Abstract
Background
Adjuvant tamoxifen therapy substantially decreases the risk of recurrence and mortality in women with hormone (estrogen and/or progesterone) receptor–positive breast cancer. Previous studies have suggested that metabolic conversion of tamoxifen to endoxifen by cytochrome P450 2D6 (CYP2D6) is required for patient benefit from tamoxifen therapy.
Methods
Tumor specimens from a subset of postmenopausal patients with hormone receptor–positive early-stage (stages I, II, and IIIA) breast cancer, who were enrolled in the randomized double-blind Arimidex, Tamoxifen, Alone or in Combination (ATAC) clinical trial, were genotyped for variants in CYP2D6 (N = 1203 patients: anastrozole [trade name: Arimidex] group, n = 615 patients; tamoxifen group, n = 588 patients) and UDP-glucuronosyltransferase-2B7 (UGT2B7), whose gene product inactivates endoxifen (N = 1209 patients; anastrozole group, n = 606 patients; tamoxifen group, n = 603 patients). Genotyping was performed using polymerase chain reaction–based TaqMan assays. Based on the genotypes for CYP2D6, patients were classified as poor metabolizer (PM), intermediate metabolizer (IM), or extensive metabolizer (EM) phenotypes. We evaluated the association of CYP2D6 and UGT2B7 genotype with distant recurrence (primary endpoint) and any recurrence (secondary endpoint) by estimating the hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) using Cox proportional hazards models. All statistical tests were two-sided.
Results
After a median follow-up of 10 years, no statistically significant associations were observed between CYP2D6 genotype and recurrence in tamoxifen-treated patients (PM vs EM: HR for distant recurrence = 1.25, 95% CI = 0.55 to 3.15, P = .64; HR for any recurrence = 0.99, 95% CI = 0.48 to 2.08, P = .99). A near-null association was observed between UGT2B7 genotype and recurrence in tamoxifen-treated patients. No associations were observed between CYP2D6 and UGT2B7 genotypes and recurrence in anastrozole-treated patients.
Conclusion
The results do not support the hypothesis that CYP2D6 genotype predicts clinical benefit of adjuvant tamoxifen treatment among postmenopausal breast cancer patients.
CONTEXT AND CAVEATS
Prior knowledge
Cytochrome P450 2D6 (CYP2D6) converts tamoxifen to the metabolically active endoxifen, and its gene polymorphisms are suggested to influence the outcome in tamoxifen-treated breast cancer patients. The question is, should patients be genotyped for CYP2D6?
Study design
Postmenopausal, hormone receptor–positive early-stage breast cancer patients from the UK population of the Arimidex (generic name anastrozole) Tamoxifen, Alone or in Combination (ATAC) Clinical Trial, who received tamoxifen, or anastrozole, were genotyped for CYP2D6 variants. UDP-glucuronosyltransferase-2B7 (UGT2B7) was also genotyped because its gene product inactivates endoxifen. Associations between genotypes and distant recurrence or any recurrence were assessed.
Contribution
CYP2D6 genotype showed no association with recurrence, which remained after adjustment for concomitant medication known to inhibit the CYP2D6 enzyme, and UGT2B7 genotype showed a near-null association with recurrence in tamoxifen-treated patients.
Implications
Reduced CYP2D6 enzyme activity was not associated with worse disease outcome. Results do not support CYP2D6 genotyping in patients considering tamoxifen because it did not predict clinical benefit of adjuvant tamoxifen treatment among postmenopausal breast cancer patients.
Limitations
Results are restricted to the UK component of the ATAC trial, and only to postmenopausal women. Circulating endoxifen levels were not measured, and compliance regarding tamoxifen intake was self-reported.
From the Editors
In hormone (estrogen and/or progesterone) receptor–positive breast cancer, 5 years of adjuvant tamoxifen therapy reduces disease recurrence by approximately half and breast cancer mortality by approximately a third (1). The parent compound tamoxifen is a relatively weak estrogen receptor (ER) antagonist but is converted in vivo into many metabolites with varying estrogenic and anti-estrogenic properties. Of these, 4-hydroxy-N-desmethyl-tamoxifen (known as endoxifen) binds ER with 100-fold greater affinity than tamoxifen and metabolite N-desmethyl-tamoxifen, and its serum concentration is 6- to 10-fold higher than the high affinity metabolite 4-hydroxy-tamoxifen (2–7). Cytochrome P450 2D6 (CYP2D6) and UDP-glucuronosyltransferase-2B7 (UGT2B7) are the primary and rate-limiting enzymes responsible for the formation and inactivation of endoxifen, respectively (3,8).
CYP2D6 genotype is associated with plasma concentrations of endoxifen (2,4), a finding that raised the hypothesis that CYP2D6 genotype may predict response to tamoxifen. We observed that the gene product of UGT2B7*2, a common genetic variant of UGT2B7 that exhibits decreased enzymatic activity (9), is associated with higher plasma endoxifen concentrations in patients receiving tamoxifen therapy (D. A. Flockhart, Indiana University, unpublished data). However, studies that tested associations between CYP2D6 genotype and benefit from tamoxifen have provided conflicting results (10,11). Some studies found that tamoxifen-treated breast cancer patients who are carriers of CYP2D6 alleles associated with reduced enzyme activity have worse outcomes compared with patients who carry the functional CYP2D6 gene (12,13). In contrast, other investigators have failed to observe a difference or have even suggested better clinical outcomes for patients with CYP2D6 genotypes associated with poor tamoxifen metabolism (14,15).
Most, if not all, previously reported studies of the association between CYP2D6 genotypes and clinical efficacy of tamoxifen have been confounded by a variety of biases and do not provide the level of evidence needed to recommend CYP2D6 genotyping for decisions regarding tamoxifen therapy (10,11,16,17). Recently, Simon et al. (18) have proposed a scale to define the level of evidence necessary for evaluation of clinical utility of tumor markers using results from archival specimens, specifically defining studies using specimens from “repurposed” prospective studies as the highest level (also designated as “prospective retrospective studies”). In this regard, the Arimidex, Tamoxifen, Alone or in Combination (ATAC) clinical trial was a prospective, randomized double-blind clinical trial to test the efficacy and safety of the aromatase inhibitor, anastrozole (trade name: Arimidex), vs tamoxifen for 5 years as initial adjuvant endocrine treatment in postmenopausal women with hormone receptor–positive early-stage breast cancer (19–26). In addition to disease outcomes, ATAC investigators collected comprehensive concomitant medication data during the 5 years of active treatment. This trial, which now has a 10-year median follow-up, provides an ideal platform for generating high-level evidence on whether CYP2D6 or UGT2B7 genotypes predict response to tamoxifen. We conducted a genetic substudy of the ATAC trial to test for associations between patient CYP2D6 and UGT2B7 genotypes with clinical outcomes in anastrozole- and tamoxifen-treated hormone receptor–positive breast cancer patients.
