Abstract
Attractant binding to bacterial chemotaxis receptors initiates a transmembrane signal that inhibits the kinase CheA bound about 300 Å away at the other end of the receptor. Chemoreceptors form large clusters in many bacterial species, and the extent of clustering has been reported to vary with signaling state. To test whether ligand binding regulates kinase activity by modulating a clustering equilibrium, we measured the effects of two-dimensional receptor concentration on kinase activity in proteoliposomes containing the purified E. coli serine receptor reconstituted into vesicles at a range of lipid-to-protein molar ratios. The IC50 of kinase inhibition was unchanged despite a 10-fold change in receptor concentration. Such a change in concentration would have produced a measurable shift in the IC50 if receptor clustering were involved in kinase regulation, based on a simple model in which the receptor oligomerization and ligand binding equilibria are coupled. These results indicate that the primary signal, ligand control of kinase activity, does not involve a change in receptor oligomerization state. In combination with previous work on cytoplasmic fragments assembled on vesicle surfaces [Besschetnova et al. (2008) Proc. Nat. Acad. Sci. USA 105, 12289–12294], this suggests that ligand binding to chemotaxis receptors inhibits the kinase by inducing a conformational change that expands the membrane area occupied by the receptor cytoplasmic domain, without changing the number of associated receptors in the signaling complex.
Two-component signaling pathways, a signal transduction motif that is widespread in prokaryotes and found in some eukaryotes, involve the autophosphorylation of histidine kinases and phosphotransfer to aspartyl groups of response regulators (1). Bacterial chemotaxis is a well-studied two-component signaling system that enables bacteria to sense chemical gradients and bias swimming toward larger attractant concentrations (2). The methyl-accepting chemotaxis receptors that provide the sensory input for this system have been the focus of numerous investigations that seek to understand the mechanism of transmembrane signaling. An intriguing property of these receptors is the formation of large receptor clusters, typically at the poles of the cell (3). These clusters are thought to be important for receptor cooperativity (4) and adaptation (5) to mediate the sensitivity, dynamic range, and integration of the chemotaxis signaling network (6–9). Clustering may also play a role in receptor activation, as observed for the EGF receptor and other receptors that are activated by ligand-induced dimerization or oligomerization (10, 11). Chemoreceptor clustering has been reported to vary with signaling state, but the evidence has been inconsistent. These data raise the question of whether a clustering equilibrium plays a role in the primary signal, ligand regulation of chemoreceptor activation of the kinase CheA.
Four chemotaxis receptors in E coli share the overall structure and interaction sites shown in Figure 1 (12). The high abundance receptors Tar and Tsr detect aspartate and serine, respectively, and the low abundance receptors Trg and Tap detect ribose/galactose and dipeptides, respectively. These chemoreceptors are transmembrane alpha-helical homodimers, based on crystal and NMR structures of the periplasmic, HAMP, and cytoplasmic domains (13–15). The membrane-distal tip of the cytoplasmic domain binds two proteins, the histidine kinase CheA and a scaffolding protein CheW. The central region of the receptor cytoplasmic domain contains 4 glutamate residues that are methylated and demethylated, which enables the receptor to adapt to an ongoing stimulus. Both the CheR methyltransferase and the CheB methylesterase that modify these adaptation sites have been shown to bind to the carboxy terminus of the high abundance receptors (16, 17). Replacing Glu with Gln at the adaptation sites mimics the effects of receptor methylation (18), and the wild type receptor is genetically encoded in an intermediate adaptation state, with two Glu and two Gln (Gln are initially deamidated to Glu by CheB).
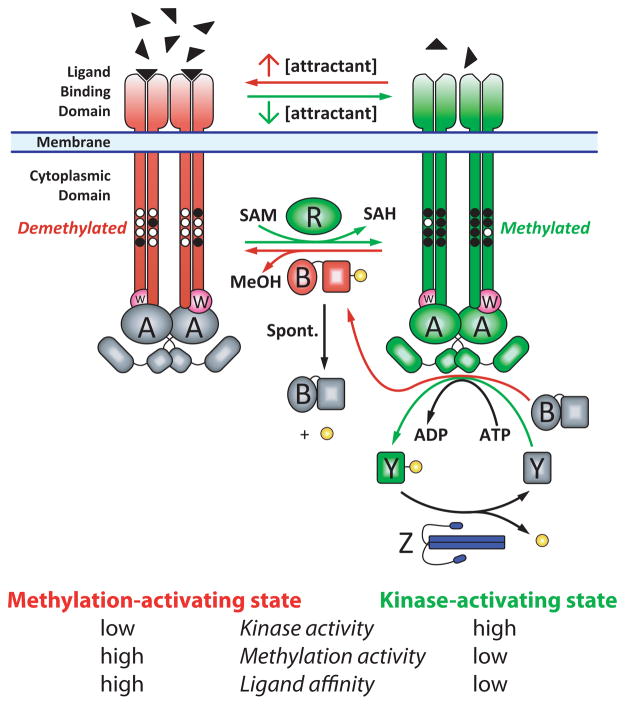
Figure 1.
The E. coli chemotaxis pathway depicted as a two state signaling system. Receptor/CheW/CheA complexes are shown in kinase-stimulating (green/magenta/green) and kinase-inhibiting (red/magenta/gray) states. The binding of attractant (filled triangles) inhibits the CheA kinase (gray A dimer) and stimulates the receptor methylation (black circles) via CheR (green R). Methylated receptors stimulate CheA (green A dimer). The two substrates of CheA, CheY (Y) and the methylesterase (B), are activated by phosphorylation (yellow). CheY-phosphate (green Y) propagates the tumble signal and CheB-phosphate (red B) provides negative feedback by demethylating (and deactivating) receptors. CheB dephosphorylates spontaneously. CheZ (blue Z dimer) dephosphorylates CheY-phosphate. Two features not represented in the figure are the packing of receptor dimers into hexagonal arrays with CheA and CheW, and the unknown receptor:CheA:CheW stoichiometry.
Although not shown in Figure 1 for simplicity, chemotaxis receptors are interconnected by CheA and CheW into hexagonal arrays that have been observed across a wide range of prokaryotes (19). Furthermore, although the stoichiometry of the proteins in these arrays is not known, recent estimates suggest they contain more receptor than CheA and CheW: based on measured stoichiometries of six Tsr per CheA and CheW, it has been suggested that a pair of receptor trimer-of-dimers is needed to activate the dimeric CheA kinase (20). Recent cryoelectron tomography studies have yielded models for the arrangement of receptor, CheA and CheW in the signaling arrays (21, 22).
Receptor signaling is usually described in terms of a two-state equilibrium, with attractant occupancy and methylation shifting the equilibrium in opposite directions (Figure 1). In the absence of attractant ligand, the receptor stimulates autophosphorylation of the histidine kinase CheA, which transfers the phosphate to either CheY or CheB. Phospho-CheY binds to the flagellar motor, causing a change from counterclockwise rotation of the flagellar bundle that propels the cell forward, to clockwise rotation that disrupts the flagellar bundle and causes the cell to tumble. Phosphorylation also activates the CheB methylesterase, which decreases the steady state level of receptor methylation that is determined by the relative activities of CheR and CheB. Binding of an attractant ligand to the receptor turns off kinase activation and thus decreases levels of phospho-CheY and tumbling frequency, so that the cell makes longer runs in the presence of attractants such as Asp or Ser. Following this rapid change in tumbling frequency, adaptation occurs on a slower timescale: decreased levels of phospho-CheB lead to increased levels of receptor methylation, shifting the equilibrium back towards the kinase-activating state. The attractant-bound receptor is also more efficiently methylated by CheR, which contributes to the adaptation shift back to the kinase-activating state.
