Abstract
Purpose
In recent years, microRNAs (miRNAs) have been reported to play important roles in a broad range of biologic processes, including oxidative stress-mediated ocular diseases. In addition, the polyphenolic compound curcumin has been shown to possess anti-inflammatory, antioxidant, anticancer, antiproliferative, and proapoptotic activities. The aim of this study was to investigate the impact of curcumin on the expression profiles of miRNAs in ARPE-19 cells exposed to oxidative stress.
Methods
MiRNA expression profiles were measured in ARPE-19 cells treated with 20 μΜ curcumin and 200 μΜ H2O2. PCR array analysis was performed using web-based software from SABiosciences. The cytotoxicity of ARPE-19 cells was determined with the CellTiter-Blue cell viability assay. The effects of curcumin on potential miRNA targets were analyzed with quantitative real-time PCR and western blotting.
Results
Curcumin treatment alone for 6 h had no effect on ARPE-19 cell viability. Incubation with H2O2 (200 µM) alone for 18 h decreased cell viability by 12.5%. Curcumin alone downregulated 20 miRNAs and upregulated nine miRNAs compared with controls. H2O2 downregulated 18 miRNAs and upregulated 29 miRNAs. Furthermore, curcumin pretreatment in cells exposed to H2O2 significantly reduced the H2O2-induced expression of 17 miRNAs. As determined with quantitative real-time PCR and western blotting, curcumin increased the expression of antioxidant genes and reduced angiotensin II type 1 receptor, nuclear factor-kappa B, and vascular endothelial growth factor expression at the messenger RNA and protein levels.
Conclusions
The results demonstrated that curcumin alters the expression of H2O2-modulated miRNAs that are putative regulators of antioxidant defense and renin-angiotensin systems, which have been reported to be linked to ocular diseases.
Introduction
Oxidative stress from reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) has been implicated in many diseases, including age-related macular degeneration (AMD), in which the retinal pigment epithelium (RPE) is considered the primary target. The RPE is the outermost layer of the retina that absorbs redundant light and processes shed photoreceptor outer segments through phagocytosis, which generates high oxidative stress [1]. Therefore, targeting oxidative damage should be considered as treating and preventing oxidative stress-mediated diseases. Microarray analysis conducted by Weigel et al. [2] and Vandenbroucke et al. [3] revealed that regulation of many genes is altered in cells treated with H2O2, mediating protective and detrimental cellular effects. Transcriptional regulation in H2O2-mediated oxidative stress has been shown by many investigators [2-4]. However, the post-transcriptional mechanism of gene expression in response to H2O2-mediated oxidative stress in RPE cells has not been thoroughly investigated.
Recently, Sun et al. [5] reported microRNA (miRNA/miR) expression profiles were altered by curcumin in pancreatic cancer cells. MiRNA expression profiling of ischemic rat hearts in the context of pretreatments with resveratrol using a quantitative real-time PCR (qRT-PCR)–based assay was conducted by Mukhopadhyay et al. [6]. Curcumin and resveratrol have been shown using oligonucleotide microarray chip and qRT-PCR-based assays to alter the expression profiles of miRNAs in human pancreatic cancer cells and the rat ischemia/reperfusion model, respectively [5,6]. Curcumin significantly protects RPE cells against H2O2-induced oxidative stress [7]. Baicalein, a naturally occurring flavonoid compound, has also been shown to protect RPE cells against oxidative stress [8].
Curcumin is a naturally occurring phenolic compound derived from the rhizome of Curcuma longa and possesses anti-inflammatory and antioxidant effects [9]. Curcumin significantly decreases lipid peroxidation, increases intracellular antioxidant, glutathione, regulates antioxidant enzymes, and scavenges ROS [10,11]. However, the mechanisms underlying the antioxidant activity of curcumin have not been completely delineated. Curcumin has also been studied as a cancer chemopreventive agent in various cancers [12].
In recent years, miRNAs have received greater attention in cancer and other research fields. These small, non-coding RNAs bind to the 3′ untranslated region of target messenger RNA (mRNA) and negatively regulate the expression of genes involved in development, differentiation, proliferation, apoptosis, and other important cellular processes. MiRNAs regulate gene expression at the post-transcriptional level by either degradation or translational repression of a target mRNA. Curcumin regulates the expression of genes involved in regulating cellular signaling pathways, including vascular endothelial growth factor (VEGF), nuclear factor-kappa B (NF-κB), protein kinase B, mitogen-activated protein kinase (MAPK), and other pathways [13], and these signaling pathways could be regulated by miRNAs. In this study, we evaluated the effects of curcumin on protecting RPE cells from H2O2-induced oxidative stress and identify a potential mechanism. The expression of miRNAs can be measured with northern blot, primer extension assay, RNase protection assay, and global profiling methods [14]. In our investigation, we used a PCR array to profile miRNA expression and to evaluate the effect of curcumin on oxidatively stressed ARPE-19 cells. We hypothesize that curcumin may play an important role in protecting RPE cells from oxidative stress by differentially modulating the expression of miRNAs that putatively regulate the expression of antioxidant, proangiogenic, proliferative, and proinflammatory genes. Our study for the first time reveals that the modulation of miRNA expression may be an important mechanism underlying the biologic effect of curcumin in human RPE, and this approach could be applied as a potential strategy for preventing and treating oxidative stress-mediated ocular diseases such as AMD and diabetic retinopathy (DR).
Methods
Cell culture
ARPE-19 cells purchased from American Type Culture Collection (ATCC; Manassas, VA) were cultured at 37 °C in 5% (v/v) of CO2 in Dulbecco’s modified Eagle’s medium and Ham’s F12 medium (DMEM/F12) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 100 U/ml of penicillin, and 100 µg/ml of streptomycin (Invitrogen, Gibco, Carlsbad, CA). The media were changed every 2–3 days. ARPE-19 cells were seeded in 12-well plates at 1.5×105 cells/well, cultured for 48 h, and then treated with curcumin (Sigma-Aldrich, St. Louis, MO) and H2O2 (Sigma-Aldrich) alone for 6 h and 18 h, respectively. The effect of curcumin on H2O2-induced oxidative stress was also assessed, in which ARPE-19 cells were treated with curcumin for 6 h before H2O2 insult for 18 h and then harvested for miRNA-enriched total RNA or protein extraction. Cells treated with dimethyl sulfoxide (DMSO) were maintained as controls.
Determination of cell viability
The CellTiter-Blue viability assay (Promega Corp, Madison, WI) was used as the index for cell survival, which measures the ability of living cells to reduce a redox dye (resazurin) into a fluorescent dye (resorufin). The assay was performed according to the manufacturer’s protocol, in which 96-well plates were seeded at 1×104 cells/well and incubated for 6 h for cells to attach to the surface. The ARPE-19 cells were then exposed to varying concentrations (1–50 μM) of curcumin for 6 h. In addition, the cell viability during various durations of exposure of 20 μM curcumin was measured. Cells were washed with phosphate buffered saline (PBS; 10 mM sodium phosphate, 150 mM sodium chloride, pH 7.8), 100 μl of DMEM-F12 without serum was added to each well, and then 20 μl CellTiter-Blue reagent was added. The plates were then incubated at 37 °C for 2 h. The absorbance was recorded at 590 nm in the Synergy 2 Multi-Mode Microplate Reader (Winooski, VT), with the CellTiter-Blue reagent without cells as the blank. The optic density (OD) of the experimental and control samples were subtracted from that of the blank. Cell viability (%) was calculated according to the following formula: Percentage cell viability=(OD of the experimental samples/OD of the control)×100.
RNA isolation, quantitative real-time polymerase chain reaction, and microRNA polymerase chain reaction arrays
MiRNA-enriched total RNA was extracted from cultured ARPE-19 cells using the QIAzol and miRNeasy kit following the manufacturer’s protocol (Qiagen, Valencia, CA). The concentration of total RNA and the RNA quality (260/280 absorbance ratio) of the samples were measured using a SmartSpec 300 Spectrophotometer (Bio-Rad, Hercules, CA). The first strand kit (Qiagen, cat # 331,401) was used to perform cDNA analysis. For each reaction, 0.8 μg of total RNA, extracted from ARPE-19 cells treated with H2O2 and with or without curcumin, was submitted to reverse transcription, following the manufacturer’s instructions (Qiagen). The RNA sample with miRNA reverse transcription (RT) enzyme mix was incubated at 37 °C for 2 h, and then the samples were heated at 95 °C for 5 min to degrade RNA and inactivate the reverse transcriptase. To measure miRNAs, the cDNA was diluted tenfold by adding RNase-free H2O. The resulting diluted cDNA was added to the RT2 Real-Time SYBR Green qPCR Master Mix (Qiagen), which contained real-time PCR buffer, a high-performance HotStart DNA Taq polymerase, nucleotides, and SYBR Green dye. The ROX and fluorescein reference dyes were also included in the PCR master mix to normalize variation from well to well.
