data. Using the analysis software, each bin of the histogram was mapped back onto the perfusion image, thus giving a map of the pixel positions for each bin, as illustrated in Figure 1. This was done for histograms of the raw and normalised rCBV data for each tumour. The pixel maps were then analysed side by side with the high-resolution T1 image representing the same anatomical region.
Figure 1.
(a) A T2 weighted image of a glioma is shown with (b) the corresponding perfusion image and superimposed rectangular region of interest (ROI). The analysis software calculates the histogram from the relative cerebral blood volume (rCBV) data within the ROI and can highlight the perfusion image pixel values corresponding to a bin (marked), as shown in (c). An observer uses these images to determine whether a pixel corresponded to signal from a vessel or tumour tissue.
Semi-automated relative cerebral blood volume analysis
An experienced neuroradiologist and a physician with little radiology experience, corresponding to our specialist and non-specialist, respectively, analysed the sections independently, determining which pixels corresponded to a vessel or artefact and which to non-vessel tumour mass. Both investigators were blinded to the grading of the tumour. The smallest bin value that contained a pixel corresponding to a vessel as well as the bin value containing the pixel with the maximum value (rCBVmax) in the non-vessel tumour region were noted.
Comparison of interobserver variation was conducted by plotting measurements of minimum vessel signal and rCBVmax in both raw and normalised tumour series for each observer. The correlation coefficient and corresponding linear equations relating the results of the two observers were calculated.
Fully automated histogram analysis
The mean, standard deviation, median, skewness and kurtosis of all data within each calculated histogram, as well as the mean and standard deviation calculated from only the top 10%, 25% and 50% rCBV values, were calculated using the software. A lower raw rCBV cut-off value of 0.1 and an upper cut-off value of 21.3 were applied, below and above which, respectively, contained only artefact or vessel signal.
Statistical analyses
Data were separated into low- and high-grade tumour groups consisting of grades II/III and grade IV, respectively, for all statistical analyses. For both rCBVmax and calculated histogram statistical values, correlation with grade was calculated using a Spearman rank correlation test, and a Mann–Whitney U-test was used to compare levels between the groups. Bonferroni correction was applied to all statistical tests. A significance level of α=0.05 was used.
Results
Semi-automated relative cerebral blood volume analysis
A strong correlation was found between the specialist (experienced neuroradiologist) and the non-specialist (inexperienced physician) with correlation coefficients of r=0.997 for both raw and normalised values of rCBVmax. Strong correlations were also present for measurements of minimum pixel value corresponding to vessel signal with values of r=0.993 and 0.959 for raw and normalised data, respectively. These results along with the linear relationship between observers are summarised in Table 2.
Table 2. The linear relationship and corresponding coefficients as calculated between specialist and non-specialist observers using semi-automated software to identify the first pixel lying within a vessel and that representing the maximum relative cerebral blood volume (rCBVmax) non-vessel tumour value.
| Analysis | Slope | Intercept | Correlation coefficient (r) |
| Raw vessel with smallest pixel value | 0.98 | 0.40 | 0.993 |
| Normalised vessel with smallest pixel value | 0.97 | 0.41 | 0.959 |
| Raw rCBVmax | 0.98 | 0.43 | 0.997 |
| Normalised nrCBVmax | 0.96 | 0.62 | 0.997 |
Data from both raw (unaltered) glioblastoma multiforme tumour series and tumour series normalised by the averaged rCBV in a region of contralateral white matter (nrCBVmax) are shown. A correlation coefficient of 1 with a gradient of 1 and intercept of 0 would correspond to absolute correlation. Values measured by the specialist can be predicted using the formula: specialist values=(slope)×(non-specialist values)+intercept.
The minimum rCBV values corresponding to vessel signal and maximum rCBV values corresponding to tumour for both raw and normalised data as measured by the non-specialist are shown in Figure 2. Similar results were obtained by the specialist and are not shown. The measured minimum rCBV values were generally much smaller than the corresponding measured rCBVmax values. This corresponds to the large gap between curves in Figure 2.