The focus of this study is to determine whether the extent of receptor clustering is different in the two signaling states depicted in Figure 1. We use the term cluster to refer to a receptor associated with other receptors, CheA, and CheW into an oligomeric multi-protein complex, with intermolecular contacts among the proteins within the cluster, as distinct from co-localized receptors that lack such contacts. Evidence from microscopy and in vivo cross-linking studies is mixed: some studies have reported that ligand binding decreases receptor clustering or methylation increases receptor clustering in cells. Libermann et al (23) observed an increase in the polar localization of CheA-containing complexes with methylation (Tar2Q2E ≈ half methylated receptor relative to Tar4E = unmethylated adaptation state), using fluorescence microscopy of E. coli expressing CheA fused to YFP (yellow fluorescent protein). However, because these changes were much smaller than the measured changes in kinase activity, they concluded that changes in assembly of CheA-containing clusters do not control the kinase (23). Based on immunoelectron microscopy of E. coli cells expressing a single type of chemotaxis receptor, Lyberger et al (24) reported that high abundance receptors are clustered independent of methylation state, but low abundance receptors are significantly less clustered in the unmethylated state. They suggested that both increases in abundance and methylation may shift the equilibrium toward a clustered state, and that such an equilibrium could also regulate kinase activation (24). Homma at al reported that attractant ligand does not decrease the polar localization of a Tar-GFP construct in E. coli, but attractant does decrease in vivo interdimer crosslinking of Tar (25). By contrast, Lamanna et al. observed that attractant ligand decreases polar clusters in both E. coli and B. subtilis, when receptors are crosslinked with paraformaldehyde and then visualized with a fluorescent antibody (26). Finally, Studdert & Parkinson reported that in vivo interdimer crosslinking of Tsr and Tar is independent of both ligand binding and methylation state (27). Limitations of these studies include the inability of microscopy to distinguish clustering (oligomerization) of receptors from co-localization, and the inability of crosslinking studies to distinguish whether changes in the extent of crosslinking result from conformational changes or dissociation of receptors.
Two in vitro studies that correlated receptor concentration with changes in kinase activity are more suggestive that a clustering equilibrium may control the kinase. Lai et al. (28) varied the overexpression level of Tsr or Tar and isolated inner membrane vesicles that contained each receptor as a variable fraction of total membrane protein. The kinase activity per receptor increased linearly with receptor fraction, up to 50% of total membrane protein in these samples (28). Besschetnova et al (29) examined simpler, more defined samples of histidine-tagged cytoplasmic fragments of Tar4E, which were assembled on the surface of liposomes with Ni-chelating lipids, along with CheA and CheW. These template-assembled receptor-signalling arrays displayed a cooperative increase in kinase activation as the 2-dimensional receptor concentration on the vesicle increased. Moreover, receptor methylation activity was observed to decrease as the two dimensional concentration (density) of receptors increased, in a manner consistent with the signaling equilibrium of Figure 1 (29). The results of these in vitro studies are consistent with a clustering equilibrium model in which high receptor concentrations favor the kinase-activating state, which would be a larger oligomeric (more clustered) state than the methylation-activating state. Such a model predicts that ligand binding would favor receptor dissociation into the kinase-inactivating state (less clustered) and thus ligand affinity (which was not measured in either study) would also vary with receptor concentration.
To test whether ligand-induced unclustering is an essential element of the mechanism of kinase regulation, we measured kinase activity and serine dose-response curves on purified E. coli Tsr reconstituted into liposomes over a range of two-dimensional concentrations of receptors. Our results indicate that the activity equilibrium does not involve a change in receptor oligomerization state. In combination with the previous template-assembly results (29), this indicates that the cytoplasmic domain of the kinase-off state has an expanded conformation.
Experimental Procedures
Purification of Serine Receptor (Tsr)
Plasmids pJL41 and pJL31 (30), coding for Tsr4E Δ4His6 and Tsr4Q Δ4His6, respectively, (and constructed analogously to pJL21 (31)) were used to express receptors with a C-terminal His-tag to facilitate purification via nickel affinity chromatography. The receptors were expressed in HCB721, an E. coli strain that lacks CheR and CheB, so the receptor adaptation state was controlled genetically by the 4E and 4Q mutations. Cells were grown at 30°C in LB containing 100 μg/mL ampicillin. Upon reaching an optical density between 0.4 and 0.6 at 600 nm, receptor expression was induced by the addition of 0.5 mM IPTG (final concentration), then grown for three hours and harvested by centrifugation at 3750 rpm (Beckman Coulter Allegra 6R Tabletop Centrifuge; GH-3.8A swinging bucket rotor) for 15 minutes at 4°C. Cell pellets were flash frozen in liquid N2 and stored at − 80°C until purification.
Cell pellets were thawed on ice and resuspended with 4 mL of resuspension buffer per gram of cells (50 mM Tris-HCl pH 8, 300 mM NaCl, 10% glycerol, 1% (w/v) freshly prepared n-octyl-β-D-glucopyranoside (OG) detergent, 10 mM imidazole). Lysozyme and PMSF (final concentrations of 0.15 mg/mL and 2 mM, respectively) were added and cells were incubated on ice for 20 minutes. Sonication was performed with a Branson Ultrasonics sonicator, using a medium-sized tip, at a duty cycle of 35 and an output control of 3.5. The cell-lysozyme suspension was sonicated on ice for three two-minute intervals separated by cooling periods of three minutes. Cell debris was removed in a Beckman L80 ultracentrifuge at 28,000 rpm (104,000g; SW28 rotor) for 1 hour at 4°C. The supernatant, containing the OG-solubilized Tsr, was loaded onto a Ni-NTA column (5 mL HisTrap HP, GE Healthcare) pre-equilibrated with resuspension buffer for separation at 4°C. The column was washed at 5 mL/min with 50 mL of resuspension buffer containing 50 mM imidazole, and Tsr was eluted at 3 mL/min with 30 mL of resuspension buffer containing 250 mM imidazole. Three-milliliter fractions were collected and evaluated by SDS-PAGE. The Tsr-containing fractions were pooled, and EDTA was added to a final concentration of 1 mM to inhibit metalloproteases. Aliquots of the protein solution were flash-frozen in liquid nitrogen and stored at −80°C.
Reconstitution of Tsr into Lipid Vesicles
E. coli polar lipid extract in chloroform (Avanti Polar Lipids) was dried to a thin film in a glass vial using N2 gas. Reconstitution buffer (25 mM Tris-HCl, 25 mM NaCl, 10% glycerol, pH 7.4) was added to generate a 10 mg/mL suspension of lipid. The suspension was extruded 21 times through a 400 nm polycarbonate filter (Avanti Polar Lipids). Purified Tsr in 1% (w/v) OG was added to the lipid vesicles at various lipid-to-receptor ratios. More OG and PMSF were added to give final concentrations of 0.7% (w/v) and 2 mM, respectively. After a one hour incubation at 25°C, the lipid-detergent-receptor mixture was diluted 2-fold with detergent-free reconstitution buffer. Biobeads were added over time to the samples (stirred slowly with microstir bars) sequentially as follows. Aliquots of biobeads, at a 2:1 (w/w) ratio of biobeads to the initial amount of detergent, were added at 0, 30, and 60 minutes at 25°C. Then at 90 minutes and 4°C an aliquot of biobeads was added at a 15:1 ratio of biobeads to the initial amount of detergent. Finally at 120 minutes an aliquot of biobeads was added at a 12:1 ratio of biobeads to the initial amount of detergent for overnight incubation (approx. 16 hours) at 4°C. Using a pipette tip cut ~0.5 cm from the end of the tip, the resulting proteoliposome suspensions were removed from the settled biobeads in each sample and centrifuged for 15–30 minutes at 14,000 rpm (16,000g). The pellets were resuspended in fresh, detergent-free reconstitution buffer. Samples were stable for about a month at 4°C.
Measurement of the Lipid-to-Receptor Ratio in Proteoliposomes
The concentration of purified Tsr in detergent micelles was determined using a BCA protein assay (Pierce Protein Research). Such samples served as a standard for measurement of Tsr concentrations in proteoliposomes by SDS-PAGE, by comparing intensities of Tsr bands using ImageJ. The amount of lipid was determined using the molybdate assay for inorganic phosphate (32). Aliquots from 0.01 to 0.10 mL of a 0.7 mM sodium phosphate (Na2HPO4) standard solution and test samples at different dilutions were placed in Pyrex 13 × 100 mm test tubes. Magnesium nitrate solution (30 μL of 10% Mg(NO3)26H2O in 95% ethanol) was added to each sample, then oven-dried overnight. The dried residue for each sample was ashed over an intense blue Bunsen flame in a fume hood until the evolution of brown NO2 gas stopped. The tubes were cooled, 0.3 mL of 0.5 N HCl was added, and the tubes were capped with a marble and heated in a boiling water bath for 15 minutes. To minimize evaporation, a stream of air was used to cool the tops of the marbles. After the tubes were cooled, 0.7 mL of a solution consisting of 1 part ascorbic acid (10% w/v) and 6 parts ammonium molybdate stock solution (0.42% w/v ammonium molybdate tetrahydrate in 1 N H2SO4) was added to each tube (and to 0.3 mL of water for the blank). After 1 hour of incubatio at 37°C, absorbance at 820 nm was measured. The phosphate concentrations in each test sample were found by comparison to a standard curve generated with Na2HPO4. The calculated [phosphate]/[Tsr] provided the actual molar lipid-to-Tsr ratio in each sample
Ternary Complex Assembly and Binding Assays
Ternary complexes of CheA, CheW and Tsr proteoliposomes were assembled at 25°C overnight in order to test their kinase activities. High and low concentrations of CheA and CheW were used to assemble complexes with 16 μM of receptor in kinase buffer (75 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, 2 mM TCEP, 5% DMSO). Low concentrations were 0.5 μM CheA and 6 μM CheW; high concentrations were 7 μM CheA and 10 μM CheW. The latter concentrations produced the greatest kinase activity with 16 μM Tsr inner membrane preparations (Adam Nelson, unpublished results). CheY was included in the overnight complex assembly for receptors assayed by [γ-32P] ATP incorporation.
