Abstract
RNA granules are structures within cells that impart key regulatory measures on gene expression. Two general types of RNA granules are conserved from yeast to mammals: stress granules (SGs), which contain many translation initiation factors, and processing bodies (P-bodies, PBs), which are enriched for proteins involved in RNA turnover. Because of the inverse relationship between appearance of RNA granules and persistence of translation, many viruses must subvert RNA granule function for replicative purposes. Here we discuss the viruses and mechanisms that manipulate stress granules and P-bodies to promote synthesis of viral proteins. Several themes have emerged for manipulation of RNA granules by viruses: (1) disruption of RNA granules at the mid-phase of infection, (2) prevention of RNA granule assembly throughout infection and (3) co-opting of RNA granule proteins for new or parallel roles in viral reproduction. Viruses must employ one or multiple of these routes for a robust and productive infection to occur. The possible role for RNA granules in promoting innate immune responses poses an additional reason why viruses must counteract the effects of RNA granules for efficient replication.
Keywords: Stress granules, P-bodies, RNA granules, Translation control
Introduction to RNA granules
A key aspect of virus host interactions is viral manipulation of cellular gene expression to maintain conditions conducive for efficient replication. Eukaryotic genes are highly regulated post-transcriptionally by evolving mRNP compositions that influence splicing, export, regulation of translation, subcellular localization and mRNA turnover. These events are often interconnected, e.g., mRNA translation is linked to poly(A) shortening and decay, and the processes share proteins (Chang et al., 2004, Shyu et al., 2008). The composition of mRNPs also determines if the mRNA constituents are translationally competent and able to access and assemble ribosomes, or translationally silenced and unable to access active ribosomal machinery. Both nuclear and cytoplasmic mRNP granules exist. Nuclear granules include cajal bodies, histone locus bodies, nuclear speckles, nuclear stress bodies and paraspeckles (Mao et al., 2011, Caudron-Herger and Rippe, 2012). The function of nuclear mRNP granules ranges from stress responsive granules to those that regulate processing of mRNAs (e.g., histone locus bodies, nuclear speckles and paraspeckles) and non-coding RNAs. This review will focus on cytoplasmic RNA granules because of the high propensity for viruses to modify these granules and recent evidence implicating cytoplasmic RNA granules in innate immunity.
Translationally silenced mRNPs can organize into two major classes of RNA granules in the cytoplasm, known as stress granules (SGs) and processing bodies (P-bodies, PBs). It has been suggested that there is a cytoplasmic mRNA cycle in which mRNPs rapidly move between active polysomes and silenced compartments of PBs and SGs. This is supported by the observation that SGs and PBs are in equilibrium with actively translating mRNPS, which is indicated by experiments using chemical and genetic blockage of multiple steps in the process of translation initiation and elongation (Mokas et al., 2009; Dang et al., 2006, Kedersha et al., 1999). Furthermore, flux between the different RNA granules has been demonstrated with experiments showing transient docking of SGs and PBs with each other, photobleaching experiments that show rapid turnover of proteins in these RNA granules and that SGs and PBs can share many protein components and specific mRNA moieties (Chang et al., 2004, Kedersha et al., 2005, Anderson and Kedersha, 2008, Shyu et al., 2008, Buchan and Parker, 2009).
Stress granules are distinguished by containing high concentrations of translation initiation factors and 40S ribosome subunits, whereas P-bodies are enriched for RNA decay machinery. However, many proteins have been described in both compartments such as Ago2, eIF4E, APOBEC3, PCBP2, TTP and others (Kedersha and Anderson, 2007, Kedersha et al., 2005). Several other types of RNA granules have been described in Caenorhabditis elegans, Drosophila, and neurons, that contain various levels of proteins uniquely found in either SG or PBs. Thus, a continuum of RNA granules has been suggested to exist in eukaryotic cells with degrees of similarity to either SG or PBs (Buchan and Parker, 2009).
Stress granules
Stress granules are dynamic structures that quickly form when external stresses are applied to cells and global translation rates decline, and disperse when translation conditions are restored. Thus, SG are mostly thought to contain stalled 43S and 48S ribosomal preinitiation complexes and are proposed to serve as temporary repositories for these complexes. In this way, largely preassembled translation complexes can be rapidly released to resume gene expression when cellular stress conditions abate. The scenario most often described for SG formation follows when oxidative, nutrient or heat stress activates one of the eIF2α kinases (heme-regulated kinase, HRI; general control non-depressible 2 kinase, GCN2; double-stranded RNA (dsRNA)-activated protein kinase R, PKR; and PKR-like endoplasmic reticulum kinase, PERK), which phosphorylate the alpha subunit of eIF2 and block translation, forcing an accumulation of the stalled 43S and 48S ribosomal preinitiation complexes. Inhibition of the function of eIF4G or eIF4A in translation initiation are also linked to SG formation (Mazroui et al., 2006) and some mechanisms of SG formation can proceed in the absence of eIF2α phosphorylation ( Fig. 1) (Dang et al., 2006, Emara et al., 2012, Reineke et al., 2012). As SGs contain hundreds of RNA-interacting proteins and an siRNA screen indicates more than 100 genes are involved in SG assembly, the mechanism of SG formation is likely very complex (Ohn et al., 2008). Yet genetically simple viruses have evolved efficient means to control their formation and function.
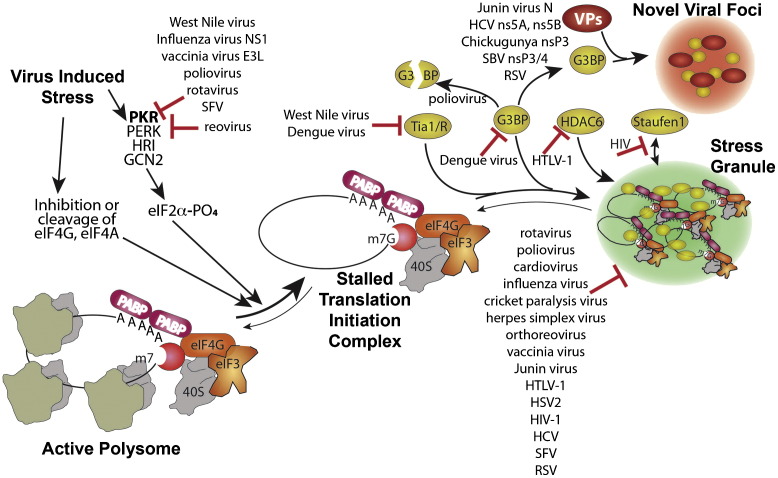
Fig. 1.
Stress granule assembly and interference by viruses. Virus infection causes stress at multiple levels that reduces host translation through activation of eIF2 kinases or other means and converts active polysome mRNPs into stalled translation initiation complex mRNPs. A complex series of events involving nucleation of several stress granule proteins such as G3BP1, Tia-1/TIAR, and HDAC6 plus transport on microtubules leads to aggregates of translation initiation complex mRNPs in stress granules. Specific points/proteins where viruses interact with and inhibit or divert the RNA granule assembly pathway are shown. Several viral proteins (VPs) interact with G3BP1 in complexes, some of which localize to novel viral foci. Note that many viruses control PKR activation; only those discussed in the text are indicated.
Constants and variables in SG composition
Since SGs contain stalled initiation complexes, canonical SGs are defined by the presence of key initiation factors (e.g., eIF4E, eIF4G, eIF4A, eIF4B, eIF3, eIF2, PABP), mRNA and the small ribosome subunit (Kedersha et al., 1999, Kedersha et al., 2005, White and Lloyd, 2011). In addition there are a host of RNA binding proteins such as FMRP, YB1, HuR, and TTP, just to name a few. It is unclear at this point, but presumably any protein that binds mRNA or interacts strongly with mRNPs may be ultimately included in SGs. Many of these are passenger proteins that may not function overtly in SG biology. However, SGs also contain key marker proteins that are linked to their formation, most notably G3BP1, TIA1 and TIAR, TDRD3, HDAC6, Caprin1, as well as others (Tourrière et al., 2003, Gilks et al., 2004, Kwon et al., 2007, Solomon et al., 2007, Goulet et al., 2008). It is important to note that not all SGs are equivalent. The composition of SGs has been found to vary depending on the mode of stress that induced them, though the majority of markers that define SG function as repositories of stalled translation complexes are consistent among all types of SG, e.g., translation initiation factors, 40S ribosome subunits, mRNA. For instance, heat shock induced stress granules (HS-SGs) contain heat shock protein 27 (hsp27), but hsp27 is not found in arsenite (Ars)-induced SGs (Kedersha et al., 1999, Gilks et al., 2004, Piotrowska et al., 2010). Virus infection produces unique types of cell stress and can induce SG to form (V-SG). Some V-SGs uniquely contain Sam68 which is not found in HS-SGs (Piotrowska et al., 2010).
Not all mRNA can be included in all SGs; ER-associated transcripts are generally excluded (Unsworth et al., 2010) and heat shock protein transcripts are omitted from HS-SGs (Nover et al., 1989). It is possible that other types of transcripts that function during cell stress, e.g., certain IRES-containing mRNAs, are preferentially excluded from SG inclusion. The mechanisms that lead to exclusion of certain transcripts from SGs remain to be determined. Thus, the overall function of aggregation of mRNPs into SGs remains unclear, but likely regulates both mRNA translation and mRNA turnover, and may be linked to signaling pathways (discussed below). Overall, these aggregate functions likely promote increased cell survival during stress conditions and rapid return to homeostasis at stress termination (Eisinger-Mathason et al., 2008).
Processing bodies
P-bodies are constitutively present in cells, and increase in size and number when translational arrest occurs. P-body constituents include decapping enzymes, exonucleases, deadenylases, RNA binding proteins involved in nonsense-mediated decay and microRNA mediated silencing ( Fig. 2). RNA decay occurs both within PBs and outside PBs and proportions of total RNA decay attributed to each compartment are controversial (Arribere et al., 2011). Recruitment of mRNA to PBs is not an occurrence of non-translation, rather requires active silencing via miRNA or RNAi mechanisms (Eulalio et al., 2007). The molecular mechanism of PB formation is thought to involve aggregation of RNA binding proteins, as well as the mRNA itself as an organizing structure. Consistent with this, PBs disperse in permeablized cells treated with RNase (Eulalio et al., 2007). The human DEAD box helicase RCK/p54 (also called DDX6) may coat and relax mRNA structures before entry into PBs (Ernoult-Lange et al., 2012). Similar to SGs, PBs can contain variable protein makeup in mammalian cell lines and Drosophila, with proteins like PCBP2, Hedls, Xrn1, defining subsets of PBs (Teixeira et al., 2005, Eulalio et al., 2007). PBs are proposed to dynamically exchange mRNP cargo with SGs and have been proposed to serve as nucleation sites for SG formation (Kedersha et al., 2005).