Figure 2.
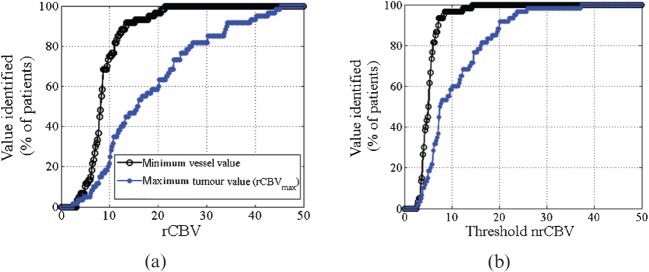
The graphs show the percentage of tumours for each relative cerebral blood volume (rCBV) value corresponding to the detection of either the smallest rCBV value of a vessel (closed circles) or the largest rCBV value of the tumour tissue, which we label rCBVmax (open circles). Raw data are shown in (a), whereas the normalised data are shown in (b). The rCBVmax line is well separated and to the right of the line representing smallest vessel rCBV values, and thus these rCBVmax values would be above any cut-off value used to separate tumour and vessel values. nrCBV, normalised rCBV.
Mean values for grade II and grade III gliomas obtained by the non-specialist using the semi-automated analysis of rCBVmax on the raw rCBV data were very close, with values of 8±2 and 7±2, respectively, with a larger value of 20±11 obtained for grade IV gliomas, as shown in Table 3 along with the corresponding normalised rCBV values. A significant correlation was found between tumour grade and rCBVmax for both raw and normalised data, with values of r=0.662 and r=0.557 where the stronger correlation is from the raw data. Grade II and grade III mean values were closely matched, as can be seen by the close clustering of points in Figure 3, and thus they were combined into a single group for significance testing. A significant difference (p<0.0001) was seen between grade II/III gliomas and grade IV gliomas for both raw and normalised series. Similar results were obtained by the specialist and are not shown.
Table 3. The mean and standard deviation of maximum relative cerebral blood volume (rCBVmax) values corresponding to raw (unaltered) glioblastoma multiforme tumour perfusion series and values normalised by the averaged rCBV in a region of contralateral white matter (nrCBVmax) for each glioma grading.
| Values | Grade II | Grade III | Grade IV | r-value | p-value |
| Mean rCBVmax | 8±2 | 7±2 | 20±11 | 0.662 | <0.0001 |
| Mean nrCBVmax | 7±5 | 4±1 | 12±7 | 0.557 | <0.0001 |
Values of Spearman rank correlation coefficients and Bonferroni corrected p-values are listed. Significance (α=0.05) is denoted by the italic font.
Figure 3.
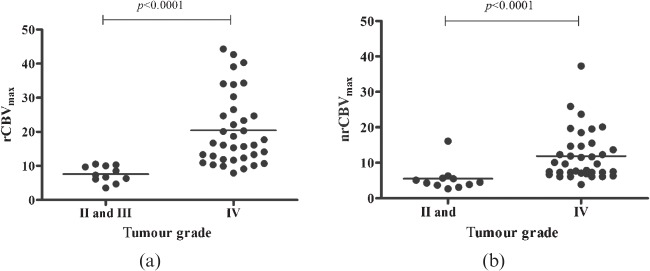
Maximum relative cerebral blood volume (rCBVmax) values for (a) raw and (b) normalised (glioblastoma multiforme) tumour perfusion series based on histological grade, either grade II and III, or grade IV. p-values are calculated using the Mann–Whitney U-test.
Fully automated histogram analysis
While the mean and standard deviation of the top 10%, 25% and 50% of values were calculated and analysed, these parameters were ultimately found to be poorer indicators of tumour grade than the mean and standard deviation of all values, and thus the results are excluded.
The mean, standard deviation, median, mode, skewness and kurtosis of the curve for all ROI values are summarized in Table 4. The mean value analyses show an increasing trend as grade increased, which was true for many of the other parameters in both raw and normalised perfusion series.