After ternary complexes were assembled and incubated at 25°C overnight, 30 μL of the sample was sedimented in a Beckmann TLX tabletop ultracentrifuge at 60,000 rpm (121000g) at 25°C for 30 minutes. The supernatant was removed and the remaining pellets, which contained the CheA bound to the receptors, were resuspended in 1X kinase buffer to a final volume of 10 μL to which was added 10 μL of 2x reducing gel loading buffer before boiling for 5 minutes. Using ImageJ, the intensities of CheA protein bands (on a 10% SDS gel) of the pellet gel samples were compared to a CheA standard series, allowing the determination of the concentration of bound CheA.
CheW binding was used to assess the accessibility of Tsr4E cytoplasmic domains in inner membrane preparations and reconstituted samples, relative to Tar4ECF assembled on liposomes, which were completely accessible (33). 30 μM receptor from each type of sample was incubated at 25°C overnight with varying amounts of CheW in kinase buffer at 25°C. Aliquots removed prior to sedimentation represented the total CheW in a sample, and aliquots removed from the supernatant after sedimentation at 25°C (for 30 min at 121000g in a Beckman TLX tabletop ultracentrifuge with a TLA-120.2 rotor) represented unbound CheW. The total and unbound CheW samples were analyzed by SDS-PAGE on a 15% gel using ImageJ to compute the fraction of receptor-bound CheW as (‘total CheW’ - ‘unbound CheW’)/‘total CheW’.
Spectrophotometric ATPase Assay
In this ATPase assay, regeneration of ATP is coupled to oxidation of NADH, which is monitored spectrophotometrically (34–36). This assay was used only for optimization of reconstitution conditions (Figure 2), using a low lipid-to-receptor ratio of 100 to 1, because it suffers from excessive noise due to light scattering for high lipid-to-receptor ratio proteoliposomes. Ternary complexes of 0.5 μM CheA, 6 μM CheW, and 16 μM receptor in proteoliposomes were assembled in kinase buffer and incubated overnight at 25°C. Following this incubation, 8 μL of complex was diluted 25-fold into a final concentration of 50 μM CheY, 4 units lactose dehydrogenase/pyruvate kinase enzymes (Sigma Aldrich), 4 mM ATP, 4 mM phosphoenolpyruvate and 0.25 mM NADH in kinase buffer. Absorbance (Abs) decrease at 340 nm was monitored immediately. Specific activity (s−1) was calculated from the slope, 6220 molar absorptivity of NADH and the concentration of CheA in the reaction (2 × 10−8 M).
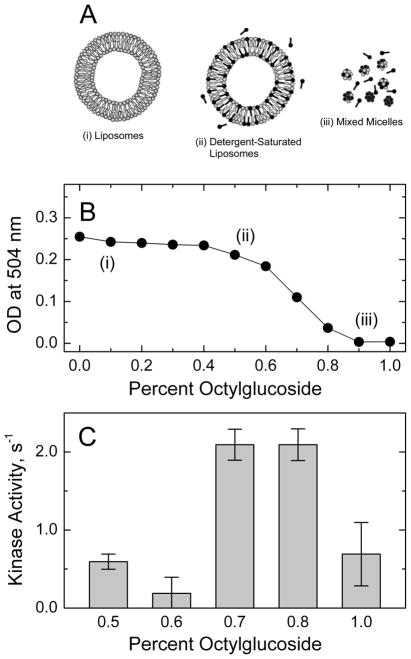
Figure 2.
Optimum conditions for Tsr proteoliposome formation. (A & B) The conversion of phospholipid liposomes (2 mg lipid/mL) into mixed micelles was monitored by light scattering at 504 nm as a function of the OG (0.0 to 1.0% w/v) added to the liposomes. OG-solubilized affinity-purified Tsr was added to the solutions and the OG was removed (as described in Materials and Methods) to yield proteoliposomes. (C) The kinase activity of receptor/CheW/CheA complexes was measured in the coupled ATPase assay using Tsr proteolipsomes prepared with different OG concentrations shown in Fig. 2B. Proteoliposomes prepared with either 0.7 or 0.8% OG (w/v) yielded the largest CheA activity
[γ-32P] ATP Incorporation Assay
To measure kinase activity by [γ-32P] ATP incorporation, ternary complexes of CheA, CheW, CheY and receptor proteoliposomes were assembled and incubated overnight in kinase buffer at 25°C. ATP fuel was prepared freshly in kinase buffer at a final concentration of 2 mM with 0.018 mCi of [γ32P]-ATP (10 mCi/mL, Perkin Elmer) and kept at 25°C. 0.5 μL of kinase buffer or serine was added to 4 μL aliquots of assembled complexes and incubated at 25°C for about 45 minutes. Phosphorylation reactions were initiated by adding 4.5 μL of the radioactive ATP, vortexed and terminated after 15 seconds with the addition of 9 μL 2x reducing SDS gel loading buffer. 3 μL of each sample (out of 18 μL total) were loaded on 16.5% SDS-PAGE gels. After drying, gels were exposed to a phosphorimager screen along with a dilution series of radioactive ATP spotted on a filter paper and sealed in a plastic bag. The phosphor screen was scanned and ImageQuant software was used to estimate phosphate incorporation from the intensities of the 32P labeled CheY bands calibrated against the [γ-32P]-ATP standards. The activity (s−1) of bound CheA was obtained by dividing the total activity per total CheA (s−1) by the fraction of bound CheA. CheA binding to the receptor was measured by SDS-PAGE using the most concentrated receptor samples (50:1 for Tsr4E and 60:1 for Tsr4Q) and applied across the series to calculate the activity per bound CheA.
Results
Vesicle Reconstitution for Control of Receptor Concentration
The strategy chosen for testing the effect of receptor concentration on its activation of CheA involves reconstitution of the purified intact receptor into vesicles at a range of lipid-to-receptor molar ratios for measurements of kinase activity and inhibition to test whether coupled ligand binding and receptor oligomerization equilibria control kinase activity (model shown in Fig 6).
Figure 6.
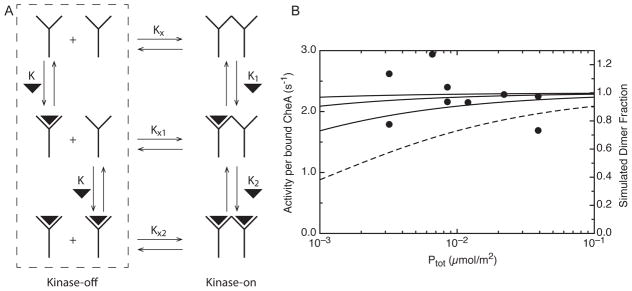
A simple ligand-modulated dimerization model for comparison with activity measurements. (A) In this model the receptors must associate to form the kinase-activating unit, here represented as a “dimer” unit for simplicity (each “monomer” unit could correspond to a trimer of receptor dimers). Binding constants for ligand association (vertical reactions) are K, K1, and K2. Binding constants for receptor association (horizontal reactions) are Kx, Kx1, and Kx2. (B) The concentration-independence of the Tsr4E receptor activity (circles, left y-axis) in the absence of ligand can be compared to the calculated dimer fraction (right y-axis) for a range of values for Kx. The dashed line for Kx = 103 m2/μmol is incompatible with the data; the solid lines for Kx = 106, 105, and 104 m2/μmol (top to bottom) are compatible, providing a lower limit of Kx ≤ 104 m2/μmol. Since the predicted ligand affinity shift with receptor concentration is not observed (Figure 5), the data suggest that the unclustered kinase-off receptor species in the dashed box (left side of Figure 6A) do not occur under physiological conditions.