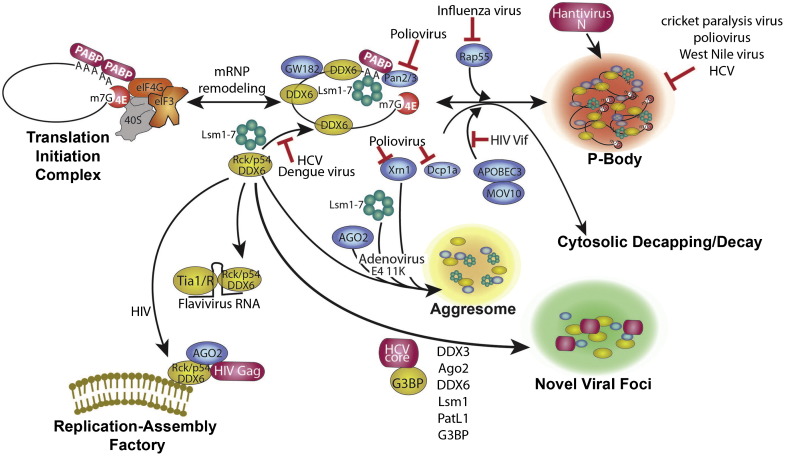
Fig. 2.
P-body assembly and interference by viruses. P-bodies form via a complex series of events involving stripping of mRNPs of initiation factors and ribosome subunits, association with GW182, undergoing Pan2/3-mediated deadenylation, MT transport, and association of other RNA decay factors (e.g., Xrn1, Dcp1a, DDX6, GW182 and Lsm components of the exosome), and final concentration in P-bodies. Decapping and decay occurs both in and outside P-bodies. The order of association of factors with mRNPs in PBs is arbitrary. For HCV, novel viral foci containing P-body components also contain some SG components including G3BP.
Relationship between RNA granules and viruses
Because SGs and PBs control the mRNA cycle, metabolism and gene expression, they become another vital point of control for viruses to manipulate. The degree and type of manipulation employed by viruses is turning out to be as variable as the replication schemes of the viruses themselves and the impact of SGs on virus replication can be wide-ranging. Virus infection induces many types of stresses on cells, even during non-lytic infections, and the perturbation of cellular homeostasis is sensed in many ways in pathways that feed directly into stress responses. An emerging theme is primordial innate immune responses and general stress responses are intimately linked, and interface at many levels. Most outcomes of stress responses serve to sharply curtail or alter host gene expression patterns, usually to the detriment of a virus. Thus, a general trend by viruses is that they block or co-opt stress responses to facilitate efficient replication. To cover the broad range of virus-RNA granule interactions, this review organizes both SG and PB interactions into roughly generalized topics according to current understanding. SG groupings are summarized in Table 1. We expect these groupings will require revision as more research emerges.
Table 1.
| Viruses | Mechanism of SG interaction | References |
|---|---|---|
| SGs are disrupted at mid-phase of infection | ||
| Mammalian orthoreovirus | Mechanism unknown, but may involve expression levels of PKR/PERK inhibitor p58IPK. | Smith et al. (2006) |
| Poliovirus | SG disruption due to G3BP cleavage polio 3C protease. | White et al. (2007) |
| Theiler's murine encephalomyelitis virus | Leader protein disrupts SG formation. | Borghese and Michiels (2011) |
| Cricket paralysis virus | Mechanism of SG disruption is unknown. | Khong and Jan (2011) |
| SGs are undetectable during infection | ||
| Influenza a virus | NS1 expression restricts PKR activity inhibiting eIF2α-mediated SGs; NS1-Rap55 complexes may also be involved in SG inhibition. | Khaperskyy et al. (2011); Mok et al. (2012) |
| Rotavirus | Mechanism of disruption is unknown, eIF2α phosphorylation induced by viral proteins Vp2, Nsp2, Nsp5. | Montero et al. (2008) |
| Human T cell leukemia virus | Viral protein Tax interacts with HDAC6, but unclear whether HDAC6 activity is impaired. | Legros et al. (2011) |
| Viruses co-opt SG proteins | ||
| Chikungunya virus (other alphaviruses?) | Viral protein Nsp3 sequesters G3BP from SGs, but function is unknown. | Fros et al. (2012) |
| Sindbis virus | Viral RdRP Nsp4 interacts with G3BP, but function is also unknown. | Cristea et al. (2010) |
| West Nile virus | Recruitment of TiaR to viral RNA replication sites, but role of TiaR in replication is unknown. | Li et al. (2002) |
| Dengue virus | G3BP, USP10 and Caprin1 interact with Dengue 3′UTR, function unknown. | Ward et al. (2011) |
| Hepatitis C virus | G3BP and others are relocalized to lipid droplets; G3BP also interacts with HCV replication complex NSP5A and NSP5B, function unknown. | Ariumi et al. (2011); Yi et al. (2011) |
| Herpes simplex virus 1 | Pbp1 (Ataxin 2 ortholog) induces mTORC1 accumulation in SGs, which inhibits mTORC1 activity, likely induces HSV1 reactivation from latency. | Takahara and Maeda (2012) |
| Herpes simplex virus 2 | Tia1 redistributed to nuclear foci, but unlikely inhibits SG, function unknown. | Finnen et al. (2012) |
| Junin virus | Junin protein N colocalizes with G3BP and replication-transcription complexes, function unknown. | Linero et al. (2011) |
| Human immunodeficiency virus | Staufen 1 is partitioned to Gag complexes and regulates virus encapsidation. | Abrahamyan et al. (2010) |
| Respiratory syncytial virus | G3BP is important in viral replication cycle, but function is unknown; Trailer region of viral RNA inhibits SG formation, but mechanism unknown. | Lindquist et al. (2010); Hanley et al. (2010) |
| Transmissible gastroenteritis virus and mouse hepatitis virus | Viruses grow worse when SGs are present, but whether translational repression or subversion of SG factors is unknown. | Raaben et al. (2007); Sola et al. (2011) |
| Vaccinia virus | Conflicting studies: (1)Replication factories contain proviral SG-like structures, and (2)Antiviral SG-like structures localize to replication factories. Function unknown. | Katsafanas and Moss (2004); Katsafanas and Moss (2007),Walsh et al. (2008); Simpson-Holley et al. (2011) |
Viruses that repress SG formation
Most viruses have been reported to modulate stress granules by suppressing their formation at some point in the infection cycle. Many variations on this theme have been reported already, but few examples exist where fully formed functional SGs (defined as containing stalled translation complexes) have been shown to co-exist within virus infected cells at mid-phase of virus replication cycle when virus gene expression is high. Typically when viruses amplify their gene expression sufficiently, SGs are absent and/or SG components have been co-opted into novel virus-specific structures or roles that are symbiotic with replication processes. Another emerging theme is that PKR is a central sensor of viral stress that drives not only stress granule responses, but also downstream innate immune functions. We have organized the discussion of virus modulation of SGs into broad groups that reflect these patterns.
Mammalian orthoreovirus (MRV) induces stress granules initially (6 hpi), which correlates with reduced translation of both cellular and viral mRNAs, and increased phosphorylation of eIF2α (Qin et al., 2011). eIF2α phosphorylation is required for MRV induction of stress granules, as is uncoating of the virus. PKR and the other individual eIF2α kinases are dispensable for induction of SGs by MRV, which indicates MRV induces SGs by signaling through multiple eIF2α kinases or possibly some other mechanism (Qin et al., 2009). Disassembly of MRV-induced SGs requires synthesis of viral proteins, and permits preferential translation of viral mRNAs even in the presence of eIF2α phosphorylation (Qin et al., 2009, Qin et al., 2011).
Smith and colleagues showed that the yield of three reoviruses strains (Dearing, c8 and c87) were reduced in mouse embryonic fibroblasts exclusively expressing the non-phosphorylatable S51A eIF2α mutant, and individual PERK and ATF4 knockout cells indicating the initial induction of stress granules improves virus production (Smith et al., 2006). The degree of virus production varied in a strain-specific manner, but was linked to variations in virus production of p58IPK, an inhibitor of eIF2α kinases PKR and PERK. The authors found that high expression of p58IPK correlated with reduced eIF2α phosphorylation and stress granule persistence later in infection (19.5 hpi) (Smith et al., 2006). ATF4 is a transcriptional regulator whose expression is regulated by eIF2α because of regulatory upstream open reading frames. Smith et al. suggested that reovirus replication depends on expression of ATF4-induced genes that augment virus replication. Using different reovirus strains, Qin et al. demonstrated that MRV restricts SG formation at a point downstream of eIF2α phosphorylation during late times post infection (24 hpi), but did not draw correlations with p58IPK levels or measure virus titers in the presence and absence of eIF2α phosphorylation. As such, there is no confirmation of the observation by Smith and colleagues that SG formation early during infection augments viral replication. However, reoviruses inhibit assembly of stress granules at a fundamental level, since even eIF4A inhibitors, which act independent of eIF2α phosphorylation, cannot induce SGs late in infection (Qin et al., 2011).
Poliovirus is also known to inhibit stress granule assembly during the late-phase of infection. The mechanism of SG disassembly involves cleavage of the stress granule protein G3BP1, which is mediated by the viral protease 3C (Fig. 1). G3BP1 cleavage separates the N-terminal protein-interacting domain from the C-terminal RNA recognition motif. Expression of a 3C protease cleavage-resistant mutant of G3BP1 rescues stress granules at late times post infection (White et al., 2007). A contradicting study showed that Tia1-containing stress granules persisted even during late times post infection (Piotrowska et al., 2010), but those granules were later shown to be devoid of the stress granule components eIF3, eIF4G and eIF4E indicating that Tia1 granules are remnants of normal stress granules and do not correlate with translational repression (White and Lloyd, 2011). Therefore, poliovirus unlinks Tia1 aggregation from aggregation of translation initiation factors in stress granules. These results reinforce the notion that mRNP granules differ in composition and function and cannot be stereotyped based on some common factors. eIF2α phosphorylation increases during the course of poliovirus infection, similar to the case with MRV. This effectively inhibits cellular protein synthesis so resources can be used for viral RNA translation. Interestingly, poliovirus translation proceeds due to cleavage of eIF5B, which bypasses the need for eIF2α during translation initiation (White et al., 2011). It is unclear whether MRV translation can persist during eIF2α phosphorylation by exploiting such a mechanism.
A related animal picornavirus, Theiler's murine encephalomyelitis virus, also blocks SG formation. In this case G3BP1 was not degraded as with poliovirus; however, the virus leader protein was determined to be responsible for blocking SG formation. Ectopic expression of the leader protein could block cells from mounting stress granule responses to arsenite and thapsigargin (Borghese and Michiels, 2011).
Cricket paralysis virus, a member of the picornavirus subgroup, is also capable of inhibiting stress granule formation at early times post infection (2 h). However, inhibition of stress granules induced by the exogenous stressors arsenite, pateamine A and heat shock does not appear until later (4 hpi) indicating that stress granule inhibition correlates with increased viral protein expression, similar to the example from poliovirus, another picornavirus. Cleavage of the G3BP paralog does not occur during infection, in contrast to poliovirus infection, and Tia1 also remains intact (Khong and Jan, 2011).