Table 4. The mean and standard deviations as calculated from relative cerebral blood volume (rCBV) histogram values of glioblastoma multiforme tumour perfusion series for grade II, III and IV gliomas.
| Parameter |
Raw |
Normalised |
||||
| Grade II | Grade III | Grade IV | Grade II | Grade II | Grade IV | |
| Mean | 3±1 | 3±1 | 5±2 | 1.7±0.6 | 3±1 | 3±1 |
| Standard deviation | 1.9±0.9 | 2±2 | 4±1 | 1.1±0.5 | 2±1 | 3±1 |
| Median | 2±1 | 2.2±0.9 | 4±2 | 1.4±0.5 | 2±1 | 3±1 |
| Mode | 0.6±0.9 | 0.9±0.8 | 1±1 | 0.8±0.4 | 1.5±0.8 | 2±4 |
| Skewness | 2±1 | 1.9±0.2 | 1.2±0.7 | 2.1±0.8 | 1.8±0.3 | 1.2±0.7 |
| Kurtosis | 15±13 | 8±2 | 5±4 | 12±7 | 7±2 | 5±3 |
Raw values represent the unchanged perfusion values, while normalised values were first normalised by the averaged rCBV value in a region of contralateral white matter.
The Spearman correlation coefficients with Bonferroni corrected p-value were calculated for all parameters for both raw and normalised data, and are summarised in Table 5. All parameters calculated from both the raw and the normalised data showed significant correlation with tumour grade except for the mode. The standard deviation as calculated from all raw data has the strongest correlation with r=0.553. However, rCBVmax, as measured in the semi-automated technique using raw data, still showed stronger correlation with a value of r=0.662. Similarly, the correlation strength obtained from the normalised rCBVmax (nrCBVmax) semi-automated technique showed stronger correlation than that obtained from all parameters calculated using the normalised data.
Table 5. Listed for each histogram parameter are the Spearman correlation coefficients assessed with Bonferroni corrected p-values along with the mean, standard deviation and corresponding p-values between low-grade (II/III) and high-grade (IV) glioblastoma multiforme tumour groups calculated using the Mann–Whitney U-test.
| Parameter |
Raw |
Normalised |
||||||||
| Correlation |
Grade II/III |
Grade IV |
p-value |
Correlation |
Grade II/III |
Grade IV |
p-value |
|||
| r-value | p-value | r-value | p-value | |||||||
| Mean | 0.4880 | 0.0156 | 3±1 | 5±2 | 0.031 | 0.4992 | 0.0104 | 2±1 | 3±1 | 0.047 |
| Standard deviation | 0.5530 | <0.0001 | 2±1 | 4±1 | 0.008 | 0.4907 | 0.0130 | 1.5±0.8 | 4±1 | >0.050 |
| Median | 0.4880 | 0.0156 | 2±1 | 4±2 | 0.023 | 0.4534 | 0.0416 | 1.7±0.8 | 3±1 | >0.050 |
| Mode | −0.0158 | >0.0500 | 0.8±0.8 | 1±1 | >0.050 | −0.1108 | >0.0500 | 1.1±0.7 | 2±4 | >0.050 |
| Skewness | −0.4890 | 0.0156 | 2.1±0.9 | 1.2±0.7 | 0.021 | −0.4496 | 0.0442 | 2.0±0.6 | 1.2±0.7 | >0.050 |
| Kurtosis | −0.5180 | 0.0052 | 12±10 | 5±4 | 0.016 | −0.4587 | 0.0338 | 10±6 | 5±3 | >0.050 |
Raw values represent the unchanged relative cerebral blood volume (rCBV) perfusion values, while normalised values were first normalised by the averaged rCBV value in a region of contralateral white matter. Significance (α=0.05) is denoted by the italic font.