Variables in the reconstitution of a membrane protein include the choice of lipid, detergent, method of detergent removal, and ratios of lipid-to-detergent and lipid-to-protein. The choice of both detergent (octyl-β-D-glucopyranoside, OG) and lipid (E. coli polar lipid extracts) was based on the fact that these were used in previous successful chemoreceptor solubilization and reconstitution studies (34, 37). Furthermore, the polar lipid extract contains phosphatidylethanolamine, phosphatidylglycerol and cardiolipin at amounts similar to those found in the inner membrane of E. coli (38). Purified Tsr was prepared by overexpression of constructs bearing a C terminal truncation (Δ34) and an appended C terminal His6 tag in HCB721 cells, for purification by nickel-NTA affinity chromatography in 1% (w/v) OG detergent. For optimization of reconstitution conditions, we tested a range of detergent-to-lipid conditions and several detergent removal methods, while maintaining the final lipid-to-receptor molar ratio at 100:1. Centrifugation followed by SDS gel electrophoresis of pellet and supernatant samples provided a rapid assay to determine the fraction of Tsr incorporated into the lipid vesicles upon detergent removal; ATP hydrolysis assays of receptors in the pellet fraction in ternary complexes with CheA and CheW provided a measure of receptor functionality.
The method developed by Rigaud and Levy (39) was used to optimize conditions for obtaining Tsr incorporated into vesicles with maximal kinase activity. A range of reconstitution conditions are prepared by adding various amounts of detergent to the pre-formed liposomes and measuring optical density to monitor solubilization as shown in Figure 2B. Light scattering remains high as OG detergent is titrated into the liposome suspension, producing detergent-destabilized liposomes. Light scattering then decreases as liposomes begin to solubilize into smaller lipid-detergent micelles, until it reaches a minimum OD close to zero when only lipid-detergent micelles are present. Purified Tsr was added to the various lipid-detergent mixtures and the resulting membrane incorporation and kinase activation by the receptor were measured.
Initial experiments indicated that (1) the highest kinase activity is achieved with detergent concentrations near the middle of the liposome solubilization curve, and (2) detergent removal with Biobeads can yield 100% incorporation of Tsr into the membrane fraction, but reconstituted Tsr shows consistently higher kinase activation when OG is diluted 2-fold before the addition of Biobeads (data not shown). Using this detergent removal method (2-fold dilution followed by Biobeads), reconstitutions were performed to find the optimum OG concentrations in the range of 0.5–1.0% by adding purified Tsr to the various lipid-detergent mixtures. As shown in Figure 2C, reconstitution using OG concentrations of 0.7–0.8% results in the greatest kinase activation; therefore 0.7% OG (2 mg/mL lipids) was chosen for reconstitutions with a series of lipid-to-receptor ratios.
The challenges of quantitative studies of membrane proteins in proteoliposomes include limited access to the vesicle interior and light scattering by the vesicles. It is important to determine whether these accessibility and scattering issues are constant over the range of receptor concentrations to be compared in this study. We found that although light scattering was not a problem in the spectrophotometric assay at the larger receptor concentrations (lipid-to-receptor ratios of 50:1 to 300:1), it resulted in unacceptably large errors for low concentration samples (40). Therefore all kinase activation measurements reported below employed a 32P incorporation assay in which light scattering was irrelevant.
The anticipated random incorporation of receptors into proteoliposomes during reconstitution would result in 50% of the intact receptors with their cytoplasmic domain exposed on the outside of the lipid vesicles. It is important to determine whether comparable amounts of cytoplasmic domains are accessible for complex formation and kinase activation across the range of samples that vary in receptor concentration. This was measured using CheW binding measurements on the low and high-concentration extremes of the reconstitution sample series. Template-assembled cytoplasmic fragments (CF) of Tar4E and inner membrane preparations (IMP) of Tsr4E, which are expected to have 100% (33) and ~90% (41, 42) cytoplasmic accessibility, respectively, served as control samples for comparison with proteoliposomes containing reconstituted Tsr4E. As shown in Figure 3A, reconstituted samples with a lipid-to-receptor ratio of 50:1 bound half the amount of CheW compared to the CF and IMP samples, as expected for random orientation with 50% cytoplasmic domains on the outside of the reconstituted proteoliposomes. Figure 3B demonstrates comparable CheW binding for the low (1000:1) and high (50:1) receptor concentration extremes of the Tsr4E proteoliposome series, demonstrating that the lipid-to-receptor ratio during the reconstitution does not alter the accessibility of the receptor cytoplasmic domain, which validates comparison of kinase activation across the sample series.
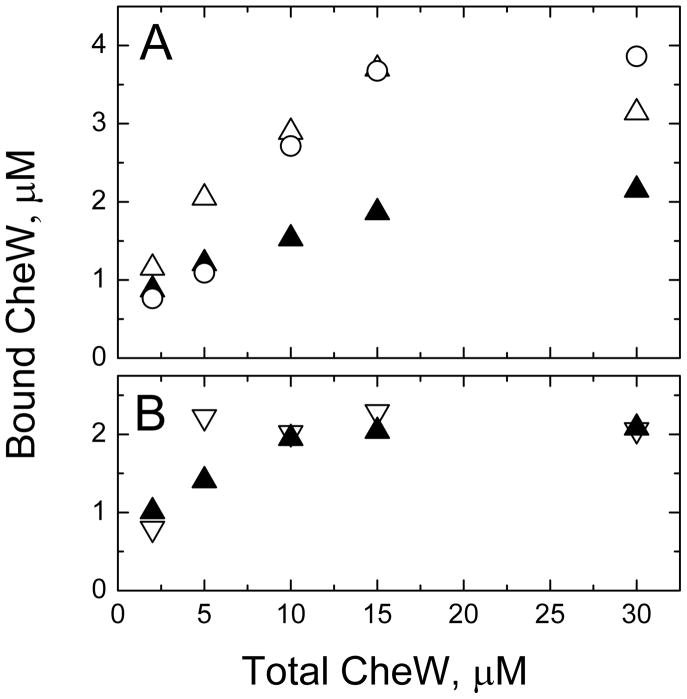
Figure 3.
Receptor accessibility measured by CheW binding. (A) 30 μM cytoplasmic domains, present either as CF assembled on liposomes (circles) or as full-length Tsr in IMPs (white triangles), served as positive controls for binding, which were assumed to have 100% and ~90% accessibility, respectively. 30 μM Tsr4E in proteoliposomes (black triangles) bound about half the amount of CheW as the two controls, consistent with a random orientation of reconstituted receptors, with 50% of the cytoplasmic domains accessible for CheW binding. (B) CheW binding to 15 μM Tsr4E reconstituted at target lipid- to-protein ratios of 50:1 and 1000:1 (white and black triangles, respectively).
Finally, Ser inhibition of kinase activation requires Ser access to the vesicle interior to bind the periplasmic domains of those receptors with their exposed cytoplasmic domains on the vesicle exterior that can form complexes with CheA and CheW. As shown below, the series of reconstituted Tsr4E proteoliposomes were nearly fully inhibited by Ser (~ 80% inhibition). This suggests that the proteoliposomes are leaky to small molecules. Such leakiness is consistent with the fact that lipid-only vesicles made via extrusion of E. coli polar lipids are leaky to Mn2+, based on broadening of 88% of the phosphorous NMR resonance of the lipid headgroups (M Harris & D Fowler, unpublished results). It is possible that such leakiness is not a property of the Tsr4Q proteoliposomes, since these receptors were not inhibited by Ser. However, in our hands inner membrane preparations of Tsr4Q are also not inhibited by Ser. Since inner membrane samples are typically leaky due to the presence of porin impurities, this suggests some other cause of the locked-on state of Tsr4Q.
Kinase Activation is Independent of Receptor Concentration
Proteoliposomes containing a range of intact Tsr concentrations were prepared using the optimized reconstitution protocol that combines OG-solubilized purified receptor with detergent-destabilized extruded vesicles (0.7% OG), followed by a 2-fold dilution and detergent removal with Biobeads. Different amounts of receptor were added to 2 mg/mL of liposomes, with target lipid-to-receptor molar ratios ranging from 50:1 to 1000:1. Measurements of the protein and phosphate concentration in the resuspended proteoliposome samples were used to compute the actual lipid-to-receptor molar ratios, which ranged from 55:1 to 603:1. Ternary complexes of each of the proteoliposome samples were assembled to contain 16 μM Tsr, 7 μM CheA and 10 μM CheW, incubated at 25°C overnight, and then subjected to 32P incorporation assays of kinase activation and SDS-PAGE assays of CheA binding.
The data in Figure 4A and Supporting Information Table S1 demonstrate that kinase activity and inhibition by serine are independent of receptor concentration in proteoliposomes containing reconstituted Tsr4E. The absence of any concentration effect on activity was observed at both high CheA/W concentrations (16 μM Tsr, 7 μM CheA, 10 μM CheW, Figure 4A) conditions under which a maximium in the total kinase signal was observed, and at lower CheA/W concentrations (16 μM Tsr, 0.5 μM CheA, 6 μM CheW, data not shown). As demonstrated in Figure 4A (white triangles), the variations in activity among the samples are all within error, and there is no trend in activity with receptor concentration. Reconstituted Tsr4E proteoliposomes show similar inhibition (~ 80%) by serine at all receptor concentrations (black triangles in Figure 4A): the average kinase activity of 2.3 s−1 decreases to 0.5 s−1 per bound CheA in the presence of serine.