SG and PKR inhibition to maintain virus translation
Unlike MRV and poliovirus, Influenza A virus (IAV) prevents stress granule formation throughout the infection. Inhibition of stress granule formation by IAV depends on expression of the viral protein NS1, as recombinant Influenza A virus expressing a variant of NS1 that does not bind dsRNA stimulates eIF2α phosphorylation and SG accumulation. Stress granule formation appears to inhibit IAV replication because accumulation of the viral protein NP is repressed when SGs persist through the infectious cycle (Khaperskyy et al., 2011). Also, PKR knockout cells do not accumulate stress granules late during infection despite the presence of NS1 mutants that induce SG formation (Khaperskyy et al., 2011) (Fig. 1). Together these data suggest that the ability of NS1 to inhibit PKR activity is critical for inhibition of SGs. Furthermore, ablation of NS1 expression in IAV results in formation of stress granules that contain many innate immunity factors including OAS, RNase L and PKR. These granules appear to be important in induction of interferon gene expression implying an importance for antiviral stress granules in priming nearby cells against infection (Onomoto et al., 2012).
The mechanism of NS1 repression of RNA granule formation involves an interaction in a complex containing cellular RNA associated protein 55 (Rap55), which is a component of both SGs and PBs. Overexpression of Rap55 induced SGs and inhibits virus replication. Viral nucleoprotein colocalized with SGs in the absence of NS1 but colocalizes with PBs during wild type virus infection. The portion of NS1 responsible for interaction with Rap55 complexes maps to the PKR-interacting domain (Mok et al., 2012).
In contrast to the study by Khaperskyy et al., another report found increased PKR activation and eIF2α phosphorylation did not significantly alter levels of the IAV NP protein despite more pronounced PKR activation during infection with an IAV NS1 deletion mutant (IAV ΔNS1) (Onomoto et al., 2012). These results indicate that not only do SGs differ in composition depending on the context (discussed above), but even the same virus can interact with SGs and/or the translational apparatus in a cell type-dependent manner (A549 cells versus HeLa). Interestingly, SGs induced from infection with IAV ΔNS1 appear to be important in induction of interferon gene expression as measured by qPCR of IFN-β mRNA in control cells or cells depleted of G3BP1, which are impaired in SG formation (Onomoto et al., 2012). The authors went on to show decreased IFN-β mRNA in cells devoid of PKR expression despite IAV infection. Although intriguing, these results leave open the possibility that either G3BP or PKR are important for the innate immune response rather than SGs themselves. Indeed, PKR has previously been shown to be required to mount a normal innate immune transcription response (Garcia et al., 2006, Steele et al., 2011, Taghavi and Samuel, 2012).
Rotavirus induces eIF2α phosphorylation that is dependent on viral proteins VP2, NSP2, and NSP5 (Montero et al., 2008). Stress granules are not assembled during the course of infection or induced in infected cells by addition of arsenite. Therefore the virus actively inhibits stress granule formation. The use of eIF2α S51A mutant mouse embryonic fibroblasts to study the significance of eIF2α phosphorylation on rotavirus replication indicates that the virus replicates better when eIF2α cannot be phosphorylated. These results are similar to the case with mouse hepatitis coronavirus which also replicate better in S51A mutant mouse embryonic fibroblasts (discussed in detail below) (Raaben et al., 2007). These examples highlight the importance of a normal innate immune response for inhibiting the spread of virus, and indicate these viruses reach a balance between eIF2α phosphorylation during the infection and reduced virus production. If SG assembly is important for innate immunity, as the IAV example suggests, rotavirus may inhibit granules to inhibit the innate immune response to increase virus production in neighboring cells.
HTLV Tax protein inhibits stress granule assembly when transiently expressed (Legros et al., 2011). Tax was shown to interact with HDAC6, whose activity was earlier shown to be required for stress granule assembly (Kwon et al., 2007). However, the study did not delineate whether Tax inhibited HDAC6 activity, or whether Tax expression during infection was even required to inhibit stress granules. It is possible that stress granules are inhibited after an initial phase of induction similar to the examples documented for MRV and poliovirus. This would be expected since Tax protein expression is unlikely to be at inhibitory levels immediately after cells are infected.
Viruses subvert SG components and enhance replication
The alphavirus Semliki Forest Virus (SFV) also inhibits stress granule formation after an initial phase of eIF2α phosphorylation and stress granule induction (McInerney et al., 2005). The translational enhancer immediately downstream of the initiating AUG codon allows SFV to escape translational repression induced by eIF2α phosphorylation. However, despite the observation that eIF2α phosphorylation favors synthesis of viral proteins over cellular proteins, it is unclear whether the stress response that accompanies eIF2α phosphorylation augments viral replication as it does with MRV.
Although the mechanism of stress granule repression during SFV infection is not known, another alphavirus, Chikungunya Virus, is capable of repressing stress granules by recruiting G3BP1 to cytoplasmic foci, a process dependent on the SH3 homology domain-binding motif in the viral protein nsp3 (Fros et al., 2012). The G3BP1/nsP3-containing foci are not canonical stress granules as they lack the SG marker eIF3. Similarly, Semliki Forest virus nsP3 also sequesters G3BP1 into viral replication complexes and SG formation. In this case a C-terminal domain of nsP3 was mapped to interact with G3BP that was distinct from the Chikungunya SH3 domain (Panas et al., 2012). Further, the efficient translation of viral mRNAs with an enhancer motif also helps disassemble SGs during infection. The precise molecular benefit of subverting G3BP1 to novel replicase complexes is unclear but deletions of the G3BP1-intereacting sequence in nsP3 reduces replication of viral replicons or virus (Fros et al., 2012, Panas et al., 2012).
With another alphavirus, Sindbis Virus (SV), nsP4, which is the RNA-dependent RNA polymerase, interacts with G3BP1 and 2 as identified using immunoprecipitation followed by mass spectrometry (Cristea et al., 2010). Previous work showed G3BP1 also interacted with nsP2 and nsP3, so this may reflect overlapping interactions with a large viral replicase complex (Gorchakov et al., 2008, Frolova et al., 2006). The role of interaction of SV nsP4 with G3BP is unclear as there are only marginal changes in SV RNA levels when both G3BP1 and 2 are depleted. Because G3BP was identified in infected cells without RNase treatment of immunocomplexes to eliminate RNA-dependent interactions, it is possible G3BP actually interacts with the SV RNA and regulates SV translation. This idea is supported by experiments indicating that SV polyprotein production is significantly increased when G3BP is depleted (Cristea et al., 2010). Furthermore, G3BP1 has been shown to regulate translation of some cellular mRNAs (Ortega et al., 2010). If G3BP1 were interacting with the SV RNA, it could be similar to subversion of the SG proteins Tia1 and TIAR by the flaviviruses, as highlighted below, although in that case Tia1 and TIAR play a positive role for Dengue and West Nile Virus. The role of G3BP in SV polyprotein production is paralleled by a 5-fold increase in virus production when G3BP 1 and 2 are depleted. Considering the similarity of Sindbis Virus to SFV and the importance of G3BP1 in SG assembly (White et al., 2007) it is likely that depletion of G3BP1 and 2 abrogates SGs during infection thereby enhancing overall virus production by eliminating the translation block coincident with SG assembly (Fig. 1). Interaction of G3BP with Sindbis Virus nsP3 has not been documented indicating that Sindbis and SFV may differentially co-opt G3BP function, which is surprising considering the similarity of these viruses.
Stress granule formation was prevented in cells infected with the flaviviruses West Nile (WNV) and Dengue Virus. Phosphorylated eIF2α did not accumulate during infection with a West Nile virus strain from lineage 1, indicating the virus is actively suppressing translational repression. In fact, arsenite induction of stress granules is impaired as viral protein expression increases during the infection (Emara and Brinton, 2007). Increased expression of viral RNA early during WNV infection from a chimeric lineage 1/2 virus W9561C has been documented to activate PKR and induce stress granules (Courtney et al., 2012). Lineage 1 WNV strain Eg101 lacks the robust early viral RNA levels observed with W9561C and has reduced or delayed activation of PKR. These data suggest WNV restricts expression of viral RNA early to avoid induction of the cellular stress response. Li et al. showed that WNV strain Eg101 replication was 6–7 fold reduced in cells depleted of TIAR, but not the highly related Tia1 gene. Therefore, native WNV strains likely prevent the SG response in order to subvert TIAR to functions that promote viral RNA replication (Li et al., 2002).
G3BP1 and 2 have been shown to bind the 3′ UTR of Dengue virus 2. Interestingly, other proteins known to interact with G3BP1 and localize to stress granules were identified to bind the Dengue virus mRNA. Among these proteins are USP10 and Caprin1 (Ward et al., 2011). However, it is unclear what influence these proteins have on Dengue virus replication because functional studies have not yet been performed. These data suggest stress granule proteins may enhance virus translation or RNA replication, or alternatively that flaviviruses recruit these stress granule proteins to prevent a strong innate immune response (discussed below).
Hepatitis C Virus also induces stress granules in a manner dependent on eIF2α phosphorylation (Garaigorta et al., 2012, Ruggieri et al., 2012). Furthermore, stress granules are more abundant in HCV infected cells treated with interferon and oscillate during the course of the infection. These oscillations are thought to be critical for viral gene expression because infected cells containing stress granules exhibit translational repression and would otherwise be incapable of expressing viral proteins necessary for chronic HCV infection. Interestingly, no viral gene is expected to induce SG oscillations during HCV infection as SG oscillation was observed with diverse viruses and even transfection of double-stranded RNA. Oscillation in SG abundance is also expected to enhance HCV virion production by liberating SG proteins involved in viral replication and egress. This prediction is based on the observation that several P-body and stress granule proteins are redistributed to lipid droplets during the course of infection, including DDX6 and G3BP1 (Ariumi et al., 2011). In fact, depletion of G3BP1, Tia1, TIAR and PABP, among others, have been described to effect different steps of the HCV lifecycle (Ariumi et al., 2011, Yi et al., 2011, Garaigorta et al., 2012). Although the significance of G3BP1 redistribution is unknown, it was found to colocalize and interact with NS5A and NS5B, two components of the HCV replication complex suggesting G3BP1 may be involved in HCV RNA replication (Yi et al., 2011). However, additional details of the mechanism of G3BP1 involvement in HCV RNA replication have not been elucidated.
There may be some role for the SG response in regulating HSV-1 latency in neurons. Pbp1 is an ortholog of Ataxin 2, which associates with PABP and enters SGs (Mangus et al., 1998, Buchan et al., 2008). Pbp1 undergoes a self-association to enter SGs, but does so in a complex that pulls TORC1 (target of rapamycin complex 1) into SGs (Takahara and Maeda, 2012). Sequestration of TORC1 in SGs inhibits TORC function, which normally regulates cap-dependent translation in response to growth signaling. Interestingly, mTORC1 controls the latency of HSV-1 in neurons and when active will suppress HSV-1 reactivation. Inhibiting TORC1 with hypoxia stress (that can induce SG that may sequester mTORC1), causes a reactivation of HSV acute replication (Kobayashi et al., 2012). In this way, latent HSV-1 may respond to SG responses to activate acute replication that then suppresses SG responses. It will be interesting to determine if hypoxic stress in cultured neurons in this system actually sequesters mTORC1, completing a linkage between the two studies.
