Abstract
The basidiomycete yeast Crytococcus neoformans is a prominent human pathogen. It primarily infects immunocompromised individuals producing a meningoencephalitis that is lethal if untreated. Recent advances in its genetics and molecular biology have made it a model system for understanding both the Basidiomycota phylum and mechanisms of fungal pathogenesis. The relative ease of experimental manipulation coupled with the development of murine models for human disease allow for powerful studies in the mechanisms of virulence and host responses. This chapter introduces the organism and its life cycle and then provides detailed step-by-step protocols for culture, manipulation of the genome, analysis of nucleic acids and proteins, and assessment of virulence and expression of virulence factors.
1. Introduction
Although members of the Ascomycota phylum, particularly Sacchromyces cerevisiae, are the most studied fungi, there are 80,000 known species of the Fungi kingdom. There is a great deal of diversity in the kingdom, ranging from small harmless unicellular yeast such as S. cerevisiae to the great plant pathogen Armillaria ostoyae, one of the largest organisms in the world. This latter species is a member of the Basidiomycota phylum, a phylum less well understood than Ascomycota.
While no basidiomycete species has been studied in as much detail as S. cerevisiae, it is a fascinating and diverse group of organisms. Basidiomycetes produce many interesting secondary metabolites used in medicine, industry, and research. Members of the phylum account for about 10% (40 species) of known human fungal pathogens (Morrow and Fraser, 2009). With the onset of the AIDS epidemic, one basidiomycete in particular, Cryptococcus neoformans, has risen from a little-known pathogen to one of the top fungal killers of immunocompromised patients.
C. neoformans is primarily found as a haploid yeast, and is widely present in the environment worldwide, including in avian excreta, soil, and tree bark. Studies have shown that humans come into frequent contact with C. neoformans: individuals with no history of cryptococcosis possess antibodies against the yeast (Chen et al., 1999), and most children appear to have been exposed by the age of five (Goldman et al., 2001). This suggests that the majority of individuals encounter C. neoformans in the environment, most likely through inhalation into the lungs. Immunocompetent individuals are usually able to control and contain the infection, often leading to an asymptomatic latent state of infection. If the patient’s immune system becomes compromised at a later date, the latent infection can reactivate. In the case of the immunocompromised individual, pulmonary infection can lead to pneumonia followed by dissemination via the bloodstream to other organs. C. neoformans is one of only a few fungal species known to cross the blood-brain barrier and infect the brain (Kim, 2006), leading to meningitis that is fatal if left untreated. When the AIDS epidemic began in the 1980s, there was a concomitant surge in cryptococcosis cases worldwide. In recent years, the increased usage of antiretroviral therapy and antifungals has reduced the overall incidences of fatal cryptococcal meningitis. Yet in areas where access to treatment is limited, C. neoformans remains an important concern in the care of the immunocompromised, including AIDS, cancer, and organ transplant patients. In addition, recent outbreaks of cryptococcosis in immunocompetent individuals in the Pacific Northwest raise concerns about the risk of cryptococcal infection even in otherwise healthy individuals (Bartlett et al., 2008; Hoang et al., 2004).
As a haploid yeast cell, C. neoformans is amenable to many of the extensive protocols that have been developed for S. cerevisiae, requiring in most cases only a few adjustments. However, having diverged from the ascomycete lineage some 400 million years ago (mya) (Taylor and Berbee, 2006), there are significant differences in its cellular machinery and life cycle (see below). Comparative genomics promises to yield rich information about the evolution of shared and diverged genes, proteins, and pathways, as well as offering insight into the differences between species that allow one yeast to exist as a benign saprophyte and another to cause lethal infection in a mammalian host.
2. Serotypes, Strains, and Sequences
C. neoformans is classified into four different serotypes based on its reactivity with monoclonal antibodies to surface capsular polysaccharide (Kabasawa et al., 1991). These serotypes have historically been further classified into three different varieties: var. neoformans (serotype D), var. grubii (serotype A), and var. gattii (serotypes B and C). However, in recent years, var. gattii has been proposed to comprise its own species as Cryptococcus gattii, based on morphological and biochemical evidence (Kwon-Chung and Varma, 2006). C. neoformans var. neoformans and var. grubii primarily infect immunocompromised individuals, with var. grubii causing ~99% of cryptococcal infections in HIV-infected patients (Mitchell and Perfect, 1995). C. gattii has the ability to infect immunocompetent individuals, as evidenced by an emergent outbreak in the Pacific Northwest that has resulted in hundreds of human and veterinary infections. Based on analysis of mutation frequency in conserved genes, it is thought that C. neoformans and C. gattii diverged about 37 mya, while C. neoformans var. neoformans and var. grubii split 18.5 mya, and within C. gattii, serotypes B and C diverged 9.6 mya (Xu et al., 2000). To date, the genomes of five strains of C. neoformans and C. gattii have been sequenced to at least 6× coverage: JEC21 (serotype D), B-3501 (serotype D), H99 (serotype A), WM276 (serotype B), and R265 (serotype B). H99 and R265 are clinical isolates, while WM276 was isolated from the environment. JEC21 and B-3501 are laboratory-derived strains, where JEC21 was derived from B-3501 through a series of crosses and backcrosses (Heitman et al., 1999), and their genomes are 99.5% identical (Loftus et al., 2005).
Online resources for C. neoformans genome sequences
The genomes of C. neoformans and C. gattii contain about 19 Mb of DNA spread over 14 chromosomes with about 7000 predicted protein-coding genes. The genomic sequence is relatively GC-rich (48% GC content) when compared to the genome of S. cerevisiae (38% GC content). Nucleic acid enzymatic protocols from S. cerevisiae laboratories that have been adapted for use with C. neoformans take this into account with the addition of DMSO (5% final concentration) or betaine (1.3 M final concentration) to resolve secondary structures resulting from the higher GC content.
Unless otherwise noted, the use of “C. neoformans” in the text of this chapter refers to C. neoformans var. neoformans and var. grubii. Although many of the same techniques are applicable to C. gattii, their usage is less well documented and may require additional adaptations.
3. Life Cycle
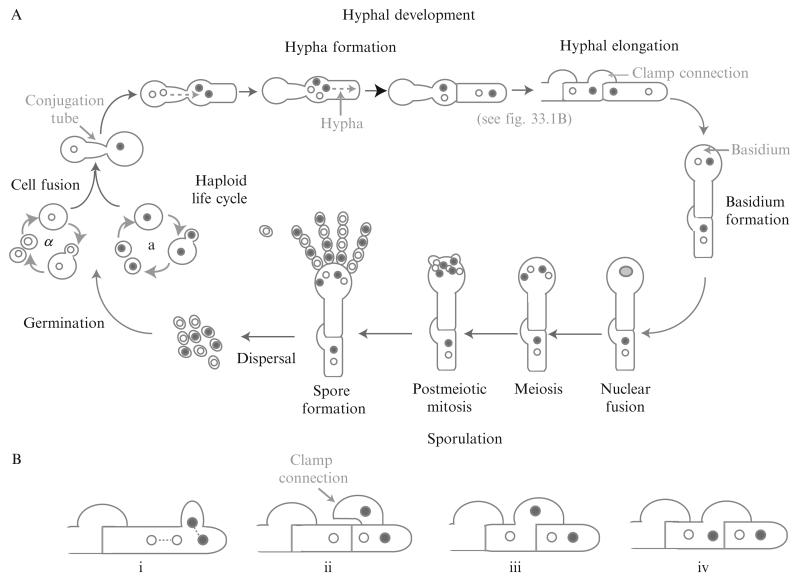
C. neoformans and C. gattii primarily exist as haploid yeast cells that reproduce asexually through budding. They also possess a bipolar mating system, with mating types a and α. The mating (MAT) locus regulates the sexual cycle and encodes for more than 20 genes, including genes for cell type identity and the production and sensing of pheromone. Similar to S. cerevisiae, MATa cells produce MFa pheromone that is sensed by MATα cells. In response to pheromone, MATα cells produce a conjugation tube (Fig. 33.1A). Likewise, MATa cells respond to the MFα pheromone produced by MATα cells, although the response of MATa cells is to form large swollen cells that can then fuse to the conjugation tubes of the MATα cells. The MATa and MATα nuclei divide, and the MATα nuclei travel through the conjugation tube into the MATa cell. MATa and MATα nuclei move into the hypha formed by the MATa cell, and a septum forms between the hypha and the MATa cell. The hypha may then elongate through cell growth and division. During hyphal elongation, the nuclei divide mitotically (Fig. 33.1B). One nucleus divides in such an orientation as to enter into a bulge in the cell wall that will later form a clamp connection (i). Septa form between the posterior cell wall, the tip of the hypha, and the clamp connection, leaving one nucleus in the posterior cell, two nuclei in the tip of the hypha, and one nucleus in the clamp (ii). The clamp fuses back to merge with the posterior cell (iii), allowing the nucleus present in the clamp to join the nucleus in the posterior cell (iv). During sporulation (Fig. 33.1A), a basidium forms at the tip of the hypha. In the basidium, the MATa and MATα nuclei fuse and undergo meiosis. The new MATa and MATα nuclei then undergo repetitive rounds of mitosis, eventually forming four chains of spores that emerge from the basidium. The spores are then dispersed, and germinate into haploid yeast cells (Bovers et al., 2008; McClelland et al., 2004).
Figure 33.1.
Life cycle of C. neoformans.
In the laboratory, mating is achieved through nitrogen starvation on V8 medium, consisting of 5% (v/v) V8 juice, 3 mM KH2PO4, 4% (w/v) agar, pH 5.0. For technical details, several excellent studies have been performed examining mating conditions; we refer you to these (Escandon et al., 2007; Nielsen et al., 2003; Xue et al., 2007).
Haploid fruiting has also been observed, where cells of one mating type become diploid and form hyphae. These monokaryotic hyphae are characterized by unfused clamp connections. Similar to mating, monokaryotic hyphae also form basidia, undergo meiosis, and sporulate (Lin et al., 2005; Tscharke et al., 2003; Wickes and Edman, 1995).