For PCR array analysis, aliquots of the mixture were placed in each well of a 96-well RT2 miRNA profiler miFinder PCR array plate (Qiagen, MAH-001A) that contained a panel of primer sets for a thoroughly researched set of 88 pathway- or disease-focused miRNAs, plus four small nuclear RNA housekeeping (SNORD 44, 47, 48, and U6) assays. The plate also contained duplicate reverse transcription controls that test the efficiency of the RT2 miRNA first strand kit (Qiagen) reaction with a primer set detecting the template synthesized from the kit’s built-in miRNA external RNA control and duplicate positive controls that tested the efficiency of the PCR reaction itself using a predispensed artificial DNA sequence and the primer set that detected it. The qRT-PCR analysis was performed in MyiQ Cycler (Bio-Rad Laboratories Inc.) with 25 μl total volume containing diluted cDNA (1 μl per well) and 2X SYBR Green PCR Master Mix. The amplification conditions were the following: 10 min at 95 °C, 40 cycles at 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. The relative amount of each miRNA in PCR array analysis was normalized to an average of four small nuclear housekeeping genes. Heatmap or cluster analysis was conducted on the expression profiles of all four groups using SABiosciences (Frederick, MD) software.
For mRNA analysis, the isolation of total RNA from ARPE-19 cells and cDNA synthesis were performed using the RNeasy kit and the QuantiTect reverse transcription kit, respectively, according to the manufacturer’s protocol (Qiagen). The qRT-PCR analysis was performed with a 25 μl total volume containing cDNA (2 μl from each sample), 1X QantiFast SYBR Green PCR Master Mix (Qiagen), and 300 nM gene-specific primers (Table 1). The amplification conditions for mRNA qRT-PCR were the following: 5 min at 95 °C, 40 cycles at 95 °C for 10 s, and 60 °C for 30 s. Each sample was assayed in duplicate, and the experimental data were normalized to the expression levels of the housekeeping gene Hprt. The absence of non-specific products was confirmed with the analysis of the melt curves and electrophoresis in 2% agarose gels.
Table 1. Primers used for quantitative real-time PCR.
| Gene | Primer sequence (5’-3’) | Amplicon size (bp) |
|---|---|---|
|
VEGF-A |
F: TGCCATCCAATCGAGACCCTG |
156 |
| |
R:GGTGATGTTGGACTCCTCAGTG |
|
|
NF-κB1 |
F:CAACCACAGATGGCACTGCC |
125 |
| |
R:GCACCAGGTAGTCCACCATG |
|
|
AT1R |
F:TGCAGATATTGTGGACACGGCC |
154 |
| |
R:GTGGGATTTGGCTTTTGGGGG |
|
|
Catalase |
F:CCATTATAAGACTGACCAGGGC |
133 |
| |
R:AGTCCAGGAGGGGTACTTTCC |
|
|
GPx-1 |
F:AGTCGGTGTATGCCTTCTCGG |
142 |
| |
R:TCGTTCATCTGGGTGTAGTCCC |
|
|
GPx-4 |
F:GAGTTTTCCGCCAAGGACATCGA |
130 |
| |
R:GGTCGACGAGCTGAGTGTAGTTT |
|
|
Hprt |
F:ACAGGACTGAACGTCTTGCTCG |
87 |
| R:TATAGCCCCCCTTGAGCACAC |
FR, fold regulation; FC, fold change. (Note: FC values greater than one indicate a positive or an upregulation, and the FR is equal to the FC. FC values less than one indicate a negative or downregulation, and the FR is the negative inverse of the FC).
The expression levels of mRNA and miRNAs were measured using the threshold cycle (Ct). The Ct is the fractional cycle number at which the fluorescence of each sample passes the fixed threshold. Briefly, the average ΔCt of each group was calculated with the following formula: ΔCt=average mRNA/miRNA Ct – average of the housekeeping genes Ct. ΔΔCt was calculated with ΔΔCt=ΔCt of the experimental group – ΔCt of the control group. The fold relationships in miRNA or gene expression among the tested samples were calculated using 2−ΔΔCt [15]. The efficiency of reverse transcription in the PCR array was calculated with ΔCt=average ΔCtRTC – ΔCtPPC. The ΔCt value of the RT control more than 5 shows evidence of poor reverse transcription efficiency.
Western blotting analysis
Protein samples were isolated from confluent ARPE-19 cells growing on 12-well plates by washing in ice-cold PBS and then lysed in RIPA buffer (50 mmol/l Tris-HCl [pH 8.0], 150 mmol/l NaCl, 100 μg/ml phenylmethylsulfonyl fluoride, 1% NP-40, 50 mmol/l NaF, 2 mmol/L EDTA), supplemented with protease inhibitor cocktail (Sigma-Aldrich). Samples were centrifuged at 600 × g for 30 min at 4 °C to remove cell debris. Protein concentrations were determined with the Lowry method [16]. Samples of 100 μg proteins mixed with loading buffer (Bio-Rad, cat# 161–0791) were boiled for 10 min, separated on 10% Bis–Tris Criterion XT precast gels (Bio-Rad), and transferred onto a polyvinylidene difluoride membrane (Millipore Co., Bedford, MA). Nonspecific binding was blocked by immersing the membrane in 5% dry milk for 3 h. Proteins were incubated with primary antibodies (anti-β-actin, Sigma-Aldrich, 1:3000, cat# A-5441; VEGF-A, Abcam, Cambridge, MA, 1:1000, ab1316; NF-κB, Santa Cruz Biotechnology Inc., Santa Cruz, CA, 1:1000, sc-8008, gift; anti-AT1R, Santa Cruz Biotechnology, 1:1000, cat# sc-1173; anticatalase, Santa Cruz Biotechnology, 1:500, cat# sc-34280; anti-GPx-1, Santa Cruz Biotechnology, 1:500, cat# sc-22145; anti-GPx-4, Santa Cruz Biotechnology, 1:500, cat# sc-50497) diluted in 5% bovine serum albumin in PBST (1x PBS, 0.05% Tween-20) overnight at 4 °C. After washing with PBST, the membrane was further incubated with horseradish peroxidase-conjugated antigoat immunoglobulin G (sc-2378, Santa Cruz Biotechnology) at 1:5,000 dilution for 1 h at room temperature. The membrane was washed three times for 15 min each with PBST, and the target proteins were detected with an enhanced chemiluminescence detection system (GE Healthcare, Buckinghamshire, England). The chemiluminescence signal was transferred on Blue Lite Autorad Film (ISC BioExpresss, Kaysville, UT), and the developed film was scanned densitometrically (Kodak Molecular Imaging, Rochester, NY). For data normalization, after the striping procedure, β-actin protein was detected on the same membrane. For β-actin and NF-κB detection, antimouse immunoglobulin G was used as a secondary antibody (sc-2005, Santa Cruz Biotechnology) at 1:5,000 dilution for 1 h at room temperature.
Statistical analysis
All data including PCR array, qRT-PCR, and immunoblotting analyses were statistically analyzed with Sigma Stat and Sigma Plot (Systat Software, Inc., Chicago IL). Cluster and volcano analyses for the PCR array were done using the PCR array analysis program from SABiosciences. Differences among the groups were analyzed with analysis of variance (ANOVA). Differences between two groups were analyzed with the Student t test. In all statistical analyses, p<0.05 was regarded as statistically significant. All values were presented as means±standard error of the mean (SEM).
Results
Effect of curcumin on cell viability
To determine the effect of curcumin on the cytotoxicity of ARPE-19, cells were grown in 96 wells, and cell viability was assessed with various concentrations (1–50 μM) of curcumin for 6 h or with various durations of exposure to 20 μM curcumin. No significant loss of viability was observed with 1–20 μM curcumin. However, treatment of cells with 50 μM curcumin for 6 h resulted in an approximate 50% decrease in cell viability (p<0.001 versus control). Therefore, 20 μM was chosen as the optimum concentration for subsequent experiments. Curcumin (20 μM) treatment for up to 6 h had no effect on cell viability as measured with CellTiter-Blue, but the CellTiter-Blue value was reduced by 18.5% (p<0.05 versus control) to 30.1% (p<0.001 versus control) of the untreated control cells after 20 µM exposure to 15 and 20 h, respectively (Figure 1). The effect of H2O2 on the viability of ARPE-19 cells using various concentrations of H2O2 was previously reported [17]. To examine the effect of H2O2-mediated oxidative stress in this study, 200 μM H2O2 was used, as the viability of the cells using this concentration was shown to be only slightly but significantly (p<0.05) reduced by 12.5% compared to control.
Figure 1.