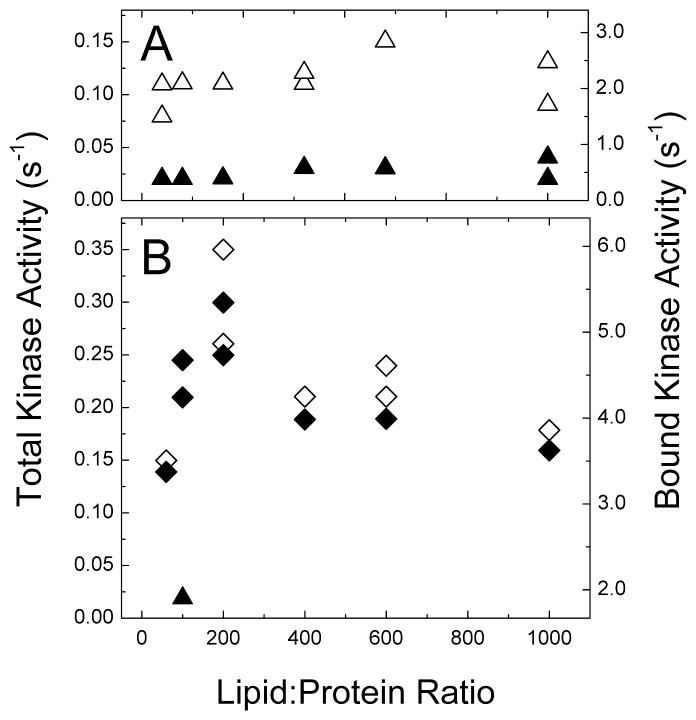
Figure 4.
Kinase activity of receptor/CheW/CheA complexes as a function of the lipid to protein ratio in proteoliposomes. Receptor/CheW/CheA complex assembly conditions: 16 μM receptor, 7 μM CheA and 10 μM CheW incubated overnight at 25°C. Kinase activities (32P incorporation assay) are scaled to the total amount of CheA added (7 μM, left y-axis) and to the amount of CheA bound in the complex (right y-axis). (A) Kinase activities of Tsr4E proteoliposomes without serine (white triangles) and with 10 mM serine (black triangles). (B) Kinase activities of Tsr4Q proteoliposomes without serine (white diamonds) and with 10 mM serine (black diamonds). In contrast to the locked-on property of Tsr4Q proteoliposomes, a 100:1 4E sample included in the same series is inhibited 90% by 10 mM serine (black triangle).
Activity assays on a similar series of proteoliposomes containing reconstituted Tsr4Q again demonstrate that kinase activation is independent of receptor concentration. The target and actual lipid-to-receptor ratios for both the Tsr4Q and Tsr4E reconstitution series (Supporting Information Tables S1 and S2) are close at high receptor concentrations but diverge at low concentrations. Tsr4Q complexes were prepared under high CheA/W conditions for kinase assays; the activity data are tabulated in Table S2 and plotted in Figure 4B. As observed for Tsr4E, there is no clear trend in kinase activation with Tsr4Q receptor concentration (white diamonds), and all activities are within a range similar to the variation between replicate measurements. The exception is the high receptor concentration sample (lipid-to- receptor ~ 60:1), which has reproducibly low activity in the Tsr4Q series.
Although Tsr4Q kinase activation is comparable to Tsr4E, a surprising result is that none of the Tsr4Q proteoliposomes are inhibited by serine (black diamonds). In contrast, a Tsr4E sample included with this series (black triangle in Figure 4B) is 90% inhibited by 10 mM Ser (see Supporting Information Table S2). Additional activity measurements on the 100:1 samples demonstrated that even 400 mM Ser did not inhibit kinase activation by Tsr4Q proteoliposomes (40). The lack of Ser inhibition of Tsr4Q might be related to the C-terminal deletion in our construct, since deletions of residues in the 502–515 range of the Asp receptor have been shown to interfere with Asp inhibition of kinase activity (43). However, our Tsr construct is intact through residue 517, and is capable of wild type levels of kinase inhibition, at least for the Tsr2Q2E (31) and Tsr4E (this study) methylation states.
Ligand Inhibition of Kinase is Independent of Receptor Concentration
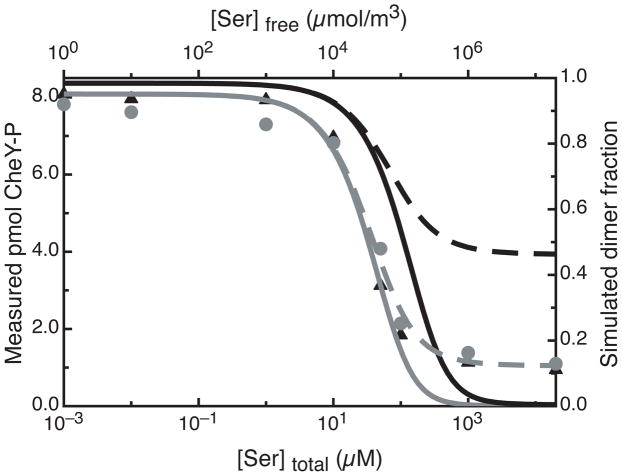
Measurements of ligand affinity can further test the clustering equilibrium model. If ligand binding turns off kinase activation by unclustering the receptor, the low concentration conditions that favor the kinase-off state should yield receptors with higher ligand affinity. An indirect measure of ligand affinity via inhibition of kinase is ideal since it measures binding only to receptors in receptor/CheA/CheW complexes. Proteoliposomes containing Tsr4E reconstituted at high and low concentrations were titrated with serine to determine the concentration of serine at half maximal kinase activity. The kinase assay results plotted in Figure 5 for target lipid-to-receptor molar ratios of 50:1 (black triangles) and 1000:1 (gray circles) demonstrate that the serine inhibition profile is not significantly different at the two concentration extremes. The titration data were fit to the Hill Equation for a quantitative comparison; the resulting parameters are listed in Table 1. [Serine]1/2, the concentration at half inhibition, shifts only slightly, from 28 μM for high- concentration receptor to 46 μM for low-concentration receptor. This is not a significant change, and is in the opposite direction from the prediction of a ligand-modulated clustering equilibrium model: if ligand binding causes unclustering, low-concentration receptors would have higher ligand affinity. Both samples also have similar Hill coefficients (nH = 1.7), indicating positive cooperativity similar to previous observations (nH = 1.5) for the Tsr4E chemoreceptor (4). The similarity of ligand inhibition at high and low receptor concentration indicates that ligand binding does not play a significant role in modulating the receptor association to control kinase activity.
Figure 5.
Apparent serine affinity (IC50) of Tsr4E is independent of receptor concentration. Data are plotted using the left y-axis & bottom x-axis: Experimentally observed amounts of CheY-32P (pmol in 3 μL gel band) generated by receptor/CheW/CheA complexes containing Tsr4E in proteoliposomes reconstituted at target lipid-to-receptor ratios of 50:1 (black triangles) and 1000:1 (gray circles) are plotted as a function of the total serine concentration. Error bars, representing the standard deviation of the CheY-P band intensities when one 32P reaction was loaded twice on the gel, are smaller than the data symbols on the plot. Simulations are plotted using the right y-axis & top x-axis: The simulated dimer fraction for a simple ligand-induced dissociation model (Figure 6A, with K = 4 × 10−4 m3/μmol and Kx = 105 m2/Mmol) is plotted as a function of free [Ser]. Gray lines represent the low receptor concentration sample (RC1000, 4 × 10−3 μmol/m2) and black lines represent the high receptor concentration sample (RC50, 4 × 10−2 Mmol/m2). Dissociation constants for the model are chosen so that ligand binding causes complete inhibition (solid lines with K1 = K2 = 4 × 10−7 m3/μmol) or partial inhibition (dashed lines with K1 = K2 = 8 × 10−6 m3/μmol). For both cases the model predicts a measurable change (gray to black) in the inhibition curve over the 10-fold change in receptor concentration tested. The data show no such change, and thus refute the ligand-induced dissociation model.
Table 1.