Methods
Study Population
Patient selection and sample collection in the ATAC trial have been described previously (19). A total of 9366 postmenopausal women from 381 centers in 21 countries were enrolled in the ATAC trial between July 12, 1996, and March 24, 2000, and were randomly assigned to the anastrozole alone (n = 3125 women), tamoxifen alone (n = 3116 women), and combined anastrozole and tamoxifen (n = 3125 women) groups. Eligible patients were postmenopausal women with histologically proven, operable invasive breast cancer, who had completed primary surgery and chemotherapy (where given) and were eligible to receive adjuvant hormonal therapy. Archival tumor blocks were requested for patients except those known to be ER negative and progesterone receptor negative according to immunohistochemical test conducted at the local institution (27). Patients were ineligible if they had metastatic disease, if chemotherapy was started more than 8 weeks after surgery, or completed more than 8 weeks before random assignment. In patients not receiving chemotherapy, surgery must have been completed less than 8 weeks before random assignment. Patients were ineligible if they had received hormonal therapy previously for breast cancer prevention or for adjuvant treatment of breast cancer.
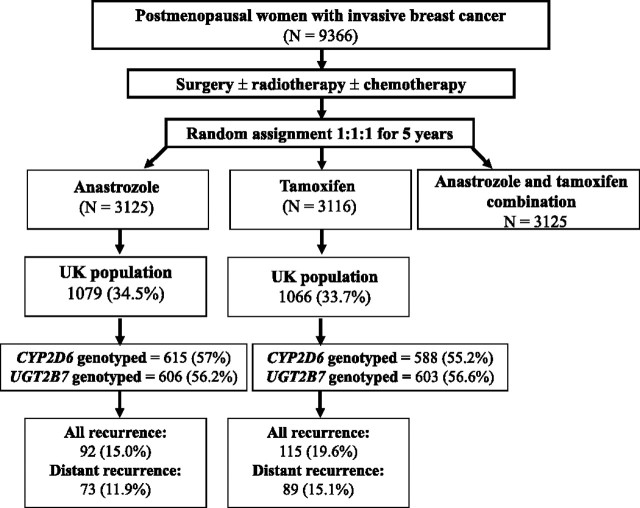
We designed a genetic substudy that represents patients enrolled in the ATAC trial only in the United Kingdom but were compared with all other patients (rest of the world [ROW]) in selected analyses. Only patients randomly assigned to either anastrazole or tamoxifen group, and none from the combined anastrazole and tamoxifen group, were included in this substudy. A CONSORT diagram (Figure 1) illustrates the selection of patients for germline genetic analyses. All patients provided consent to undergo genetic analyses, and this study was approved by the English National Research Ethics Service (via the Lewisham Local Ethics Committee).
Figure 1.
CONSORT diagram showing the number of patients from the Arimidex, Tamoxifen, Alone or in Combination (ATAC) clinical trial included in the genetic substudy. CYP2D6 = cytochrome P450 2D6; UGT2B7 = UDP-glucuronosyltransferase-2B7.
Genotype Analysis
DNA was extracted from formalin-fixed paraffin-embedded tumor specimens (n = 1327), as described previously (28). All single-nucleotide polymorphisms (SNPs) (Supplementary Table 1, available online) were determined using the polymerase chain reaction (PCR)–based TaqMan Allelic Discrimination Assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions, as described previously (28). All genotyping reactions were prepared in a vertical AirClean 600 laminar flow hood with HEPA filtration (AirClean Systems, Raleigh, NC), and 15% of tumor specimens from anastrozole and tamoxifen groups were randomly selected for duplicate determinations of SNPs resulting in 100% concordance. Researchers were blinded to the groups while genotyping.
CYP2D6 Genotyping.
Formalin-fixed, paraffin-embedded tissue-extracted DNA was genotyped for known SNPs in CYP2D6 using a TaqMan Allelic Discrimination Assay, as described previously (28). The following SNPs were genotyped: 1846G>A (rs3892097), T1707del (rs5030655), A2549del (rs35742686), 2988G>A (rs28371725), 100C>T (rs1065852), and 4180G>C (rs1135840). Genotyping for these SNPs allows the identification of the most common alleles of CYP2D6: ancestral or wild-type (WT) allele CYP2D6*1, and variant alleles CYP2D6*2, CYP2D6*3, CYP2D6*4, CYP2D6*6, CYP2D6*10, and CYP2D6*41. PCR reactions were carried out to 60 cycles to allow amplification of sub-nanogram quantities of DNA. The PCR conditions were as follows: 95°C for 10 minutes followed by 60 cycles of denaturation at 92°C for 15 seconds and annealing and extension for 1 minute. The temperature for the annealing and extension step was optimized for each individual SNP and ranged from 57°C to 60°C.
UGT2B7 Genotyping.
Metabolic inactivation of endoxifen is mediated by UGT2B7 (8). We found that the common genetic variant of UGT2B7, UGT2B*2, was associated with higher endoxifen concentrations in patients receiving tamoxifen (D. A. Flockhart, Indiana University, unpublished data). Tumor specimens were genotyped for ancestral or WT allele UGT2B7*1 and variant allele UGT2B7*2 (rs7439366) using a TaqMan Allelic Discrimination Assay, as described previously (28). The PCR conditions were as follows: 95°C for 10 minutes followed by 60 cycles of denaturation at 92°C for 15 seconds and annealing and extension at 59°C for 1 minute.
Phenotypic Scoring of CYP2D6
We assigned a CYP2D6 “activity score” to patients based on their CYP2D6 genotype, using the method described by Blake et al. (29) with minor modifications as described previously (30). The CYP2D6 activity score is an approach to predict most accurately a patient's CYP2D6 metabolic phenotype based on their genotype (31). Briefly, each CYP2D6 allele was assigned a value from 0 (for nonfunctional alleles) to 1 (for fully functioning alleles) based on the relative catalytic activity of its gene product for substrate O-demethylation of dextromethorphan (29). Each patient's activity score represents the sum total of their individual CYP2D6 alleles, and patients can be categorized into one of the five groups of scores (0, 0.5, 1.0, 1.5, or 2.0); score 0 representing the poor metabolizer (PM) phenotype; scores 0.5, 1.0, and 1.5 representing intermediate metabolizer (IM) phenotype; and score 2.0 representing the extensive metabolizer (EM) phenotype of CYP2D6. The EM phenotype is also the CYP2D6 homozygous WT phenotype.
