Since the discovery of Systemin in tomatoes in 1991, many additional peptide hormones have been discovered in plants. Plant peptide hormones typically start as small precursor proteins (~100a.a.), which include N-terminal secretory or anchor sequences, and are post-translationally processed into a smaller hormonally active peptide (on the range of 5–18a.a.). To date, peptide hormones have been shown to have diverse functions in plants, including nearly every aspect of development and morphogenesis. Here, we discuss recent reports regarding the CLV3/ESR-RELATED (CLE) and ROOT GROWTH FACTOR (RGF) families of peptide hormones.
CLE FAMILY
The CLAVATA3 (CLV3) and EMBRYO SURROUNDING REGION (ESR) peptide hormones share a common peptide sequence with several other proteins in plants (32 so far in Arabidopsis), and were collectively named the CLE family (Cock and McCormick, 2001). Mature CLE peptide hormones are 12–13a.a. long, and are proteolytically released from their precursors by a serine protease (Ni et al., 2011). Receptor kinases are known receptors for CLE peptides, and several of the WUSCHEL-RELATED HOMEBOX (WOX) transcription factors have been implicated as downstream targets in CLE signaling pathways. Expression domains and/or posttranslational modifications to the CLE peptides are likely to be the critical factors for determining their unique functions (Ito et al., 2006).
CLV3 is produced in and excreted from the Shoot Apical Meristem (SAM) central zone (CZ), which contains the stem cells. CLV3 signals through its receptors to repress the expression of WUSCHEL (WUS) in the organizing center (OC) of the SAM (Schoof et al., 2000). WUS acts non-cell autonomously to promote the accumulation of stem cells in the CZ (Schoof et al., 2000). The CLV3/WUS signaling pathway regulates SAM stem cell number through a negative feedback loop (Schoof et al., 2000). Despite the obvious enlarged meristem phenotype of clv3 mutants (Fletcher et al., 1999), none of the other CLE genes was initially identified through mutagenesis.
The root apical meristem (RAM) contains a set of stem cells, called initials, which surround an organizing core called the quiescent center (QC). Initial cells divide to produce one cell for development and one cell to remain as an initial—a system reminiscent of animal stem cells. SAMs also provide new cells to developing tissues, but the identity of those cells is determined later in development. It is intriguing that a CLE signaling pathway was also discovered to function in the regulation of the RAM. CLE40 is expressed in differentiating cells in the root, and the CLE40 peptide acts through the receptor-like kinase CRINKLY4 (ACR4) to restrict and position the expression of WOX5 (Stahl et al., 2009). WOX5 is expressed in the QC and promotes RAM activity by preventing the premature differentiation of the distal stem cells. CLV3 and CLE40, and WUS and WOX5, are functionally equivalent if expressed in the other’s domains (Stahl et al., 2009). Thus, the two primary meristems in plants, which are functionally, morphologically, and evolutionarily quite distinct, both utilize CLE signaling to regulate their size, position, and function.
Tracheary Element Differentiation Inhibitory Factor (TDIF) is a 12-a.a. peptide hormone produced from the CLE41/44 precursors in Arabidopsis (Ito et al., 2006). TDIF is excreted from phloem cells (and their neighbors), and promotes the division and maintenance of vascular cambium cells and also prevents them from differentiating into xylem. TDIF signals through PHLOEM INTERCALATED WITH XYLEM (PXY/TDR), a LRR-receptor kinase, and up-regulates the expression of WOX4 in vascular cambium cells (Hirakawa et al., 2008). WOX4 was shown to be required for the maintenance of the vascular cambium, but not the prevention of differentiation into xylem (Hirakawa et al., 2010), suggesting that TDIF prevents xylem differentiation through a second mechanism. That a secondary meristem also utilizes a CLE signaling pathway suggests that all meristematic tissues in plants may utilize CLE signaling to maintain a proper balance between differentiation and maintenance of stem cells.