Herpes simplex virus 2 (HSV2) does not induce stress granules during acute infection. In fact, stress granule formation induced by oxidative stress is also inhibited, similar to the observation with MRV, SFV and PV. However, eIF2α phosphorylation does accumulate 10 hpi without induction of SGs (Finnen et al., 2012). Strikingly, stress granules containing G3BP1, Tia1 and PABP can be induced in infected cells with the eIF4A inhibitor pateamine A. However, instead of Tia1 localizing to cytoplasmic SG as seen in RNA-virus infected cells, it primarily localizes to discrete nuclear foci that contain Sam68. Even in the presence of pateamine A-induced stress granules, Tia1 predominantly localizes to Sam68-containing nuclear foci (Finnen et al., 2012). The role of the Tia1 nuclear foci is unclear, but unlike examples of G3BP1 redistribution where cells are resistant to SG induction with exogenous stressors, HSV-2-infected cells with Tia1 nuclear foci are still capable of forming SGs. Therefore, it is unlikely that sequestration of Tia1 in nuclear foci is responsible for inhibition of SG formation during infection. These data indicate that Tia1 may be augmenting HSV-2 viral replication or transcription similar to the case for Dengue and West Nile viruses.
The Arenavirus Junin virus does not cause induction of stress granules. These results are consistent with a lack of accumulation of eIF2α phosphorylation. Interestingly, expression of viral proteins N and glycoprotein precursor are sufficient to inhibit stress granule formation, while the matrix protein Z cannot inhibit SG assembly (Fig. 1) (Linero et al., 2011). Another study showed that Junin virus infection subverts some components of stress granules into replication-transcription complexes. G3BP1 colocalizes with the viral protein N, which are thought to augment the infection. However, neither PABP nor Tia1 enter the same foci, which are novel virus-specific foci but not SG. Interestingly, some initiation factors including eIF4G and eIF4A and large and small ribosomal subunit proteins L10a and S6 are also present in replication-transcription complexes. Although initiation factors and the ribosome may participate in translation of the viral RNA in replication-transcription complexes, it is unclear how G3BP may function. It is possible that G3BP1 is recruited to sequester it and disassemble stress granules to enhance Junin virus translation (Baird et al., 2012).
HIV1 is capable of inhibiting stress granules despite addition of arsenite, which normally induces eIF2α phosphorylation and stress granules. However, it is unclear whether SG inhibition happens early or later during infection as HIV1 infection was accomplished through transfection of proviral DNA so kinetics of SG formation have not been dissected. The mechanism of SG inhibition appears to be related to sequestration of the stress granule protein Staufen1 to RNPs containing the viral protein Gag and HIV1 viral RNA (Fig. 1) (Abrahamyan et al., 2010). Interestingly, depletion of Staufen1 co-depletes Gag from infected cells and results in enhanced encapsidation of viral mRNA and residual Staufen1. Furthermore, levels of the nonsense-mediated decay factor Upf1 and large ribosomal subunit protein L7 were increased in purified virus assembled in Staufen1 depleted cells. These results indicate that HIV1 prevents stress granule assembly to subvert Staufen1 to alternative RNPs that regulate viral encapsidation. It is unclear whether the virions assembled in Staufen1 depleted cells, which differ in composition from normal virus, are equally infectious.
Respiratory Syncytial Virus (RSV) was shown to induce stress granules during the course of infection (Lindquist et al., 2010, Lindquist et al., 2011). Lindquist et al. showed a striking increase in viral protein N production in cells containing stress granules. In cells that were genetically deficient for SG formation by knockdown of G3BP1, there was a 10-fold reduction in RSV titer, consistent with the observation that viral protein production is impaired in cells lacking stress granules. These results are reminiscent of results from the vaccinia virus system where, under some conditions, SG-like aggregates promote virus replication (discussed below) (Katsafanas and Moss, 2007). However, these viruses are clearly from different families and no association between RSV viral inclusion bodies and stress granules was observed in these studies (Lindquist et al., 2010, Lindquist et al., 2011). A contradictory study using the RSV system showed that stress granule formation was inhibited specifically by the trailer region of the viral genome (Hanley et al., 2010). However, this was consistent with earlier studies with Sendai virus, which is similar to RSV, and its trailer blocks SG formation during infection (Iseni et al., 2002). Previous work has documented a role for the trailer region in synthesis of progeny genomes. Hanley et al. showed that replacement of the trailer with a sequence complimentary to the 5′ leader resulted in robust SG formation. However, an unspecified eIF3 subunit was used as the only marker of stress granule formation, leaving the possibility that SG-like proviral structures form, which lack that eIF3 subunit, in a trailer-dependent manner.
The coronaviruses Transmissible Gastroenteritis (TGEV) and Mouse Hepatitis Coronavirus (MHC) both form stress granules as the infection progresses (Raaben et al., 2007, Sola et al., 2011). Appearance of stress granules during MHC infection correlates with an increase in eIF2α phosphorylation at 6 h post infection (Raaben et al., 2007). Consistent with those findings, infection with TGEV correlates with the appearance of stress granules at later times post infection (Sola et al., 2011). Appearance of TGEV-induced SGs is evident between 16 and 48 h post infection depending on the cell line being examined. There is no evidence that these granules disassemble during the infection for either TGEV or MHC. However, SGs were not examined at late time points in infection with MHC (Raaben et al., 2007).
Stress granules are unlikely to augment infection by TGEV or MHC, since depletion of PTB (TGEV), which may disrupt SGs as observed with other SG nucleating proteins (Tourrière et al., 2003, Gilks et al., 2004; Wilczynska et al., 2005; De Leeuw et al., 2007; Stoeklin et al. 2004), and growth of virus in mouse embryonic fibroblasts expressing the nonphosphorylatable eIF2α S51A mutant (MHC) both result in higher virus titers. These results indicate that, although the viruses continue to replicate under SG conditions, they do so less efficiently and benefit when SGs are ablated. The coronavirus examples presented here are in stark contrast to the RSV example where SGs appear to promote virus replication in at least some studies (Lindquist et al., 2010, Lindquist et al., 2011).
Vaccinia virus (VV) forms large viral replication factories that contain several components of stress granules including G3BP1, Caprin1, eIF4E, eIF4G and poly(A) mRNAs. Localization of these components of stress granules was suggested to result from coordinated transcription and translation in the replication factory rather than assembly of bona fide stress granules in the vicinity of the replication factory (Katsafanas and Moss, 2007). Caprin1 and G3BP1 interact with each other and regulate transcription of the VV genome (Katsafanas and Moss, 2004, Solomon et al., 2007). Coordinated transcription and translation agrees with recent data indicating eIF4G is recruited to single-stranded viral DNA by the action of the viral protein I3 (Zaborowska et al., 2012). In a separate study, aggregation of translation initiation factories in and around virus factories were shown to be devoid of the translational suppressor Tia1 (Walsh et al., 2008). These results indicate VV subverts several components of stress granules for proviral roles in replication factories.
A recent study has documented the colocalization of SG components (Tia1, eIF3b, G3BP1 and Usp10) in foci surrounding VV factories using cells infected with ΔE3L mutant vaccinia virus and suggested the SG components in “antiviral” granules can block virus replication in some circumstances (Simpson-Holley et al., 2011). E3L inhibits PKR activation and therefore prevents translational repression due to eIF2α phosphorylation; which is consistent with the observation that PKR activation, eIF2α phosphorylation and global translation arrest are necessary for formation of “antiviral” granules with E3L mutant virus (Simpson-Holley et al., 2011). These granules differ from canonical SGs in that they lack ribosomal subunits and are not dispersed by cycloheximide, but like SGs, they are formed in response to eIF2α phosphorylation. Because of the different properties they were termed “antiviral granules” (Simpson-Holley et al., 2011). Further, these authors did not find G3BP1-foci in wild type VV infected cells, but only in cells with translation and replication inhibition (E3L mutant virus). These results present a discrepancy with the previous study suggesting proviral roles for SG components in viral factories (Katsafanas and Moss, 2007). The authors explain the conflicting results by saying that the proviral granules previously documented (i) lacked Tia1, (ii) were present within viral factories whereas “antiviral” granules form surrounding virus factories, and (iii) “antiviral” granules require eIF2α phosphorylation whereas the proviral SG aggregates correlate with ongoing translation. Although a clear distinction has been made regarding the nature of proviral and “antiviral” granules, several questions remain. First, are the granules observed in each study the same, and why were not proviral granules observed in infections by wild type vaccinia virus by Simpson–Holley et al.? Second, is translation occurring in either proviral aggregates or the “antiviral” stress granule-like structures surrounding VV replication factories? These questions could be addressed using new technologies to visualize active translation at the single-cell level (Reineke et al., 2012, David et al., 2011, Dieterich et al., 2006). Third, if these mRNPs are modified stress granule structures and not sites of active translation, is translation repressed throughout the cell when “antiviral” granules are present as suggested by studies with ΔE3L mutant VV? And finally, are “antiviral” granules really distinct from stress granules or just morphologically distinct because of virus-induced alterations in cellular processes?
Viruses that modulate P-bodies
Compared to a large, rapidly growing literature on virus-stress granule interactions, less is known about how viruses interact with P-bodies and their constituents. This is an understudied area given that RNA viruses must regulate RNA decay processes/machinery that are concentrated within these RNA granules to prevent degradation of virus genomes, templates and mRNAs. However, early interest in the relationship between P-body components and some viruses has occurred because gene expression is closely linked to these structures. As one might expect, the emerging picture of the relationship between viruses and P-bodies appears as varied as with stress granules. We have loosely grouped the relationships into broad categories that will likely require revision as more research findings are produced ( Table 2). Fig. 2 illustrates some of the virus-PB relationships that are known.