All concomitant medications were documented by self-report at each follow-up visit and recorded in the database. Concomitant medication usage was collected for the 5 years of active treatment, permitting further refinement of CYP2D6 activity scores based on known inhibition of CYP2D6 enzymatic activity (4). Concomitant medications were categorized for strong, moderate, or weak/no inhibition of CYP2D6 as described previously (30). Patients were considered to have taken inhibitor drugs if prescription and/or use of the drug was recorded at any time during their endocrine therapy. In patients taking more than one drug, only the strongest CYP2D6 inhibitor, such as paroxetine, fluoxetine, quinidine, and bupropion, was factored into the CYP2D6 scoring. Duloxetine, diphenydramine, thioridazine, amiodarone, sertraline, and cimetidine partially inhibit CYP2D6 and were considered moderate inhibitors. Based on these designations, 2 points were subtracted from each patient's CYP2D6 metabolism score for strong inhibitors, 1 point for moderate inhibitors, and 0 points for the weak inhibitor and no inhibitors. Adjusted scores less than 0.5 (0, −0.5, −1.0, −1.5, and −2.0) were assigned a value of 0 as they all correspond with lack of CYP2D6 activity.
Statistical Analysis
In this genetic substudy, the primary endpoint was distant recurrence, and any recurrence was also evaluated as secondary endpoints. The association between CYP2D6 and UGT2B7 genotype and recurrence was determined by estimating the hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) using Cox proportional (approximate proportionality was verified by visual inspection of the Kaplan–Meier curves) hazards regression model, both with and without adjustment for age, tumor size, grade, and nodal status. Proportionality was based on the normal approximations of the distribution of test of equality of proportions. P values were based on a normal approximation to the partial likelihood score test. A trend test for CYP2D6 used the metabolic score as a covariate and a partial likelihood ratio score test (32). Time-to-recurrence curves were constructed using the Kaplan–Meier method (33). Patients were censored at last follow-up or death from a non-breast cancer cause as verified by a death certificate. Comparisons of baseline characteristics were based on normal approximations of the distribution of tests of equality of proportions. Hardy–Weinberg equilibrium was tested by a goodness-of-fit χ2 test with allele frequencies estimated from the same data (1 df). The allele frequencies of CYP2D6 and UGT2B7 were tested for consistency with those expected in a predominantly white population of European descent and their distributions tested for Hardy–Weinberg equilibrium. All tests were two-sided, and all P values less than .05 were considered statistically significant. All calculations were performed using STATA (Version 11) (StataCorp LP, College Station, TX). “Methods” and “Results” are reported according to the REMARK criteria (34). Baseline characteristics to compare genotyped vs non-genotyped groups were as follows: hysterectomy (yes vs no), ever smoker (yes vs no), previous hormone replacement therapy use (yes vs no), age (≤60 vs 60–69 vs ≥70 years), body mass index (<25, 25–30, >30 kg/m2), radiotherapy (yes vs no), chemotherapy (yes vs no), and mastectomy (yes vs no). Tumor characteristics were defined as follows: grade (well vs intermediate vs poorly differentiated vs unknown and/or could not be assessed), nodal status (positive vs negative vs unknown), hormone receptor status (positive vs negative vs unknown), and tumor size (<20 vs 20–50 vs >50 mm). Recurrence was defined either as any or distant recurrence, and cause of death (as verified by death certificate) was either any breast cancer or other than breast cancer death. The comparisons of these groups were based on simple comparison of proportions.
Results
Clinical Characteristics of the ATAC Trial Patients Included in the Genetic Substudy
Of the 2145 postmenopausal women with invasive breast cancer included from the UK component of the randomized ATAC trial, and with a median follow-up time of 10 years, tumor specimens were obtained from 1327 women, and genotyping was completed for CYP2D6 and UGT2B7 in 1203 and 1209 patients, respectively (Figure 1). The allele frequencies for the seven most common CYP2D6 (ancestral CYP2D6*1, and variants CYP2D6*2, CYP2D6*3, CYP2D6*4, CYP2D6*6, CYP2D6*10, and CYP2D6*41), and UGT2B7 (ancestral UGT2B7*1 and variant UGT2B7*2) alleles were in Hardy–Weinberg equilibrium in this population (Supplementary Table 1, available online). The allele frequencies were consistent with those reported for a predominantly white population of European descent (9,35).
Of the 1203 patients genotyped for CYP2D6, 615 and 588 patients were from the anastrozole and tamoxifen groups, respectively, representing approximately 20% of the total patients accrued to each of these treatment groups in the ATAC trial, and 56% of the UK ATAC patients (Figure 1). Of the 1209 patients genotyped for UGT2B7, 606 and 603 patients were from the anastrozole and tamoxifen groups, respectively (Figure 1). Overall, the characteristics of the patients genotyped for both CYP2D6 and UGT2B7 (the genotyped UK population) from the tamoxifen subgroup were similar to the non-genotyped UK ATAC population with regards to hysterectomy, ever smokers, previous hormone replacement therapy, age, and body mass index (Table 1). However, the genotyped patients treated with tamoxifen had higher rates of radiotherapy, lower rates of chemotherapy and mastectomy, and exhibited slight differences in tumor grade and nodal status compared with the non-genotyped patient population from the ROW. Most UK patients (544 [92.5%] of 588) in the tamoxifen group who were genotyped for CYP2D6 had hormone receptor–positive cancers. Tumors less than 20 mm in size were more frequent in the genotyped patients in the tamoxifen group compared with the frequency in non-genotyped ROW tamoxifen group (404 [68.7%] of 588 vs 1253 [61.1%] of 2050 patients; P < .001). Proportions of distant recurrence were similar in the genotyped vs non-genotyped UK population or non-genotyped ROW tamoxifen-treated patients (15.1% vs 18.2% or 17.5%). However, more deaths from any cause were observed in the non-genotyped vs genotyped UK population (P = .004). Similar baseline distributions between genotyped and non-genotyped patients were seen for the UK population in the anastrozole group (data not shown). Side effects in the tamoxifen and anastrozole groups in the genotyped UK population were similar to those in the non-genotyped ROW (data not shown).
Table 1.