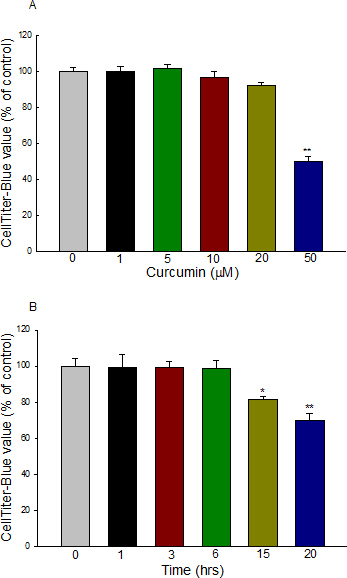
Viability effect of curcumin on ARPE-19 cells at different concentrations and time points. A: Dose effect of curcumin on cells treated with different concentrations of curcumin for 6 h. Curcumin at concentrations 1–20 μM did not affect the viability of the ARPE-19 cells. B: Time effect of curcumin on cells treated with 20 μM curcumin at different time points. Cells treated with curcumin (20 μM) for 15 h and 20 h induced cell death, compared with the control. Cell survival was determined with CellTiter-Blue assay (absorbance at 560/590 nm). Data represent mean±SEM from five separate samples. *p<0.05 versus control, **p<0.001 versus control.
Global microRNA expression profile with polymerase chain reaction array analysis
To study the responses of miRNAs to curcumin and H2O2, global miRNA expression profiling using the RT2 Profiler miFinder PCR array (SABiosciences) was conducted with miRNA-enriched total RNAs extracted from ARPE-19 cells treated with DMSO (control), curcumin, and H2O2 in the following ways as shown in Figure 2: 1) DMSO for 6 h; 2) 20 µM curcumin for 6 h; 3) 200 µM H2O2 for 18 h; and 4) in the curcumin+H2O2 group, 20 μM curcumin for 6 h before H2O2 (200 μM) exposure for 18 h. Three experimental assays were performed independently resulting in three PCR array replicates for each condition. In addition, the influence of curcumin was evaluated by comparing the values of the treatment groups with respective non-treated cells exposed to H2O2 only. Two major bioinformatic approaches were used to analyze the expression of miRNAs and their changes in H2O2- and curcumin-treated cells.
Figure 2.
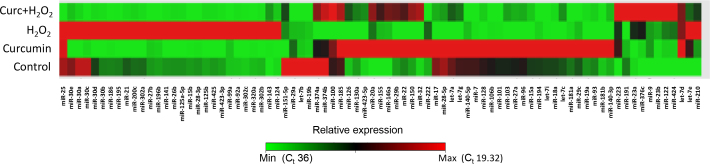
Clustergram or heatmap analysis: Curcumin alters microRNAs (miRNAs) expression profiling in ARPE-19 cells. This clustergram or heatmap represents the expression levels of 81 microRNAs (miRNAs) in four conditions (control, curcumin, hydrogen peroxide [H2O2], and curc+H2O2) relative to an average of four housekeeping genes. ARPE-19 cells were treated with 20 μM curcumin and 200 μM H2O2 for 6 and 18 h, respectively. In the curc+H2O2 group, the cells were treated with curcumin for 6 h before H2O2 insult for 18 h. Cells with vehicle DMSO were used as control. Each vertical line in the heatmap represents a single miRNA, and the rows represent groups. Samples clustered according to the condition. Red represents high expression, while green represents low expression. The average Ct values for miRNAs with minimum (Min) and Maximum (Max) expression are given in parentheses. Based on clustering, curcumin and H2O2 alone changed the expression of several miRNAs compared with the control. In addition, curcumin pretreatment altered H2O2-induced miRNA expression. There were three samples (n=3) in each condition.
Heatmap/clustering analysis
Out of 88 miRNAs surveyed in this array, the qRT-PCR detected 81 miRNAs in ARPE-19 cells. Several observations can be made from Figure 2. In the PCR array, either curcumin or H2O2 alone altered the expression of several miRNAs that are clustered as red and green, respectively, when compared to the controls. Curcumin and H2O2 up- or downregulated several miRNAs. Significantly, curcumin pretreatment altered the expression profile of H2O2-modulated miRNA expression. Five miRNAs (miR-20a, miR-126, miR-146, miR-150, and miR-155) that target VEGF-A, platelet-derived growth factor β (PDGFβ), NF-κB, endothelin 1, p53, and AT1R were tightly clustered together in curcumin-treated samples, and their expression was significantly (p<0.05) different compared to the controls. In addition, a tight cluster of nine miRNAs (miR-223, miR-191, miR-23a, miR-376c, miR-9, miR-23b, miR-122, miR-424, and let-7d) was significantly (p<0.05) induced by curcumin compared with the controls. All five members of the miR-30 family (miR-30a-e) were upregulated and downregulated by H2O2 and curcumin, respectively. However, only miR-30b and miR-30d were differentially expressed more than twofold after H2O2 treatment.
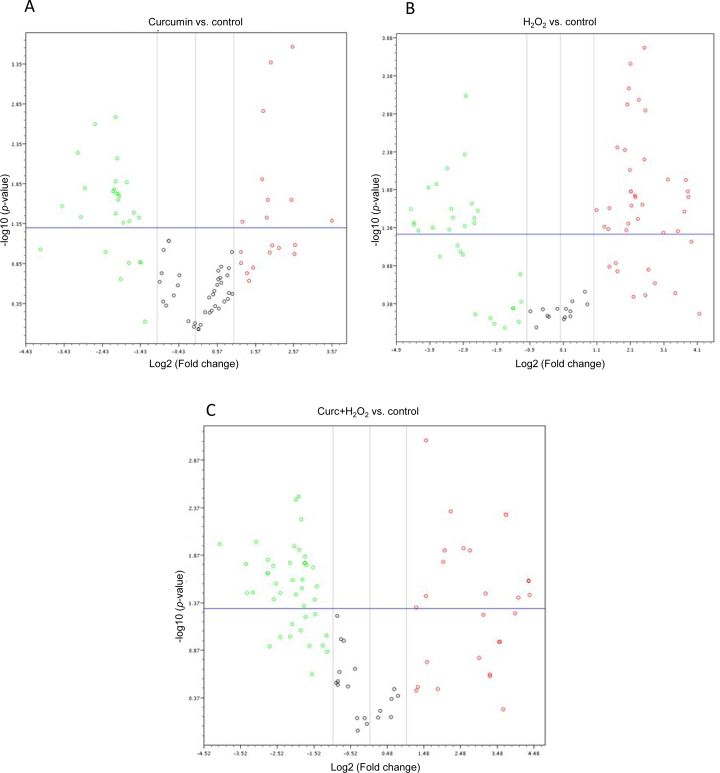
Volcano analysis
To compare the miRNA expression levels between two different conditions (curcumin-treated samples to control, H2O2-treated samples to control, and curcumin+H2O2-treated samples to control) following the criteria of twofold change in expression and statistical significance (Student t test, p<0.05), a volcano plot was generated (Figure 3). The volcano plot also demonstrated that the curcumin pretreatment had altered several H2O2-modulated miRNAs. In the plot, the first (horizontal) dimension (x-axis) is the fold change (FC) between the two groups (on a log2 scale, so that up- and downregulation appear symmetric), and the second (vertical) axis represents the p value for a Student t test of differences between samples (on a negative log scale, so smaller p values appear higher up). The first axis indicates the biologic impact of the change, and the second indicates the statistical evidence, or reliability of the change. For each miRNA, this plot demonstrated the log2 of the FC in the average expression of the two groups (e.g., control versus curcumin-treated samples) as plotted against −log10(p). P represents the probability value for a given miRNA associated with the Student t test comparison of the two groups of samples. MiRNAs with statistically significant differential expression were found above the horizontal threshold line of 1.3 (-log of p value=0.05). MiRNAs with 2- or more than 2 FC values would lie to the left (downregulated genes) or right (upregulated genes) of a vertical threshold line. Therefore, significantly upregulated or downregulated miRNAs identified with Student t tests would be located in the upper left or upper right parts of the plot. The volcano plot serves as a useful tool for presenting statistically significant results when two groups of samples were compared.
Figure 3.
Volcano plot of significance against the relative expression differences between the control and treated groups (A–C). Each dot represents one of the 81 microRNAs (miRNAs) that was filtered and had detectable expression in either treatment. The X-axis displays log2-transformed signal intensity differences between the control group and the experimental group; the Y axis is the log-odds calculated according to the moderated Student t statistic test for differential expression between the control group and the treated group. The horizontal dashed line and the vertical lines represent significance threshold log-odds=2 and twofold expression differences, respectively. All spots above the horizontal dashed line are miRNAs that were identified as showing significant differential expression between the two treatments. MiRNAs positioned in the left and right upper-lateral quadrants represent downregulation and upregulation, respectively.