New research by Fiume and Fletcher (2012) identifies the function of CLE8 in embryogenesis in Arabidopsis. CLE8 is expressed in the endosperm and in the early developmental stages of the apical portion of the embryo. One role of the CLE8 peptide is to function as a signal to the suspensor, where it acts to promote the expression of WOX8. In their study, ~15% of cle8 mutant embryos were morphologically defective; however, all of the cle8 embryos that did develop a normal morphology produced a smaller-than-normal seed/embryo. CLE8 expression is maintained for the longest period of time in the region of the endosperm responsible for nutrient acquisition from the maternal sporophyte, and CLE8 overexpression produced larger-than-normal seeds/embryos. In addition to its role in embryo development, the authors suggest a role for CLE8 in promoting proliferation of endosperm and preventing the premature differentiation of endosperm cells. Their data indicate that CLE8 promotes the proper expression of WOX8 in the embryo-surrounding region of the endosperm. Consistently with this, a wox8 mutant was able to suppress the seed size increase caused by CLE8 overexpression. Interestingly, this research implicates yet another CLE signaling pathway in a developmental decision between differentiation and proliferation.
RGF FAMILY
The RGF peptides were discovered through a mutant in tyrosylprotein sulfotransferase (TPST) in Arabidopsis, which caused a severe short root phenotype (Matsuzaki et al., 2010). tpst mutants have both a reduced number of meristematic root cells and an increased number of QC cells. Application of the known sulfated peptide hormones Phytosulfokine (PSK) and PLANT PEPTIDE CONTAINING SULFATED TYROSINE1 (PSY1) to the growth media failed to rescue the tpst phenotype. Another family of proteins containing a tyrosine sulfation motif (Asp-Tyr) was identified (named RGF) and, when exogenously applied as a sulfated peptide, one member (RGF1) restored ~70% of the meristem size of tpst mutants, and reduced the number of QC cells by a similar amount. Eight out of the nine sulfated RGF peptides that Matsuzaki et al. tested restored at least some root growth. Sulfated RGF1 in combination with PSK and PSY1 fully restored the size of tpst mutant root meristems. The strongest expression of RGF genes in the roots was observed in the RAM stem cell niche, and the PLETHORA transcription factors, which are known to promote RAM activity, were shown to be up-regulated by sulfated RGF peptides.
Recently, Meng et al. (2012) reported on their independent discovery of the RGF peptides (which they called CLE-Like (CLEL) due to existence of an RGF domain in the CLE18 precursor in addition to its CLE domain). They reported that RGF genes are also expressed in tissues outside of the root, suggesting that they have additional roles outside of the RAM. Overexpression of some RGF genes led to enlarged RAMs that produced longer roots, confirming the Matsuzaki et al. report that the RGF peptide response is dose-dependent. Additionally, overexpression of several of the RGF genes drastically reduced lateral root formation and increased root waviness. Interestingly, Meng et al. reported that exogenously applied un-sulfated RGF peptides increased the root length of wild-type plants. Matsuzaki et al. reported that the un-sulfated form of RGF1 peptide had little or no ability to recover root growth in tpst mutants, but their data did show a small, potentially significant improvement in tpst root length with the application of 100nM un-sulfated RGF1. Meng et al. applied higher concentrations of the unmodified RGF peptides than did Matsuzaki et al. (1µM versus 100nM, respectively), so it is possible that tyrosine sulfation acts to enhance the potency of the RGF peptides. It will be interesting to learn the exact role of tyrosine sulfation in the function of RGF peptides, and whether sulfated and un-sulfated RGF peptides play different roles in plant development. The roles of RGF peptides outside of the root also remain to be determined.
Conclusions
The importance of peptide hormones in plant development has grown exponentially over the last decade. CLE peptide signaling functions in both primary and secondary meristems, and also in embryogenesis (Figure1). CLE signaling is likely an ancient pathway involved in the stem cell proliferation versus differentiation decision, and was repeatedly utilized as new meristems evolved. The newly discovered RGF/CLEL peptides may prove to have developmental functions as equally ubiquitous to the CLE family. Determining the functions of the additional CLE and RGF members will be important for our overall understanding of plant development and evolution. Additionally, the manipulation of peptide hormones in plants may provide a relatively simple means by which to modify plants for human benefit; a clear example of this is the seed enlargement caused by CLE8 overexpression.