Serine binding parameters for Tsr4E proteoliposomes.a
| Proteoliposome sample | 50:1 (RC50) | 1000:1 (RC1000) |
|---|---|---|
| [CheY-P] (pmol) | 8.05 ± 0.04 | 7.30 ± 0.06 |
| Δ[CheY-P] (pmol) | −6.93 ± 0.05 | −6.20 ± 0.06 |
| Hill Coefficient (nH) | 1.65 ± 0.09 | 1.73 ± 0.2 |
| [serine] 1/2 (μM) | 28 ± 1 | 46 ± 3 |
The measured kinase activation properties of Tsr4E at a range of concentrations were compared to a simplified model of a ligand-modulated dimerization equilibrium, based on the treatment by Wofsy et al (44) and using pro Fit (Quantum Soft) to simulate the active dimer fraction of receptors. This model (Figure 6A) assumes that dimerization of some receptor unit (such as dimerization of two receptor trimer of dimers) is necessary to create the kinase-activating complex. Kinase activation is then proportional to the concentration of this active dimer fraction. Vertical reactions in the figure represent ligand binding, with association constants K, K1, and K2, (in three dimensional units of m3/μmol) and horizontal reactions represent receptor dimerization into the kinase-activating complex, with association constants Kx, Kx1, and Kx2 (in two dimensional units of m2/μmol). Note that only 4 of these 6 constants are independent, since Kx1 = KxK1/K and Kx2 = KxK1K2/K2.
The high receptor activity in the absence of ligand at all tested concentrations (physiological range, see Discussion) suggests that the ligand-free dissociated kinase-inactive receptor (top left in Figure 6A) does not occur under physiological conditions. As shown in Figure 6B, the concentration independence of kinase activation in the absence of Ser provides a lower limit for the receptor-receptor affinity in this kinase-activating unit: comparison of the measured activity (same data as plotted in Figure 4A) with simulated curves representing the predicted active dimer fraction for various values of Kx demonstrates that Kx ≥ 104 m2/μmol in order to see similar kinase activation throughout the concentration series. A value of Kx = 105 is used in simulations of the dimer fraction as a function of ligand concentration in Figure 5. In this model, ligand-induced dissociation of the active dimer occurs when the ligand affinity of the inactive “monomer” is greater than the ligand affinity of the active dimer, K > K1 and K2. Simulations can be used to predict the magnitude of the shift in [Ser]1/2 for the receptor inactivation transition, for comparison with the data. The simulated curves plotted in Figure 5 show that ligand binding would fully dissociate (and thus inactivate) the receptor dimer for both the high (solid black line) and low (solid gray line) receptor concentration extremes when K = 4 × 10−4 m3/μmol, and K1 = K2 = 4 × 10−7 m3/μmol. Lower values of K1 and K2 (i.e. 4 × 10−8 m3/μmol) give superimposable curves, and higher values (i.e. K1 = K2 = 8 × 10−6 m3/μmol) give incomplete inhibition (dashed lines). Kx = 1 × 105 was chosen for the simulations so the receptor would be close to fully dimerized and maximally active. For either complete (solid lines) or partial inhibition (dashed lines), the simulations predict a significant shift in [Ser]1/2 for the high vs low receptor concentration samples (Figure 5 black vs gray, respectively), which is not observed. These simulations demonstrate that the 10-fold change in receptor concentration in these two samples would have significantly shifted [Ser]1/2 for a ligand-induced receptor dissociation model and thus these data are sufficient to disprove this mechanism for ligand control of kinase activation.
Discussion
Proteoliposomes Mimic Physiological Conditions to Test Clustering Mechanisms
We have prepared proteoliposomes containing purified bacterial chemotaxis receptors in order to control the receptor concentration and test the role of receptor oligomerization in ligand control of kinase activation. Our results demonstrate that both kinase activation and ligand affinity are independent of receptor concentration. Simulations indicate that we tested a large enough change in receptor concentration to predict a measureable change in ligand affinity, thus demonstrating that modulation of an oligomerization or clustering equilibrium is not the means by which ligand binding controls kinase activation.
Proteoliposomes provide a good mimic of physiological conditions to test the role of receptor clustering in the mechanism of kinase activation. By incorporating purified receptor into proteoliposomes, we avoided the variable amounts of protein impurities that result if receptor overexpression level is varied, to generate a series of samples that differ only in the receptor concentration. As discussed below, the series of receptor proteoliposome samples spans a physiologically relevant range of lipid-to-protein ratios and exhibits kinase activities comparable to those reported for other in vitro receptor studies. The least physiological aspect of the proteoliposome sample is the mixed orientation of the receptors. The only purified receptor system that avoids this issue is the vesicle-templated CF (36), but this system lacks the ligand-binding domain and thus cannot be used to investigate receptor concentration effects on ligand affinity. Finally, proteoliposomes are advantageous over nanodiscs (20) for a study of the effects of receptor clustering because the small nanodiscs (~10 nm diameter) cannot accommodate physiologically relevant clusters which are observed in cells to extend over 200 nm (45).
The Tsr proteoliposomes contain receptors in the concentration range expected for bacteria. An approximate physiological lipid-to-receptor molar ratio can be estimated as follows (46). Since the receptors are found predominantly at the poles of the cells (3, 24), the area of the hemispherical cap is calculated as ½(4πr2) = 0.98 μm2, for a cell diameter of 0.79 μm (r = radius = 0.395 μm). Using a membrane surface area per lipid of 70 Å2 = 7 × 10−7 μm2 (47), the number of lipids at the cell poles = (0.98 μm2 total area/7 × 10−7 μm2 area per lipid) x 2 membrane leaflets x 2 cell poles = 5.6 × 106 lipids. This value for the number of polar lipids divided by reported estimates of 3,600 – 41,000 receptors per cell (48), yields a lipid-to-receptor molar ratio range of 1556:1 to 137:1. Super-resolution light microscopy measurements suggest that about two thirds of Tar receptors localize to the poles (49), which would increase the lipid-to-receptor molar ratio range at the poles to 2334:1 to 206:1. To calculate the effective lipid-to-receptor molar ratio in the Tsr proteoliposomes, the measured ratio ranging from 603:1 to 55:1 (see Table S1 in the Supporting Information), is corrected by a factor of two because only half of the receptors are oriented with the cytoplasmic domain accessible for assembly of complexes with CheA and CheW. So the experimental range of ≈ 1200:1 – 100:1 is similar to the estimated physiological range of ≈ 2300:1 to 200:1 at the cell poles.
It is important to note that an effective system for ligand control of receptor dissociation would likely position the coupled equilibrium for maximal ligand-induced change at physiological concentrations and densities. For example, simulations of the ligand-induced dissociation model described above can position the on/off transitions for the ligand-free and ligand-saturated receptor on either side of the physiological concentration range (transitions at Ptot values of 10−5 Mmol/m3 and 1 Mmol/m3, as shown in Figure S1 in Supplemental Information). Thus throughout the physiological concentration range the receptor would exhibit maximal (and constant) kinase-activation and maximal inhibition by ligand binding. This suggests that the best way to detect such a coupled equilibrium is through its effects on ligand affinity (as shown in Figure 5) rather than effects on maximal activity, which are likely to be negligible.
Kinase activities achieved with the optimized reconstitution protocol compare favorably with previously reported activities. Activities are difficult to compare because of differences in conditions for assembly of ternary complexes resulting in different amounts of bound CheA. Furthermore, specific activity per total or per bound CheA is not always reported. The activity of Tsr4E proteoliposomes observed in this study ranges from ≈ 2 s−1 per total CheA (Figure 2, measured with the spectrophotometric assay with 0.5 μM CheA) to ≈ 0.1 s−1 per total CheA ≈ 2 s−1 per bound CheA (Figure 4A, measured with the 32P incorporation assay with 7 μM CheA). These values are similar to those obtained in the original reconstitution study of Ninfa at al (3 μM ATP/min with 0.2 μM CheA, equivalent to 0.25 s−1 per total CheA) for Tar2Q2E (34). Similar activities in the range of 0.15 – 1.5 s−1 per total CheA have also been reported for native membrane vesicle preparations of Tsr2Q2E (4, 28) or Tar2Q2E (50). Cytoplasmic fragment constructs (TarCF2Q2E) yield somewhat higher activities in the ≈ 12 s−1 range, when either bound to templating vesicles (29) or dimerized by leucine zippers (51). Ligand binding curves for the proteoliposomes display comparable cooperativity to previous studies of receptors in native membrane vesicles, with Hill coefficients of ≈ 1.5 (4) or 1.7 (this study) for Tsr4E, and ≈ 1.8 for Tsr2Q2E (28) and Tar2Q2E (52). However values of the serine concentration at half maximal inhibition vary widely, with 35 μM for Tsr4E in proteoliposomes (this study) found between values previously observed for inner membrane vesicles: ≈ 0.1 μM and 5 μM for Tsr4E and Tsr2Q2E, respectively (4), to ≈ 260 μM for Tsr2Q2E (28). With the exception of the locked-on state of Tsr4Q proteoliposomes observed in this study, which was not observed for Tsr4Q in inner membrane vesicles (4), the proteoliposomes appear to have captured the physiological behavior of Tsr as well as previous in vitro preparations of intact chemotaxis receptors, while achieving control of receptor purity and concentration. In a different, in vivo, setting Tar4E had a Kd of 38 MM and nH of 1.2 (7).