Out of 81 miRNAs screened, based on these criteria (statistical significance and [less than or equal to]-2 or [greater than or equal to] +2 FC values), treating cells with curcumin or H2O2 alone downregulated 20 (24.69%) and 18 (22.22%) miRNAs, and upregulated nine (11.11%) and 29 (35.8%) miRNAs, respectively, compared to the controls. Pretreatment with curcumin followed by addition of H2O2 down- and upregulated 29 (35.8%) and 14 (17.28%) miRNAs, respectively, when compared to controls. Out of 29 miRNAs downregulated by curcumin pretreatment, 17 miRNAs were induced by H2O2 alone based on two criteria, i.e., statistical significance and 2-FC. The volcano analysis for the effect of curcumin on miRNA expression has been arranged into two categories.
Curcumin-downregulated microRNAs
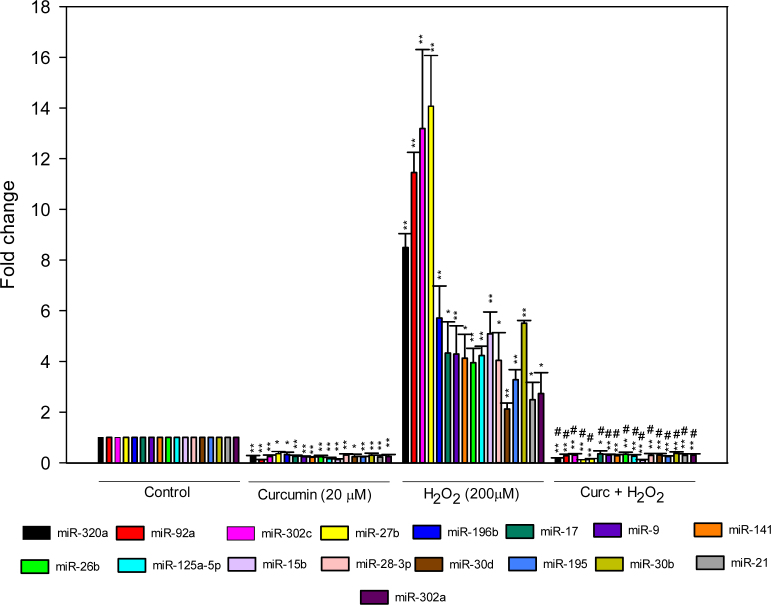
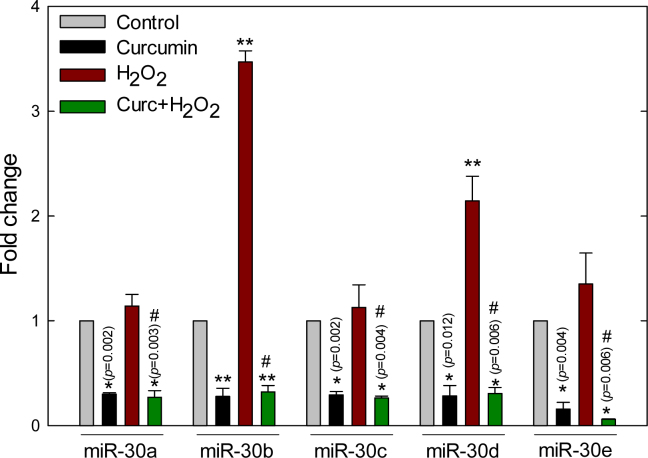
Based on statistical significance (p<0.05) and 2-FC, curcumin pretreatment attenuated the H2O2-induced expression of 17 miRNAs (miR-15b, miR-17, miR-21, miR-26b, miR-27b, miR-28–3p, miR-30b, miR-30d, miR-92a, miR-125a-5p, miR-141, miR-196b,, miR-195, miR-302a, miR-302c, miR-320a, and miR-9), which were also significantly reduced by the curcumin treatment alone (Figure 4, Table 2). Out of 17 H2O2-induced miRNAs, the maximum miRNA expression induced by H2O2 was observed for miR-124 (14.23 FC, p=0.017), and the most downregulated miRNA expression by curcumin pretreatment was observed for miR-30e (−17.16 FC, p=0.014). In the array, miR-23b, another member of the miR-23–27–24 clusters, was also significantly up- and downregulated by H2O2 and curcumin, respectively. Two members of the miR-30 family (miR-30b and miR-30d), putative regulators of the genes implicated in oxidative stress-mediated ocular diseases, were significantly (miR-30b: p=0.001, FC 3.52; miR-30d: p=0.001, FC 2.14) upregulated under the oxidative environment, and curcumin alone significantly (p<0.05) reduced the expression of all five members of the miR-30 family, compared to the controls, and significantly reduced induction by H2O2 (Figure 5).
Figure 4.
Curcumin attenuated hydrogen peroxide-induced microRNA expression in ARPE-19 cells. In the polymerase chain reaction (PCR) array, cells were treated with 20 μM curcumin for 6 h and 200 μM hydrogen peroxide (H2O2) for 18 h. In the curcumin pretreatment group, the cells were treated with 20 μM curcumin for 6 h first, and after washing, the cells were insulted with 200 μM H2O2 for 18 h. The curcumin pretreatment significantly suppressed the H2O2-induced expression of 17 microRNAs (miRNAs) compared to controls. Data represent mean±SEM from three separate samples. *p<0.05 versus control, **p<0.001 versus control, #p<0.001 versus H2O2, FC≥ or ≤2.
Table 2. List of miRNAs altered by curcumin and H2O2.
| Control versus curcumin | Control versus H2O2 | Control versus H2O2+curcumin | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Downregulated |
Downregulated |
Downregulated |
||||||||||
| miRNA (miR) |
P value |
FR (FC) |
miRNA (miR) |
P value |
FR (FC) |
miRNA (miR) |
P value |
FR (FC) |
||||
| miR-302c |
0.033 |
−4.26 (0.24) |
miR-15a |
0.041 |
−13.98 (0.07) |
miR-7 |
0.011 |
−4.18 (0.24) |
||||
| miR-30e |
0.016 |
−7.37 (0.14) |
let-7a |
0.031 |
−5.91 (0.17) |
miR-30d |
0.014 |
−3.40 (0.29) |
||||
| miR-30d |
0.019 |
−4.19 (0.24) |
miR-126 |
0.045 |
−19.01 (0.05) |
miR-140–5p |
0.017 |
−10.42 (0.10) |
||||
| miR-30b |
0.02 |
−3.97 (0.30) |
miR-27a |
0.024 |
−22.15 (0.05) |
miR-141 |
0.031 |
−3.62 (0.28) |
||||
| miR-141 |
0.017 |
−4.46 (0.22) |
miR-29a |
0.036 |
−20.81 (0.05) |
miR-92a |
0.025 |
−3.60 (0.28) |
||||
| miR-92a |
0.006 |
−8.39 (0.12) |
miR-29b |
0.038 |
−20.86 (0.05) |
miR-26b |
0.018 |
−2.94 (0.34) |
||||
| miR-27b |
0.037 |
−2.78 (0.36) |
miR-22 |
0.024 |
−9.57 (0.10) |
miR-196b |
0.015 |
−6.84 (0.15) |
||||
| miR-30a |
0.007 |
−3.33 (0.30) |
Let-7d |
0.001 |
−7.09 (0.14) |
miR-30a |
0.006 |
−3.67 (0.27) |
||||
| miR-26b |
0.013 |
−4.26 (0.24) |
let-7i |
0.036 |
−9.51 (0.17) |
miR-30b |
0.029 |
−2.72 (0.37) |
||||
| miR-17 |
0.023 |
−4.06 (0.25) |
miR-106b |
0.04 |
−7.22 (0.14) |
miR-423–3p |
0.04 |
−6.15 (0.16) |
||||
| miR-15b |
0.037 |
−7.94 (0.13) |
let-7g |
0.005 |
−7.23 (0.14) |
let-7i |
0.027 |
−5.91 (0.17) |
||||
| miR-302a |
0.019 |
−4.06 (0.25) |
miR-28–5p |
0.011 |
−12.99 (0.08) |
miR-106b |
0.034 |
−9.15 (0.11) |
||||
| miR-30c |
0.014 |
−3.46 (0.29) |
miR-374a |
0.044 |
−9.93 (0.10) |
miR-17 |
0.04 |
−2.84 (0.35) |
||||
| miR-9 |
0.007 |
−4.13 (0.24) |
miR-100 |
0.007 |
−10.4 (0.10) |
miR-15b |
0.034 |
−10.18 (0.10) |
||||
| miR-196b |
0.032 |
−3.05 (0.33) |
miR-103 |
0.02 |
−6.27 (0.16) |
miR-302a |
0.017 |
−3.30 (0.30) |
||||
| miR-125a-5p |
0.003 |
−6.16 (0.16) |
miR-128 |
0.012 |
−15.4 (0.06) |
miR-29c |
0.021 |
−6.95 (0.14) |
||||
| miR-195 |
0.002 |
−4.26 (0.24) |
miR-150 |
0.031 |
−9.29 (0.11) |
miR-186 |
0.016 |
−4.51 (0.22) |
||||