Table 2.
| Viruses | Mechanism of PB interaction | References |
|---|---|---|
| P-bodies are dispersed or redistributed during infection | ||
| Poliovirus and Coxsackievirus | Disruption of P-bodies possibly through cleavage of Pan3, Xrn1 and Dcp1a. | Dougherty et al. (2011) |
| Adenovirus | P-bodies are diminished because E4 11K redistributes P-body components to aggresomes. | Greer et al. (2011) |
| Influenza A virus | Inhibition of P-bodies at mid-phase of infection through interaction between NS1 and Rap55. | Mok et al. (2012) |
| P-body components are co-opted to augment infection | ||
| West Nile virus | Disperses P-bodies and relocalizes Lsm1, GW182, DDX3, DDX6 and Xrn1 to viral replication centers. | Emara and Brinton (2007); Chahar et al. (2012) |
| Yellow fever virus, dengue virus, Kunjin Virus, West Nile virus | Xrn1 is necessary for production of viral sfRNA, but the consequences for P-body assembly are unknown. | Silva et al. (2010); Moon et al. (2012); Schnettler et al. (2012) |
| HIV1 | Conflicting studies: (1) HIV RNA does not localize to P-bodies, and (2) HIV RNA does localize to P-bodies. Also, Gag complexes with DDX6 and Ago2 to promote virion assembly. | Phalora et al. (2012); Nathans et al. (2009); Reed et al. (2012) |
| Hepatitis C virus | P-body proteins DDX6 (Rck/p54), Lsm1, Xrn1, PATL1 and Ago2 are redistributed to lipid droplets where viral production factories function. | Ariumi et al. (2011) |
| Hantavirus | Protein N protects the 5′ cap structure of cellular mRNAs in P-bodies to prime viral mRNA synthesis. | Mir et al. (2008) |
| Brome mosaic virus | Lsm1p, Pat1p and Dhh1p (Rck/p54) regulate translation of viral RNA and packaging at the cell membrane in yeast. | Noueiry et al. (2003) |
| Gamma herpes virus | SOX nuclease generates uncapped host RNAs for degradation by Xrn1. Effect on PBs unknown. | Covarrubias et al. (2011) |
PBs are dispersed or redistributed by virus infection
Several viruses appear to disperse PBs and PB components during the course of infection, however details of mechanisms involved are sparse. The flavivirus West Nile virus, which sequesters Tia1 on viral RNA, causes a progressive decline in the numbers of PBs in cells (Emara and Brinton, 2007). WNV also relocalizes many P-body components, including Lsm1, GW182, DDX3, DDX6 and Xrn1, to viral replication centers (Chahar et al., 2012). Localization of at least some of these components to WNV replication centers is important for RNA replication because siRNA-mediated depletion of Ago2, DDX3, Dicer, Lsm1 and GW182 all result in coincident reductions in viral RNA (Chahar et al., 2012). Localization of these P-body components to WNV replication centers may be due to interaction with viral genomic RNA. Indeed, with another flavivirus, Dengue virus genomic RNA binds DDX6 (Rck/p54) at a conserved stem loop structure in the 3′ UTR. Interaction of Dengue RNA with stress granule proteins G3BP1, USP10 and caprin1 occurs at an adjacent unstructured region of the 3′ UTR (Ward et al., 2011). The interaction with DDX6 is expected to be important since DDX6 knockdown reduced virus replication presumably through this interaction. Thus, Dengue and possibly West Nile virus can co-opt critical PB proteins for virus replication, and may interfere with their role in PB assembly.
Recent evidence indicates that the sfRNA generated by flaviviruses may be an important component of the viral lifecycle (Pijlman et al., 2008). During Yellow Fever Virus (YFV) infection, the sfRNA corresponds to a portion of the 3′ UTR of the genomic transcript generated by stalling of 5′–3′ exonunucleolytic decay by Xrn1 at pseudoknot 3 (Silva et al., 2010). Previous evidence indicates that the sfRNA does colocalize with Xrn1 in P-bodies, and its production is important for cytopathogenicity of the flavivirus Kunjin Virus (Pijlman et al., 2008). Although, the effect of the sfRNA on P-body integrity during infection was not examined for YFV, disruption of pseudoknot 3 and subsequent depletion of the sfRNA did not effect YFV viral titers (Silva et al., 2010). Furthermore, the consequences of depletion of Xrn1 on virus yield were not examined for YFV. More recent results indicate that sfRNA inhibits Xrn1 activity, resulting in accumulation of uncapped cellular mRNAs in cells (Moon et al., 2012). sfRNA also exhibits RNAi suppressor activity and can inhibit cleavage of dsRNA by Dicer (Schnettler et al., 2012). Thus, sfRNA has emerging roles in inhibiting host nucleases involved in gene regulation and innate immunity. These roles may indirectly affect RNA granule function.
Infection with the cytolytic enteroviruses poliovirus and Coxsackievirus B3 results in complete disruption of PB foci by the mid-phase of the replication cycle. Coincident with PB dispersal, three factors involved in mRNA turnover are quickly degraded, Xrn1 and Pan3, and Dcp1a may be directly cleaved by viral proteinase 3C (Fig. 2) (Dougherty et al., 2011). Deadenylation of mRNA has been linked to PB formation as a requirement, thus loss of Pan3, which regulates Pan2-mediated deadenylation mRNA, may be sufficient to block incorporation of mRNA into foci (Zheng et al., 2008). Alternatively, Dcp1a has also been linked to PB control and its cleavage may be key or contribute to PB loss (Rzeczkowski et al., 2011), but further work is required to test these hypotheses.
Adenovirus, a lytic DNA virus, has been shown to relocalize several P-body components, including Lsm-1, Ge-1, Ago2, Xrn1 and DDX6, to aggresomes where they are presumably slated for rapid proteolytic destruction. P-bodies were found to decrease in number by about a third in conjunction with this relocalization, but did not disappear. The adenovirus E4 11k protein drives this reorganization and was found to interact with DDX6 (Rck/p54), which may facilitate this process (Greer et al., 2011). E4 11k has many roles in controlling innate immunity and also relocalizes promyelocytic leukemia protein (PML) from nuclear PML bodies to unique elongated structures associated with virus replication centers. Thus, this virus protein can be seen to reorganize both cytoplasmic and nuclear structures while controlling host responses to infection (Greer et al., 2011).
Influenza virus causes disruption of PBs and SGs during mid-phase of the replication cycle in a process involving association of virus NS1 with the PB protein Rap55 (discussed above). One function of the NS1-Rap55 complex formation is to keep viral nucleoprotein from entering PBs (Mok et al., 2012).
Viruses co-opt P-body components to augment infection
Considerable attention within the HIV field has been directed at PBs due to the observation that certain APOBEC3 (apolipoprotein B mRNA editing enzyme catalytic polypeptide like 3) family members, that catalyze cytidine deamination, were included within PBs. The APOBEC3 family provides innate immune functions against HIV and contains seven proteins, four of which have anti-HIV1 functions and are targeted by viral Vif protein for degradation (Fig. 2). Two APOBEC3 members, APOBEC3F and APOBEC3G, interact with many RNA binding proteins, including the RISC protein Ago2, Dcp1a, Dcp2 and DDX6, and thus enter PBs (Kozak et al., 2006, Gallois-Montbrun et al., 2007). Argonaute proteins involved in miRNA repression also localize to PBs and interact with APOBEC3F and APOBEC3G (Gallois-Montbrun et al., 2007, Gallois-Montbrun et al., 2008). Antiviral activities of APOBEC3 proteins take place at multiple stages of the viral replication cycle, including promiscuous packaging of APOBEC3 into virions where its deaminase function damages viral genomes, thus inhibiting the next cycle of infection (Wang et al., 2008). Inclusion of APOBEC3 into PBs was not required for this antiviral packaging step indicating APOBEC3 family members do not alter HIV-1 virion production in a P-body-dependent manner.
Repression of HIV infection has been reported by a human miRNA miR-29a (Nathans et al., 2009). However, the antiviral effect of APOBEC3G and other APOBEC family members was not attributed to regulation of miRNA-mediated repression of HIV-1 translation (Nathans et al., 2009). As APOBEC proteins have been shown to confer antiviral activity, interact with P-body proteins and localize to P-bodies, interaction of HIV-1 mRNA with P-bodies was examined (Nathans et al., 2009, Phalora et al., 2012). While neither HIV-1 Gag RNA nor Gag protein could be detected in P-bodies in one study (Phalora et al., 2012), HIV-1 mRNA was shown to interact with P-body proteins APOBEC3G, Ago2 and DDX6 and colocalize P-bodies in another (Nathans et al., 2009). This discrepancy may be due to artifacts associated with tethering of MS2-GFP fusion proteins to the 3′ UTR of the Gag mRNA. Phalora and colleagues showed depletion of Lsm1 and DDX6 did not rescue the antiviral effect of APOBEC3G expression. However, Nathans et al. showed ablation of expression of Lsm1 and DDX6 caused an increase in viral production and infectivity by 2–3 fold in the absence of APOBEC3G overexpression. These results indicate APOBEC3G may act downstream of Lsm1 and DDX6 in the HIV-1 lifecycle. Despite the discrepancy between these results, both studies showed that knockdown of Ago proteins increase viral infectivity. Nathans et al. went on to show knockdown of other components of the siRNA pathway including Dicer and Drosha resulted in increased HIV-1 viral infectivity. It remains unclear whether the results observed in these studies are due to decreased function of miRNAs on the virus, as suggested by Drosha, Dicer and miR-29a depletion, or due to the depletion of P-bodies and associated functions from the cell as suggested by DDX6, Lsm1 or Ago proteins (Phalora et al., 2012, Nathans et al., 2009).
Finally, investigation of the role of APOBEC in HIV biology focused attention on Mov10, a putative RNA helicase protein that associates with APOBEC in RISC complexes and is found in PBs. Like APOBEC, Mov10 can be incorporated into virions and was found to inhibit HIV replication at multiple stages and also associated with HIV RNA (Burdick et al., 2010). Another report mostly concurred, but found overexpression inhibits retrovirus replication, primarily at the reverse transcriptase stage, for both HIV and murine leukemia virus. Curiously, depletion of Mov10 also inhibited replication modestly (Burdick et al., 2010, Furtak et al., 2010). Virus inhibitory activity of Mov10 was mapped to the N-terminus of Mov10 rather than the putative helicase domain indicating the helicase activity of Mov10 is required. No specific role for Mov10 in PB biology has yet been linked to HIV replication.
HIV Gag protein was shown to co-opt a cellular complex containing DDX6 and Ago 2 (Reed et al., 2012). This complex is thought to promote virion assembly. Consistent with that hypothesis, Ago2 and DDX6 colocalize with Gag at assembly sites at the plasma membrane (Fig. 2). A role for DDX6 in HIV assembly is supported by knockdown of DDX6, which results in a marked reduction in virion assembly. However, these results are inconsistent with those of the aforementioned studies showing an increase in viral titer when DDX6 is depleted. It is unclear what the discrepancy is between these studies, but they require resolution to understand the function of P-body proteins in the HIV-1 lifecycle.
Several lines of evidence suggest HCV interact with PBs or PB proteins. Two reports indicated that the HCV core protein forms complexes with DDX3 and colocalizes in cytoplasmic punctae similar to PBs (Mamiya and Worman, 1999, You et al., 1999). Knockdown of DDX3 results in repression of HCV replication, implying an important role in replication (Ariumi et al., 2007). However a subsequent report utilized mutagenesis of DDX3 to abrogate interaction with HCV core protein to determine that the requirement of HCV for DDX3 did not require interaction with the core protein (Angus et al., 2010).
As discussed above, more recent work suggests HCV co-opts SG components into novel viral-induced foci that block SG formation. In conjunction with this, HCV also reorganizes the distribution of PB components DDX6 (Rck/p54), Lsm1, Xrn1, PATL1 and Ago2 into viral production factories containing HCV core protein that organize around lipid droplets, the same factories that gain SG components G3BP1, ataxin-2 and PABP (Ariumi et al., 2011). Interestingly, not all PB components are co-opted into these new structures, e.g., Dcp2 remained in foci reminiscent of endogenous PBs and did not colocalize with new viral structures (Ariumi et al., 2011). Although HCV is clearly interfering with canonical PBs, retention of Dcp2 suggests an alternate vestigial structure may be retained, analogous to Tia1 pseudo-SG foci that persist in poliovirus infected cells (White and Lloyd, 2011). These vestigial foci may provide clues about the minimal requirements for foci formation.