Models for Ligand Control of Kinase Activation: Ligand Binding Expands the Receptor Structure without Changing the Number of Associated Receptors
This study was motivated by a variety of results in the published literature that suggest a possible correlation between receptor clustering and receptor signaling state. Two studies in particular yielded data that strongly suggested that high receptor concentration favors the kinase-activating state. Lai et al. prepared native membrane vesicles using a range of overexpression levels of intact receptors and found that kinase activation increased linearly with the receptor fraction of total protein (28). However, since the lipid and total protein content of the vesicles was not measured, it is not known how the receptor concentration varied among the samples. It is also possible that the receptor orientation in the isolated vesicles varied with receptor overexpression level. Native membrane vesicles isolated from high receptor overexpression conditions are known to contain receptors oriented predominantly inside-out (accessible cytoplasmic domain). If low overexpression yields random orientation, then the increased activity observed with increased overexpression could be due to an increasing fraction of receptors oriented with accessible kinase-binding cytoplasmic domains.
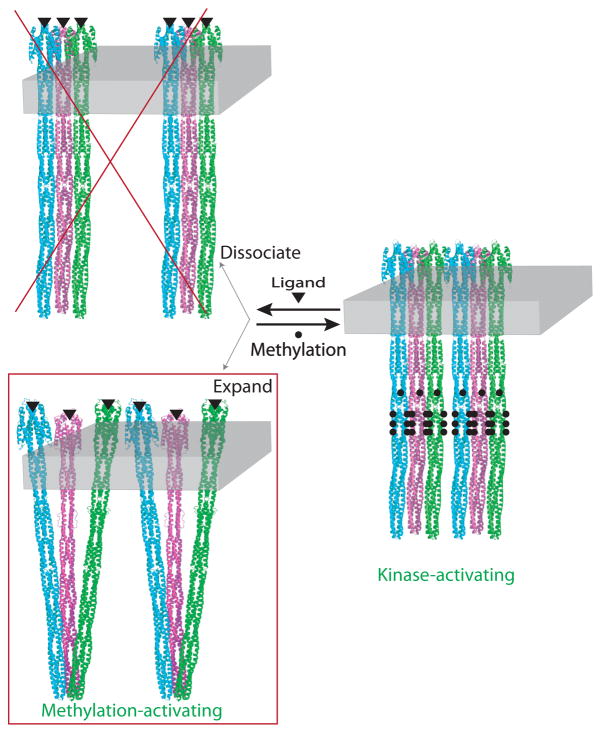
Besschetnova et al (29) employed a simplified system with greater control of receptor concentration and orientation: purified receptor cytoplasmic fragments were bound via N-terminal His tags to vesicles bearing lipids with Ni-chelating headgroups. Thus all receptors are bound to the outside of the vesicle, accessible for kinase binding, and receptor concentration can be controlled precisely. A cooperative increase in kinase activity was observed with increasing receptor density on the vesicles, suggesting that the kinase-activating state is either (i) an oligomerized structure or (ii) a more compact structure, both of which would be favored by high receptor density. Results on intact receptors reconstituted into proteoliposomes now rule out model (i). Figure 7 summarizes this insight into the signaling mechanism: the kinase-activating state (right) forms the methylation-activating state either by dissociating (top) or by expanding (bottom). The dissociation model is eliminated by the current observations that receptor concentration does not alter kinase activity (Figure 4) or ligand affinity (Figure 5), which suggests that ligand binding induces an expanded receptor conformation.
Figure 7.
Model for kinase control: Ligand binding induces an expanded receptor conformation. Six receptor dimers are shown associated as two trimers of dimers bound to a membrane slab of constant size (each dimer in a trimer is depicted in a different color). Cytoplasmic fragments assembled on vesicles are forced into the kinase-activating state by high density (29), which would drive either an oligomerization or contraction of the receptor. The ligand-induced dissociation model is ruled out by the results reported above, the concentration independence of both kinase activity (Figure 4) and ligand affinity (Figure 5). Therefore ligand-binding induces an expanded receptor conformation.
Like chemoreceptors in cells, the intact Ser receptor (Tsr4E) adopts the kinase-activating state when reconstituted into proteoliposomes, and is inhibited by Ser binding. In contrast, binding of the cytoplasmic fragment (TarCF4E) to membrane vesicles is not sufficient to create a kinase-activating state; high density is needed to drive it into the kinase-activating conformation. As noted previously, the density transition for TarCF4E mimics the 2-state equilibrium shown in Figure 1: density of the CF on vesicles, like ligand binding to intact receptors, causes inverse effects on methylation and kinase activities (29). Reconstitutions in the current study all employed receptor densities lower than those used to drive the CF transition to the kinase-activating state (see Supporting Information), and thus would not be expected to trap Tsr4E in a locked-on state, insensitive to inhibition by Ser. It is interesting that the 4Q receptor is found locked-on in both studies, with high kinase activity at both low density (vesicle- templated TarCF4Q) and high ligand concentration (Tsr4Q proteoliposomes).
Results of other studies have also suggested that the kinase-inactivating state of the receptor adopts an expanded conformation. Vaknin and Berg (53) showed that ligand binding to receptor-YFP constructs causes a 10% increase in the distance between the YFP fluorophores at the receptor C-termini. In a cryo-electron tomography study, Khursigara et al (54) observed that ligand binding shifts the receptor population toward a conformation with an expanded HAMP domain. Both of these studies were performed on chemoreceptors in the absence of CheA and CheW. Our current results, interpreted in light of the template-assembled CF study (29), suggest a similar conclusion for chemoreceptors in active complexes with CheA and CheW: attractant ligand binding to the receptor periplasmic domain turns off kinase activation by inducing an expanded receptor conformation that increases the membrane area occupied by each receptor cytoplasmic domain. In contrast to the results of Khursigara et al, this lateral expansion is not restricted to the HAMP domain, since it occurs in TarCF4E that lack the HAMP domain. One possible model for the structural expansion is pictured in Figure 7: the expanded methylation-activating state, made by superimposing intact receptor models onto the trimer of dimers crystal structure of the cytoplasmic domain (1qu7 (15)), consists of tilted receptors that touch at their cytoplasmic tips, consistent with in vivo crosslinking results of Parkinson and coworkers that suggest such contacts occur in bacteria (27). The contracted kinase-activating state, pictured as a parallel bundle of receptors, lacks the trimer of dimers contact at the tip, consistent with NMR measurements on the kinase-activating state (55).
Further insight into the magnitude of the receptor expansion can be drawn from a recent electron cryotomography study of chemoreceptor arrays in Caulobacter crescentus. Briegel et al showed that indistinguishable ≈12 nm hexagonal arrays are observed in the presence and absence of galactose attractant (56). Thus attractant does not dissociate the hexagonal arrays in C crescentus, consistent with our in vitro demonstration that attractant does not inhibit CheA kinase by dissociating E coli chemoreceptors. Assuming E coli and C crescentus chemoreceptors operate similarly, the magnitude of the attractant-induced expansion of the cytoplasmic domain is below the detection limit of the published electron cryotomography study. Our work may also suggest that the overall expansion of the intact receptor is small, since we did not oberve any change in ligand affinity with changes in receptor concentration and density in proteoliposomes.
In summary, we have demonstrated that kinase activation and ligand affinity of bacterial chemoreceptors are independent of concentration, which provides insight into both the role of clustering and the mechanism of transmembrane signaling. Clustering changes may play a role in tuning the sensitivity of the receptor to the conditions of the cell: according to a recent study, E. coli grown under different environmental conditions (minimal vs rich media) exhibited differences in both the receptor arrays (density and/or order) and the sensitivity (steepness) of the chemotaxis response curve (9). The observed change in the response curve is not likely to be due to changes in overall receptor density, since our in vitro data demonstrate that changes in receptor density do not alter the cooperativity of the receptor. Regarding the transmembrane signaling mechanism, we have shown that clustering changes are not involved in the primary signal, ligand inhibition of the kinase. The primary signal occurs when ligand binding to the receptor induces a conformational change without changing the number of associated receptors in the signaling complex. This conformational change expands the lateral area of the membrane occupied by the receptor cytoplasmic domain. Further studies are needed to establish the structural details of the ligand-induced expansion of the cytoplasmic domain, how it is coupled via the HAMP domain to the ligand-induced piston that has been shown to occur in the periplasmic and transmembrane domains (57), and how it inhibits the kinase activity of the associated CheA.