| miR-423–3p |
0.027 |
−11.23(0.09) |
miR-7c |
0.024 |
−5.52 (0.18) |
miR-21 |
0.017 |
−3.45 (0.29) |
||||
| miR-21 |
0.017 |
−4.35 (0.23) |
Upregulated |
miR-302c |
0.047 |
−3.46 (0.29) |
||||||
| miR-28–3p |
0.041 |
−3.30 (0.30) |
miR-30d |
0.001 |
2.14 |
miR-9 |
0.016 |
−3.39 (0.30) |
||||
|
Upregulated |
miR-23a |
0.014 |
4.32 |
miR-30c |
0.012 |
−3.80 (0.26) |
||||||
| miR-18a |
0.042 |
2.35 |
miR-142–3p |
0.021 |
4.36 |
miR-320a |
0.034 |
−5.42 (0.18) |
||||
| miR-142–3p |
0.022 |
3.71 |
miR-143 |
0.037 |
10.83 |
miR-125a-5p |
0.003 |
−3.84 (0.26) |
||||
| miR-146a |
0.037 |
3.64 |
miR-124 |
0.017 |
14.23 |
miR-195 |
0.004 |
−4.06 (0.25) |
||||
| miR-20a |
0.002 |
3.4 |
miR-122 |
0.004 |
3.82 |
miR-28–3p |
0.036 |
−4.06 (0.25) |
||||
| miR-126 |
0.041 |
11.9 |
miR-125b |
0.032 |
5 |
miR-128 |
0.018 |
−6.17 (0.16) |
||||
| miR-29b |
0.001 |
5.79 |
miR-141 |
0.001 |
4.13 |
miR-27b |
0.009 |
−8.62 (0.12) |
||||
| miR-22 |
0.001 |
3.91 |
miR-9 |
0.001 |
4.28 |
miR-30e |
0.014 |
−17.16 (0.06) |
||||
| miR-150 |
0.022 |
5.7 |
miR-92a |
0.046 |
11.45 |
miR-143 |
0.021 |
−6.89 (0.15) |
||||
| miR-155 |
0.012 |
3.33 |
miR-27b |
0.014 |
14.06 |
Upregulated |
||||||
| miR-26b |
0.044 |
3.95 |
miR-22 |
0.012 |
20.1 |
|||||||
| miR-96 |
0.016 |
4.69 |
let-7b |
0.025 |
20.1 |
|||||||
| miR-21 |
0.04 |
2.49 |
miR-424 |
0.035 |
8.93 |
|||||||
| miR-23b |
0.009 |
9.34 |
miR-99a |
0.012 |
6.66 |
|||||||
| miR-17 |
0.014 |
4.33 |
let-7d |
0.038 |
16.63 |
|||||||
| miR-15b |
0.001 |
5.09 |
miR-150 |
0.005 |
4.59 |
|||||||
| miR-302a |
0.043 |
2.73 |
miR-20a |
0.001 |
2.89 |
|||||||
| miR-186 |
0.037 |
4.11 |
miR-23b |
0.011 |
5.89 |
|||||||
| miR-223 |
0.017 |
4.76 |
miR-29a |
0.012 |
4.12 |
|||||||
| miR-320a |
0.048 |
8.46 |
miR-29b |
0.016 |
4.02 |
|||||||
| miR-196b |
0.005 |
5.71 |
miR-155 |
0.049 |
2.4 |
|||||||
| miR-302b |
0.001 |
5.85 |
miR-146a |
0.037 |
2.89 |
|||||||
| miR-125a-5p |
0.007 |
4.23 |
miR-126 |
0.006 |
4.35 |
|||||||
| miR-195 |
0.004 |
3.28 |
miR-142–3p |
0.036 |
20.57 |
|||||||
| miR-28–3p |
0.001 |
4.04 |
||||||||||
| miR-302c |
0.026 |
13.19 |
||||||||||
| miR-423–3p |
0.001 |
5.71 |
||||||||||
| miR-30b |
0.001 |
3.52 |
||||||||||
| miR-30d | 0.024 | 2.12 | ||||||||||
Figure 5.
Curcumin alters hydrogen peroxide-induced miR-30 expression in ARPE-19 cells. As measured with the quantitative reverse-transcription polymerase chain reaction (qRT-PCR)–based array, cells treated with 200 μM hydrogen peroxide (H2O2) for 18 h significantly induced miR-30b and miR-30d expression, compared to controls. However, curcumin and curcumin pretreatments for 6 h significantly reduced all five members of miR-30, when compared with controls. In the curcumin pretreated group, the cells were treated with curcumin for 6 h first, and then the cells were incubated for 18 h with H2O2. Data represent mean±SEM from three separate samples. *p<0.05 versus control, **p<0.001 versus control, #p<0.001 versus H2O2, FC≥ or ≤2.
Curcumin-induced microRNAs
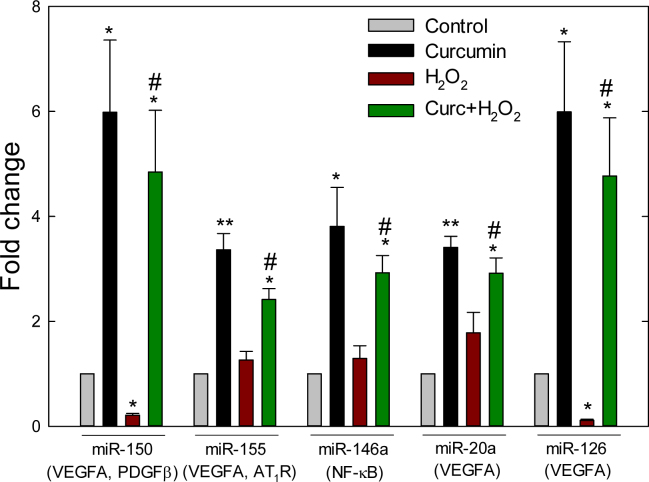
Based on statistical significance (p<0.05) and 2-FC, curcumin alone significantly increased the expression of nine miRNAs (miR-18a, miR-22, miR-20a, miR-29b, miR-126, miR-142–3p, miR-146a, miR-150, and miR-155). However, compared to controls, curcumin pretreatment upregulated 14 H2O2-modulated miRNAs of which seven miRNAs (miR-20a: p=0.001, FC=2.89; miR-126: p=0.006, FC=4.35; miR-146a: p=0.037, FC=2.89; miR-150: p=0.005, FC=4.59; miR-155: p=0.049, FC=2.40; miR-29b: p=0.016, FC=4.02; and miR-142–3p: p=0.036, FC=20.57) were also significantly upregulated by curcumin treatment alone. Five miRNAs (miR-150: p=0.031, FC=−9.29; miR-126: p=0.045, FC=−19.01; miR-29a: p=0.036, FC=−20.81; miR-29b: p=0.038, FC=−20.86; let-7d: p=0.001, FC=−7.09) out of 14 miRNA induced by curcumin pretreatment were significantly downregulated by H2O2 treatment (Figure 6, Table 2).
Figure 6.
Curcumin-induced microRNAs in ARPE-19 cells. Following two criteria of statistical significance (p<0.05) and fold change (FC) (≥ or ≤2), the quantitative real-time polymerase chain reaction (qRT-PCR) array revealed five microRNAs (miRNAs) that were upregulated in cells treated with 20 μM curcumin for 6 h. The miRNAs have been reported to target angiotensin II type 1 receptor (AT1R), nuclear factor-kappa B (NF-κB), platelet-derived growth factor β (PDGFβ), and vascular endothelial growth factor (VEGF). Data represent mean±SEM from three separate samples. Data represent mean±SEM from three separate samples. *p<0.05 versus control, **p<0.001 versus control, #p<0.05 versus H2O2.
In addition to two major groups, PCR revealed a third group in which miRNAs were downregulated by H2O2 and curcumin. Out of 81 miRNAs examined and followed by two criteria (statistical significance and 2-FC), the expression of three H2O2-downregulated miRNAs (let-7i, miR-106b, and miR-128) was significantly downregulated by curcumin pretreatment.