Clinical characteristics of genetic substudy patients in the tamoxifen group compared with non-genotyped tamoxifen-treated patients in the United Kingdom and rest of the world (ROW)*
| Baseline characteristics | Genotyped UK tamoxifen group (n = 588), No. (%) | Non-genotyped UK tamoxifen group (n = 478), No. (%) | Non-genotyped ROW tamoxifen group (n = 2050), No. (%) | P† (genotyped vs non-genotyped UK tamoxifen group) | P† (genotyped UK tamoxifen vs non-genotyped ROW tamoxifen) |
| Hysterectomy | 142 (24.2) | 109 (22.8) | 614 (30.0) | .61 | .006 |
| Ever smoker | 295 (50.2) | 266 (55.7) | 712 (34.7) | .07 | <.001 |
| Previous HRT | 213 (36.2) | 167 (34.9) | 723 (35.3) | .66 | .66 |
| Age, y | |||||
| ≤60 | 209 (35.5) | 185 (38.7) | 710 (34.6) | .28 | .68 |
| 60–69 | 232 (39.5) | 176 (36.8) | 749 (35.5) | .37 | .19 |
| ≥70 | 147 (25.0) | 117 (24.5) | 591 (28.8) | .84 | .07 |
| BMI, kg/m2 | |||||
| <25 | 205 (34.9) | 156 (32.6) | 683 (33.3) | .44 | .48 |
| 25–30 | 202 (34.4) | 196 (41.0) | 706 (34.4) | .03 | .96 |
| >30 | 136 (23.1) | 96 (20.1) | 596 (29.1) | .23 | .005 |
| Unknown | 45 (7.7) | 30 (6.3) | 65 (3.2) | .38 | <.001 |
| Radiotherapy | 410 (69.7) | 282 (59.0) | 1255 (61.2) | <.001 | <.001 |
| Chemotherapy | 25 (4.3) | 40 (8.4) | 582 (28.4) | .005 | <.001 |
| Mastectomy | 224 (38.1) | 226 (47.3) | 1024 (50.0) | .003 | <.001 |
| Tumor characteristics | |||||
| Grade‡ | |||||
| Well differentiated | 134 (22.8) | 84 (17.6) | 420 (20.5) | .04 | .22 |
| Moderately differentiated | 323 (54.9) | 224 (46.9) | 941 (45.9) | .009 | <.001 |
| Poorly differentiated | 110 (18.7) | 138 (28.9) | 467 (22.8) | <.001 | .04 |
| Unknown or could not be assessed | 31 (3.6) | 32 (6.7) | 210 (10.2) | .32 | <.001 |
| Nodal status | |||||
| Positive | 167 (28.4) | 126 (26.4) | 754 (36.8) | .45 | <.001 |
| Negative | 398 (67.7) | 327 (68.4) | 1193 (58.2) | .80 | <.001 |
| Unknown | 23 (3.9) | 25 (5.2) | 103 (5.0) | .30 | .26 |
| Hormonal receptor status | |||||
| Positive | 544 (92.5) | 270 (56.5) | 1784 (87.0) | <.001 | <.001 |
| Negative | 2 (0.3) | 122 (25.5) | 149 (7.3) | <.001 | <.001 |
| Unknown | 42 (7.1) | 86 (18.0) | 117 (5.7) | <.001 | .19 |
| Tumor size, mm | |||||
| <20 | 404 (68.7) | 303 (63.4) | 1253 (61.1) | .07 | <.001 |
| 20–50 | 174 (29.6) | 164 (34.3) | 727 (35.5) | .09 | .008 |
| >50 | 8 (1.4) | 5 (1.1) | 56 (2.7) | .64 | .06 |
| Unknown | 2 (0.3) | 6 (1.3) | 14 (0.7) | .09 | .34 |
| Recurrence | |||||
| Any | 115 (19.6) | 107 (22.4) | 483 (23.6) | .25 | .04 |
| Distant | 89 (15.1) | 87 (18.2) | 359 (17.5) | .18 | .17 |
| Death | |||||
| Any | 117 (19.9) | 131 (27.4) | 470 (22.9) | .004 | .12 |
| Breast cancer | 70 (11.9) | 78 (16.3) | 279 (13.6) | .04 | .28 |
| Other cause | 47 (8.0) | 53 (11.1) | 191 (9.3) | .08 | .32 |
Eligible patients in the randomized, double-blind Arimidex, Tamoxifen, Alone or in Combination (ATAC) clinical trial were postmenopausal women with histologically proven, operable invasive breast cancer who had completed primary surgery and chemotherapy (where given) and were eligible to receive adjuvant hormonal therapy. Patients were randomly assigned to the anastrozole [trade name: Arimidex] alone, tamoxifen alone, and combined anastrozole and tamoxifen groups. The genetic substudy represents patients enrolled only in the United Kingdom and were compared with patients from the ROW for some analysis. Patients were genotyped for CYP2D6 and UGT2B7 using ABI Taqman assays. BMI = body mass index; HRT = hormone replacement therapy.
The comparisons of these groups were based on proportions using two-sided χ2 test, and P values were calculated using a two-sided Fisher's exact test.
Tumor grade (well vs intermediate vs poorly differentiated vs unknown and/or could not be assessed) was based on local institutional guidelines.
Association Between CYP2D6 Genotype and Recurrence Rate
We compared the rates of distant recurrence for patients according to CYP2D6 phenotypes—PM (phenotypic score 0), IM (phenotypic scores 0.5, 1.0, and 1.5), and EM (phenotypic score 2.0); scores were based on CYP2D6 genotypes (Figure 2, A). For tamoxifen-treated patients, no statistically significant difference in the rates of distant recurrence was observed between CYP2D6 homozygous WT and metabolic variants (PM vs IM [score 0.5], HR of distant recurrence = 2.80, 95% CI = 0.93 to 8.46, P = .068; PM vs IM [score 1.0], HR of distant recurrence = 1.31, 95% CI = 0.49 to 3.48, P = .58; PM vs IM [score 1.5], HR of distant recurrence = 0.76, 95% CI = 0.20 to 2.84, P = .68; PM vs EM [score 2], HR of distant recurrence = 1.25, 95% CI = 0.50 to 3.15, P = .64). Test for trend across the CYP2D6 genotypes for distant recurrence was not statistically significant (Ptrend = .66). We also compared rates of any recurrence for tamoxifen-treated patients according to CYP2D6 status. No statistically significant differences in any recurrence rates were observed between CYP2D6 homozygous WT and metabolic variants (PM vs IM [score 0.5], HR of any recurrence = 2.15, 95% CI = 0.85 to 5.40, P = .10; PM vs IM [score 1.0], HR of any recurrence = 0.94, 95% CI = 0.43 to 2.08, P = .88; PM vs IM [score 1.5], HR of any recurrence = 0.68, 95% CI = 0.23 to 1.96, P = .47; PM vs EM [score 2], HR of any recurrence = 0.99, 95% CI = 0.48 to 2.08, P = .99) (Figure 2, B). A multivariable analysis adjusted for clinical pathological factors including tumor size, grade, nodal status, and age did not influence the lack of association between CYP2D6 genotypes and recurrence rates in the tamoxifen group (data not shown).
Figure 2.