4. Techniques for Basic Culture
C. neoformans is classified as a Biosafety Level 2 (BSL-2) organism, and as such does not require elaborate biohazard safety facilities. Current precaution recommendations include the use of a Class I or Class II biological safety cabinet for manipulation of environmental samples or spore forms. Incidences of infection in laboratory personnel are rare, limited in the literature to skin puncture accidents with needles heavily contaminated with C. neoformans (Casadevall et al., 1994).
C. neoformans may be cultured using similar medium to that used in S. cerevisiae cultivation. Common media used include YPAD and YNB. For some assays (e.g., see Sections 6.1.6 and 6.3.2), Sabouraud dextrose medium is used for culturing for historic reasons and its promotion of yeast growth over bacterial growth.
| Composition | |
|---|---|
| YPAD (yeast peptone adenine dextrose) | |
| 1% bacto yeast extract (Becton Dickinson, Cat. No. 212720) |
10 g |
| 2% bacto peptone (Beckton Dickinson, Cat. No. 211820) |
20 g |
| 2% glucose | 20 g |
| 0.73 mM L-tryptophan (Sigma, Cat. No. T8941) | 0.15 g |
| 0.27 mM adenine (Sigma, Cat. No. A2786) | 0.037 g |
| Water | to 1 l |
| YNB (yeast nitrogen base) | |
| 0.15% YNB w/o amino acids, w/o dextrose, w/o ammonium sulfate (BIO 101, Cat. No. 4027-032) |
3 g |
| 75 mM ammonium sulfate | 10 g |
| 2% glucose | 20 g |
| Water | to 1 l |
| Sabouraud dextrose | |
| 3% Sabouraud dextrose broth (Becton Dickinson, Cat. No. 238210) |
30 g |
| Water | to 1 l |
Standard growth is performed in YPAD medium, typically at 30 °C, with the alternative use of the defined medium YNB. During logarithmic growth in YPAD medium at 30 °C, the doubling time of wild-type C. neoformans is approximately 110 min. Consistent with its role as a human pathogen, C. neoformans also grows robustly at 37 °C. Unlike S. cerevisiae, C. neoformans does not perform fermentation, and therefore requires a minimal amount of oxygen for growth. C. neoformans is sensitive to alkaline pH, growing poorly at pH 9. However, it is insensitive to acidic pH, exhibiting normal doubling times in conditions as low as pH 3.
Frozen stocks of C. neoformans can be maintained in 15% glycerol solution at −80 °C. These stocks may be revived following transfer by sterile applicator stick to a YPAD plate.
4.1. Dominant drug selection markers
Our laboratory and others have used resistance to nourseothricin (NAT), G418, and hygromycin for selection in C. neoformans.
In the plasmids pHL001-STM-# and pJAF1, the genes encoding for proteins conferring resistance to NAT and G418 have been inserted in between the promoter element of C. neoformans ACT1 and the terminator element of C. neoformans TRP1 (both of these sequences were derived from the H99 strain) (Table 33.1). In the plasmid pHYG7-KB1, the gene encoding for resistance to hygromycin was inserted between the promoter element of C. neoformans ACT1 and the untranslated region (UTR) of C. neoformans GAL7 (where the ACT1 sequence was derived from H99 and the GAL7 sequence was derived from JEC21).
Table 33.1.
Dominant drug selection markers
| Drug selection | Plasmid name | Structure | Reference |
|---|---|---|---|
| Nourseothricin | pHL001-STM-# |
CnACT1 promoter-NATR -CnTRP1 term |
Gerik et al. (2005) |
| G418 | pJAF1 |
CnACT1 promoter-NEOR -CnTRP1 term |
Fraser et al. (2003) |
| Hygromycin | pHYG7-KB1 |
CnACT1 promoter-HYGR -CnGAL7 UTR |
Hua et al. (2000) |
For selection of yeast containing the appropriate drug resistance cassette, we use YPAD agar plates made with 0.1 mg/ml nouseothricin (clonNAT, Werner BioAgents), 0.2 mg/ml G418 (VWR, Cat. No. 45000-626), and/or 0.3 mg/ml hygromycin (Sigma, Cat. No. H7772).
5. Basic Molecular Biology Techniques
5.1. Fusion polymerase chain reaction
Manipulation of the genomic sequence of a species is a powerful tool for analyzing the importance of specific genes in the function of the organism. Homologous recombination, or the integration of an exogenous DNA construct into the genome, is a crucial step in the site-directed mutagenesis of a target gene. C. neoformans performs homologous recombination at relatively low frequencies when compared with other fungi (1–4% as compared to nearly 100% in S. cerevisiae) but transformation with linear constructs flanked by a significant amount (0.3–1 kb) of sequence homologous to the genome creates stable integrants reproducibly (Davidson et al., 2000; Nelson et al., 2003). For example, a linear construct to target a gene for deletion might contain an antibiotic resistance cassette flanked on the 5′- and 3′-ends with 1 kb sequences homologous to the 5′- and 3′-ends of the targeted gene.
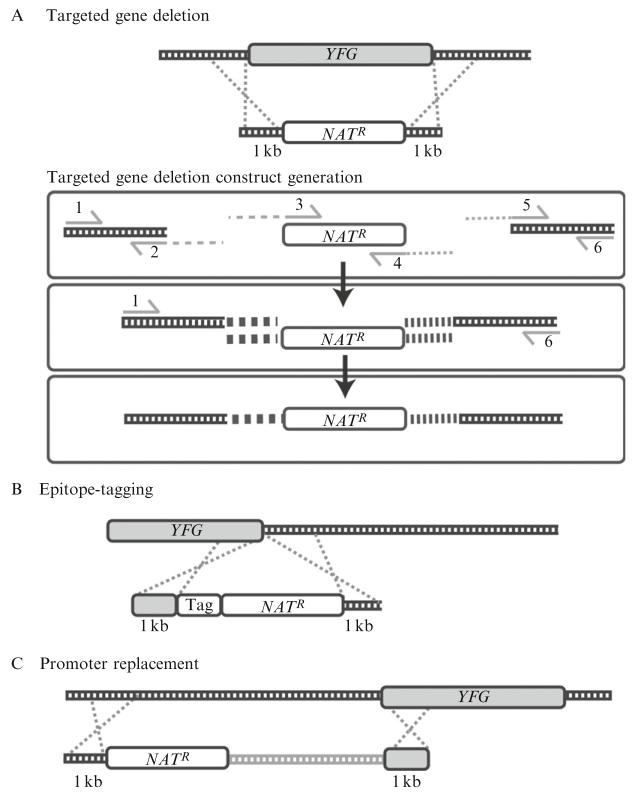
Construction of linear constructs for homologous recombination uses a procedure known as fusion PCR or PCR overlap (Davidson et al., 2002). In this process, two or more DNA fragments are joined together during the polymerase chain reaction (PCR) by virtue of a shared region of homology. This region of homology is engineered during previous PCR steps using primers containing linker sequences that are then shared between the two fragments to be fused together (Fig. 33.2).
Figure 33.2.
(A) Targeted gene deletion: The construct contains an antibiotic resistance cassette flanked on the 5′- and 3′-ends with 1 kb regions of homology upstream and downstream of the targeted gene. (B) Epitope-tagging: The construct contains the epitope tag and an antibiotic resistance cassette flanked on the 5′-end with a 1-kb region of homology to the 3′-end of the targeted gene, and on the 3′-end with a 1-kb region of homology to the region immediately downstream of the targeted gene. (C) Promoter replacement: The construct contains an antibiotic resistance cassette and the desired promoter region flanked on the 5′-end with a 1-kb region homologous to the sequence upstream of the promoter to be replaced, and on the 3′-end with a 1-kb region homologous to the 5′-end of the targeted gene. NATR, nourseothricin resistance cassette; YFG, targeted gene.
5.1.1. Fusion PCR for targeted gene deletion
As mentioned previously, a linear construct for targeted gene deletion is designed to contain an antibiotic resistance cassette flanked by 1 kb sequences homologous to the targeted sequence. To create this construct, an antibiotic resistance cassette, such as resistance to NAT, is first amplified with primers containing 22 bp of homology to the 5′ and 3′ ends of the cassette, and 21 bp of linker sequence which is different for the 5′ and 3′ primers (these primers are designated primers 3 and 4 in Table 33.2 and Fig. 33.2A). Then, from genomic DNA we amplify 1 kb of sequence upstream and downstream of the ORF targeted for deletion. We term these sequences the 5′ and 3′ flanks for the targeted gene deletion construct. For the 5′ flank, the forward primer (1) is 22 bp of exact homology to the genomic sequence. The reverse primer (2) is 21 bp of linker sequence that is antiparallel to the linker sequence in the forward primer (3) for amplifying the antibiotic resistance cassette, followed by 22 bp of homology to the genomic sequence. For the 3′ flank, the forward primer (5) is 21 bp of linker sequence that is antiparallel to the linker sequence in the reverse primer (6) for amplifying the antibiotic resistance cassette, followed by 22 bp of homology to the genomic sequence. The reverse primer is 22 bp of exact homology to the genomic sequence. Table 33.2 contains the linker sequences we use to design these primers.
Table 33.2.
Primers for construction of targeted gene deletion construct by fusion PCR
| Primer | Sequencea,b |
|---|---|
| 1 | Forward primer to 5′ flank: 22 bp of sequence 1 kb upstream of ORF |
| 2 | Reverse primer to 5′ flank: CACGGCGCGCCTAGCAGCGGA-22 bp of sequence immediately upstream of ORF |
| 3 | Forward primer to antibiotic resistance cassette: CCGCTGCTAGGCGCGCCGTGA-22 bp of sequence at 5′ end of antibiotic resistance cassette |
| 4 | Reverse primer to antibiotic resistance cassette: GCAGGGATGCGGCCGCTGACA-22 bp of sequence at 3′ end of antibiotic resistance cassette |
| 5 | Forward primer to 3′ flank: GTCAGCGGCCGCATCCCTGCA-22 bp of sequence immediately downstream of ORF |
| 6 | Reverse primer to 3′ flank: 22 bp of sequence 1 kb downstream of ORF |
The linker sequences are not exactly antiparallel with each other; you will note that all linker sequences in primers 2′–5′ end in an adenine prior to the 22 bp of homologous sequence. Taq polymerase exhibits terminal transferase activity, which adds an additional adenosine onto the 3′ ends of PCR products. Therefore, the extra adenine in the primers allows for perfect homology between the linker sequences during the actual fusion PCR that fuses the three fragments together into the targeted gene deletion construct. The sequences for these primers are listed 5′–3′.