For each parameter, the mean, standard deviation and significance determined using the Mann–Whitney U-test, calculated after combining grade II and III tumours into a single group and comparing them with grade IV tumours, are also summarised in Table 5. Significant differences were found for all histogram parameters based on the raw data except for the mode. For the normalised data, a significant difference was obtained only for the mean.
Discussion
We have presented semi-automated and automated techniques based on MRI perfusion scans and custom-designed GUI-based software to grade intra-axial neoplasms. Our work builds on previous work to both calculate rCBVmax values [15,22] and perform a histogram analysis [16,17], but tries to solve the problems that limit these techniques: time-consuming and observer-dependent measurements, and need for highly trained neuroradiologist observers.
We have made our software freely available under a standard open source licence in an effort to maximise the availability, promoting further research and use of these techniques. The liberal nature of the licence also enables others to build easily on our work, perhaps enabling a fully automated calculation of rCBVmax values.
Marking a departure from previous studies, we used raw (non-normalised) rCBV values to compare gliomas in addition to normalised values. This was undertaken as correlation was found to be better using raw data; our data, perhaps counterintuitively, indicate that normalisation may mask differences between gliomas. One reason for this may be concurrent contralateral hemisphere perfusion changes. This difference between raw and normalised data should be taken into account and further explored in future studies.
Semi-automated relative cerebral blood volume analysis
Our semi-automated analysis is performed by rapidly scrolling through all pixels in the perfusion series, from the largest to the smallest, and determining the pixel with the maximal rCBV (i.e. rCBVmax) that lies outside a vessel. Our method is comparable to other techniques that analyse colour perfusion maps for the highest signal [23,24]. However, our method is not prone to the wide variation reported for these techniques, as noted by the strong correlation between our observers. For example, Wetzel et al [24] showed a best interobserver correlation coefficient of 0.71, compared with our better correlation values that were all >0.9. The reason for this may include the ease of use of the software and the use of a one-pixel ROI analysis for rCBVmax. This obviates observer selection of small ROIs within the tumour, thereby eliminating interobserver variability; the only decision to be made by the observer is whether a pixel lies within a vessel or not. It is also important to note that our measurements were carried out under ideal conditions in which the observers were not limited by time or presented with distractions. In clinical settings, an observer may not be able to dedicate the same amount of time and/or concentration to finding the maximum value, resulting in pixel selections that do not represent the true rCBVmax. We would expect this to result in an increase in both inter- and intra-observer variability.
The strong agreement between specialist and non-specialist suggests that use of the software not only helps eliminate the observer dependence of the measurements, but also successfully enables non-specialists to make the measurements. This could enable glioma tumour grading that is both quicker and potentially even less expensive as it could be done by general physicians or even technologists.
One of our hypotheses was that rCBV values corresponding to vessels would be generally larger and separated from those corresponding to the tumour. Thus, use of a “vessel cut-off” (threshold) value was expected to avoid the potential confounding effect of tumour vessels on MRI-based glioma grading [21]. However, this hypothesis was not supported by our data, and vessel signals were consistently smaller than rCBVmax measurements for both raw and normalised perfusion series. Therefore, if a vessel cut-off value were generated from our data, it would exclude many larger tumour tissue rCBV values, leading to an underestimation of rCBVmax and thus preventing an automated calculation of rCBVmax and nrCBVmax. A possible explanation for this may be that small vessel sizes and/or partial voluming effects resulting when signal from surrounding tumour is averaged with rCBV values of the vessels cause the measured value to be underestimated.