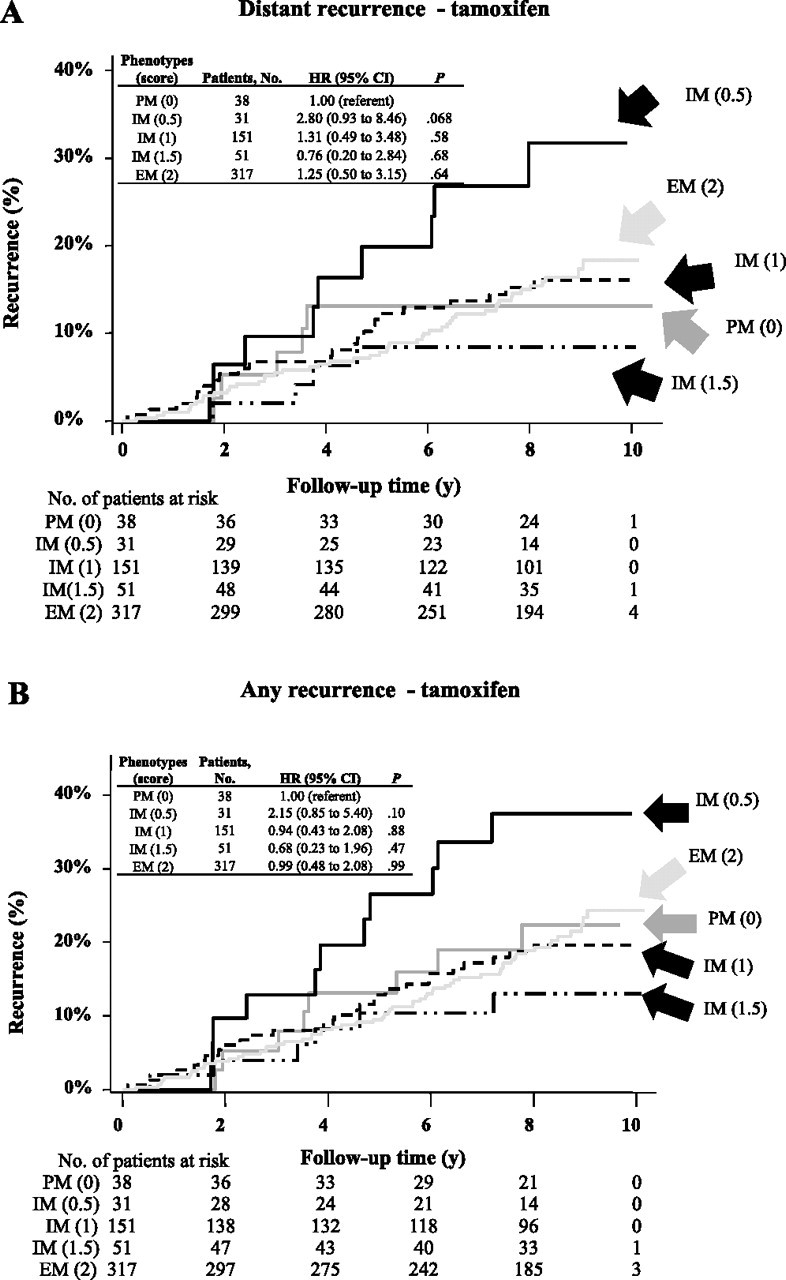
Probability of recurrence in patients enrolled in the Arimidex, Tamoxifen, Alone or in Combination (ATAC) trial according to cytochrome P450 2D6 (CYP2D6) phenotype. Recurrence was assessed in patients who were randomly assigned to the tamoxifen group using a Cox proportional hazard model. A CYP2D6 activity score was assigned to each patient, which most accurately predicts a patient's CYP2D6 metabolic phenotype based on her genotype (31). The phenotypic scores are indicated in parentheses. Two-sided P values were calculated using an adjusted Cox proportional hazard model. A) Distant recurrence. B) Any recurrence. CI = confidence intervalHR = hazard ratio; EM = extensive metabolizer; IM = intermediate metabolizer; PM = poor metabolizer.
Some of the previous studies that assessed the associations between CYP2D6 genotype and benefit from tamoxifen classified patients into only one of the three metabolic phenotypes based on the knowledge of homozygosity and heterozygosity of the most common nonfunctional CYP2D6 allele, CYP2D6*4 [see review (11)]. Therefore, we compared rates of recurrence for tamoxifen-treated patients according to CYP2D6*4 alone. No statistically significant difference in the rate of distant recurrence was observed between the homozygous WT CYP2D6 (CYP2D6*1/*1, ie, WT/WT) with heterozygous (CYP2D6*4/WT) or homozygous (CYP2D6*4/*4) genotypes in patients from the tamoxifen group (WT/WT vs CYP2D6*4/WT, HR of distant recurrence = 1.18, 95% CI = 0.75 to 1.88, P = .48; WT/WT vs CYP2D6*4/*4, HR of distant recurrence = 0.71, 95% CI = 0.26 to 2.00, P = .51) (Supplementary Figure 1, A, available online ). Similarly, there was no association between CYP2D6*4 genotype alone and rates of any recurrence (Supplementary Figure 1, B, available online).
Association Between CYP2D6 Genotype and Concomitant Medication and Recurrence Rate
Of the 588 tamoxifen-treated patients genotyped for CYP2D6, 19 (3.2%) and 13 (2.2%) patients had been prescribed potent and moderate CYP2D6 inhibitors, respectively. Incorporating concomitant medications to adjust the CYP2D6 score did not alter the lack of observed association between CYP2D6 genotypes (based on phenotypic scores as described earlier) and rates of either distant recurrence or any recurrence. After adjusting for concomitant medications, no statistically significant difference in rate of distant recurrence was observed between CYP2D6 phenotypes (PM and IM [score 1.0], HR of distant recurrence = 0.74, 95% CI = 0.40 to 1.37, P = .34; PM vs IM [score 1.5] phenotype, HR of distant recurrence = 0.49, 95% CI = 0.16 to 1.44, P = .19; PM vs EM [score 2], HR of distant recurrence = 0.78, 95% CI = 0.45 to 1.35, P = .38) (Table 2). No statistically significant differences in the rate of any recurrence between PM and IM or EM phenotypes were noted (Table 2).
Table 2.
Rates of distant recurrence and any recurrence according to CYP2D6 phenotype and concomitant medication usage*
| CYP2D6 phenotype | Distant recurrence |
P† | Any recurrence |
P† | ||
| Tamoxifen group, No. (%) | HR (95% CI) | Tamoxifen group, No. (%) | HR (95% CI) | |||
| PM (phenotypic score = 0) | 19 (18.5) | 1.00 (referent) | 24 (23.3) | 1.00 (referent) | ||
| IM (phenotypic score = 1.0) | 22 (14.3) | 0.74 (0.40 to 1.37) | .34 | 27 (17.5) | 0.67 (0.39 to 1.17) | .20 |
| IM (phenotypic score = 1.5) | 4 (8.3) | 0.49 (0.16 to 1.44) | .19 | 6 (12.5) | 0.58 (0.23 to 1.41) | .21 |
| EM (phenotypic score = 2.0) | 44 (15.6) | 0.78 (0.45 to 1.35) | .38 | 58 (20.5) | 0.82 (0.51 to 1.32) | .44 |
The relative catalytic activity of CYP2D6 for substrate O-demethylated dextromethorphan was used to calculate an “activity score”; all ATAC UK genetic substudy patients were assigned a phenotypic score based on their CYP2D6 genotype. The CYP2D6 activity score is an approach to predict most accurately a patient's CYP2D6 metabolic phenotype based on their genotype (31). The scores were adjusted for concomitant CYP2D6 inhibitors during active treatment. Cox proportional hazard model was used for multivariable analysis adjusted for tumor size (<20 mm, 20–50 mm, >50 mm, unknown), grade (well differentiated, moderately differentiated, poorly differentiated, unknown), nodal status (positive, negative, unknown), and age. CI = confidence interval; EM = extensive metabolizer; HR = hazard ratio; IM = intermediate metabolizer; PM = poor metabolizer.