The linker sequences were adapted from previous work Reid et al. (2002).
Primers 1–6 are used first for amplification of the 5′- and 3′-flanks and the antibiotic resistance cassette. Then primers 1 and 6 are used in the fusion PCR to amplify the full-length linear construct.
5.1.1.1. Conditions for amplification of 5′ and 3′ flanks and antibiotic resistance cassette
(50 μl final volume): 400 nM each primer, 0.25 mM dNTPs, 20 mM Tris–HCl (pH 8.8), 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% (v/v) Triton X-100, 0.01% (w/v) BSA (98% electrophoresis grade, Sigma, Cat. No. A7906), 5% (v/v) DMSO, 2.5 U Pfu polymerase, 0.5 μl of template DNA (genomic DNA at 1 μg/μl or plasmid bearing antibiotic resistance cassette at 30 ng/μl).
We maintain at 4 °C a 10× stock of PCR buffer that contains 200 mM Tris–HCl (pH 8.8), 20 mM MgSO4, 100 mM KCl, 100 mM (NH4)2SO4, 1% (v/v) Triton X-100, 0.1% (w/v) BSA. We then add the primers, DMSO, dNTPs, Pfu, and template DNA separately.
PCR conditions, performed on a PTC-200 Peltier Thermal Cycler (MJ Research): 93 °C for 3 min, followed by 35 cycles of (93 °C for 30 s, 45 °C for 30 s, 72 °C for 3.5 min or appropriate amount of time for the length of your antibiotic resistance cassette), followed by 72 °C for 5 min.
Purify the PCR products by running the PCR out on a 0.8% agarose gel. Cut out the appropriate size band in the gel, and purify using a QIAquick Gel Extraction kit (Qiagen, Cat. No. 28704), following the manufacturer’s instructions. In the final step, elute from the column with 30 μl of elution buffer (EB), letting the column stand for 1 min then centrifuging for 1 min at 13,000 rpm.
5.1.1.2. Conditions for fusion PCR
(50 μl final volume): 400 nM each primer (primers 1 and 6), 0.25 mM dNTPs, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1.3 M betaine, 1 U Taq polymerase, 0.25 U Pfu polymerase, 50 nmol each of 5′ flank, 3′ flank, and antibiotic resistance cassette (roughly equal to 2 μl of the eluted volume from the QIAquick Gel Extraction).
We maintain at 4 °C a 10× stock of PCR buffer that contains 100 mM Tris–HCl (pH 8.3), 500 mM KCl, 20 mM MgCl2. We then add the betaine, dNTPs, Pfu, and Taq polymerases separately. Betaine is maintained as a 5 M stock at 4 °C.
PCR conditions, performed on a PTC-200 Peltier Thermal Cycler (MJ Research): 72 °C for 10 min, 92.5 °C for 3.5 min, followed by 35 cycles of (92.5 °C for 12 s, 52 °C for 12 s, 72 °C for 7 min or appropriate amount of time for the length of the full targeted gene deletion construct), followed by 72 °C for 5 min.
Purify the PCR product by running the PCR out on a 0.8% agarose gel. Cut out the appropriate size band in the gel, and purify the DNA in the gel slice using a QIAquick Gel Extraction kit, following the manufacturer’s instructions. In the final step, elute from the column with 50 μl of EB, letting the column stand for 1 min, then centrifuging the column for 1 min at 13,000 rpm. Add another 30 μl of EB to the column, let stand for 1 min, then centrifuge for 1 min at 13,000 rpm.
With modifications, fusion PCR may be utilized for a variety of genetic manipulations. By selecting different sequences for amplification, we have successfully used fusion PCR to introduce epitope tags into the 5′- and 3′-ends of genes (Fig. 33.2B), and to replace the promoters of genes (e.g., for overexpression of genes or placing genes under the control of inducible promoters) (Fig. 33.2C).
5.2. Transformation
The preferred method for transformation into C. neoformans is biolistic delivery. While studies have shown that C. neoformans is transformable by electroporation, these transformations have been low efficiency, resulting in some stable ectopic transformants but also many unstable transformants harboring extrachromosomal DNA material (Edman, 1992; Edman and Kwon-Chung, 1990). In addition, electroporation of different C. neoformans strains has varying degrees of success; strain H99 (serotype A) is much less tractable to transformation in this way than strain B-3501 (serotype D). In contrast, both serotypes A and D are readily transformed with relatively high efficiency by biolistic delivery (Davidson et al., 2000; Toffaletti et al., 1993).
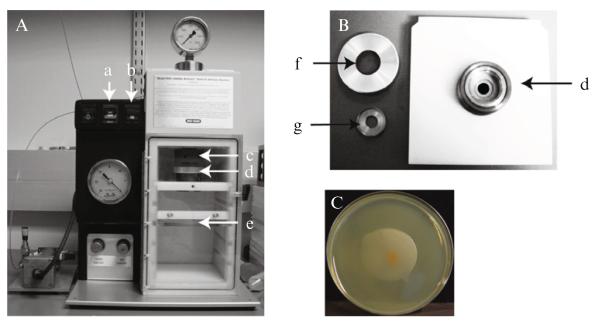
In biolistic delivery, DNA is introduced into the yeast cell using a biolistic particle delivery system (PDS-1000/He, Bio-Rad) hooked up to a vacuum pump (Maxima C Plus M6C, Fisher Scientific) and compressed helium tank (Fig. 33.3). The targeted gene deletion constructs generated by fusion PCR (see above) are deposited onto gold bead microcarriers that are then positioned on a macrocarrier disk in the main chamber of the biolistic PDS. The air in the biolistic PDS is removed by a vacuum pump, and helium is pumped into a small chamber (termed the gas acceleration tube) positioned above the macrocarrier disk, separated from the main chamber by a pressure-calibrated rupture disk. At a high enough pressure, the rupture disk breaks and the helium blasts into the main chamber of the biolistic PDS, propelling the macrocarrier disk downward against a metal stopping screen. The force of the impact against the stopping screen propels the DNA-coated microcarriers off the macrocarrier disk at high velocity and into C. neoformans cells that have been plated onto an agar plate and positioned below.
Figure 33.3.
(A) Example PDS-1000/He biolistic particle delivery system (Bio-Rad): (a) “VENT/HOLD/VAC” toggle switch, (b) “FIRE” toggle switch, (c) gas acceleration tube/retaining cap, (d) microcarrier launch assembly, and (e) plate holder. (B) Close-up of microcarrier launch assembly for biolistic particle delivery system: (d) microcarrier launch assembly, (f) Top to microcarrier launch assembly, (g) Macrocarrier holder. (C) Example of a YPAD plate immediately following biolistic transformation. Note the scattering of microcarriers in the center of the patch of C. neoformans cells.
5.2.1 Protocol for biolistic transformation
Preparation of constructs (may be done anytime prior to the day of transformation)
Transfer 30 μl of the purified construct from fusion PCR (see above) into a microcentrifuge tube or into one well of a 96-well skirted PCR plate (Fisher, Cat. No. 055068).
Concentrate the DNA to the bottom of the tube by removing all moisture by SpeedVac.
Add 2.5 μl water, pipetting up and down and around the walls of the tube/well multiple times to resuspend the DNA.
Add 12.5 μl of microcarriers (0.6 μm gold beads, Bio-Rad, Cat. No. 1652262, resuspended in water to 60 mg/ml and maintained at 4 °C).
Add 12.5 μl of 2.5 M CaCl2 (maintained at 4 °C). Pipette up and down to mix.
Add 5 μl of 0.1 M spermidine (1 M stocks are maintained at −80 °C and diluted to 0.1 M in water prior to use).
Mix on a vortexer at low speed for 4 min. We use a Vorex Genie 2 (Fisher, Cat. No.12-812) with a platform attachment, set to Vortex level 1. If using a skirted PCR plate, cover with a plastic plate seal (Qiagen, Cat. No. 1018104).
Collect the microcarriers by centrifugation at 500 rpm for 10 s.
Remove the supernatant by pipette.
Wash microcarriers by adding 50 μl 70% ethanol and immediately removing by pipette, being careful to not disturb the microcarrier pellet.
Add 50 μl 100% ethanol and immediately remove by pipette.
Resuspend the DNA-coated microcarriers in 12.5 μl 100% ethanol by pipetting up and down.
Transfer all 12.5 μl of microcarriers onto the center of a macrocarrier disk (Bio-Rad, Cat. No. 1652335) deposited in a 6-well culture dish (Falcon, Cat. No. 35-3224).
Dry the disk until all ethanol has evaporated, leaving a dark gold residue on the surface of the macrocarrier. We dry the disk by placing the 6-well culture dish in a desiccator hooked up to the house vacuum.
Day one
Inoculate C. neoformans from plate stock into 50 ml liquid YPAD medium in a 250 ml flask. Grow with aeration at 30 °C for 2-3 days.
Day three
Collect C. neoformans culture by centrifugation at 3000 rpm for 10 min.
Resuspend cell pellet in 5 ml regeneration medium (see recipe below).
Pipette 140 μl of resuspended cells onto the center of a YPAD plate, one plate for each transformation to be performed. Use a spreader device (Marsh Brand, Cat. No. KG-5P) to spread the cells into a circular patch, 4–5 cm in diameter.