Effect of curcumin on gene expression
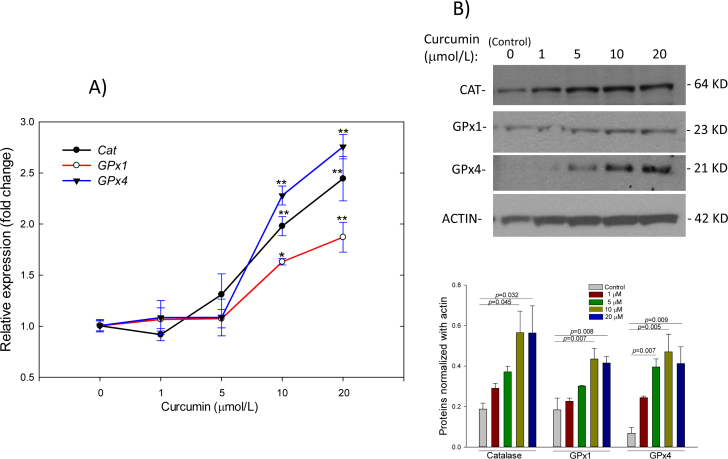
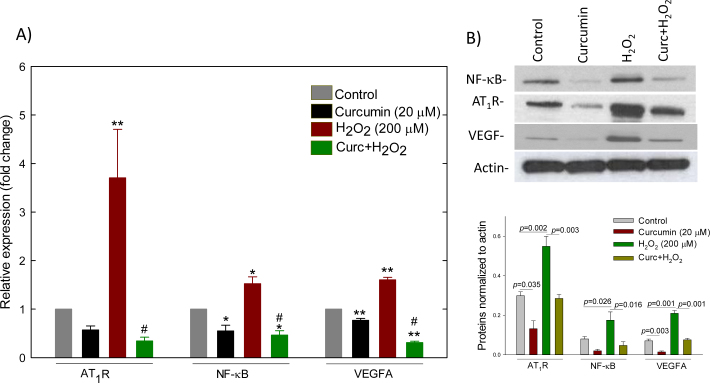
We evaluated the effect of curcumin on the expression of catalase, GPx-s, AT1R, NF-κB, and VEGF-A at the mRNA and protein levels. Exposure of ARPE-19 cells to various concentrations of curcumin (1–20 μM) for 6 h resulted in a concentration-dependent increase in catalase and GPx-s expression at the mRNA and protein levels (Figure 7). The increase in catalase and GPx-s expression at concentrations of 10 μM and above was significantly different from the vehicle (ethanol)-treated cells (control, p<0.001). Compared with the control, a sublethal concentration of H2O2 (200 μM) significantly induced (p<0.001) the expression of AT1R, NF-κB, and VEGF-A at the mRNA and protein levels (Figure 8). Curcumin (20 μM) significantly reduced the expression of NF-κB (p<0.05) and VEGF-A (p<0.001) at the mRNA and protein levels. In addition, curcumin pretreatment significantly (p<0.001) attenuated the H2O2-induced expression of AT1R, NF-κB, and VEGF-A at the mRNA and protein levels (Figure 8), indicating that the activation of AT1R, NF-κB, and VEGF-A is mediated by a prooxidant mechanism.
Figure 7.
Dose effect of curcumin on catalase and Gpx-s messenger RNA (A) and protein (B) levels in ARPE-19 cells. Cells treated with various concentrations of curcumin (1–20 μΜ) for 6 h dose-dependently increased catalase, GPx-1, and GPx-4 at the messenger RNA (mRNA) and protein levels, compared with controls. Representative western blots of catalase (CAT), GPx-1, GPx-4, actin, and densitometric analysis of protein levels are shown in the upper and lower panels, respectively. Data represent mean±SEM from four separate samples. *p<0.05 versus control, **p<0.001 versus control.
Figure 8.
Curcumin attenuated hydrogen peroxide (H2O2)-induced expression of angiotensin II type 1 receptor, nuclear factor-kappa B, and vascular endothelial growth factor at the mRNA (A) and protein (B) levels in ARPE-19 cells. The mRNA and protein measurements were done with quantitative real-time polymerase chain reaction (qRT-PCR) and western blotting, respectively. Cells were treated with curcumin and hydrogen peroxide (H2O2) for 6 and 18 h, respectively. In the curc+H2O2 group, cells were treated with 6 h followed by insult with 200 μM H2O2 for 18 h; cells were washed with fresh media in-between the curcumin and H2O2 treatments. Representative western blots of angiotensin II type 1 receptor (AT1R), nuclear factor-kappa B (NF-κB), and actin, and densitometric analysis of the protein levels are shown in the upper and lower panels, respectively (B). Data represent mean±SEM from three (for western) or four (for qRT-PCR) separate samples. *p<0.05 versus control, **p<0.001 versus control, #p<0.001 versus H2O2.
Discussion
This study explored the potential modulation of miRNAs by curcumin in ARPE-19 cells using PCR array and identified several H2O2-modulated miRNAs whose expression was altered by curcumin. Dietary polyphenolic components such as curcumin have been implicated in many biologic pathways involved in development, differentiation, apoptosis, proliferation, and cellular stress signaling [9,18,19]. These processes have been reported to be regulated by miRNAs [20-22]. Bioinformatic analysis showed that a single miRNA is capable of modulating the expression of more than 100 mRNA targets and more than 50% of human protein coding genes could be regulated by miRNAs [23]. Therefore, to investigate the functional aspects of miRNAs, array-based miRNA surveys and other high-throughput approaches are becoming increasingly popular in biologic sciences. To date, 1,527 human mature miRNAs have been reported (miRBase 18). However, the exact number of ocular miRNAs expressed in the human retina or RPE is not yet known. For the first time, we report the effect of curcumin on the expression profiles of miRNAs in ARPE-19 cells, a cellular model for human retinal pigment epithelium.
Curcumin or H2O2 treatment significantly affected the levels of many miRNAs in ARPE-19 cells. In general, more miRNAs were upregulated than downregulated in response to H2O2 treatment, while curcumin treatment primarily downregulated expression. Of the miRNAs that were affected by both treatments, the direction (up- or downregulation) was opposite in all cases except one, miR-142–3p. In addition, curcumin counteracted the upregulation of miR expression by H2O2 treatment.
H2O2 treatment significantly upregulated miR-30b and miR-30d, two members of the miR-30 family, which is consistent with our previous results [17], and all five members of the family were downregulated by the curcumin treatments. The mechanism of ROS-mediated gene regulation of miR-30b and miR-30d seems to be different from that of the other three members of the family. In silico analysis suggests that the expression of miR-30b and miR-30d, but not the other three members of the family, is regulated by the promoter of the zinc finger and AT hook domain–containing (ZFAT) gene. The epigenetic [2] and transcription factor-mediated regulation [3] of miRNA genes may underlie the molecular mechanism of ROS-mediated regulation of miRNAs in ARPE-19 cells.
Curcumin and other dietary components have been reported to alter the expression profiles of miRNAs in other tissues. In the human pancreatic cell line, curcumin has been shown to significantly up- and downregulate 11 and 18 miRNAs, respectively [5]. Consistent with those data, in our study, curcumin upregulated miR-103, miR-22, and miR-23b and downregulated miR-195, miR-15b, miR-196, and miR-92. In the human multidrug-resistant adenocarcinoma cell line A549/DDP, curcumin altered miRNA expression and significantly downregulated the expression of miR-186 [24], a negative regulator of the proapoptotic purinergic P2X7 receptor [25], which is also consistent with our result.
Curcumin was shown to significantly downregulate the H2O2-induced expression of miR-302 cluster in ARPE-19 cells. MiR-302 has been reported to inhibit several epigenetic regulators, including AOF1/2, methyl-CpG binding proteins 1 and 2, and DNA (cytosine-5-)-methyltransferase 1, that induce global DNA demethylation and subsequently activate transcription factors Oct4, Sox2, and Nanog [26]. The treatment of ARPE-19 cells with H2O2 in our experiment induced miR-26b, miR-15b, and miR-9, and that induction was significantly suppressed by curcumin. The oxidant-induced expression of these three miRNAs showed consistency with the result shown in ARPE-19 cells treated with a retinoic acid derivative (4HPR), which induces ROS generation [27]. MiR-21 has been shown to protect cardiac myocytes against H2O2-induced injury via targeting the programmed cell death protein 4 and activator protein-1 pathway [28]. Our data also showed that miR-21 was sensitive to H2O2 stimulation, and expression of miR-21 was significantly downregulated by curcumin pretreatment. The miR-17–92 cluster is expressed in human retinoblastoma, and upon deletion of Rb family members, miR-17–92 overexpression leads to explosive development of retinoblastoma [29]. In our investigation, miR-17 and miR-92 of the cluster were induced by H2O2-mediated stress, whereas curcumin treatments significantly downregulated the expression of the cluster.