Two-sided P values were calculated using an adjusted Cox proportional hazard model.
Association Between UGT2B7 Genotype and Recurrence Rate
No statistically significant difference in the rate of distant recurrence was observed between the homozygous WT UGT2B7 (UGT2B7*1/*1, ie, WT/WT) with heterozygous (UGT2B7*2/WT) or homozygous (UGT2B7*2/*2) genotypes in patients from the tamoxifen group (WT/WT vs UGT2B7*2/WT, HR of distant recurrence = 1.03, 95% CI = 0.59 to 1.79, P = .92; WT/WT vs UGT2B7*2/*2, HR of distant recurrence = 1.10, 95% CI = 0.61 to 2.00, P = .74) (Supplementary Figure 2, A, available online). Similarly, there was no association between UGT2B7*2 genotype and rates of any recurrence (Supplementary Figure 2, B, available online). A multivariable analysis adjusted for clinical pathological factors including tumor size, grade, nodal status, and age did not influence the lack of association between UGT2B7 genotypes and recurrence rates in the tamoxifen group (Table 3).
Table 3.
Rates of distant recurrence and any recurrence according to UGT2B7 genotype*
| UGT2B7 genotype | Distant recurrence |
P† | Any recurrence |
P† | ||
| Tamoxifen group, No. (%) | HR (95% CI) | Tamoxifen group, No. (%) | HR (95% CI) | |||
| UGT2B7*1/*1 (WT/WT) | 18 (14.9) | 1.00 (referent) | 21 (17.5) | 1.00 (referent) | ||
| UGT2B7*2/WT | 42 (15.3) | 1.12 (0.64 to 1.95) | .69 | 60 (22.1) | 1.29 (0.79 to 2.09) | .18 |
| UGT2B7*2/*2 | 28 (16.1) | 1.11 (0.62 to 2.08) | .71 | 33 (19.2) | 1.11 (0.65 to 1.90) | .64 |
Patients from ATAC UK genetic substudy were genotyped for UGT2B7*2 (rs7439366) using the polymerase chain reaction–based TaqMan Allelic Discrimination assay. Cox proportional hazard model was used for multivariable analysis adjusted for tumor size (<20 mm, 20–50 mm, >50 mm, unknown), grade (well differentiated, moderately differentiated, poorly differentiated, unknown), nodal status (positive, negative, unknown) and age. CI = confidence interval; HR = hazard ratio.
Two-sided P values were calculated using an adjusted Cox proportional hazard model.
Association Between CYP2D6 and UGT2B7 Genotypes and Recurrence Rates in Patients Treated With Anastrozole
Because CYP2D6 and UGT2B7 enzymes are not involved in the metabolism of anastrozole (36), the patients in this group served as an internal control group. As expected, CYP2D6 and UGT2B7 genotypes were not associated with rates of distant recurrence or any recurrence in patients treated with anastrozole. No statistically significant difference in the rates of distant recurrence was observed between CYP2D6 homozygous WT and metabolic variants (PM vs IM [score 0.5], HR of distant recurrence = 1.30, 95% CI = 0.22 to 7.76, P = .78; PM vs IM [score 1.0], HR of distant recurrence = 1.55, 95% CI = 0.45 to 5.31, P = .49; PM vs IM [score 1.5], HR of distant recurrence = 0.89, 95% CI = 0.18 to 4.43, P = .89; PM vs EM [score 2], HR of distant recurrence = 1.90, 95% CI = 0.59 to 6.09, P = .28) (Supplementary Figure 3, A, available online). Similarly, there was no association between CYP2D6 genotype and rates of any recurrence (Supplementary Figure 3, B, available online). No statistically significant difference in the rate of distant recurrence was observed between the homozygous WT UGT2B7 (WT/WT) with heterozygous (UGT2B7*2/WT) or homozygous (UGT2B7*2/*2) genotypes in patients from the anastrozole group (WT/WT vs UGT2B7*2/WT, HR = 0.63, 95% CI = 0.35 to 1.12, P = .12; WT/WT vs UGT2B7*2/*2, HR = 0.68, 95% CI = 0.34 to 1.23, P = .23) (Supplementary Figure 4, A, available online). Similarly, there was no association between UGT2B7*2 genotype and rates of any recurrence (Supplementary Figure 4, B, available online).
Discussion
In this retrospective genetic substudy of patients enrolled in the prospective ATAC Trial, which is one of the seminal randomized clinical trials comparing adjuvant endocrine treatments of breast cancer, we genotyped patients who were randomly assigned to either the tamoxifen or anastrozole groups for the common variants in CYP2D6 and UGT2B7. Our results failed to confirm previously reported associations between patient CYP2D6 genotype, or pharmacological inhibition of CYP2D6 enzymatic activity, and clinical benefit from tamoxifen in postmenopausal patients. Therefore, although variants in CYP2D6 and the use of CYP2D6 inhibitors correlate with decreased plasma concentrations of endoxifen, these differences do not appear to influence benefit from tamoxifen. Furthermore, our results do not suggest an association between UGT2B7 genotype, which is also expected to influence plasma endoxifen concentrations and clinical outcomes on tamoxifen.
Early reports of an apparently strong association of CYP2D6 genotype with clinical outcomes in women with hormone receptor–positive early-stage breast cancer treated with adjuvant tamoxifen created enthusiasm to use CYP2D6 genetic testing to guide selection of adjuvant endocrine therapy (37–39). These results led to a Food and Drug Administration Advisory Committee recommending changing the label of tamoxifen (40) and stimulated a large central pharmacy service to recommend CYP2D6 genetic testing for enrolled patients for whom tamoxifen has been prescribed (41). Furthermore, a study modeling the effects of CYP2D6 concluded that patients with homozygous WT CYP2D6 genotype would have lower rates of relapse when treated with tamoxifen compared with letrozole (42). However, published results addressing CYP2D6 genetic testing in the adjuvant and preventive settings have been highly mixed (10,11,16,17). Most results reported to date have been generated in studies of convenience, limited by relatively small numbers of patients, lack of comprehensive genotype data, lack of detailed clinical outcome data, and patient selection biases. In contrast, the data from this study are from a large double-blind clinical trial, in which CYP2D6 and UGT2B7 genotype information, as well as concomitant medication which could have affected tamoxifen metabolism was collected and was completely blinded to disease outcome and treatment allocation. Furthermore, the anastrozole group, in which CYP2D6 and UGT2B7 genotypes also showed no statistically significant association with outcome, served as an internal control group. The conduct of this work within a registration standard clinical trial leads to enhanced confidence in recording of clinicopathologic, co-medication, and outcomes data.