Let the plates dry with lids ajar at 30 °C for 20–30 min.
- Perform transformation with biolistic PDS.
- Open valve of compressed helium tank.
- Turn on vacuum pump and biolistic delivery system.
- Unscrew retaining cap at the end of the gas acceleration tube.
- Dip rupture disk briefly (2–3 s) in 70% isopropanol, to sterilize the disk and aid in its retention in the retaining cap following rupture.
- Place the rupture disk in the retaining cap of the biolistic delivery system, screw retaining cap back into place.
- Press microcarrier-coated macrocarrier (microcarrier-side up) in the macrocarrier holder.
- Place stopping screen at bottom of the microcarrier launch assembly of the biolistic delivery system.
- Place the macrocarrier holder into the microcarrier launch assembly, above the stopping screen (microcarrier-side down). Screw top on microcarrier launch assembly.
- Load microcarrier launch assembly in the first slot from the top in the main chamber of the biolistic delivery system.
- Load plate holder in the third slot from the top in the main chamber of the biolistic delivery system.
- Place a YPAD plate with patch of C. neoformans cells from step 3 on the plate holder. Remove its lid.
- Close the door of the biolistic delivery system, flipping the switch of the biolistic delivery system to “VAC” to start drawing air out of the chamber.
- When the pressure gauge reads more than 27 in. Hg vacuum, flip the switch of the biolistic delivery system to “HOLD.”
- Hold down the “FIRE” button until you hear the rupture disk break—it will sound like a loud pop—and the helium pressure drops down to zero.
- Release the “FIRE” button and flip the switch of the biolistic delivery system to “VENT” to release the vacuum from the chamber.
Incubate transformed plates at 30 °C for 4 h to allow for recovery.
Resuspend cells in 800 μl phosphate-buffered saline (PBS) using a spreader device, then transfer by pipette to a plate containing selective medium. Spread the cells over the surface of the plate using a spreader device.
Dry the plates at 30 °C with the lids ajar for 30 min or until dry.
Cover plates and incubate 2–3 days. Colonies should be visible by the end of the next day and of a pickable size (~0.5 mM diameter) by the second day. These colonies should be picked and patched out onto selective medium plates for confirmation of their genotype by PCR. We typically patch out the colonies for a verification of the 5′-junction. The colonies that show successful integration of the construct at the 5′-junction are streaked out to single colonies on selective medium, and new colonies are patched out for verification of the 3′-junction.
Notes
For concentrating the transformation construct, if using microcentrifuge tubes we run them for 30–45 min in a Savant SC100 SpeedVac on high drying rate. If using a skirted PCR plate, we use a Savant AES2010 SpeedVac outfitted with plate holders, set to run for 6 h with 45 min of radiant cover heating on high drying rate.
Sterilize the macrocarriers and stopping screens prior to use by washing in 70% ethanol and drying in a sterile environment. We find a 15-cm Petri dish to work well for this purpose.
We use rupture disks rated between 1100 and 1350 psi (Bio-Rad, Cat. Nos. 1652329 and 1652330). Both have given good transformation results.
Following successful transformation, it is usually possible to see a spattering of gold beads embedded into the YPAD plate (Fig. 33.3C). If this is not visible, it is likely that not enough gold beads were used in the macrocarrier setup.
We find it best to pick the colonies by the end of the second day, because one obtains a higher rate at that time of successful transformants that test positive for the integration of the drug selection cassette at the targeted locus. Waiting until the third day or later allows for false positive colonies to catch up in size with true positives. In general, we find it best to pick the largest colonies on the plate, although for disruptions in genes that positively regulate cell growth, these knockouts can be slower growing than some false positives. We typically pick and patch out 6-8 colonies per transformation, although more may be picked for transformations with a lower success rate.
We have observed varying transformation success rates among strains that are theoretically genetically identical (i.e., H99 strains from different laboratory sources). We hypothesize that in the process of passaging these strains, mutations have been acquired that affect homologous recombination efficiency.
| Regeneration medium | Composition |
|---|---|
| 0.9% YNB w/o ammonium sulfate w/o dextrose w/o ammonium sulfate (BIO 101, Cat. No. 4027-032) |
9 g |
| 1 M sorbitol | 182 g |
| 1 M mannitol | 182 g |
| 2.6% glucose | 26 g |
| 0.267% bacto yeast extract (Becton Dickinson, Cat. No. 212720) |
0.27 g |
| 0.054% bacto peptone (Beckton Dickinson, Cat. No. 211820) |
0.54 g |
| 0.133% Gelatin (Sigma, Cat. No. G-8150) | 1.33 g |
| Water | to 1 l |
5.3. Colony PCR
The genotype of the transformed strain is verified through PCR-based detection of the expected 5′- and 3′-junctions of the resistance marker with the genomic DNA. While verifying the genotypes of many transformations, it is easiest and fastest to perform colony PCR.
Patch out colonies from the transformation into 48- or 96-well grid format on plates containing selective medium.
row at 30 °C for 2 days.
Using a 48- or 96-well pin replicator, transfer a generous amount of cells into 7 μl of water in each well of a PCR plate.
Seal the PCR plate with PCR plate thermal adhesive sealing film.
Flash freeze the PCR plate in liquid nitrogen, then immediately transfer to a PCR block set at 100 °C. Incubate for 2–5 min.
Perform PCR as below.
(50 μl final volume): 400 nM each primer, 0.25 mM dNTPs, 20 mM Tris–HCl (pH 8.8), 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% (v/v) Triton X-100, 0.01% (w/v) BSA (98% electrophoresis grade, Sigma, Cat. No. A7906), 5% (v/v) DMSO, 0.15 U Pfu polymerase, 0.5 U Taq polymerase.
We maintain at 4 °C a 10× stock of PCR buffer that contains 200 mM Tris–HCl (pH 8.8), 20 mM MgSO4, 100 mM KCl, 100 mM (NH4)2SO4, 1% (v/v) Triton X-100, 0.1% (w/v) BSA. We then add the primers, DMSO, dNTPs, Pfu, and Taq polymerases separately.
PCR conditions, performed on a PTC-200 Peltier Thermal Cycler (MJ Research): 92.5 °C for 3 min, followed by 35 cycles of 92.5 °C for 15 s, 45 °C for 15 s, 72 °C for 1 min 45 s or appropriate amount of time for the length of targeted amplicon), followed by 72 °C for 5 min.
Notes
For adequate DNA recovery, there should be a visible amount of cells in the 7 μl of water in the PCR plate prior to flash freezing.
We use a fixed solid pin replicator (V&P Scientific, Cat. No. VP 408H) both to mark the selection medium plates on which colonies are patched and for the transfer of cells into a PCR plate. We use thin well PCR plates from RPI (Research Products International, Cat. No. 141314) and TempPlate Sealing Film (USA Scientific, Cat. No. 2921-000) for the PCR.
Colony PCR may also be performed in single tube reactions, using a sterile toothpick in this case to transfer an appropriate number of cells into 7 μl water in a PCR tube.
To verify successful gene deletion, we use primers designed to amplify DNA sequences of approximately 1 kb. The verification primers target the sequences outside of the region amplified as 5′- and 3′-flanks for the targeted gene deletion construct, and are paired with common primers internal to the gene encoding for the drug resistance.
As an additional test for successful gene replacement, it is often useful to perform a PCR to the ORF of the targeted gene to confirm its absence in the transformed strain.
5.4. Genomic DNA extraction
Inoculate 50 ml of YPAD with a C. neoformans. Culture at 30 °C until saturation (1–2 days).
Harvest cells by centrifugation in a 50-ml Falcon tube at 3000 rpm for 10 min.
Remove supernatant, add 30 ml water.
Vortex to mix, then harvest cells by centrifugation at 3000 rpm for 10 min.
Remove supernatant and flash freeze cell pellet in liquid nitrogen.
Transfer the conical tube containing the cell pellet into a lyophilizer vessel.
Attach to a lyophilizer (FreeZone 4.5 Liter Benchtop Freeze Dry System, Labconco) connected to a vacuum pump (Maxima C Plus M6C, Fisher Scientific).
Lyophilize cell pellet overnight, or until all the liquid has sublimated and a dry powdery pellet is left.
Add 3–5 ml of 3 mM glass beads and vortex vigorously until a fine powder is created.
Add 10 ml CTAB extraction buffer (100 mM Tris (pH 7.5), 0.7 M NaCl, 10 mM EDTA, 1% (w/v) CTAB (hexadecyltrimethylammonium bromide, Sigma, Cat. No. H6269), 1% (v/v) beta-mercaptoethanol) and mix.
Incubate at least 30 min at 65 °C.
Add an equal volume of chloroform and mix gently.
Pellet cell debris to the interphase by centrifugation at 3000 rpm for 10 min.
Transfer aqueous phase to a fresh tube.
Add an equal volume of isopropanol and mix gently.
Pellet DNA by centrifugation at 3000 rpm for 10 min.
Wash DNA pellet with 70% ethanol.
Aspirate out supernatant. Invert tube and allow pellet to dry overnight.
Resuspend DNA in 500 μl TE (100 mM Tris (pH 7.5), 1 mM EDTA) and transfer to a 1.5-ml microcentrifuge tube.
Add 1 μl RNase (1 mg/ml stock solution), and incubate at least 30 min at 37 °C.
Add 5 μl proteinase K (20 mg/ml stock solution), and incubate 2 h at 55 °C.
Add 500 μl equilibrated phenol (Sigma, Cat. No. P4557). Separate phases by centrifugation at 14,000 rpm for 10 min.
Transfer aqueous phase to a new 1.5-ml microcentrifuge tube. Add 500 μl chloroform. Vortex briefly to mix. Separate phases by centrifugation at 14,000 rpm for 10 min.
Transfer aqueous phase to a new 1.5-ml microcentrifuge tube. Add 1/10 volume 3 M NaOAc and 2–3 volumes 100% EtOH. Briefly vortex or flick to mix. Incubate at −20 °C for 2 h or −80 °C for 0.5–1 h.
