Abstract
A high-copy-number plasmid genomic library was screened for genes that when overexpressed down-regulate Ras protein activity in Saccharomyces cerevisiae. We report on the structure and characterization of one such gene, RPI1, which potentially encodes a novel 46-kDa negative regulator of the Ras-cyclic AMP pathway. Three lines of evidence suggest that the RPI1 gene product operates upstream to negatively regulate the activity of normal but not mutationally activated Ras proteins: (i) overexpressed RPI1 lowers cyclic AMP levels in wild-type yeast cells but not in yeast cells carrying the RAS2Val-19 mutation, (ii) overexpressed RPI1 suppresses the heat shock sensitivity phenotype induced by overexpression of normal RAS2 but does not suppress the same phenotype induced by RAS2Val-19, and (iii) disruption of RPI1 results in a heat shock sensitivity phenotype which can be suppressed by mutations that lower normal Ras activity. Thus, RPI1 appears to encode an inhibitor of Ras activity that shares a common feature with Ras GTPase-activating proteins in that it fails to down-regulate activated RAS2Val-19 function. We present evidence that the down-regulatory effect of RPI1 requires the presence of one of the two Ras GTPase activators, IRA1 and IRA2.
Full text
PDF

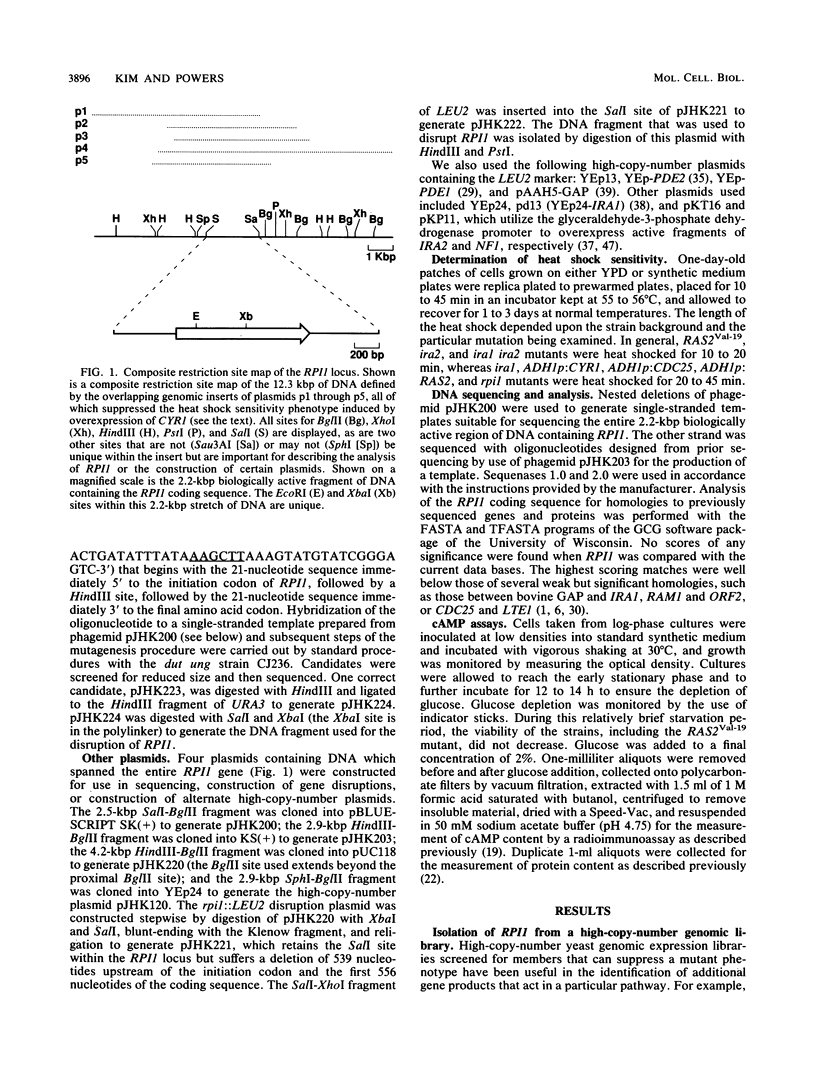

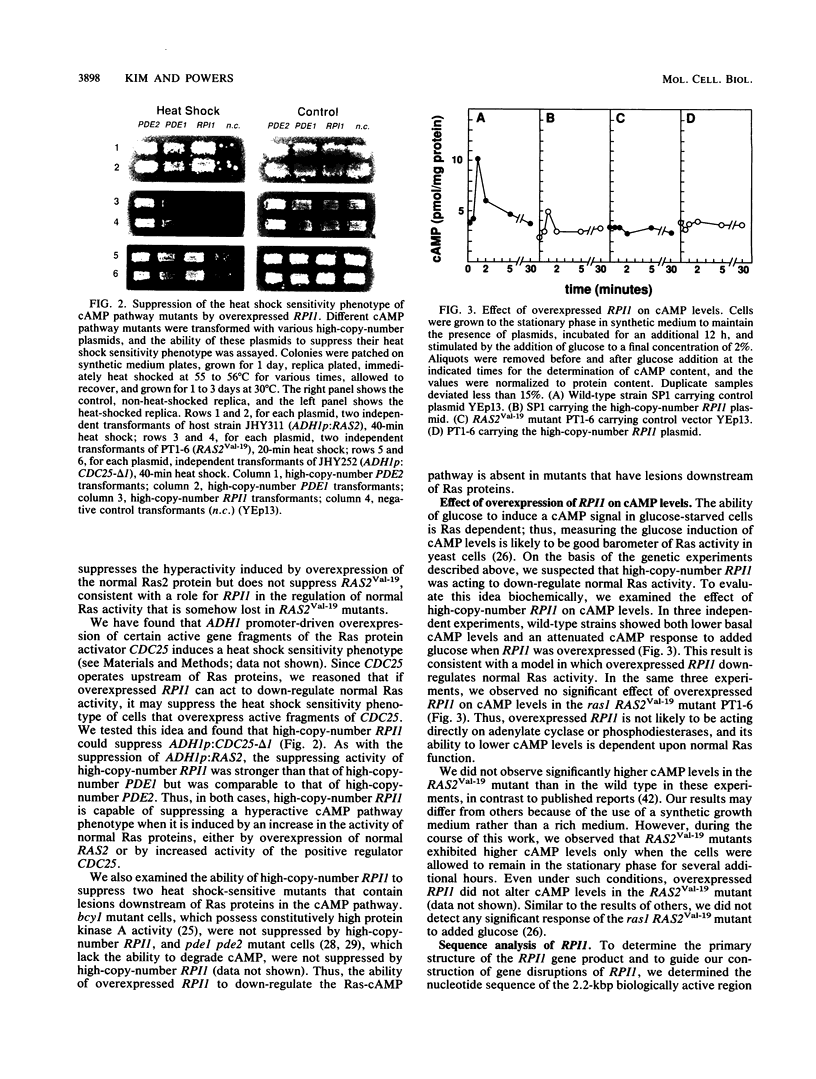

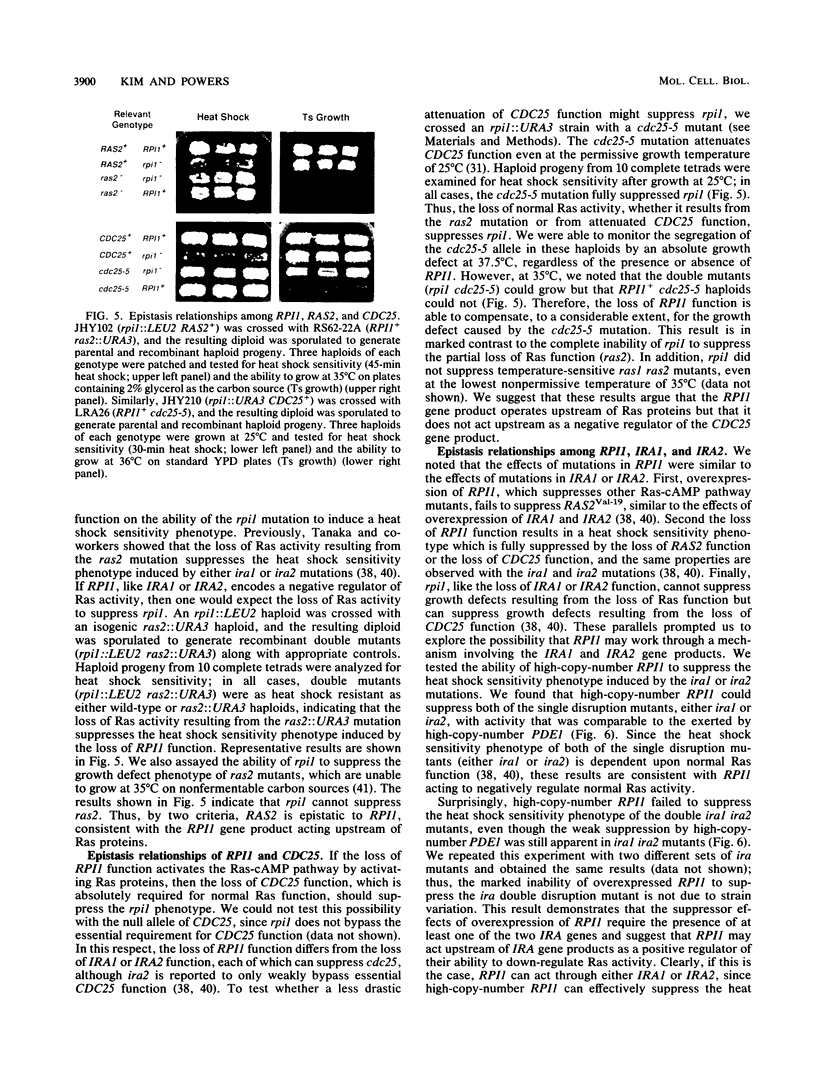




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballester R., Marchuk D., Boguski M., Saulino A., Letcher R., Wigler M., Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990 Nov 16;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- Ballester R., Michaeli T., Ferguson K., Xu H. P., McCormick F., Wigler M. Genetic analysis of mammalian GAP expressed in yeast. Cell. 1989 Nov 17;59(4):681–686. doi: 10.1016/0092-8674(89)90014-7. [DOI] [PubMed] [Google Scholar]
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990 Nov 8;348(6297):125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte E., Damak F., Camonis J., Garreau H., Jacquet M. The C-terminal part of a gene partially homologous to CDC 25 gene suppresses the cdc25-5 mutation in Saccharomyces cerevisiae. Gene. 1989 Apr 15;77(1):21–30. doi: 10.1016/0378-1119(89)90355-7. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Deschenes R. J. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Créchet J. B., Poullet P., Mistou M. Y., Parmeggiani A., Camonis J., Boy-Marcotte E., Damak F., Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990 May 18;248(4957):866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- Downward J., Graves J. D., Warne P. H., Rayter S., Cantrell D. A. Stimulation of p21ras upon T-cell activation. Nature. 1990 Aug 23;346(6286):719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- Downward J., Riehl R., Wu L., Weinberg R. A. Identification of a nucleotide exchange-promoting activity for p21ras. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5998–6002. doi: 10.1073/pnas.87.15.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg D., Simchen G., Levitzki A. In vitro reconstitution of cdc25 regulated S. cerevisiae adenylyl cyclase and its kinetic properties. EMBO J. 1990 Mar;9(3):641–651. doi: 10.1002/j.1460-2075.1990.tb08156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freissmuth M., Casey P. J., Gilman A. G. G proteins control diverse pathways of transmembrane signaling. FASEB J. 1989 Aug;3(10):2125–2131. [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Schaber M. D., Marshall M. S., Scolnick E. M., Sigal I. S. Identification of guanine nucleotides bound to ras-encoded proteins in growing yeast cells. J Biol Chem. 1987 Aug 5;262(22):10426–10429. [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Richardson H. E., de Barros Lopes M., Reed S. I. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. The cellular functions of small GTP-binding proteins. Science. 1990 Aug 10;249(4969):635–640. doi: 10.1126/science.2116664. [DOI] [PubMed] [Google Scholar]
- Harper J. F., Brooker G. Femtomole sensitive radioimmunoassay for cyclic AMP and cyclic GMP after 2'0 acetylation by acetic anhydride in aqueous solution. J Cyclic Nucleotide Res. 1975;1(4):207–218. [PubMed] [Google Scholar]
- Hughes D. A., Fukui Y., Yamamoto M. Homologous activators of ras in fission and budding yeast. Nature. 1990 Mar 22;344(6264):355–357. doi: 10.1038/344355a0. [DOI] [PubMed] [Google Scholar]
- Iida H., Yahara I. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J Cell Biol. 1984 Apr;98(4):1185–1193. doi: 10.1083/jcb.98.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Pringle J. R., Hartwell L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977 Mar 1;105(1):79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Marshall M. S., Hill W. S., Ng A. S., Vogel U. S., Schaber M. D., Scolnick E. M., Dixon R. A., Sigal I. S., Gibbs J. B. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J. 1989 Apr;8(4):1105–1110. doi: 10.1002/j.1460-2075.1989.tb03480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. A., Viskochil D., Bollag G., McCabe P. C., Crosier W. J., Haubruck H., Conroy L., Clark R., O'Connell P., Cawthon R. M. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990 Nov 16;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Genetic analysis of the role of cAMP in yeast. Yeast. 1985 Sep;1(1):15–24. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- Mbonyi K., Beullens M., Detremerie K., Geerts L., Thevelein J. M. Requirement of one functional RAS gene and inability of an oncogenic ras variant to mediate the glucose-induced cyclic AMP signal in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1988 Aug;8(8):3051–3057. doi: 10.1128/mcb.8.8.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J., Cameron S., Toda T., Ferguson K. M., Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987 Nov;1(9):931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- Nikawa J., Sass P., Wigler M. Cloning and characterization of the low-affinity cyclic AMP phosphodiesterase gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3629–3636. doi: 10.1128/mcb.7.10.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Bjørn S., Harrington T. R., Friesen J. D. An essential gene in Saccharomyces cerevisiae shares an upstream regulatory element with PRP4. Yeast. 1990 Jul-Aug;6(4):345–352. doi: 10.1002/yea.320060407. [DOI] [PubMed] [Google Scholar]
- Petitjean A., Hilger F., Tatchell K. Comparison of thermosensitive alleles of the CDC25 gene involved in the cAMP metabolism of Saccharomyces cerevisiae. Genetics. 1990 Apr;124(4):797–806. doi: 10.1093/genetics/124.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers S., O'Neill K., Wigler M. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):390–395. doi: 10.1128/mcb.9.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson L. C., Gibbs J. B., Marshall M. S., Sigal I. S., Tatchell K. CDC25: a component of the RAS-adenylate cyclase pathway in Saccharomyces cerevisiae. Science. 1987 Mar 6;235(4793):1218–1221. doi: 10.1126/science.3547648. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Kikuchi A., Araki S., Hata Y., Isomura M., Kuroda S., Takai Y. Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A, a ras p21-like GTP-binding protein. J Biol Chem. 1990 Feb 5;265(4):2333–2337. [PubMed] [Google Scholar]