Figure 1.
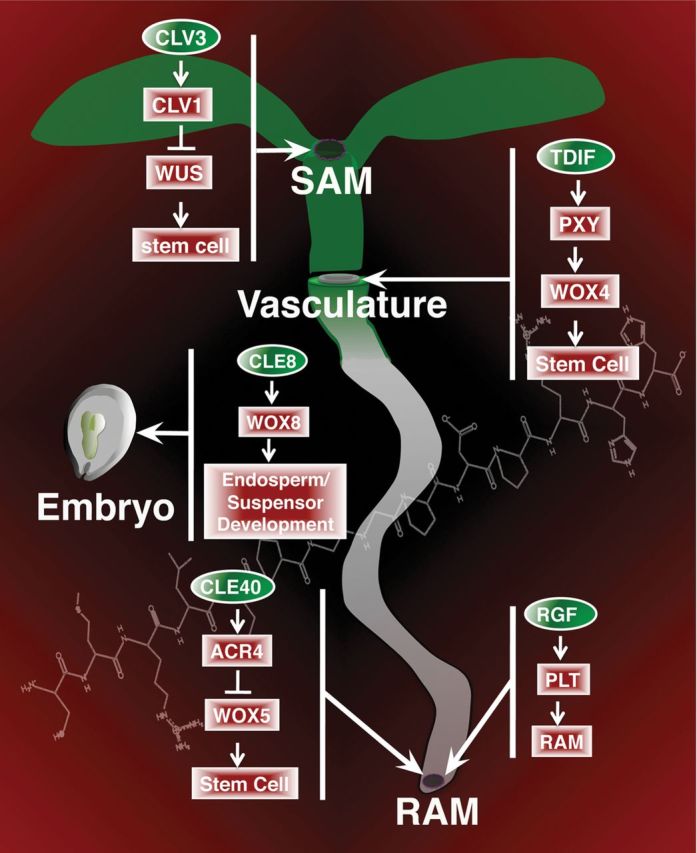
Roles of CLE and RGF Peptide Hormone Signaling Pathways in Arabidopsis.CLV3, TDIF, and CLE40 regulate the proper balance of stem cell proliferation versus differentiation in various meristems. CLE8 has a similar role in regulating embryo and endosperm developmental decisions. The RGF peptides are critical for promoting RAM function and root growth.
FUNDING
This work was supported by the National Institute of General Medical Sciences (NIH Award Number SC1GM095462). No conflict of interest declared.
References
- Cock J.M, McCormick S. (2001). A large family of genes that share homology with CLAVATA3 Plant Physiol 126 939–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiume E, Fletcher J.C. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8 Plant Cell 24 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J.C, Brand U, Running M.P, Simon R, Meyerowitz E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot apical meristems Science 283 1911–1914 [DOI] [PubMed] [Google Scholar]
- Hirakawa Y, Kondo Y, Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis Plant Cell 22 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y, Shinohara H, Kondo Y, Inoue A, Nakanomyo I, Ogawa M, Sawa S, Ohashi–Ito K, Matsubayashi Y, Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system Proc. Natl Acad. Sci. U S A 105 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Nakanomyo I, Motose H, Iwamoto K, Sawa S, Dohmae N, Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation Science 313 842–845 [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa–Ohnishi M, Mori A, Matsubayashi Y. (2010). Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis Science 329 1065–1067 [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan B.B, Feldman L.J, Luan S. (2012). CLE-Like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis Proc. Natl Acad. Sci. U S A 109 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Guo Y, Jin H, Hartsell J, Clark S.E. (2011). Characterization of a CLE processing activity Plant. Mol. Biol 75 67–75 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer K.F, Jürgens G, Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes Cell 100 635–644 [DOI] [PubMed] [Google Scholar]
- Stahl Y, Wink R.H, Ingram G.C, Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems Curr. Biol 19 909–914 [DOI] [PubMed] [Google Scholar]