To the best of our knowledge, this work is the first demonstration that rCBVmax as measured from a single pixel correlates with tumour grade. All previous studies using ROI analysis have used ROIs of at least 10 pixels, such as the work of Danchaivijitr et al [22], who used 15–20 pixels, and Lev et al [15], who used 16 pixels. Clearly, even with one pixel, it is not possible to exclude the inclusion of vessels beyond the resolution of the T2 weighted imaging used as a comparator; however, this is an inevitable failing of any analysis of perfusion imaging. The correlations calculated in this study, shown in Table 3, are weaker than some previously reported, such as the correlation values between 0.7 and 0.8 achieved by Law, Young and colleagues [19,20]. However, this may be explained by our tumour grade grouping compared with the majority of previous studies, which grouped grades I/II and III/IV together [6,13,15,19,20,22,23,25-27], as a larger difference may be expected between these groups than between groups of grade II/III and grade IV. A second explanation is that this may be a result of our use of rectangular ROIs, encompassing the tumour, compared with ROIs defined by free-line selection of tumour boundaries used in previous studies. Although this may lead to slightly lower correlations, and therefore a lower likelihood of differentiating between tumour grades, it is also one of the strengths of our analysis as it enables a non-specialist user to perform the analysis quickly and eliminates the inherent differences caused by ROI positioning choice, as shown by Young et al [20].
While significant differences (p<0.0001) were found between the grade II/III and grade IV gliomas in both raw and normalised tumour series, as shown in Table 3, a decreasing trend in mean rCBVmax is seen in grade III tumours compared with grade II tumours. This is an unexpected finding as previous studies have suggested that the grade III group should contain a subset with a higher vascularity, and therefore larger rCBVmax values, likely to transform into grade IV tumours [22]. However, it should be noted that only a small number (11) of histology-verified neoplasms in the lower glioma grades (II and III) were found. This prevented meaningful statistical analysis between groups, perhaps explaining these findings, and restricted us to a comparison between grades II/III and grade IV. A prospective study with greater numbers will help address whether the grouping of rCBVmax values into grades II and III vs grade IV is a valid means of quickly calculating the management and prognosis of patients in the clinical setting. Alternatively, use of just one contrast bolus, rather than the two used by many groups to minimise contrast extravasation effects [28], may have exaggerated differences between grade III and IV tumours, also explaining the results.
Fully automated histogram analysis
The mean and standard deviation of the top 10%, 25% and 50% of histogram data were calculated and analysed in addition to all histogram values. However, following analysis, it was seen that these values either did not show significance or showed poorer correlation to tumour grade, and thus the values were not shown or discussed further. Our results show no advantage to calculating these parameters over the mean and standard deviation of all histogram values.
Numerous normalised histogram parameters that correlate with tumour grade and show significant differences between grade II and grades III/IV tumours, grouped as low and high grade in these studies, respectively, have been reported by other groups [16,17,19,20]. Our study differs in that we compared tumour grades II/III and grade IV. As previously mentioned, this was because of the limited number and similar values measured for our grade II and III tumours. We are the first to show significance between tumour grades using these groupings, and our study concurs with previous studies, finding differences among many similar histogram parameters for the glioma grades, including the work by Law, Young and colleagues [19,20]. However, as with the semi-automated analysis, better results were obtained using raw rather than normalised tumour data, a comparison not previously investigated. Indeed, only a single calculated histogram parameter, the mean, showed significance when using normalised data.
Our results show that the standard deviation as measured using the raw data best correlates with tumour grade, and larger standard deviations correspond to a high tumour grade. This result may reflect the greater vasculisation that would be expected in higher grade tumours, thus creating a larger spread of rCBV values within the ROI. However, it should be noted that the rCBVmax value as measured using the semi-automated technique shows better correlation to tumour grade, and thus we recommend its use when trying to maximise grading accuracy. In situations in which observer time is more limited and rCBVmax cannot be measured, this measurement of standard deviation provides the best alternative.
Conclusions
Our study produced the following main findings:
rCBVmax, as measured using the semi-automated analysis and raw perfusion data, gave the best correlation with tumour grade.
rCBVmax values can be determined from a single pixel of the tumour perfusion scans.
rCBV values corresponding to vessels cannot be separated from those corresponding to tumour based on an rCBV cut-off value, preventing fully automated calculation of rCBVmax.
Use of a rectangular ROI enables a non-specialist user to perform the analysis but results in a weaker correlation between grades than was previously reported.
Standard deviation as measured from raw perfusion data provides the best automated correlation with tumour grade, but semi-automated rCBVmax values provide better correlation.