Selected authors on this study have previously published data modeling the expected saturation levels of ER by tamoxifen and its metabolites in breast cancer in postmenopausal women as more than 99.9% (43). The estimates were based on published data on the plasma concentration of tamoxifen and its metabolites and on their affinities for ER. Importantly, this modeling was conducted before the suggestion that endoxifen might play a critical role in breast cancer. This metabolite was therefore excluded from the modeling. More recent modeling suggests that inclusion of endoxifen predicts even greater ER saturation levels (44). While these estimates are subject to substantial error, if they approximate to the truth, the argument for a link based on degree of saturation between benefit from tamoxifen and different degrees of metabolism of tamoxifen to endoxifen is difficult to sustain. In premenopausal women, however, tamoxifen's anti-estrogenic effect at the hypothalamic–pituitary axis leads to increased gonadotrophin secretion and to markedly elevated plasma estradiol levels that are up to 100-fold higher than those in postmenopausal women (45). Premenopausal women usually receive the same dose of tamoxifen as postmenopausal women, and in these circumstances, saturation of around 97% in the absence of endoxifen can be calculated. Given the potential for error in this estimate, and the absence of premenopausal women in the ATAC trial, there remains room for a pharmacogenetic interaction between CYP2D6 and outcome on tamoxifen in premenopausal women.
This study has a few limitations. Traditionally, genotyping for pharmacogenetic studies has been performed using fresh and/or frozen germline DNA collected from leukocytes or buccal smears, whereas in our study, we used formalin-fixed paraffin-embedded cancer tissue. However, after we first reported the technical and biological feasibility of genotyping in the latter specimen type, subsequent studies have validated this methodology (46,47). Although our study represents one of the largest overall studies of the association of CYP2D6 and adjuvant tamoxifen, the confidence intervals for distant recurrence for those with CYP2D6 EM phenotype range from 0.45 to 1.35. In this regard, a 35% increase in risk for PM compared with EM is still possible. In our study, the number of patients taking CYP2D6 inhibitors was low. However, this observation strengthens our results since inhibitors could not be a major confounding factor in our genotype-alone analyses. Furthermore, although patient-reported information on compliance with their tamoxifen therapy was collected at each follow-up visit, blood samples were not collected, which prohibited measurement of circulating endoxifen levels and objective assessment of compliance. These results pertain only to the UK patients in ATAC and only to postmenopausal women. However, genotype frequencies in the United Kingdom are similar to the rest of Europe and North America, and there is no reason to suspect a systematic bias in UK women compared with others who participated in ATAC (as noted in Table 1). Finally, our study did not have a “no-treatment” group to test whether patient genotype was predictive vs prognostic. But the anastrozole arm in this study served as a surrogate for no tamoxifen treatment.
In conclusion, in this large, randomized double-blind clinical trial, in which all assays were conducted without knowledge of outcome or treatment allocation, we were unable to detect a statistically significant association between either CYP2D6 or UGT2B7 status and breast cancer outcomes for patients with hormone receptor–positive early-stage breast cancer treated with adjuvant tamoxifen. The CYP2D6 results from ATAC are very similar to those recently reported in a separate study performed by investigators of another registration trial, the Breast International Group (BIG) trial BIG1-98 study (48), in which patients were assigned to tamoxifen or letrozole. Taken together, these represent a high level of evidence demonstrating that CYP2D6 genotyping should not be recommended for such patients and that there is no need to avoid CYP2D6 inhibitors in postmenopausal patients taking tamoxifen. Data of equivalent quality are needed to provide greater certainty for premenopausal patients.
Funding
This work was supported in part by The Breast Cancer Research Foundation (BCRF) (grant numbers N003173 to JMR and DFH and 1RO1GM099143 to JMR), Fashion Footwear Charitable Foundation of New York/QVC Presents Shoes on Sale (to DFH), Breakthrough Breast Cancer and the National Institute Health Research Royal Marsden Biomedical Research Centre (MD, SD, BH) and BCRF (MD, BH), and Cancer Research United Kingdom program grants (C569-A10404 and C569-A11449 to JC).
Footnotes
Conflict of interest declarations: J. M. Rae is a Scientific Advisor for Olema Pharmaceuticals, has received speaking honoraria from GTx Pharmaceuticals, and research funding from Pfizer, Inc. D. F. Hayes has research grants from Pfizer, Inc, Novartis, Inc, and AstraZeneca, Inc. V. Stearns has research funding from Novartis, Inc, and Pfizer, Inc, honoraria from AstraZeneca, Inc, and consults for Otsuka, Inc. M. Dowsett has research grants, given lectures, on the advisory board and given legal advice to AstraZeneca, Inc. J. Cuzick has a research grant and consulted for AstraZeneca. The authors are solely responsible for the study design, data collection, analysis and interpretation of the data, writing the article, and decision to submit the article for publication.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 2.Stearns V, Johnson MD, Rae JM, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95(23):1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 3.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310(3):1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 4.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 5.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy- N -desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85(2):151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 6.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Lim YC, Li L, Desta Z, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318(2):503–512. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- 8.Sun D, Sharma AK, Dellinger RW, et al. Glucuronidation of active tamoxifen metabolites by the human UDP-glucuronosyltransferases (UGTs) Drug Metab Dispos. 2007;35(11):2006–2014. doi: 10.1124/dmd.107.017145. [DOI] [PubMed] [Google Scholar]
- 9.Bhasker CR, McKinnon W, Stone A, et al. Genetic polymorphism of UDP-glucuronosyltransferase 2B7 (UGT2B7) at amino acid 268: ethnic diversity of alleles and potential clinical significance. Pharmacogenetics. 2000;10(8):679–685. doi: 10.1097/00008571-200011000-00002. [DOI] [PubMed] [Google Scholar]
- 10.Cronin-Fenton DP, Lash TL. Clinical epidemiology and pharmacology of CYP2D6 inhibition related to breast cancer outcomes. Expert Rev Clin Pharmacol. 2011;4(3):363–377. doi: 10.1586/ecp.11.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seruga B, Amir E. Cytochrome P450 2D6 and outcomes of adjuvant tamoxifen therapy: results of a meta-analysis. Breast Cancer Res Treat. 2010;122(3):609–617. doi: 10.1007/s10549-010-0902-3. [DOI] [PubMed] [Google Scholar]