Knockdown and interaction experiments indicate several PB constituents may be required at some level to support HCV replication, including DDX3, DDX6, Lsm1 and PatL1 (Ariumi et al., 2011). DDX6, PatL1 and the Lsm1–7 heptameric ring play essential roles in the HCV life cycle at the translational and RNA replication levels (Scheller et al., 2009, Jangra et al., 2010). Lsm1 and DDX6 interact with HCV viral RNA directly, but given the interplay of PB components with HCV replication, it is unclear if PB foci per se inhibit or influence HCV replication. Recent work suggests they do not inhibit HCV as PB knockdown by siRNA depletion of Rap55 did not alter HCV replication or influence HCV protein levels (Pérez-Vilaró et al., 2012). HCV may hijack DDX6 for RNA packaging, which has been observed previously for the spumaretrovirus foamy virus (Yu et al. 2011). Together these data suggest that HCV co-opts certain PB constituents for replicative functions; however, there is no requirement for PB foci in HCV replication (Fig. 2). The converse experiment to examine the whether stabilization of PBs during infection has an inhibitory effect on replication has not been tested. Finally, unlike more rapidly growing and lytic enteroviruses, HCV does not seem to rely on cleavage and degradation of PB components by virus proteases.
Like the well-characterized Orthomyxovirus influenza, the ambisense segmented RNA virus family Bunyaviridae, also initiate viral transcription by “cap-snatching,” to acquire 5′ m7-guanosine capped oligonucleotides from cellular mRNAs. Unlike influenza, Bunyaviruses such as Hantavirus replicate in the cytoplasm of infected cells. To facilitate this process, Hantavirus nucleocapsid protein (N) binds tightly to the 5′ cap of cellular mRNAs and accumulates in PBs to prevent decapping/degradation from Dcp1a/Dcp2 complexes (Fig. 2). The sequestered and protected 5′ caps are then utilized to prime virus mRNA synthesis (Mir et al., 2008). An important feature is that PBs naturally can exchange/release bound mRNP cargo back to the cytoplasm. This is required for Hantavirus transcripts to reenter the soluble cytoplasmic milieu to engage ribosomes to translate virus proteins. It will be interesting to determine if Hantavirus accelerates PB content flux to facilitate release of virus transcripts. The ability of Hantavirus N protein to bind caps, thus excluding eIF4E/eIF4F interaction, may play a role in preferential transport of virus transcripts out of PBs (Mir and Panganiban, 2008). This mechanism would involve subverting P-body function, rather than specific PB components, for generating capped viral mRNAs that could then be preferentially translated.
Gammaherpesvirus promotes turnover of cellular RNAs to promote translation of viral transcripts. The viral protein SOX was recently shown to induce endonucleolytic cleavage of cellular RNAs at TGAAG motifs (Covarrubias et al., 2011). This cleavage event generates unprotected 5′ end intermediates that are then turned over by the P-body component Xrn1. Cleavage intermediates accumulate in 40S translation preinitiation complexes indicating that SOX may bind or be activated in these complexes. However, neither movement of the ribosome over the SOX cleavage site nor ribosome subunit joining are believed to be required for activity of SOX on its substrates (Covarrubias et al., 2011). Therefore, a detailed mechanism of SOX activity is still needed. One point for clarification is whether all cellular mRNAs contain enough of a consensus motif to trigger endonucleolytic cleavage by SOX or if degradation of the most highly translated cellular mRNAs is sufficient “host shutoff” for ongoing viral RNA translation. It will also be important to investigate the proportion of Xrn1 involved in SOX-induced degradation of cellular mRNAs and whether removal of Xrn1 from P-bodies during gammaherpesvirus infection results in P-body disassembly.
Viruses of non-mammalian animals and plants
The insect dicistrovirus Cricket paralysis virus also moderately disperses insect PBs foci marked with certain GFP-tagged proteins by late times in infection. Foci tagged with GFP-GW182 and GFP-DCP1 diminished; however, AGO1 or AGO2 foci tagged with GFP did not, suggesting that PB constituents are modified during infection resulting in alternate PB-like foci of undetermined function (Khong and Jan, 2011).
The plant virus-RNA granule relationships are poorly studied. However, emerging evidence indicates plant cells utilize RNA granules for posttranscriptional gene control similar to the mammalian cell example. PB structures containing Dcp1, Dcp2 and Xrn4 have been identified in Arabidopsis and tobacco protoplasts, and SG-like structures were identified by eIF4E, UBP1, PABP and small ribosome subunit proteins (Weber et al., 2008, Pomeranz et al., 2010). In Arabadopsis, a tandem zinc finger protein, AtTZF1, shuttles into both PB-like structures and SG-like structures. TZF proteins recruit and activate mRNA decay machinery in mammalian cells (Fenger-Grøn et al., 2005; Lykke-Andersen and Wagner, 2005), and may nucleate PBs in conjunction with silencing of mRNAs containing AU-rich elements. AtTZF1 may fill a similar role in plant cells.
We are not aware of studies examining plant virus effects directly on plant RNA granules; however brome mosaic virus (BMV), which belongs to the alphavirus-like superfamily, has been studied during its replication cycle in yeast. PB constituent proteins affect BMV replication in two ways. First, genetic screens indicated the Lsm1p-7p complex, Pat1p and Dhh1p (Rck/p54, DDX6) were all required for efficient translation of all the virus mRNAs. Second, the same proteins were also required for entry of viral RNA into replication complexes on membranes (Noueiry et al., 2003). Since these proteins are generally associated with translation repression, it is unclear how they stimulate BMV translation; however; a role in the switch from active translation to RNA replication is possible, which requires clearing viral RNA templates of ribosomes. Interestingly, viral RNA and the viral RNA-dependent RNA polymerase, which complexes with Lsm1p, was observed to enter PBs (Beckham et al., 2007). Also, some PBs can associate with membranes where viral replication complexes are built (Wang et al., 2005, Beckham et al., 2007). It will be interesting to determine if PBs interact at any level with membranes such as autophagic vesicles that are co-opted by RNA viruses to anchor RNA replication complexes.
Interfaces between RNA granules, stress responses and innate immunity
All cells process stress responses through regulation of gene expression at translation and RNA decay levels. Plus strand RNA viruses must also transition their genomes from active translation to translational repression to allow RNA replication on the same template. Thus, it is not surprising that many RNA regulatory proteins discussed above are linked to virus replication schemes. However, virus infection induces host stress responses at multiple levels, thus cellular stress sensors may be part of virus sensing mechanisms used to activate innate immune functions. Indeed emerging evidence indicates innate immunity and cell stress responses, even SG and PB function, are linked at many levels. These interfaces may principally signal through PKR, and G3BP1 may also play a central role.
Several links between PKR and either stress or nutrient sensing exist. PKR was shown to coordinate pathogen sensing with cellular stress and metabolic homeostasis (Nakamura et al., 2010). PKR is also important for regulation of c-Jun N-terminal kinase (JNK) activation that is involved in stress responses, and PKR has been implicated in insulin activity and metabolism by phosphorylating the insulin receptor substrate IRS1 (Yang et al., 2010). Thus, the nutrient and pathogen response systems may be integrated through PKR.
On a different level, PKR is involved in inflammasome activation in macrophages in response to dsRNA, adjuvant and bacterial infection that results in release of cytokines IL-1b and HMGB1 (Lu et al., 2012). It has also been demonstrated that G3BP1 can induce stress granules to form in the absence of applied stress or infection, which can activate PKR thereby inducing eIF2-dependent translational repression. This provides a link between the formation of a stress granule alone and PKR activation (Reineke et al., 2012). It is unclear how PKR is activated by stress granules and whether other components of the PKR signaling axis are also activated. Nuclear factor kappa B (NF-kB) is a transcription factor involved in immune and inflammatory responses (Ghosh et al., 1998). PKR activity has been linked to NF-kB translocation to the nucleus and subsequent transcriptional activation (Gil et al., 2001). Consistent with those results, PKR directly interacts with the IkB kinase complex, which phosphorylates IkB and induces dissociation of IkB from NF-kB thereby inducing NF-kB transcriptional activation (Gil et al., 2001, Bonnet et al., 2000).
RIG-I like receptors (RIG-I, MDA-5, LGP2) (RLRs) that sense viral RNA can enter SGs after arsenite induction and virus infection with an NS1 deletion mutant influenza virus (Onomoto et al., 2012). This indicates that cells naturally concentrate interferon response activating proteins together with stress granule proteins. Functional linkage of the granule-based sensing mechanism to the interferon response was shown by a loss of IFNβ mRNA production by depletion of PKR or G3BP1, the latter of which depletes SGs. PKR was also found to enter stress granules, bringing together, in a concentrated fashion, many components of the innate immune response and viral RNA, which entered granules in the IAV ΔNS1 virus. PKR has also been shown to enter P-bodies during human papilloma virus infection (Hebner et al., 2006). Further, PKR was shown to have a critical role in the formation of SG in influenza infection as SGs did not form in PKR knockout MEFs, a phenotype observed with many other viruses (Khaperskyy et al., 2011, Onomoto et al., 2012). One feature of the influenza-induced antiviral stress granules in this study was the inclusion of virus RNA in the granule, but this only occurred with IAV ΔNS1 virus, not wild type IAV (Onomoto et al., 2012). Studies with wild type poliovirus indicate that viral RNA does not enter virus-induced stress granules (Piotrowska et al., 2010); thus, inclusion of viral RNA in these structures is variable depending on the virus and is likely counteracted by viral proteins. Together, these studies raise the possibility that activation of other stress signaling pathways is mediated by stress granules. Indeed, the stress-responsive MAPK JNK is activated in a noncanonical manner during stress granule formation (Wasserman et al., 2010).
Perspectives
As the field of virus-RNA granule interactions is moving from its infancy to adolescence, much work and several key questions remain. Although it is clear that RNA granules negatively regulate many viruses, the scope of viruses regulated by RNA granules is not completely understood. Additionally, the number of mechanisms employed by viruses to subvert RNA granule function appears nearly as great as the number of different virus families, but details of the mechanisms involved remain murky. Several virus systems described above may sequester RNA granule components, but in all cases, the real role of those components in RNA granule assembly and function is unclear. As such, viruses are excellent probes of cellular biology and regulation of RNA granules by viruses poses an opportunity to understand more about mechanisms behind RNA granule biology. Furthermore, all stress granules are not identical, so the persistence of one type of granule during infection does not mean that a functional SG or PB is present in the cell. Thus it is not only important to follow multiple components of each granule during infection, but to examine the functional consequences of RNA granule persistence, e.g., translational repression for SGs and RNA stability for PBs. Finally, the emerging concept that SG formation signals downstream stress signals that activate innate antiviral mechanisms as part of an integrated stress response should receive more attention. As stress responses and innate immunity likely crosstalk at multiple levels, it is possible that RNA granule biology may be exploited as a broad spectrum antiviral strategy.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant R01 AI AI50237 to R.E.L. and an NCI Cancer Center Support Grant (P30CA125123). L.C.R. was supported in part by National Institutes of Health Training Grant T32 AI07471. No conflicts of interest exist between the authors and the subject matter.