Centrifuge at 14,000 rpm for 10 min. Aspirate out the supernatant.
Dry the pellet in a SpeedVac concentrator for 2 min.
Resuspend DNA in 500 μl TE.
Note
Lyophilization greatly enhances recovery of nucleic acid from C. neoformans. Dessication may weaken the structure of the polysaccharide capsule and cell wall, allowing greater disruption in later steps.
5.5. RNA extraction
Culture C. neoformans cells in the conditions desired for harvesting RNA.
Harvest cultures by centrifugation.
Remove medium and flash freeze cell pellet in liquid nitrogen.
Lyophilize cell pellet until dry.
Resuspend cell pellet in 1 ml TRIzol Reagent (Invitrogen, Cat. No. 15596018) and transfer to a 2-ml screw-cap microcentrifuge tube (Sarstedt, Cat. No. 72.693.005) containing ~200 μl volume of 0.5 mM zirconia/silica beads (Bio-Spec Products, Cat. No. 11079105z).
Bead-beat at least twice for 2.5-min intervals in a Mini-BeadBeater-8 (BioSpec Products).
Centrifuge samples at 12,000×g for 10 min at 4 °C.
Transfer cleared lysate to a 1.5-ml microcentrifuge tube and add 200 μl chloroform.
Vortex for 15 s to mix.
Centrifuge samples at 12,000×g for 10 min at 4 °C.
Transfer the aqueous phase to a new 1.5-ml microcentrifuge tube and add 500 μl isopropanol.
Briefly vortex and allow to sit at 4 °C for at least 15 min.
Centrifuge samples at 12,000×g for 10 min at 4 °C.
Remove supernatant and wash pellets with 1 ml 75% ethanol (prepared with RNase-free water).
Vortex to mix and centrifuge sample 10,000×g for 5 min at 4 °C.
Remove supernatant and dry pellet by spinning in a SpeedVac concentrator (Savant, Model SC100) for 2 min.
Resuspend pellet in 100 μl RNase-free water if performing DNase treatment, or 500 μl if not DNase-treating the sample.
- Optional: You may DNase-treat the RNA at this step to remove contaminating DNA.
- Add 10 μl of 10× DNase buffer (0.1 M Tris (pH 7.5), 25 mM MgCl2, 5 mM CaCl2, made with RNase-free water and stored at −20 °C), and 5 μl (50 U) of DNase I (Roche, Cat. No. 047 716 728 001)
- Incubate at 37 °C for 30 min.
- Incubate at 75 °C for 5 min to heat-inactivate the DNase I.
- Add 400 μl RNase-free water.
- Add 500 μl acid-equilibrated phenol:chloroform (Sigma, Cat. No. P1944). Vortex 15 s to mix.
- Let stand at room temperature until phases have separated (~10 min), then spin at 14,000 rpm for 10 min at 4 °C.
- Transfer aqueous phase to a new microcentrifuge tube.
- Add 500 μl chloroform. Vortex 1 min to mix.
- Spin at 14,000 rpm for 10 min at 4 °C.
- Transfer aqueous phase to a new tube.
Add 500 μl chloroform to the samples.
Vortex for 1 min.
Centrifuge samples at 12,000×g for 10 min at 4 °C.
Transfer the aqueous phase to a new microcentrifuge tube and add 15 μl 3 M NaOAc and 900 μl isopropanol. Briefly vortex and allow to sit at −20 °C for at least 15 min.
Centrifuge samples at 12,000×g for 10 min at 4 °C.
Remove supernatant and wash pellets with 1 ml 75% ethanol (prepared with RNase-free water).
Vortex to mix and centrifuge sample 10,000×g for 5 min at 4 °C.
Remove supernatant and dry pellet in a SpeedVac concentrator for 2 min.
Resuspend pellet in 100 μl RNase-free water.
Notes
Depending on the growth conditions being assayed, it may be necessary to increase the number and length of intervals in the Mini-BeadBeater-8. For example, conditions of increased capsule synthesis require upward of five 10-min intervals. Experimentation may be required in order to determine optimal durations for your growth conditions.
DNase treatment is optional but highly recommended, especially if the RNA will later be reverse-transcribed for use in quantitative PCR (qPCR). This step appears to be less critical for microarray analysis of transcript level, but is nonetheless recommended.
The second round of chloroform extractions (step 19 onward) has in our hands led to cleaner RNA extractions that offer greater yields of cDNA following reverse transcription.
5.6. Protein extraction for SDS–PAGE
Harvest cells in mid-logarithmic growth phase corresponding to OD600 = 2 (e.g., if cells are at OD = 0.5, harvest 4 ml) by centrifugation.
Remove medium and resuspend cells in 500 μl ice-cold H2O.
Transfer to 2 ml screw-cap microcentrifuge tube (Sarstedt, Cat. No. 72.693.005).
Centrifuge samples at 12,000×g for 5–10 min.
Remove supernatant and flash freeze cell pellet in liquid nitrogen.
Lyophilize until pellet is dry.
Resuspend pellet in 1 ml ice-cold water.
Add 150 μl NaOH/beta-mercaptoethanol mixture (1.85 N NaOH, 7.5% (v/v) beta-mercaptoethanol) to each sample.
Incubate on ice with occasional vortexing for 30 min.
Add 150 μl trichloracetic acid (TCA, 55% (w/v) in water kept at 4 °C in a foil-wrapped bottle) to each sample.
Incubate on ice with occasional vortexing for 30 min.
Centrifuge samples at 12,000×g for 10–20 min at 4 °C.
Remove most of the supernatant.
Optional (when harvesting >1 OD of cells): Add 100 μl ice-cold acetone to optimize removal of residual TCA.
Centrifuge samples at 12,000×g for 1 min at 4 °C.
Remove the remaining supernatant.
17. Resuspend pellet in 50 μl HU buffer (200 mM sodium phosphate buffer (pH 6.8), 8 M urea, 5% (w/v) SDS, 1 mM EDTA, bromophenol blue. Store at −20 °C and add 100 mM DTT immediately before use).
Notes
To load samples in HU buffer, care should be taken not to boil them. Instead, the samples should be heat-denatured at 65–70 °C for 10–15 min or at 37 °C for 30 min.
If HU buffer in the resuspended protein pellet turns yellow due to residual TCA, add 10–20 μl of 1 M Tris (pH 6.8).
6. Methods for Assaying Pathogenesis
6.1. Murine model of infection
Mice are relatively susceptible to C. neoformans infection, when compared with other mammalian hosts such as rats and rabbits. Immunocompetent murine strains will succumb to pulmonary infection, and will experience dissemination to other organs including the brain, similar to human cryptococcosis. A murine model of cryptococcal infection offers several advantages over other species, including the consistency of susceptibility within a given strain, the availability of genetically modified strains (useful for examining host factors that may be involved in infection), as well as their small size and low cost. Our laboratory utilizes two routes of infection for introducing C. neoformans into a murine model of infection: intranasal and intravenous.
6.1.1. Considerations of murine strain and age
Inbred mouse strains may vary in their susceptibility to C. neoformans infection. For example, in some studies, BALB/c mice have been demonstrated to be more resistant to C. neoformans infection than C57BL/6 mice, as evidenced by both fungal load in the lungs following infection (Chen et al., 2008; Huffnagle et al., 1998) and degree of dissemination to other organs (Chen et al., 2008), although in some survival curve analyses, C57BL/6 mice survive slightly longer than BALB/c mice following infection with C. neoformans (Nielsen et al., 2005). There are varying theories as to the source of the differences in susceptibility in mouse strains to C. neoformans: studies have linked relative resistance of a mouse strain to the production of Th1-type cytokines, where their production is associated with pulmonary clearance (Huffnagle et al., 1998), or the presence of complement protein C5 (Rhodes et al., 1980). Our laboratory performs murine infections with 5- to 6-week-old A/J mice, which are C5-deficient and therefore slightly more susceptible to C. neoformans infection than C5-sufficient mouse strains (e.g., BALB/c and C57BL/6 mice) (Nielsen et al., 2005; Wormley et al., 2007). It bears noting that the inocula listed below have been determined by our laboratory for use with our strain of H99 C. neoformans in A/J mice. Use of other mouse strains may necessitate adjustment of the inocula to a higher or lower dosage of C. neoformans cells. Additionally, derivatives of the H99 strain (i.e., H99 stocks maintained by different laboratories) appear to have varying levels of virulence.
Studies have also shown that the age of the mice may affect their relative susceptibility to infection. Older C57BL/6 mice (e.g., 17-week-old) are better able to clear an intratracheal infection from their lungs, brains, and spleens than younger (e.g., 5-week-old) mice (Blackstock and Murphy, 2004). We have found it best to infect mice of a consistent age to reduce variability in our data.