The actions of curcumin and resveratrol, a structurally-related polyphenolic compound, are similar in some respects [30,31]. Curcumin and resveratrol target many of the same signaling molecules, including NF-κB, B-cell lymphoma 2, B-cell lymphoma-extra large, Bim, and survivin [32]. Curcumin and resveratrol have antioxidant effects that protect ARPE-19 cells from cytotoxicity [7,33,34]. Although the effects of resveratrol on miRNA expression in RPE cells have not been reported to our knowledge, resveratrol affects miRNA expression in several other cell types, and miRNA regulation is increasingly thought of as a means of delivering the beneficial effects of resveratrol [35]
A major challenge for retinal miRNA studies is identifying relevant genes and their downstream targets that regulate angiogenesis and increased vascular permeability, the major factors for wet AMD and DR [36]. AMD and DR are the leading causes of blindness. Oxidative stress-mediated increases of VEGF, vascular endothelial growth factor receptor, Ang II, AT1R, NF-κB, and transforming growth factor beta promote angiogenesis and increased vascular permeability; these are the well-recognized regulatory factors for wet AMD and proliferative DR. Our qRT-PCR and immunoblotting data showed that the expression of AT1R, VEGF, and NF-κB was strongly upregulated with H2O2-mediated oxidative stress at the mRNA and protein levels. However, curcumin not only reversed the H2O2-mediated expression but also significantly decreased their expression compared with control. In our investigation, curcumin significantly induced the expression of five miRNAs (miR-146a, miR-150, miR-155, miR-20a, miR-22, and miR-126) that target downstream molecules such as VEGF, NF-κB, PDGFβ, and endothelin 1. The mechanism of action of curcumin on the modulation of miRNA expression is not well understood. The altered expression of miRNAs can occur via several molecular mechanisms such as transcriptional regulation, post-transcriptional processing, genomic abnormalities [37], and regulation by epigenetic factors [38]. MiR-146a was shown to be transactivated by NF-κB, but also to inhibit NF-κB activation, showing negative feedback regulation on NF-κB activation [39]. VEGF was reported to induce miR-20a and miR-155 in human umbilical vein endothelial cells [40]. However, miR-20a was also reported to target VEGF, indicating negative feedback regulation of miR-20a on VEGF [41]. MiR-150 and miR-155 were also reported to regulate PDGFβ [42] and AT1R [43], respectively. The miR-23–27–24 clusters enhance angiogenesis and choroidal neovascularization in mice by repressing sprouty2 and Sema6a proteins, which negatively regulate MAPK and VEGFR2 signaling in response to angiogenic factors [44]. In our analysis, curcumin downregulated two members of the clusters, miR-23b and miR-27b, which were upregulated by H2O2-mediated oxidative stress.
Curcumin has been known to target several biochemical and molecular signaling cascades either through direct binding to proteins or through modulation of gene expression [45]. Curcumin has been shown to physically interact with 33 proteins and is known to modulate various molecular targets, including cytokines, transcription factors, growth factors and their receptors, and genes controlling cell proliferation and apoptosis [46]. The inhibitory effect of curcumin on carcinogenic, angiogenic, inflammatory, and proliferative properties is mediated through suppression of a host of cell-signaling molecules, including activator protein-1 [46], NF-κB [46], early growth response-1 factor [47], tumor necrosis factor alpha [48,49], cytokines [49], IκB kinase phosphorylation [50], Janus kinase-2 phosphorylation [51], c-Jun N-terminal kinase [52], phosphoinositide 3-kinase/protein kinase B phosphorylation [53,54] and p38 MAPK [54,55], extracellular signal-regulated kinase phosphorylation [56], epidermal growth factor receptor [54], tyrosine kinase, serine/threonine and tyrosine protein kinase [46,56], matrix metalloprotease-9 [57], nitric oxide synthase [55], PDGF and epidermal growth factor [58], transforming growth factor beta-1, connective tissue growth factor, VEGF, and vascular endothelial growth factor receptor [56,59,60]. Curcumin-mediated reduction of NF-κB protein in our experiments could be through direct interaction with the protein, as curcumin was shown to directly bind to NF-κB and the binding was reversed by glutathione [61].
In summary, we evaluated the effect of curcumin on the expression levels of miRNA in ARPE-19 cells in the presence of an oxidative environment. For the first time, we have shown that this polyphenolic compound can alter the expression profiles of H2O2-modulated miRNAs in this human RPE culture system. Modulation of miRNA expression may be an important mechanism in the pathogenesis of AMD and DR, and curcumin may provide a therapeutic approach for preventing and treating these oxidative stress-mediated diseases.
Acknowledgments
The research was supported by an unrestricted departmental award from Research to Prevent Blindness (RPB), Inc., and NIH grants R01EY004864, and P30EY006360. PMI is a recipient of a Senior Scientific Investigator Award from RPB. The authors thank Hana Kim, Jane abbey, and Fazila Aseem for technical assistance, and Dr. Purnachandra Ganji (Emory University) for donating NF-κB antibody. The authors declare no conflict of interest with respect to the research reported herein.
References
- 1.Tate DJ, Jr, Miceli MV, Newsome DA. Phagocytosis and H2O2 induce catalase and metallothionein gene expression in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1995;36:1271–9. [PubMed] [Google Scholar]
- 2.Weigel AL, Handa JT, Hjelmeland LM. Microarray analysis of H2O2-, HNE-, or tBH-treated ARPE-19 cells. Free Radic Biol Med. 2002;33:1419–32. doi: 10.1016/s0891-5849(02)01082-1. [DOI] [PubMed] [Google Scholar]
- 3.Vandenbroucke K, Robbens S, Vandepoele K, Inze D, Van de Peer Y, Van Breusegem F. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–16. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 4.Irani K. Oxidant signaling in vascular cell growth, death, and survival: a review of the roles of reactive oxygen species in smooth muscle and endothelial cell mitogenic and apoptotic signaling. Circ Res. 2000;87:179–83. doi: 10.1161/01.res.87.3.179. [DOI] [PubMed] [Google Scholar]
- 5.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–73. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay P, Mukherjee S, Ahsan K, Bagchi A, Pacher P, Das DK. Restoration of altered microRNA expression in the ischemic heart with resveratrol. PLoS ONE. 2010;5:e15705. doi: 10.1371/journal.pone.0015705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo JM, Shin DY, Lee SJ, Joe Y, Zheng M, Yim JH, Callaway Z, Chung HT. Curcumin protects retinal pigment epithelial cells against oxidative stress via induction of heme oxygenase-1 expression and reduction of reactive oxygen. Mol Vis. 2012;18:901–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JH, Wann H, Chen MM, Pan WH, Chen YC, Liu CM, Yeh MY, Tsai SK, Young MS, Chuang HY, Chao FP, Chao HM. Baicalein significantly protects human retinal pigment epithelium cells against H(2)O(2)-induced oxidative stress by scavenging reactive oxygen species and downregulating the expression of matrix metalloproteinase-9 and vascular endothelial growth factor. J Ocul Pharmacol Ther. 2010;26:421–9. doi: 10.1089/jop.2010.0063. [DOI] [PubMed] [Google Scholar]
- 9.Epstein J, Sanderson IR, Macdonald TT. Curcumin as a therapeutic agent: the evidence from in vitro, animal and human studies. Br J Nutr. 2010;103:1545–57. doi: 10.1017/S0007114509993667. [DOI] [PubMed] [Google Scholar]
- 10.Strasser EM, Wessner B, Manhart N, Roth E. The relationship between the anti-inflammatory effects of curcumin and cellular glutathione content in myelomonocytic cells. Biochem Pharmacol. 2005;70:552–9. doi: 10.1016/j.bcp.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 11.Osawa T, Kato Y. Protective role of antioxidative food factors in oxidative stress caused by hyperglycemia. Ann N Y Acad Sci. 2005;1043:440–51. doi: 10.1196/annals.1333.050. [DOI] [PubMed] [Google Scholar]
- 12.Shao ZM, Shen ZZ, Liu CH, Sartippour MR, Go VL, Heber D, Nguyen M. Curcumin exerts multiple suppressive effects on human breast carcinoma cells. Int J Cancer. 2002;98:234–40. doi: 10.1002/ijc.10183. [DOI] [PubMed] [Google Scholar]