- 12.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23(36):9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 13.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25(33):5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 14.Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9(1):R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lash TL, Cronin-Fenton D, Ahern TP, et al. CYP2D6 inhibition and breast cancer recurrence in a population-based study in Denmark. J Natl Cancer Inst. 2011;103(6):489–500. doi: 10.1093/jnci/djr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes DF, Stearns V, Rae J, Flockhart D. A model citizen? Is tamoxifen more effective than aromatase inhibitors if we pick the right patients? J Natl Cancer Inst. 2008;100(9):610–613. doi: 10.1093/jnci/djn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lash TL, Rosenberg CL. Evidence and practice regarding the role for CYP2D6 inhibition in decisions about tamoxifen therapy. J Clin Oncol. 2010;28(8):1273–1275. doi: 10.1200/JCO.2009.26.7906. [DOI] [PubMed] [Google Scholar]
- 18.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101(21):1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baum M, Budzar AU, Cuzick J, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early breast cancer: first results of the ATAC randomised trial. Lancet. 2002;359(9324):2131–2139. doi: 10.1016/s0140-6736(02)09088-8. [DOI] [PubMed] [Google Scholar]
- 20.Howell A, Cuzick J, Baum M, et al. Results of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial after completion of 5 years’ adjuvant treatment for breast cancer. Lancet. 2005;365(9453):60–62. doi: 10.1016/S0140-6736(04)17666-6. [DOI] [PubMed] [Google Scholar]
- 21.Baum M, Buzdar A, Cuzick J, et al. Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early-stage breast cancer: results of the ATAC (Arimidex, Tamoxifen Alone or in Combination) trial efficacy and safety update analyses. Cancer. 2003;98(9):1802–1810. doi: 10.1002/cncr.11745. [DOI] [PubMed] [Google Scholar]
- 22.Buzdar A, Howell A, Cuzick J, et al. Comprehensive side-effect profile of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: long-term safety analysis of the ATAC trial. Lancet Oncol. 2006;7(8):633–643. doi: 10.1016/S1470-2045(06)70767-7. [DOI] [PubMed] [Google Scholar]
- 23.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9(1):45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 24.Cuzick J, Sestak I, Baum M, et al. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 10-year analysis of the ATAC trial. Lancet Oncol. 2010;11(12):1135–1141. doi: 10.1016/S1470-2045(10)70257-6. [DOI] [PubMed] [Google Scholar]
- 25.Fallowfield L, Cella D, Cuzick J, Francis S, Locker G, Howell A. Quality of life of postmenopausal women in the Arimidex, Tamoxifen, Alone or in Combination (ATAC) Adjuvant Breast Cancer Trial. J Clin Oncol. 2004;22(21):4261–4271. doi: 10.1200/JCO.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 26.Cuzick J, Sestak I, Cella D, Fallowfield L. Treatment-emergent endocrine symptoms and the risk of breast cancer recurrence: a retrospective analysis of the ATAC trial. Lancet Oncol. 2008;9(12):1143–1148. doi: 10.1016/S1470-2045(08)70259-6. [DOI] [PubMed] [Google Scholar]
- 27.Dowsett M, Allred C, Knox J, et al. Relationship between quantitative estrogen and progesterone receptor expression and human epidermal growth factor receptor 2 (HER-2) status with recurrence in the Arimidex, Tamoxifen, Alone or in Combination trial. J Clin Oncol. 2008;26(7):1059–1065. doi: 10.1200/JCO.2007.12.9437. [DOI] [PubMed] [Google Scholar]
- 28.Sikora MJ, Thibert JN, Salter J, Dowsett M, Johnson MD, Rae JM. High-efficiency genotype analysis from formalin-fixed, paraffin-embedded tumor tissues. Pharmacogenomics J. 2010;11(15):348–358. doi: 10.1038/tpj.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blake MJ, Gaedigk A, Pearce RE, et al. Ontogeny of dextromethorphan O- and N-demethylation in the first year of life. Clin Pharmacol Ther. 2007;81(4):510–516. doi: 10.1038/sj.clpt.6100101. [DOI] [PubMed] [Google Scholar]
- 30.Rae JM, Sikora MJ, Henry NL, et al. Cytochrome P450 2D6 activity predicts discontinuation of tamoxifen therapy in breast cancer patients. Pharmacogenomics J. 2009;9(4):258–264. doi: 10.1038/tpj.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83(2):234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 32.Cox DR. Regression models and life-tables. J R Stat Soc. 1972;68(10):187–220. [Google Scholar]
- 33.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 34.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23(36):9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 35.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 36.Kamdem LK, Liu Y, Stearns V, et al. In vitro and in vivo oxidative metabolism and glucuronidation of anastrozole. Br J Clin Pharmacol. 2010;70(6):854–869. doi: 10.1111/j.1365-2125.2010.03791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes FA, Liticker JD. Pharmacogenomics of tamoxifen in a nutshell-and who broke the nutcracker? J Oncol Pract. 2005;1(4):155–159. doi: 10.1200/jop.2005.1.4.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goetz MP, Kamal A, Ames MM. Tamoxifen pharmacogenomics: the role of CYP2D6 as a predictor of drug response. Clin Pharmacol Ther. 2008;83(1):160–166. doi: 10.1038/sj.clpt.6100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302(13):1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phan MT, Venitz J. Summary Minutes of the Advisory Committee Pharmaceutical Science Clinical Pharmacology Subcommittee. 2006. http://www.fda.gov/ohrms/dockets/ac/06/minutes/2006-4248m1.pdf. [Google Scholar]
- 41.Medco Health Solutions, Inc. CYp2D6 Testing for Tamoxifen. 2011. http://www.medcohealth.com/medco/corporate/home.jsp?ltSess=y&articleID=CorpPM_Tamoxifen. [Google Scholar]
- 42.Punglia RS, Burstein HJ, Winer EP, Weeks JC. Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst. 2008;100(9):642–648. doi: 10.1093/jnci/djn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowsett M, Haynes BP. Hormonal effects of aromatase inhibitors: focus on premenopausal effects and interaction with tamoxifen. J Steroid Biochem Mol Biol. 2003;86(3–5):255–263. doi: 10.1016/s0960-0760(03)00365-0. [DOI] [PubMed] [Google Scholar]
- 44.Lash TL, Lien EA, Sorensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection? Lancet Oncol. 2009;10(8):825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravdin PM, Fritz NF, Tormey DC, Jordan VC. Endocrine status of premenopausal node-positive breast cancer patients following adjuvant chemotherapy and long-term tamoxifen. Cancer Res. 1988;48(4):1026–1029. [PubMed] [Google Scholar]
- 46.Schneider BP, Wang M, Radovich M, et al. Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol. 2008;26(28):4672–4678. doi: 10.1200/JCO.2008.16.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss JR, Baer MR, Ambrosone CB, et al. Concordance of pharmacogenetic polymorphisms in tumor and germ line DNA in adult patients with acute myeloid leukemia. Cancer Epidemiol Biomarkers Prev. 2007;16(5):1038–1041. doi: 10.1158/1055-9965.EPI-06-0964. [DOI] [PubMed] [Google Scholar]
- 48.Regan MM, Leyland-Jones B, Bouzyk M, et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 Trial. J Natl Cancer Inst. 2012;104(6):441–451. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]