Normalisation of perfusion values to the contralateral white matter may mask results and cause a loss of correlation.
Footnotes
S N Friedman and P J Bambrough are joint first coauthors.
References
- 1.Al-Okaili RN, Krejza J, Wang S, Woo JH, Melhem ER. Advanced MR imaging techniques in the diagnosis of intraaxial brain tumors in adults. Radiographics 2006;26Suppl. 1:S173–89 [DOI] [PubMed] [Google Scholar]
- 2.Wintermark M, Sesay M, Barbier E, Borbély K, Dillon WP, Eastwood JD, et al. Comparative overview of brain perfusion imaging techniques. Stroke 2005;36:e83–99 [DOI] [PubMed] [Google Scholar]
- 3.Yang D, Korogi Y, Sugahara T, Kitajima M, Shigematsu Y, Liang L, et al. Cerebral gliomas: prospective comparison of multivoxel 2D chemical-shift imaging proton MR spectroscopy, echoplanar perfusion and diffusion-weighted MRI. Neuroradiology 2002;44:656–66 [DOI] [PubMed] [Google Scholar]
- 4.Roberts HC, Roberts TP, Brasch RC, Dillon WP. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol 2000;21:891–9 [PMC free article] [PubMed] [Google Scholar]
- 5.Petrella JR, Provenzale JM. MR perfusion imaging of the brain: techniques and applications. AJR Am J Roentgenol 2000;175:207–19 [DOI] [PubMed] [Google Scholar]
- 6.Provenzale JM, Wang GR, Brenner T, Petrella JR, Sorensen AG. Comparison of permeability in high-grade and low-grade brain tumors using dynamic susceptibility contrast MR imaging. AJR Am J Roentgenol 2002;178:711–16 [DOI] [PubMed] [Google Scholar]
- 7.Barbier EL, Lamalle L, Decorps M. Methodology of brain perfusion imaging. J Magn Reson Imaging 2001;13:496–520 [DOI] [PubMed] [Google Scholar]
- 8.Donahue KM, Krouwer HG, Rand SD, Pathak AP, Marszalkowski SC, Censky SC, et al. Utility of simultaneously acquired gradient-echo and spin-echo cerebral blood volume and morphology maps in brain tumor patients. Magn Reson Med 2000;43:845–53 [DOI] [PubMed] [Google Scholar]
- 9.Ostergaard L. Principles of cerebral perfusion imaging by bolus tracking. J Magn Reson Imaging 2005;22:710–17 [DOI] [PubMed] [Google Scholar]
- 10.Weber MA, Zoubaa S, Schlieter M, Jüttler E, Huttner HB, Geletneky K, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology 2006;66:1899–906 [DOI] [PubMed] [Google Scholar]
- 11.Law M, Yang S, Wang H, Babb JS, Johnson G, Cha S, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24:1989–98 [PMC free article] [PubMed] [Google Scholar]
- 12.Aronen HJ, Gazit IE, Louis DN, Buchbinder BR, Pardo FS, Weisskoff RM, et al. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology 1994;191:41–51 [DOI] [PubMed] [Google Scholar]
- 13.Jackson A, Kassner A, Annesley-Williams D, Reid H, Zhu XP, Li KL. Abnormalities in the recirculation phase of contrast agent bolus passage in cerebral gliomas: comparison with relative blood volume and tumor grade. AJNR Am J Neuroradiol 2002;23:7–14 [PMC free article] [PubMed] [Google Scholar]
- 14.Cha S, Tihan T, Crawford F, Fischbein NJ, Chang S, Bollen A, et al. Differentiation of low-grade oligodendrogliomas from low-grade astrocytomas by using quantitative blood-volume measurements derived from dynamic susceptibility contrast-enhanced MR imaging. AJNR Am J Neuroradiol 2005;26:266–73 [PMC free article] [PubMed] [Google Scholar]
- 15.Lev MH, Ozsunar Y, Henson JW, Rasheed AA, Barest GD, Harsh GR, 4th, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol 2004;25:214–21 [PMC free article] [PubMed] [Google Scholar]