Supplementary Material
Acknowledgments
Funding
This research was supported by U.S. Public Health Service Grant GM47601 to LKT and GM085288 to LKT and RMW, and also supported in part by the U. S. Army Research Office under grant number 54635CH and a Faculty Research Grant from the University of Massachusetts, Amherst. FCS was partially supported by a Fellowship from the University of Massachusetts as part of the Chemistry-Biology Interface Training Program (National Research Service Award T32 GM08515).
The authors thank Jiayin Li for construction of the plasmids pJL31 and pJL41, Abdalin Asinas for help with the initial activity assays, Seena Koshy for proteins used in some kinase assays, Dan Fowler and Michael Harris for NMR measurements of vesicle leakiness, and Adam Nelson for measurement of CheA and CheW levels that maximize kinase activity for inner membrane preparations.
ABBREVIATIONS
- CF
aspartate receptor cytoplasmic fragment
- DMSO
dimethyl sulfoxide
- EDTA
ethylenediaminetetraacetic acid
- EGF
epidermal growth factor
- GFP
green fluorescent protein
- HAMP
histidine kinases, adenylyl cyclases, methyl-accepting proteins and phosphatases
- IMP
inner membrane preparation of overexpressed receptors in native membrane vesicles
- IPTG
Isopropyl β-D-1- thiogalactopyranoside
- LB
Luria-Bertani
- OD
optical density
- OG
n-octyl-β-D-glucopyranoside
- PMSF
phenylmethylsulfonyl fluoride
- RC
reconstituted proteoliposome
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Ser
serine
- Tar
asparate receptor
- Tap
dipeptide receptor
- TCEP
tris(2-carboxyethyl)phosphine
- Trg
ribose/galactose receptor
- Tsr
serine receptor
- YFP
yellow fluorescent protein
Footnotes
Comparison of receptor density in proteoliposomes (this study) and vesicle-templated CF,(29) and Tables S1 and S2 listing kinase activity of proteoliposomes. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes:
The authors declare no competing interests.
References
- 1.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Lai WC. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 4.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 6.Duke TA, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci U S A. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci U S A. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 9.Khursigara CM, Lan G, Neumann S, Wu X, Ravindran S, Borgnia MJ, Sourjik V, Milne J, Tu Y, Subramaniam S. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E. coli chemotaxis response. Embo J. 2011;30:1719–1729. doi: 10.1038/emboj.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 11.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 12.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeh JI, Biemann HP, Pandit J, Koshland DE, Kim SH. The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J Biol Chem. 1993;268:9787–9792. [PubMed] [Google Scholar]
- 14.Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, Coles M. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126:929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 15.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 16.Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 17.Barnakov AN, Barnakova LA, Hazelbauer GL. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci U S A. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunten P, Koshland DE., Jr Tuning the responsiveness of a sensory receptor via covalent modification. J Biol Chem. 1991;266:1491–1496. [PubMed] [Google Scholar]
- 19.Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ, Zhulin IB, Jensen GJ. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci U S A. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci U S A. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci U S A. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Hu B, Morado DR, Jani S, Manson MD, Margolin W. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci U S A. 2012;109:E1481–1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liberman L, Berg HC, Sourjik V. Effect of chemoreceptor modification on assembly and activity of the receptor-kinase complex in Escherichia coli. J Bacteriol. 2004;186:6643–6646. doi: 10.1128/JB.186.19.6643-6646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lybarger SR, Nair U, Lilly AA, Hazelbauer GL, Maddock JR. Clustering requires modified methyl-accepting sites in low-abundance but not high-abundance chemoreceptors of Escherichia coli. Mol Microbiol. 2005;56:1078–1086. doi: 10.1111/j.1365-2958.2005.04593.x. [DOI] [PubMed] [Google Scholar]
- 25.Homma M, Shiomi D, Kawagishi I. Attractant binding alters arrangement of chemoreceptor dimers within its cluster at a cell pole. Proc Natl Acad Sci U S A. 2004;101:3462–3467. doi: 10.1073/pnas.0306660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamanna AC, Ordal GW, Kiessling LL. Large increases in attractant concentration disrupt the polar localization of bacterial chemoreceptors. Mol Microbiol. 2005;57:774–785. doi: 10.1111/j.1365-2958.2005.04728.x. [DOI] [PubMed] [Google Scholar]
- 27.Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci U S A. 2004;101:2117–2122. doi: 10.1073/pnas.0308622100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai RZ, Manson JM, Bormans AF, Draheim RR, Nguyen NT, Manson MD. Cooperative signaling among bacterial chemoreceptors. Biochemistry. 2005;44:14298–14307. doi: 10.1021/bi050567y. [DOI] [PubMed] [Google Scholar]
- 29.Besschetnova TY, Montefusco DJ, Asinas AE, Shrout AL, Antommattei FM, Weis RM. Receptor density balances signal stimulation and attenuation in membrane-assembled complexes of bacterial chemotaxis signaling proteins. Proc Natl Acad Sci U S A. 2008;105:12289–12294. doi: 10.1073/pnas.0802868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J. PhD. Molecular and Cellular Biology Graduate Program, University of Massachusetts; Amherst: 1996. Seeking the primary sense: A biochemical and biophysical study of the signaling mechanism of bacterial chemotaxis; p. 147. [Google Scholar]
- 31.Li J, Li G, Weis RM. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 32.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 33.Asinas AE, Weis RM. Competitive and cooperative interactions in receptor signaling complexes. J Biol Chem. 2006;281:30512–30523. doi: 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- 34.Ninfa EG, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 35.Norby JG. Coupled assay of Na+, K+-ATPase activity. Methods Enzymol. 1988;156:116–119. doi: 10.1016/0076-6879(88)56014-7. [DOI] [PubMed] [Google Scholar]
- 36.Shrout AL, Montefusco DJ, Weis RM. Template-directed assembly of receptor signaling complexes. Biochemistry. 2003;42:13379–13385. doi: 10.1021/bi0352769. [DOI] [PubMed] [Google Scholar]
- 37.Bogonez E, Koshland DE., Jr Solubilization of a vectorial transmembrane receptor in functional form: aspartate receptor of chemotaxis. Proc Natl Acad Sci U S A. 1985;82:4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morein S, Andersson A, Rilfors L, Lindblom G. Wild-type Escherichia coli cells regulate the membrane lipid composition in a “window” between gel and non-lamellar structures. J Biol Chem. 1996;271:6801–6809. doi: 10.1074/jbc.271.12.6801. [DOI] [PubMed] [Google Scholar]
- 39.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 40.Consolacion FC. PhD. Chemistry Department, University of Massachusetts; Amherst: 2011. Clustering independence of ligand affinity and kinase activity of reconstituted bacterial chemoreceptors: Insight into signaling mechanisms; p. 115. [Google Scholar]
- 41.Gegner JA, Graham DR, Roth AF, Dahlquist FW. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 42.Levit MN, Grebe TW, Stock JB. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem. 2002;277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 43.Lai RZ, Bormans AF, Draheim RR, Wright GA, Manson MD. The region preceding the C-terminal NWETF pentapeptide modulates baseline activity and aspartate inhibition of Escherichia coli Tar. Biochemistry. 2008;47:13287–13295. doi: 10.1021/bi8013399. [DOI] [PubMed] [Google Scholar]
- 44.Wofsy C, Goldstein B, Lund K, Wiley HS. Implications of epidermal growth factor (EGF) induced egf receptor aggregation. Biophys J. 1992;63:98–110. doi: 10.1016/S0006-3495(92)81572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci U S A. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asinas AE. PhD. Chemistry Department, University of Massachusetts; Amherst: 2007. Protein interactions and kinase regulation in template-assembled complexes of chemotaxis receptor fragments; p. 172. [Google Scholar]
- 47.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochimica et biophysica acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signaling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y, Levit M, Lurz R, Surette MG, Stock JB. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. Embo J. 1997;16:7231–7240. doi: 10.1093/emboj/16.24.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaknin A, Berg HC. Physical responses of bacterial chemoreceptors. J Mol Biol. 2007;366:1416–1423. doi: 10.1016/j.jmb.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci U S A. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fowler DJ, Weis RM, Thompson LK. Kinase-active signaling complexes of bacterial chemoreceptors do not contain proposed receptor-receptor contacts observed in crystal structures. Biochemistry. 2010;49:1425–1434. doi: 10.1021/bi901565k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26:257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.