- Sass P., Field J., Nikawa J., Toda T., Wigler M. Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9303–9307. doi: 10.1073/pnas.83.24.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Lin B. K., Wood D. R., Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Matsumoto K., Toh-E A. IRA1, an inhibitory regulator of the RAS-cyclic AMP pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Feb;9(2):757–768. doi: 10.1128/mcb.9.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Satoh T., Marshall M. S., Gibbs J. B., Matsumoto K., Kaziro Y., Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990 Mar 9;60(5):803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Nakafuku M., Tamanoi F., Kaziro Y., Matsumoto K., Toh-e A. IRA2, a second gene of Saccharomyces cerevisiae that encodes a protein with a domain homologous to mammalian ras GTPase-activating protein. Mol Cell Biol. 1990 Aug;10(8):4303–4313. doi: 10.1128/mcb.10.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K. RAS genes and growth control in Saccharomyces cerevisiae. J Bacteriol. 1986 May;166(2):364–367. doi: 10.1128/jb.166.2.364-367.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985 Jan;40(1):27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987 Oct 23;238(4826):542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Tsai M. H., Yu C. L., Wei F. S., Stacey D. W. The effect of GTPase activating protein upon ras is inhibited by mitogenically responsive lipids. Science. 1989 Jan 27;243(4890):522–526. doi: 10.1126/science.2536192. [DOI] [PubMed] [Google Scholar]
- Ueda T., Kikuchi A., Ohga N., Yamamoto J., Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990 Jun 5;265(16):9373–9380. [PubMed] [Google Scholar]
- Wolfman A., Macara I. G. A cytosolic protein catalyzes the release of GDP from p21ras. Science. 1990 Apr 6;248(4951):67–69. doi: 10.1126/science.2181667. [DOI] [PubMed] [Google Scholar]
- Xu G. F., Lin B., Tanaka K., Dunn D., Wood D., Gesteland R., White R., Weiss R., Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990 Nov 16;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- Xu G. F., O'Connell P., Viskochil D., Cawthon R., Robertson M., Culver M., Dunn D., Stevens J., Gesteland R., White R. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990 Aug 10;62(3):599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]