6.1.2. Intranasal infection
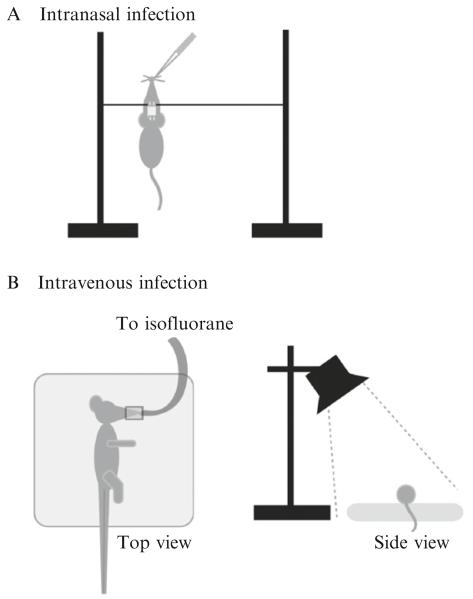
An intranasal infection is thought to more closely mimic the natural course of infection, beginning with the inhalation of C. neoformans cells leading to pulmonary disease followed by dissemination to other organs. The mice are first anesthetized with a mixture of ketamine hydrochloride (Orion Pharma Animal Health) and medetomidine hydrochloride (Domitor®, Orion Pharma Animal Health) via intraperitoneal injection. For all murine injections, we use ½ cc insulin syringes with 28G½ needles (Becton Dickinson, 329461). The anesthetic is mixed to contain 18.75 mg/ml ketamine hydrochloride and 0.625 mg/ml medetomidine hydrochloride. We administer 30–50 μl of this formulation to each mouse (10–15 g), leading to doses of 50–60 mg/kg ketamine hydrochloride and 1.5–2.0 mg/kg medetomidine hydrochloride. The mice usually succumb to the anesthesia after 5–10 min, at which point they are weighed, their ears notched for later identification, and ointment (Artifical Tears, Webster Veterinary, Cat. No. 07-841-4071) applied to their eyes to prevent them from drying out. A silk thread (50 Denier Weight, obtainable from a sewing supply store) is strung between two supports—we use ring stands for this purpose. The mice are then suspended by their incisors upon this silk thread (Fig. 33.4A). The inoculumof C. neoformans cells (5×105 cells/mouse in 50 μl) is slowly pipetted directly into one nare using a pipette fitted with a filter tip. Take care to allow for complete dispension of the inoculum into the nare; if signs of struggling are seen in the mouse, pipetting should be suspended until the mouse no longer shows signs of discomfort. We typically anesthetize and inoculate batches of five mice at a time. Following completion of inoculation, the mice remain suspended for 10 min, to allow for complete aspiration into the lungs, before being lowered and the anesthesia reversed via intraperitoneal injection of atipamezole hydrochloride (Antisedan®, Orion Pharma Animal Health). We administer 40–50 μl of 1 mg/ml atipamezole hyrochloride per mouse, leading to a dose of 2.5–3.5 mg/kg. It typically takes 10–15 min following injection with atipamezole hydrochloride to see signs of stirring in the mice, and another 15–20 min before the mice begin to walk around again.
Figure 33.4.
(A) Intranasal infection. A silk thread is tied across two supports (such as ring stands). The anesthetized mouse is suspended from its front incisors on the thread. The inoculum of yeast cells is pipetted down one nare. (B) Intravenous infection. The mouse is anesthetized with isofluorane administered by face mask (top view), while the tail vein is dilated through a combination of a sodium acetate heating pad from below and a heating lamp from above (side view). When the mouse is laid on its side, the lateral tail vein of the mouse will be at the top of the tail (top view).
6.1.3. Inocculum preparation
Inocula are prepared by growing C. neoformans in liquid YPAD overnight at 30 °C. Cells are counted by hemocytometer and, for an intranasal infection, 1×107 cells are washed twice with PBS and resuspended in 1 ml of PBS. Fifty microliters of this inoculum are used per mouse (5×105 cells). For an intravenous infection, 2×107 cells are washed in PBS and resuspended in 1 ml of PBS. One hundred microliters of this inoculum is used per mouse (2×106 cells). Inocula concentrations are confirmed by plating appropriate dilutions onto YPAD plates and counting the colony forming units (CFU) after 2 days growth at 30 °C.
6.1.4. Intravenous infection
An intravenous infection, via the lateral tail vein, leads to more uniform dissemination to the organs. The mice are weighed prior to infection and marked by ear notching for later identification. They are anesthetized via inhalation of 3% isofluorane in oxygen, administered by face mask, then remain on a sodium acetate rechargeable heating pad (Heat Solution, Prism Enterprises) beneath a heating lamp during the procedure (see Fig. 33.4B) in order to dilate the vein so that it is more visible for easier injection. The inoculum (2×106 cells in 100 μl PBS) is injected into the lateral tail vein. Following successful inoculation, the mice are immediately removed to their cage where they will rapidly recover from the anesthesia.
6.1.5. Monitoring disease progression
Mice are weighed prior to infection, and then monitored every 2–3 days postinfection. Signs of disease progression include hunched posture, abnormal gait, weight loss, and decreased grooming as indicated by ruffled fur. Our laboratory uses two endpoints for assessing time of survival: the point at which the mouse has lost 15% of its initial weight, or 25% of its peak weight. We find the latter to be more consistent when the mice were infected at a younger age (e.g., close to 4 weeks in age) and are hence smaller at the initial time point.
6.1.6. Murine infection evaluations
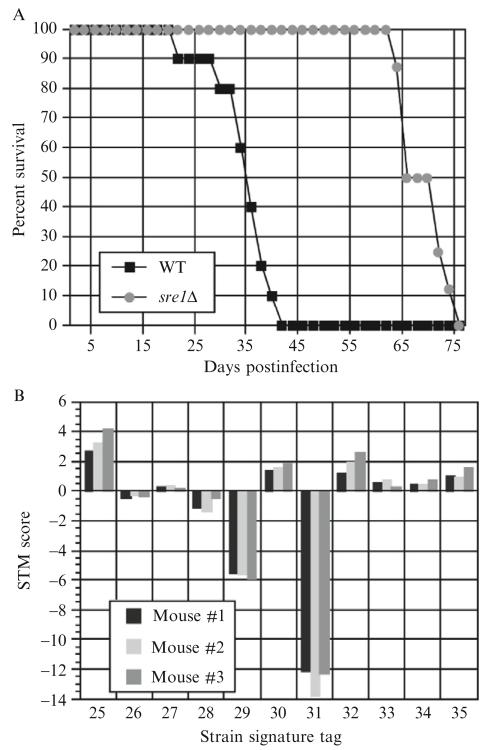
“Time-to-endpoint” survival curve analysis monitors the infection of 8–10 mice with a single strain of C. neoformans, until their endpoints (as defined above). In this manner, mice infected with less virulent strains of C. neoformans survive longer than mice infected with more virulent strains (Fig. 33.5A).
Figure 33.5.
(A) Example of a survival curve. Mice were inoculated via tail-vein injection with 2×105 cells/mouse of either WT (H99) or sre1Δ strains of C. neoformans. On average, mice infected with sre1Δ survived 30 days longer, indicating sre1Δ that is a hypovirulent strain. (B) Example of STM score data. Forty-eight signature-tagged strains were grown individually in liquid YPAD medium in a 96-well deep pocket plate, then pooled together to generate the inoculum. Three mice were inoculated with 5×105 cells/mouse via intranasal infection. The mice were monitored to the disease endpoint, at which point they were sacrificed. Shown are a subset of the data from the lungs, following qPCR and calculation of the STM score for each signature tag in each mouse.
This analysis gives a gross determination of the virulence of a single strain on the entire host system. A more specific analysis might address questions such as the initial rate of colonization to a specific organ, the rate of proliferation and/or rate of killing by the host immune cells, or the rate of dissemination to other organs. This additional analysis may be performed by assessing fungal load in the organs at various time points following infection. We typically examine fungal loads in the lungs, brain and spleen, although we have also examined the liver and kidneys. To measure organ loads after the animal is euthanized, the selected organs are removed by dissection, and placed on ice in 17×100 mM polypropylene sterile tubes (Evergreen Scientific, Cat. No. 222-2393-080), one tube per organ per mouse. Take care to wash the dissecting tools in water and ethanol between organs to eliminate carryover of yeast from organ to organ. Each organ is homogenized in 5 ml sterile PBS (we use a PRO200 tissue homogenizer, PRO Scientific, Oxford, CT), then serial dilutions in PBS are plated on Sabouraud dextrose agar plates (made with Sabouraud dextrose agar, Becton Dickinson, Cat. No. 211661) containing 40 μg/ml gentamycin and 50 μg/ml carbenicillin to discourage bacterial growth. CFU are assessed, and comparisons can be made for a single strain in different organs, rate of growth in different organs over time, or between multiple strains for relative fitness.
6.1.7. Signature-tagged mutagenesis screening
Evaluation of infectivity and virulence for a large number of C. neoformans strains through single-strain infections as described above can quickly add up in terms of both time and cost, as many mice must be used for each strain. Pooling mutant strains into a single infection allows rapid assessment of multiple strains in a single mouse. This can be easily and effectively performed using a technique known as signature-tagged mutagenesis (STM) screening. Each mutant contains a signature tag, or a unique sequence similar to a barcode, in its DNA. When pooled together in a group, individual strains can still be identified through qPCR of pooled genomic DNA using signature-tag-specific primers. By identifying relative representation in the pool of genomic DNA before and after infection, relative rates of infectivity can be assessed rapidly and reproducibly for multiple mutants in a single infection. Using 48 unique signature tag sequences, this technique has been employed by our laboratory for the production and quantitative analysis of a library containing ~1200 targeted gene deletion strains (available without restriction from the Fungal Genetic Stock Center or the American Type Culture Collection (ATCC)) (Liu et al., 2008).
In detail, to analyze a group of 48 signature-tagged strains, the group is first grown up in liquid YPAD in 96-well deep-pocket plates (Grenier Bio-One, Cat. No 780270), one strain per well, at 30 °C without shaking for 3 days. Two hundred microliters of each culture is pooled together, and the number of cells assessed by hemocytometer. 2×107 cells (for tail vein injection) or 1×107 cells (for intravenous infection) are washed twice in sterile PBS and resuspended in 1 ml sterile PBS. This pool is used as the inoculum to infect three mice, either by intranasal infection (5×105 cells/mouse) or tail vein injection (2×106 cells/mouse). Fifty microliters (5×105 cells) of this pool is also plated in triplicate on Sabouraud dextrose agar plates containing 40 μg/ml gentamycin and 50 μg/ml carbenicillin, which are then incubated at 30 °C for 2 days. The resulting colonies are scraped off each plate, resuspended in water, flash frozen in liquid nitrogen and then lyophilized. Genomic DNA is prepared from these samples as described above. This DNA constitutes the “input DNA” for later analysis.
After monitoring and sacrifice of the animals, the organs of interest are removed and homogenized in 5 ml sterile PBS. Serial dilutions in triplicate are made in sterile PBS and plated on Sabouraud dextrose agar plates containing 40 μg/ml gentamycin and 50 μg/ml carbenicillin. These plates are incubated at 30 °C for 2 days. The resulting colonies are scraped off each plate, resuspended in water, flash frozen in liquid nitrogen, and then lyophilized. The genomic DNA that is prepared from these samples constitutes the “output DNA” of the experiment.