References
- Abrahamyan L.G., Chatel-Chaix L., Ajamian L., Milev M.P., Monette A., Clément J.-F., Song R., Lehmann M., DesGroseillers L., Laughrea M., Boccaccio G., Mouland A.J. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Angus A.G.N., Dalrymple D., Boulant S., Mcgivern D.R., Clayton R.F., Scott M.J., Adair R., Graham S., Owsianka A.M., Targett-Adams P., Li K., Wakita T., McLauchlan J., Lemon S.M., Patel A.H. Requirement of cellular DDX3 for hepatitis C virus replication is unrelated to its interaction with the viral core protein. J. Gen. Virol. 2010;91:122–132. doi: 10.1099/vir.0.015909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Abe K.-I., Dansako H., Ikeda M., Wakita T., Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariumi Y., Kuroki M., Kushima Y., Osugi K., Hijikata M., Maki M., Ikeda M., Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011;85:6882–6892. doi: 10.1128/JVI.02418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere J.A., Doudna J.A., Gilbert W.V. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell. 2011;44:745–758. doi: 10.1016/j.molcel.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N.L., York J., Nunberg J.H. Arenavirus infection induces discrete cytosolic structures for RNA replication. J. Virol. 2012;86:11301–11310. doi: 10.1128/JVI.01635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham C.J., Light H.R., Nissan T.A., Ahlquist P., Parker R., Noueiry A. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 2007;81:9759–9768. doi: 10.1128/JVI.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet M.C., Weil R., Dam E., Hovanessian A.G., Meurs E.F. PKR stimulates NF-kappaB irrespective of its kinase function by interacting with the IkappaB kinase complex. Mol. Cell. Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese F., Michiels T. The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 2011;85:9614–9622. doi: 10.1128/JVI.00480-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Muhlrad D., Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick R., Smith J.L., Chaipan C., Friew Y., Chen J., Venkatachari N.J., Delviks-Frankenberry K.A., Hu W.-S., Pathak V.K. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J. Virol. 2010;84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron-Herger M., Rippe K. Nuclear architecture by RNA. Curr. Opin. Genet. Dev. 2012;22:179–187. doi: 10.1016/j.gde.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Chahar H.S., Chen S., Manjunath N. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology. 2012 doi: 10.1016/j.virol.2012.09.041. Epub Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.-C., Yamashita A., Chen C.-Y.A., Yamashita Y., Zhu W., Durdan S., Kahvejian A., Sonenberg N., Shyu A.-B. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Gene. Dev. 2004;18:2010–2023. doi: 10.1101/gad.1219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias S., Gaglia M.M., Kumar G.R., Wong W., Jackson A.O., Glaunsinger B.A. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. Plos Pathogen. 2011;7:e1002339. doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney S.C., Scherbik S.V., Stockman B.M., Brinton M.A. West nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J. Virol. 2012;86:3647–3657. doi: 10.1128/JVI.06549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea I.M., Rozjabek H., Molloy K.R., Karki S., White L.L., Rice C.M., Rout M.P., Chait B.T., MacDonald M.R. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J. Virol. 2010;84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- David A., Netzer N., Strader M.B., Das S.R., Chen C.Y., Gibbs J., Pierre P., Bennink J.R., Yewdell J.W. RNA binding targets aminoacyl-tRNA synthetases to translating ribosomes. J. Cell Biol. 2011;286:20688–20700. doi: 10.1074/jbc.M110.209452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leeuw F., Zhang H., Wauquier C., Huez G., Kruys V., Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Dieterich D.C., Link A.J., Graumann J., Tirrell D.A., Schuman E.M. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal moncanonical amino acid tagging (BONCAT) Proc. Nat. Acad. Sci. U.S.A. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty J.D., White J.P., Lloyd R.E. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011;85:64–75. doi: 10.1128/JVI.01657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason T.S.K., Andrade J., Groehler A.L., Clark D.E., Muratore-Schroeder T.L., Pasic L., Smith J.A., Shabanowitz J., Hunt D.F., Macara I.G., Lannigan D.A. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell. 2008;31:722–736. doi: 10.1016/j.molcel.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M.M., Brinton M.A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Nat. Acad. Sci. U.S.A. 2007;104:9041–9046. doi: 10.1073/pnas.0703348104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara M.M., Fujimura K., Sciaranghella D., Ivanova V., Ivanov P., Anderson P. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem. Biophys. Res. Commun. 2012;423:763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M., Baconnais S., Harper M., Minshall N., Souquere S., Boudier T., Benard M., Andrey P., Pierron G., Kress M., Standart N., le Cam E., Weil D. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–1715. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Grøn M., Fillman C., Norrild B., Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Finnen R.L., Pangka K.R., Banfield B.W. Herpes simplex virus 2 infection impacts stress granule accumulation. J. Virol. 2012;86:8119–8130. doi: 10.1128/JVI.00313-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova E., Gorchakov R., Garmashova N., Atasheva S., Vergara L.A., Frolov I. Formation of nsP3-specific protein complexes during Sindbis virus replication. J. Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fros J.J., Domeradzka N.E., Baggen J., Geertsema C., Flipse J., Vlak J.M., Pijlman G.P. Chikungunya Virus nsP3 Blocks Stress Granule Assembly by Recruitment of G3BP into Cytoplasmic Foci. J. Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak V., Mulky A., Rawlings S.A., Kozhaya L., Lee K., Kewalramani V.N., Unutmaz D. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS ONE. 2010;5:e9081. doi: 10.1371/journal.pone.0009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Holmes R.K., Swanson C.M., Fernández-Ocaña M., Byers H.L., Ward M.A., Malim M.H. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J. Virol. 2008;82:5636–5642. doi: 10.1128/JVI.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C.M., Byers H., Lynham S., Ward M., Malim M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaigorta U., Heim M.H., Boyd B., Wieland S., Chisari F.V. Hepatitis C Virus Induces the Formation of Stress Granules whose Proteins Regulate HCV RNA Replication, Virus Assembly and Egress. J. Virol. 2012;86:11043–11056. doi: 10.1128/JVI.07101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. Impact of Protein Kinase PKR in Cell Biology: from Antiviral to Antiproliferative Action. Microbiol. Mol. Biol. Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., May M.J., Kopp E.B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Gil J., Rullas J., Garcia M.A., Alcamí J., Esteban M. The catalytic activity of dsRNA-dependent protein kinase, PKR, is required for NF-kappaB activation. Oncogene. 2001;20:385–394. doi: 10.1038/sj.onc.1204109. [DOI] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchakov R., Garmashova N., Frolova E., Frolov I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 2008;82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I., Boisvenue S., Mokas S., Mazroui R., Côté J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet. 2008;17:3055–3074. doi: 10.1093/hmg/ddn203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer A.E., Hearing P., Ketner G. The adenovirus E4 11k protein binds and relocalizes the cytoplasmic P-body component Ddx6 to aggresomes. Virology. 2011;417:161–168. doi: 10.1016/j.virol.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley L.L., Mcgivern D.R., Teng M.N., Djang R., Collins P.L., Fearns R. Roles of the respiratory syncytial virus trailer region: effects of mutations on genome production and stress granule formation. Virology. 2010;406:241–252. doi: 10.1016/j.virol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebner C.M., Wilson R., Rader J., Bidder M., Laimins L.A. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol. 2006;87:3183–3193. doi: 10.1099/vir.0.82098-0. [DOI] [PubMed] [Google Scholar]
- Iseni F., Garcin D., Nishio M., Kedersha N., Anderson P., Kolakofsky D. Sendai virus trailer RNA binds TIAR, a cellular protein involved in virus-induced apoptosis. EMBO J. 2002;19:5141–5150. doi: 10.1093/emboj/cdf513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra R.K., Yi M., Lemon S.M. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J. Virol. 2010;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas G.C., Moss B. Vaccinia virus intermediate stage transcription is complemented by Ras-GTPase-activating protein SH3 domain-binding protein (G3BP) and cytoplasmic activation/proliferation-associated protein (p137) individually or as a heterodimer. J. Biol. Chem. 2004;279:52210–52217. doi: 10.1074/jbc.M411033200. [DOI] [PubMed] [Google Scholar]
- Katsafanas G.C., Moss B. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe. 2007;2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Meth. Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy D.A., Hatchette T.F., McCormick C. Influenza a virus inhibits cytoplasmic stress granule formation. FASEB J. 2011 doi: 10.1096/fj.11-196915. [DOI] [PubMed] [Google Scholar]
- Khong A., Jan E. Modulation of stress granules and P bodies during dicistrovirus infection. J. Virol. 2011;85:1439–1451. doi: 10.1128/JVI.02220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Wilson A.C., Chao M.V., Mohr I. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Gene. Dev. 2012;26:1527–1532. doi: 10.1101/gad.190157.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak S.L., Marin M., Rose K.M., Bystrom C., Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- Kwon S., Zhang Y., Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Gene. Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros S., Boxus M., Gatot J.S., Van Lint C., Kruys V., Kettmann R., Twizere J.C., Dequiedt F. The HTLV-1 Tax protein inhibits formation of stress granules by interacting with histone deacetylase 6. Oncogene. 2011;30:4050–4062. doi: 10.1038/onc.2011.120. [DOI] [PubMed] [Google Scholar]
- Li W., Li Y., Kedersha N., Anderson P., Emara M., Swiderek K.M., Moreno G.T., Brinton M.A. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 2002;76:11989–12000. doi: 10.1128/JVI.76.23.11989-12000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M.E., Lifland A.W., Utley T.J., Santangelo P.J., Crowe J.E. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 2010;84:12274–12284. doi: 10.1128/JVI.00260-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M.E., Mainou B.A., Dermody T.S., Crowe J.E. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology. 2011 doi: 10.1016/j.virol.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linero F.N., Thomas M.G., Boccaccio G.L., Scolaro L.A. Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2α phosphorylation. J. Gen. Virol. 2011;92:2889–2899. doi: 10.1099/vir.0.033407-0. [DOI] [PubMed] [Google Scholar]
- Lu B., Nakamura T., Inouye K., Li J., Tang Y., Lundbäck P., Valdes-Ferrer S.I., Olofsson P.S., Kalb T., Roth J., Zou Y., Erlandsson-Harris H., Yang H., Ting J.P.Y., Wang H., Andersson U., Antoine D.J., Chavan S.S., Hotamisligil G.S., Tracey K.J. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 2012;488:670–674. doi: 10.1038/nature11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J., Wagner E., Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Gene. Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N., Worman H.J. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J. Biol. Chem. 1999;274:15751–15756. doi: 10.1074/jbc.274.22.15751. [DOI] [PubMed] [Google Scholar]