- 16.Emblem KE, Nedregaard B, Nome T, Due-Tonnessen P, Hald JK, Scheie D, et al. Glioma grading by using histogram analysis of blood volume heterogeneity from MR-derived cerebral blood volume maps. Radiology 2008;247:808–17 [DOI] [PubMed] [Google Scholar]
- 17.Emblem KE, Scheie D, Due-Tonnessen P, Hald JK, Scheie D, Borota OC, et al. Histogram analysis of MR imaging-derived cerebral blood volume maps: combined glioma grading and identification of low-grade oligodendroglial subtypes. AJNR Am J Neuroradiol 2008;29:1664–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maia AC, Jr, Malheiros SM, da Rocha AJ, da Silva CJ, Gabbai AA, Ferraz FA, et al. MR cerebral blood volume maps correlated with vascular endothelial growth factor expression and tumor grade in nonenhancing gliomas. AJNR Am J Neuroradiol 2005;26:777–83 [PMC free article] [PubMed] [Google Scholar]
- 19.Law M, Young R, Babb J, Pollack E, Johnson G. Histogram analysis versus region of interest analysis of dynamic susceptibility contrast perfusion MR imaging data in the grading of cerebral gliomas. AJNR Am J Neuroradiol 2007;28:761–6 [PMC free article] [PubMed] [Google Scholar]
- 20.Young R, Babb J, Law M, Pollack E, Johnson G. Comparison of region-of-interest analysis with three different histogram analysis methods in the determination of perfusion metrics in patients with brain gliomas. J Magn Reson Imaging 2007;26:1053–63 [DOI] [PubMed] [Google Scholar]
- 21.Caseiras GB, Thornton JS, Yousry T, Benton C, Rees J, Waldman AD, et al. Inclusion or exclusion of intratumoral vessels in relative cerebral blood volume characterization in low-grade gliomas: does it make a difference? AJNR Am J Neuroradiol 2008;29:1140–1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danchaivijitr N, Waldman AD, Tozer DJ, Benton CE, Brasil Caseiras G, Tofts PS, et al. Low-grade gliomas: do changes in rCBV measurements at longitudinal perfusion-weighted MR imaging predict malignant transformation? Radiology 2008;247:170–8 [DOI] [PubMed] [Google Scholar]
- 23.Shin JH, Lee HK, Kwun BD, Kim JS, Kang W, Choi CG, et al. Using relative cerebral blood flow and volume to evaluate the histopathologic grade of cerebral gliomas: preliminary results. AJR Am J Roentgenol 2002;179:783–9 [DOI] [PubMed] [Google Scholar]
- 24.Wetzel SG, Cha S, Johnson G, Lee P, Law M, Kasow DL, et al. Relative cerebral blood volume measurements in intracranial mass lesions: interobserver and intraobserver reproducibility study. Radiology 2002;224:797–803 [DOI] [PubMed] [Google Scholar]
- 25.Lam WW, Chan KW, Wong WL, Poon WS, Metreweli C. Pre-operative grading of intracranial glioma. Acta Radiol 2001;42:548–54 [DOI] [PubMed] [Google Scholar]
- 26.Schmainda KM, Rand SD, Joseph AM, Lund R, Ward BD, Pathak AP, et al. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. AJNR Am J Neuroradiol 2004;25:1524–32 [PMC free article] [PubMed] [Google Scholar]
- 27.Sugahara T, Korogi Y, Kochi M, Ushio Y, Takahashi M. Perfusion-sensitive MR imaging of gliomas: comparison between gradient-echo and spin-echo echo-planar imaging techniques. AJNR Am J Neuroradiol 2001;22:1306–15 [PMC free article] [PubMed] [Google Scholar]
- 28.Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology 2008;249:601–13 [DOI] [PMC free article] [PubMed] [Google Scholar]