The input and output DNA are analyzed using qPCR using a common primer targeted to the drug resistance marker that has replaced the targeted gene, coupled with signature tag-specific primers.
6.1.7.1. STM qPCR conditions
(50 μl final volume): 400 nM each primer, 0.25 mM dNTPs, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 1.3 M betaine, 1 U Taq polymerase, 0.25 U Pfu polymerase, 1–4 μg genomic DNA, 2 μl 2× Sybr Green I (Molecular Probes, Cat. No. S-7563).
We maintain at 4 °C a 10× stock of PCR buffer that contains 100 mM Tris–HCl (pH 8.3), 500 mM KCl, 20 mM MgCl2. We then add the betaine, dNTPs, Sybr Green I, Pfu, and Taq polymerases separately. Betaine is maintained as a 5 M stock at 4 °C. Sybr Green I is kept at −20 °C as a 100× stock in DMSO, and diluted 1:50 in TE buffer to 2× stock immediately prior to addition to the PCR mix.
PCR conditions, performed on a DNA Engine Opticon (MJ Research): 93 °C for 4 min, followed by 40 cycles of (93 °C for 45 s, 52 °C for 25 s, 72 °C for 1 min, then a plate read by the machine), followed by 72 °C for 5 min.
6.1.7.2. Calculating STM score
The threshold cycle (CT), or cycle number where the amplified target reaches a fixed threshold, of each primer pair is used to calculate an STM score, using a variation of the 2−ΔΔCT method for quantitation analysis (Livak and Schmittgen, 2001). For each signature tag, a ΔCT is calculated by subtracting the CT of the specific primer pair from the median CT for all 48 pooled strains to (ΔCT = CT-median − CT-tag). The ΔCT values for each of the three independent input DNA samples are averaged to calculate the ΔCT-input value. The ΔCT values for each of the three independent output DNA samples is similarly calculated by subtracting the median CT for all 48 pooled strains to the CT of the specific primer pair. However, ΔCT values (ΔCT-output) for the three independent output DNA samples are not averaged. The value ΔΔCT is then calculated, where (ΔΔCT = ΔCT-output − ΔCT-input). The STM score is then equal to ΔΔCT. The STM scores from the three mice (i.e., each of the three output DNA samples) are then averaged to determine a final STM score for each mutant. Strains with reduced levels of persistence in the organ have STM scores less than 0, while strains with increased levels of persistence have STM scores greater than 0 (Fig. 33.5B). The STM score correlates with the relative fold change in persistence of a strain with respect to wild type.
This method of analysis makes the basic assumption in its normalization of the data that most of the signature-tagged strains in the pooled infection will have phenotypes similar to wild type. If you desire to assay a significant number of strains that you believe to have different survival rates than wild-type C. neoformans in the mouse, you may need to also seed the inoculum pool with signature-tagged strains that are known to have a wild-type phenotype to prevent skewing during the normalization process. We frequently use knockouts in the gene SXI1 (CNAG_06814 in the H99 sequence database of the Broad Institute) in this manner, as SXI1 is required for mating but dispensable for virulence (Hull et al., 2004).
It is important to note that this screen assays for relative persistence of a strain within a particular organ. It is not a true test of virulence per se, as it is conceivable that a strain may persist in large numbers in a tissue but fail to cause disease in the host. However, in our experience (Liu et al., 2008), the STM screen, when used to assay persistence of mutant strains in the lungs of 5-week-old A/J mice following intranasal infection, has resulted in STM scores that are both reproducible from mouse-to-mouse and pool-to-pool, but are also to a certain degree quantitative, by which we mean that the relative value of the STM score correlates with relative hypo- and hypervirulent phenotypes of the strains when assayed by survival curve analysis.
STM screens also cannot avoid in trans effects from mixing of strains; theoretically a wild-type strain may complement the phenotype of a mutant strain, allowing for a false negative result. Single-strain infections bypass this limitation of the STM screen approach.
6.2. Tissue culture
Although analysis of virulence in the host organismal level offers obvious correlations between a particular genotype and its efficacy at disease development, it is often problematic to determine the specific host-pathogen interactions responsible for a certain virulence phenotype. It is therefore useful to examine in closer detail the interaction of C. neoformans with a particular host tissue or cell type.
Many studies of the virulence of C. neoformans have focused on its interactions with the immune system, and, in particular, its interactions with macrophages. Alveolar macrophages are thought to be the first line of defense against pulmonary cryptococcal infection. Macrophages and macrophage-derived cells have been observed in the periphery of cryptococcal-containing granuloma formations in the lungs during latent infection of immunocompetent hosts. Additionally, depletion of macrophages from the murine host through the administration of silica has proven to be detrimental to fungal clearance (Monga, 1981). C. neoformans mutants that are more susceptible to killing by macrophages are hypovirulent in “time-to-endpoint” survival curves (e.g., FHB1 which encodes for flavohemoglobin; de Jesus-Berrios et al., 2003). For these and other reasons, it is of interest to examine the interaction of macrophages and macrophage-like cells with C. neoformans yeast.
Unopsonized C. neoformans cells are rarely taken up by macrophages in the absence of activation by cytokines such as IFN-γ or potent antigens such as lipopolysaccharide (LPS). Therefore, phagocytosis assays and assessments of killing by macrophages are commonly done in the presence of both opsonins (such as anti-C. neoformans antibodies or murine or human sera) and activating agents. We most commonly use the murine macrophage-like cell line RAW264.7 (American Type Culture Collection, No. TIB-71), and have had better success using anti-C. neoformans antibody than sera as an opsonizing agent.
6.2.1. Assay for killing of C. neoformans by macrophages
Seed RAW264.7 macrophages overnight into 96-well tissue culture plates (Corning, Cat. No. 3598) in 200 μl RAW cell medium (high-glucose DMEM (UCSF Cell Culture Facility, Cat. No. CCFAA005), 20 mM HEPES/NaOH buffer (pH 7.4) (UCSF Cell Culture Facility, Cat. No. CCFGL001), 20 mM glutamine (UCSF Cell Culture Facility, Cat. No. CCFGB002)) with IFN-γ (100 U/ml, Millipore, Cat. No. 005) at a density of 5×105 cells/well.
Culture the strain(s) of C. neoformans in 5 ml YPAD medium overnight at 30 °C.
The following day, wash an aliquot of the overnight C. neoformans culture 3× in sterile PBS.
Resuspend the C. neoformans cells to a concentration of 107 cells/ml.
Remove the RAW medium from the macrophages and replace with 200 μl of fresh RAW medium with 30 ng/ml LPS (Sigma, Cat. No. L4391), 100 U/ml IFN-γ, and anti-C. neoformans antibody.
Add 10 μl of the C. neoformans cells (105 cells) to the macrophages, and 10 μl to a well containing only 200 μl of RAW medium. Incubate for 24 h.
Remove supernatant from the wells, and retain for plating.
Lyse the macrophages by adding 0.01% SDS to each well. Wait 15 min, then remove and add to the supernatant previously removed. Repeat at least three times.
Check for complete lysis of the macrophages via a microscope.
Dilute the collected supernatants and plate for CFU. Determine the rate of killing by the macrophages by comparing the CFU from the wells with macrophages to the CFU from the wells without macrophages.
6.3. Assays for characterized virulence factors
C. neoformans has a number of characteristics previously shown to be involved in its virulence. These include (1) ability to grow at 37 °C (2) melanization, thought to aid in resistance to host killing (Nosanchuk and Casadevall, 2003), and (3) polysaccharide capsule formation, thought to be involved in host immune system evasion (Del Poeta, 2004), (Monari et al., 2006). Below are methods to test the relative efficiency of a strain for production of the virulence factors melanin and capsule.
6.3.1. Melanization
Melanization, or the ability for the yeast to form dark pigment compounds from catecholamine substances such as l-DOPA (3,4-dihydroxy-l-phenylalanine) by the enzyme laccase (Lac1), has long been associated with C. neoformans virulence. It has been hypothesized that melanin protects the yeast from oxidative or nitrosative damage originating from the host cells. To test strains for melanization, we utilize plates containing 100 ng/ml l-DOPA.
| l-DOPA plates | Composition |
|---|---|
| 2% Difco Bacto Agar (Becton-Dickinson, Cat. No. 214030) |
20 g |
| 7.6 mM l-asparagine monohydrate | 1 g |
| 5.6 mM glucose | 1 g |
| 22 mM KH2PO4 | 3 g |
| 1 mM MgSO4·7H2O | 250 mg |
| 0.5 mM l-DOPA (Sigma, Cat. No. D9628) | 100 mg |
| 0.3 mM thiamine–HCl | 1 mg |
| 20 nM biotin | 5 μg |
| Water | to 1 l |
To make 1 l of l-DOPA plate medium, autoclave 20 g of Difco Bacto Agar in 900 ml water so that it dissolves. In 100 ml water, add l-asparagine, glucose, KH2PO4, MgSO4·7H2O, and l-DOPA in the amounts indicated in the above recipe. Add phosphoric acid to the medium to pH 5.6, then add thiamine–HCl and biotin. Mix with the dissolved agar, and pour into plates.
6.3.1.1. Melanization test protocol
Inoculate cultures into YPAD from colonies on a plate for growth overnight.
Measure the optical density (OD) by spectrophotometer for each culture to be tested.
Dilute the cultures to the equivalent of OD600 = 0.6 with PBS and array in a 96-well assay plate.
Spot 4–6 μl of each diluted strain onto an l-DOPA plate.