- 13.Sarkar FH, Li Y. Cell signaling pathways altered by natural chemopreventive agents. Mutat Res. 2004;555:53–64. doi: 10.1016/j.mrfmmm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 14.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 17.Haque R, Chun E, Howell JC, Sengupta T, Chen D, Kim H. MicroRNA-30b-Mediated Regulation of Catalase Expression in Human ARPE-19 Cells. PLoS ONE. 2012;7:e42542. doi: 10.1371/journal.pone.0042542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Incalci M, Steward WP, Gescher AJ. Use of cancer chemopreventive phytochemicals as antineoplastic agents. Lancet Oncol. 2005;6:899–904. doi: 10.1016/S1470-2045(05)70425-3. [DOI] [PubMed] [Google Scholar]
- 19.Kowluru RA, Kanwar M. Effects of curcumin on retinal oxidative stress and inflammation in diabetes. Nutr Metab (Lond) 2007;4:8. doi: 10.1186/1743-7075-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410–5. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–73. doi: 10.1016/j.tig.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Li SC, Tang P, Lin WC. Intronic microRNA: discovery and biological implications. DNA Cell Biol. 2007;26:195–207. doi: 10.1089/dna.2006.0558. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Du Y, Wu C, Ren X, Ti X, Shi J, Zhao F, Yin H. Curcumin promotes apoptosis in human lung adenocarcinoma cells through miR-186* signaling pathway. Oncol Rep. 2010;24:1217–23. doi: 10.3892/or_00000975. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Qi X, Potashkin JA, Abdul-Karim FW, Gorodeski GI. MicroRNAs miR-186 and miR-150 down-regulate expression of the pro-apoptotic purinergic P2X7 receptor by activation of instability sites at the 3′-untranslated region of the gene that decrease steady-state levels of the transcript. J Biol Chem. 2008;283:28274–86. doi: 10.1074/jbc.M802663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SL. Concise review: Deciphering the mechanism behind induced pluripotent stem cell generation. Stem Cells. 2011;29:1645–9. doi: 10.1002/stem.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutty RK, Samuel W, Jaworski C, Duncan T, Nagineni CN, Raghavachari N, Wiggert B, Redmond TM. MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol Vis. 2010;16:1475–86. [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25:1734–45. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Chen LX, Ouyang L, Cheng Y, Liu B. Plant natural compounds: targeting pathways of autophagy as anti-cancer therapeutic agents. Cell Prolif. 2012;45:466–76. doi: 10.1111/j.1365-2184.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recio MC, Andujar I, Rios JL. Anti-inflammatory agents from plants: progress and potential. Curr Med Chem. 2012;19:2088–103. doi: 10.2174/092986712800229069. [DOI] [PubMed] [Google Scholar]
- 32.Vinod BS, Maliekal TT, Anto RJ. Phytochemicals As Chemosensitizers: From Molecular Mechanism to Clinical Significance. Antioxid Redox Signal. 2012;9:9. doi: 10.1089/ars.2012.4573. [DOI] [PubMed] [Google Scholar]
- 33.King RE, Kent KD, Bomser JA. Resveratrol reduces oxidation and proliferation of human retinal pigment epithelial cells via extracellular signal-regulated kinase inhibition. Chem Biol Interact. 2005;151:143–9. doi: 10.1016/j.cbi.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Pintea A, Rugina D, Pop R, Bunea A, Socaciu C, Diehl HA. Antioxidant effect of trans-resveratrol in cultured human retinal pigment epithelial cells. J Ocul Pharmacol Ther. 2011;27:315–21. doi: 10.1089/jop.2010.0144. [DOI] [PubMed] [Google Scholar]
- 35.Lançon A, Kaminski J, Tili E, Michaille JJ, Latruffe N. Control of MicroRNA expression as a new way for resveratrol to deliver its beneficial effects. J Agric Food Chem. 2012;60:8783–9. doi: 10.1021/jf301479v. [DOI] [PubMed] [Google Scholar]
- 36.Witmer AN, Vrensen GF, Van Noorden CJ, Schlingemann RO. Vascular endothelial growth factors and angiogenesis in eye disease. Prog Retin Eye Res. 2003;22:1–29. doi: 10.1016/s1350-9462(02)00043-5. [DOI] [PubMed] [Google Scholar]
- 37.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 38.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52:4402–9. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- 40.Suárez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105:14082–7. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16:1208–16. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin MM, Lee EJ, Buckenberger JA, Schmittgen TD, Elton TS. MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts. J Biol Chem. 2006;281:18277–84. doi: 10.1074/jbc.M601496200. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci USA. 2011;108:8287–92. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kunnumakkara AB, Anand P, Aggarwal BB. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008;269:199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: the Indian solid gold. Adv Exp Med Biol. 2007;595:1–75. doi: 10.1007/978-0-387-46401-5_1. [DOI] [PubMed] [Google Scholar]
- 47.Pendurthi UR, Rao LV. Suppression of transcription factor Egr-1 by curcumin. Thromb Res. 2000;97:179–89. doi: 10.1016/s0049-3848(99)00148-6. [DOI] [PubMed] [Google Scholar]
- 48.Singh S, Aggarwal BB. Activation of transcription factor NF-kappa B is suppressed by curcumin (diferuloylmethane) J Biol Chem. 1995;270:24995–5000. doi: 10.1074/jbc.270.42.24995. corrected. [DOI] [PubMed] [Google Scholar]
- 49.Cho JW, Lee KS, Kim CW. Curcumin attenuates the expression of IL-1beta, IL-6, and TNF-alpha as well as cyclin E in TNF-alpha-treated HaCaT cells; NF-kappaB and MAPKs as potential upstream targets. Int J Mol Med. 2007;19:469–74. [PubMed] [Google Scholar]
- 50.Singh S, Khar A. Biological effects of curcumin and its role in cancer chemoprevention and therapy. Anticancer Agents Med Chem. 2006;6:259–70. doi: 10.2174/187152006776930918. [DOI] [PubMed] [Google Scholar]
- 51.Bharti AC, Donato N, Aggarwal BB. Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol. 2003;171:3863–71. doi: 10.4049/jimmunol.171.7.3863. [DOI] [PubMed] [Google Scholar]
- 52.Chen YR, Tan TH. Inhibition of the c-Jun N-terminal kinase (JNK) signaling pathway by curcumin. Oncogene. 1998;17:173–8. doi: 10.1038/sj.onc.1201941. [DOI] [PubMed] [Google Scholar]
- 53.Weir NM, Selvendiran K, Kutala VK, Tong L, Vishwanath S, Rajaram M, Tridandapani S, Anant S, Kuppusamy P. Curcumin induces G2/M arrest and apoptosis in cisplatin-resistant human ovarian cancer cells by modulating Akt and p38 MAPK. Cancer Biol Ther. 2007;6:178–84. doi: 10.4161/cbt.6.2.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Yu S, Shi W, Ge L, Yu X, Fan J, Zhang J. Curcumin Inhibits the Migration and Invasion of Mouse Hepatoma Hca-F Cells Through Down-regulating Caveolin-1 Expression and Epidermal Growth Factor Receptor Signaling. IUBMB Life. 2011;63:775–82. doi: 10.1002/iub.507. [DOI] [PubMed] [Google Scholar]
- 55.Camacho-Barquero L, Villegas I, Sanchez-Calvo JM, Talero E, Sanchez-Fidalgo S, Motilva V, Alarcon de la Lastra C. Curcumin, a Curcuma longa constituent, acts on MAPK p38 pathway modulating COX-2 and iNOS expression in chronic experimental colitis. Int Immunopharmacol. 2007;7:333–42. doi: 10.1016/j.intimp.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Soetikno V, Watanabe K, Sari FR, Harima M, Thandavarayan RA, Veeraveedu PT, Arozal W, Sukumaran V, Lakshmanan AP, Arumugam S, Suzuki K. Curcumin attenuates diabetic nephropathy by inhibiting PKC-alpha and PKC-beta1 activity in streptozotocin-induced type I diabetic rats. Mol Nutr Food Res. 2011;55:1655–65. doi: 10.1002/mnfr.201100080. [DOI] [PubMed] [Google Scholar]
- 57.Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB. Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res. 2007;67:3853–61. doi: 10.1158/0008-5472.CAN-06-4257. [DOI] [PubMed] [Google Scholar]
- 58.Lin J, Chen A. Activation of peroxisome proliferator-activated receptor-gamma by curcumin blocks the signaling pathways for PDGF and EGF in hepatic stellate cells. Lab Invest. 2008;88:529–40. doi: 10.1038/labinvest.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arbiser JL, Klauber N, Rohan R, van Leeuwen R, Huang MT, Fisher C, Flynn E, Byers HR. Curcumin is an in vivo inhibitor of angiogenesis. Mol Med. 1998;4:376–83. [PMC free article] [PubMed] [Google Scholar]
- 60.Gururaj AE, Belakavadi M, Venkatesh DA, Marme D, Salimath BP. Molecular mechanisms of anti-angiogenic effect of curcumin. Biochem Biophys Res Commun. 2002;297:934–42. doi: 10.1016/s0006-291x(02)02306-9. [DOI] [PubMed] [Google Scholar]
- 61.Anand P, Sung B, Kunnumakkara AB, Rajasekharan KN, Aggarwal BB. Suppression of pro-inflammatory and proliferative pathways by diferuloylmethane (curcumin) and its analogues dibenzoylmethane, dibenzoylpropane, and dibenzylideneacetone: role of Michael acceptors and Michael donors. Biochem Pharmacol. 2011;82:1901–9. doi: 10.1016/j.bcp.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]