- Mangus D.A., Amrani N., Jacobson A. Pbp1p, a factor interacting with saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell Biol. 1998;18:7383–7396. doi: 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Zhang B., Spector D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M.-E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., Pelletier J. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 2006;17:4212–4219. doi: 10.1091/mbc.E06-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney G.M., Kedersha N.L., Kaufman R.J., Anderson P., Liljeström P. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell. 2005;16:3753–3763. doi: 10.1091/mbc.E05-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Duran W.A., Hjelle B.L., Ye C., Panganiban A.T. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Nat. Acad. Sci. U.S.A. 2008;105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.A., Panganiban A.T. A protein that replaces the entire cellular eIF4F complex. EMBO J. 2008;27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok B.W.-Y., Song W., Wang P., Tai H., Chen Y., Zheng M., Wen X., Lau S.-Y., Wu W.L., Matsumoto K., Yuen K.-Y., Chen H. The NS1 protein of influenza a virus interacts with cellular processing bodies (P-bodies) and stress granules through RNA-associated protein 55 (RAP55) during virus infection. J. Virol. 2012 doi: 10.1128/JVI.00647-12. epub Sept 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokas S., Mills J.R., Garreau C., Fournier M.-J., Robert F., Arya P., Kaufman R.J., Pelletier J., Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell. 2009;20:2673–2683. doi: 10.1091/mbc.E08-10-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero H., Rojas M., Arias C.F., López S. Rotavirus infection induces the phosphorylation of eIF2alpha but prevents the formation of stress granules. J. Virol. 2008;82:1496–1504. doi: 10.1128/JVI.01779-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.L., Anderson J.R., Kumagi Y., Wilusz C.J., Akira S., Khromykh A.A., Wilusz J. A nondoing RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA. 2012;18:2029–2040. doi: 10.1261/rna.034330.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Furuhashi M., Li P., Cao H., Tuncman G., Sonenberg N., Gorgun C.Z., Hotamisligil G.S. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R., Chu C.-Y., Serquina A.K., Lu C.-C., Cao H., Rana T.M. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noueiry A.O., Díez J., Falk S.P., Chen J., Ahlquist P. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell Biol. 2003;23:4094–4106. doi: 10.1128/MCB.23.12.4094-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Scharf K.D., Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onomoto K., Jogi M., Yoo J.-S., Narita R., Morimoto S., Takemura A., Sambhara S., Kawaguchi A., Osari S., Nagata K., Matsumiya T., Namiki H., Yoneyama M., Fujita T. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS ONE. 2012;7:e43031. doi: 10.1371/journal.pone.0043031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A.D., Willers I.M., Sala S., Cuezva J.M. Human G3BP1 interacts with {beta}-F1-ATPase mRNA and inhibits its translation. J. Cell Sci. 2010;123:2685–2696. doi: 10.1242/jcs.065920. [DOI] [PubMed] [Google Scholar]
- Panas M.D., Varjak M., Lulla A., Eng K.E., Merits A., Hedestam G.B., McInerney G.M. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 2012 doi: 10.1091/mbc.E12-08-0619. Epub Oct 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Vilaró G., Scheller N., Saludes V., Díez J. Hepatitis C virus infection alters p-body composition but is independent of p-body granules. J. Virol. 2012;86:8740–8749. doi: 10.1128/JVI.07167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalora P.K., Sherer N.M., Wolinsky S.M., Swanson C.M., Malim M.H. HIV-1 replication and APOBEC3 antiviral activity are not regulated by P-bodies. J. Virol. 2012;86:11712–11724. doi: 10.1128/JVI.00595-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijlman G.P., Funk A., Kondratieva N., Leung J., Torres S., van der Aa L., Liu W.J., Palmenberg A.C., Shi P.-Y., Hall R.A., Kromykh A.A. A highly structured, nuclease-resistant noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–591. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Piotrowska J., Hansen S.J., Park N., Jamka K., Sarnow P., Gustin K.E. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 2010;84:3654–3665. doi: 10.1128/JVI.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz M.C., Hah C., Lin P.-C., Kang S.G., Finer J.J., Blackshear P.J., Jang J.-C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010;152:151–165. doi: 10.1104/pp.109.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Carroll K., Hastings C., Miller C.L. Mammalian Orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2{alpha} phosphorylation and PKR. J. Virol. 2011;85:8798–8810. doi: 10.1128/JVI.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q., Hastings C., Miller C.L. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J. Virol. 2009;83:11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Groot Koerkamp M.J.A., Rottier P.J.M., de Haan C.A.M. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 2007;9:2218–2229. doi: 10.1111/j.1462-5822.2007.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.C., Molter B., Geary C.D., McNevin J., McElrath J., Giri S., Klein K.C., Lingappa J.R. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., Dougherty J.D., Pierre P., Lloyd R.E. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 2012;23:3499–3510. doi: 10.1091/mbc.E12-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggieri A., Dazert E., Metz P., Hofmann S., Bergeest J.-P., Mazur J., Bankhead P., Hiet M.-S., Kallis S., Alvisi G., Samuel C.E., Lohmann V., Kaderali L., Rohr K., Frese M., Stoecklin G., Bartenschlager R. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe. 2012;12:71–85. doi: 10.1016/j.chom.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeczkowski K., Beuerlein K., Müller H., Dittrich-Breiholz O., Schneider H., Kettner-Buhrow D., Holtmann H., Kracht M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J. Cell Biol. 2011;194:581–596. doi: 10.1083/jcb.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller N., Mina L.B., Galão R.P., Chari A., Giménez-Barcons M., Noueiry A., Fischer U., Meyerhans A., Díez J. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Nat. Acad. Sci. U.S.A. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnettler E., Sterken M.G., Leung J.Y., Metz S.W., Geertsema C., Goldbach R.W., Vlak J.M., Kohl A., Kromykh A.A., Pijlman G.P. Non-coding flavivirus RNA displays RNAi suppressor activity in insect and mammalian cells. J.Virol. 2012 doi: 10.1128/JVI.01104-12. Epub Oct 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P.A., Pereira C.F., Dalebout T.J., Spaan W.J., Bredenbeek P.J., Silva AN RNA pesedoknot is required for production fo yellow fever virus subgenomic RNA by the host nuclease XRN1. J. Virol. 2010;84:11395–11406. doi: 10.1128/JVI.01047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu A.-B., Wilkinson M.F., van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M., Kedersha N., Dower K., Rubins K.H., Anderson P., Hensley L.E., Connor J.H. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J. Virol. 2011;85:1581–1593. doi: 10.1128/JVI.02247-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.A., Schmechel S.C., Raghavan A., Abelson M., Reilly C., Katze M.G., Kaufman R.J., Bohjanen P.R., Schiff L.A. Reovirus induces and benefits from an integrated cellular stress response. J. Virol. 2006;80:2019–2033. doi: 10.1128/JVI.80.4.2019-2033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Galán C., Mateos-Gómez P.A., Palacio L., Zúñiga S., Cruz J.L., Almazán F., Enjuanes L. The polypyrimidine tract-binding protein affects coronavirus RNA accumulation levels and relocalizes viral RNAs to novel cytoplasmic domains different from replication-transcription sites. J. Virol. 2011;85:5136–5149. doi: 10.1128/JVI.00195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., Schrader J.W. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell Biol. 2007;27:2324–2342. doi: 10.1128/MCB.02300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele L., Errington F., Prestwich R., Ilett E., Harrington K., Pandha H., Coffey M., Selby P., Vile R., Melcher A. Pro-inflammatory cytokine/chemokine production by reovirus treated melanoma cells is PKR/NF-κB mediated and supports innate and adaptive anti-tumour immune priming. Mol. Cancer. 2011;10:20. doi: 10.1186/1476-4598-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G., Stubbs T., Kedersha N., Wax S., Rigby W.F.C., Blackwell T.K., Anderson P. MK2-induced tristetraprolin: 14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi N., Samuel C.E. Protien kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology. 2012;427:208–216. doi: 10.1016/j.virol.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara T., Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell. 2012;47:242–252. doi: 10.1016/j.molcel.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Unsworth H., Raguz S., Edwards H.J., Higgins C.F., Yagüe E. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 2010;24:3370–3380. doi: 10.1096/fj.09-151142. [DOI] [PubMed] [Google Scholar]
- Walsh D., Arias C., Perez C., Halladin D., Escandon M., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Mohr I. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell Biol. 2008;28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Tian C., Zhang W., Sarkis P.T.N., Yu X.-F. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J. Mol. Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Wang X., Lee W.-M., Watanabe T., Schwartz M., Janda M., Ahlquist P. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 2005;79:13747–13758. doi: 10.1128/JVI.79.21.13747-13758.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward A.M., Bidet K., Yinglin A., Ler S.G., Hogue K., Blackstock W., Gunaratne J., Garcia-Blanco M.A. Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3′ UTR structures. RNA Biol. 2011;8:1173–1186. doi: 10.4161/rna.8.6.17836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell. 2010;21:117–130. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C., Nover L., Fauth M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008;56:517–530. doi: 10.1111/j.1365-313X.2008.03623.x. [DOI] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- White J.P., Lloyd R.E. Poliovirus unlinks TIA1 aggregation and mRNA stress granule formation. J. Virol. 2011;85:12442–12454. doi: 10.1128/JVI.05888-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Reineke L.C., Lloyd R.E. Poliovirus switches to an eIF2-independent mode of translation during infection. J. Virol. 2011;85:8884–8893. doi: 10.1128/JVI.00792-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Yang X., Nath A., Opperman M.J., Chan C. The double-stranded RNA-dependent protein kinase differentially regulates insulin receptor substrates 1 and 2 in HepG2 cells. Mol. Biol. Cell. 2010;21:3449–3458. doi: 10.1091/mbc.E10-06-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z., Pan T., Wu X., Song W., Wang S., Xu Y., Rice C.M., MacDonald M.R., Yuan Z. Hepatitis C virus co-opts ras-GTPase-activating protein-binding protein 1 for its genome replication. J. Virol. 2011;85:6996–7004. doi: 10.1128/JVI.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L.R., Chen C.M., Yeh T.S., Tsai T.Y., Mai R.T., Lin C.H., Lee Y.H. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J. Virol. 1999;73:2841–2853. doi: 10.1128/jvi.73.4.2841-2853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S.F., Lujan P., Jackson D.L., Emerman M., Linial M. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. Plos. Pathog. 2011 doi: 10.1371/journal.ppat.1002303. Oct;7(10):e1002303. Epub 2011 Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowska I., Kellner K., Henry M., Meleady P., Walsh D. Recruitment of host translation initiation factor eIF4G by the vaccinia virus ssDNA-binding protein I3. Virology. 2012;425:11–22. doi: 10.1016/j.virol.2011.12.022. [DOI] [PubMed] [Google Scholar]
- Zheng D., Ezzeddine N., Chen C.-Y.A., Zhu W., He X., Shyu A.-B. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]