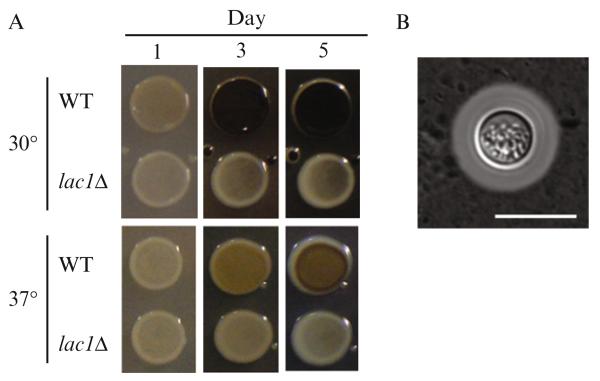
Incubate for 2–5 days at 30 or 37 °C, under observation (Fig. 33.6A).
Figure 33.6.
(A) Melanin assay: The kinetics of melanization varies depending on the incubation temperature. lac1Δ is deficient in the primary laccase enzyme responsible for melanization in C. neoformans. (B) Capsule formation assay: WT (H99) cell grown under capsule inducting conditions (DMEM, 37 °C, 5% CO2) and visualized with India ink. Bar denotes 10 μm.
Note
We have observed that the kinetics of melanization differ between growth at 30 and 37 °C, with a greater range of phenotypes visible at 30 °C. If screening a large numbers of strains, growth at 37 °C may be useful to highlight the mutants with more extreme defects in melanization. In addition, it may be useful to monitor the degree of melanization at early time points, as we have observed some strains that begin to melanize later than wild type, but reach a similar final level of melanization after 3 days.
6.3.2. Capsule formation
Secretion of a polysaccharide capsule is one of the major virulence factors of C. neoformans. When C. neoformans is mixed with India ink, the particles of the ink are excluded by a network of capsule fibers, thereby producing a characteristic halo around the yeast cell (Fig. 33.6B). Capsule is produced at low levels in typical YPAD culture—for best visualization, capsule production must be induced. Capsule can be induced through two different methods, described as follows.
Capsule induction via low nutrient conditions:
Inoculate C. neoformans from a colony on a plate into liquid Sabouraud dextrose medium.
Grow overnight at 30 °C.
Dilute the culture 1/100 in 10% Sabouraud dextrose medium buffered to pH 7.3 with 50 mM MOPS.
Grow cultures at 30 °C for 2 days in a rotating drum.
Capsule induction via carbon dioxide exposure:
Inoculate C. neoformans from a colony on a plate in liquid YNB medium.
Grow overnight at 30 °C.
Count cells on hemocytometer.
Wash 2×107 cells three times with PBS.
Resuspend the cells in 2.5 ml DMEM in a 6-well tissue culture dish (Falcon, Cat. No. 35-3224).
Culture cells for 24 h at 37 °C with 5% CO2.
Visualization of capsule by India ink staining:
Collect cells grown in capsule-inducing conditions. Concentrate the cells by centrifugation for ease of viewing if necessary.
Add 4 μl India ink (obtainable from a stationery store) to 20 μl of culture.
Drop 2 μl onto a microscope slide, mount with coverslip glass.
Visualize on a microscope with 60×–100× objective.
7. Concluding Remarks
While the methods described here are by no means all inclusive for what can be accomplished with C. neoformans, we hope they provide a guide for working with this basidiomycete, as well as a starting point for adapting other techniques.
ACKNOWLEDGMENTS
We thank Joseph Heitman, Jennifer Lodge, Gary Cox, Tamara Doering, John Perfect, Peter Williamson, Christina Hull, Andrew Alspaugh, James Kronstad, Thomas Kozel, Arturo Casadevall, June Kwon-Chung, and Suzanne Noble for sharing strains, protocols, reagents, and general expertise. We thank Jessica Brown and Oliver Liu for critical reading of this manuscript. The work was supported by an Opportunity Grant from the Herb and Marion Sandler Foundation and a grant from the NIAID (R01AI065519). C. D. C. was supported by a National Science Foundation Predoctoral Fellowship.
REFERENCES
- Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr. Infect. Dis. Rep. 2008;10:58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- Blackstock R, Murphy JW. Age-related resistance of C57BL/6 mice to Cryptococcus neoformans is dependent on maturation of NKT cells. Infect. Immun. 2004;72:5175–5180. doi: 10.1128/IAI.72.9.5175-5180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Boekhout T. Diversity of the Cryptococcus neoformans–Cryptococcus gattii species complex. Rev. Iberoam. Micol. 2008;25:S4–S12. doi: 10.1016/s1130-1406(08)70019-6. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Mukherjee J, Yuan R, Perfect J. Management of injuries caused by Cryptococcus neoformans—Contaminated needles. Clin. Infect. Dis. 1994;19:951–953. doi: 10.1093/clinids/19.5.951. [DOI] [PubMed] [Google Scholar]
- Chen LC, Goldman DL, Doering TL, Pirofski L, Casadevall A. Antibody response to Cryptococcus neoformans proteins in rodents and humans. Infect. Immun. 1999;67:2218–2224. doi: 10.1128/iai.67.5.2218-2224.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GH, McNamara DA, Hernandez Y, Huffnagle GB, Toews GB, Olszewski MA. Inheritance of immune polarization patterns is linked to resistance versus susceptibility to Cryptococcus neoformans in a mouse model. Infect. Immun. 2008;76:2379–2391. doi: 10.1128/IAI.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D’Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr. Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- Del Poeta M. Role of phagocytosis in the virulence of Cryptococcus neoformans. Eukaryot. Cell. 2004;3:1067–1075. doi: 10.1128/EC.3.5.1067-1075.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC. Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 1992;12:2777–2783. doi: 10.1128/mcb.12.6.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol. Cell. Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escandon P, Ngamskulrungroj P, Meyer W, Castaneda E. In vitro mating of Colombian isolates of the Cryptococcus neoformans species complex. Biomedica. 2007;27:308–314. [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: Implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Donlin MJ, Soto CE, Banks AM, Banks IR, Maligie MA, Selitrennikoff CP, Lodge JK. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 2005;58:393–408. doi: 10.1111/j.1365-2958.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, Casadevall A. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics. 2001;107:E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- Heitman J, Allen B, Alspaugh JA, Kwon-Chung KJ. On the origins of congenic MATalpha and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet. Biol. 1999;28:1–5. doi: 10.1006/fgbi.1999.1155. [DOI] [PubMed] [Google Scholar]
- Hoang LM, Maguire JA, Doyle P, Fyfe M, Roscoe DL. Cryptococcus neoformans infections at Vancouver Hospital and Health Sciences Centre (1997-2002): Epidemiology, microbiology and histopathology. J. Med. Microbiol. 2004;53:935–940. doi: 10.1099/jmm.0.05427-0. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyer JD, Lodge JK. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin. Diagn. Lab. Immunol. 2000;7:125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, Boyd MB, Street NE, Lipscomb MF. IL-5 is required for eosinophil recruitment, crystal deposition, and mononuclear cell recruitment during a pulmonary Cryptococcus neoformans infection in genetically susceptible mice (C57BL/6) J. Immunol. 1998;160:2393–2400. [PubMed] [Google Scholar]
- Hull CM, Cox GM, Heitman J. The alpha-specific cell identity factor Sxi1alpha is not required for virulence of Cryptococcus neoformans. Infect. Immun. 2004;72:3643–3645. doi: 10.1128/IAI.72.6.3643-3645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabasawa K, Itagaki H, Ikeda R, Shinoda T, Kagaya K, Fukazawa Y. Evaluation of a new method for identification of Cryptococcus neoformans which uses serologic tests aided by selected biological tests. J. Clin. Microbiol. 1991;29:2873–2876. doi: 10.1128/jcm.29.12.2873-2876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS. Microbial translocation of the blood-brain barrier. Int. J. Parasitol. 2006;36:607–614. doi: 10.1016/j.ijpara.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland CM, Chang YC, Varma A, Chung KJ. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 2004;12:208–212. doi: 10.1016/j.tim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS-100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monari C, Kozel TR, Paganelli F, Pericolini E, Perito S, Bistoni F, Casadevall A, Vecchiarelli A. Microbial immune suppression mediated by direct engagement of inhibitory Fc receptor. J. Immunol. 2006;177:6842–6851. doi: 10.4049/jimmunol.177.10.6842. [DOI] [PubMed] [Google Scholar]
- Monga DP. Role of macrophages in resistance of mice to experimental cryptococcosis. Infect. Immun. 1981;32:975–978. doi: 10.1128/iai.32.3.975-978.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow CA, Fraser JA. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 2009;9:161–177. doi: 10.1111/j.1567-1364.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Nelson RT, Pryor BA, Lodge JK. Sequence length required for homologous recombination in Cryptococcus neoformans. Fungal Genet. Biol. 2003;38:1–9. doi: 10.1016/s1087-1845(02)00510-8. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Litvintseva AP, Mylonakis E, Malliaris SD, Benjamin DK, Jr., Giles SS, Mitchell TG, Casadevall A, Perfect JR, Heitman J. Cryptococcus neoformans alpha strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 2005;73:4922–4933. doi: 10.1128/IAI.73.8.4922-4933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell. Microbiol. 2003;5:203–223. doi: 10.1046/j.1462-5814.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- Reid RJ, Lisby M, Rothstein R. Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Methods Enzymol. 2002;350:258–277. doi: 10.1016/s0076-6879(02)50968-x. [DOI] [PubMed] [Google Scholar]
- Rhodes JC, Wicker LS, Urba WJ. Genetic control of susceptibility to Cryptococcus neoformans in mice. Infect. Immun. 1980;29:494–499. doi: 10.1128/iai.29.2.494-499.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Berbee ML. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia. 2006;98:838–849. doi: 10.3852/mycologia.98.6.838. [DOI] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ. Haploid fruiting in Cryptococcus neoformans is not mating type alpha-specific. Fungal Genet. Biol. 2003;39:230–237. doi: 10.1016/s1087-1845(03)00046-x. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol. Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- Wormley FL, Jr., Perfect JR, Steele C, Cox GM. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect. Immun. 2007;75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Vilgalys R, Mitchell TG. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol. Ecol. 2000;9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]