Abstract
Cytokines are small proteins secreted by leukocytes in blood in response to infections, thus offering valuable diagnostic information. Given that the same cytokines may be produced by different leukocyte subsets in blood, it is beneficial to connect production of cytokines to specific cell types. In this paper, we describe integration of antibody (Ab) microarrays into a microfluidic device to enable enhanced cytokine detection. The Ab arrays contain spots specific to cell-surface antigens as well as anti-cytokine detection spots. Infusion of blood into a microfluidic device results in the capture of specific leukocytes (CD4 T-cells) and is followed by detection of secreted cytokines on the neighboring Ab spots using sandwich immunoassay. The enhancement of cytokine signal comes from leveraging the concept of reconfigurable microfluidics. A three layer polydimethylsiloxane microfluidic device is fabricated so as to contain six microchambers (1 mm × 1 mm × 30 μm) in the ceiling of the device. Once the T-cell capture is complete, the device is reconfigured by withdrawing liquid from the channel, causing the chambers to collapse onto Ab arrays and enclose cell/anti-cytokine spots within a 30 nl volume. In a set of proof-of-concept experiments, we demonstrate that ∼90% pure CD4 T-cells can be captured inside the device and that signals for three important T-cell secreted cytokines, tissue necrosis factor-alpha, interferon-gamma, and interleukin-2, may be enhanced by 2 to 3 folds through the use of reconfigurable microfluidics.
INTRODUCTION
Numbers and functions of leukocytes in blood are important indicators of infections or malignancies. In the case of HIV/AIDS, numbers of the leukocyte subset called CD4 T-cells are monitored routinely to assess compromised immunity.1, 2 The ratio of CD4 T-cells to CD8 T-cells is also frequently assessed in the case of HIV/AIDS. There also have been numerous reports attempting to describe the speed of progression from HIV to AIDS (slow vs. fast) in terms of cytokine profiles but to date these studies have not translated into diagnostic assays.3, 4, 5, 6 The best example of cytokine production being used for diagnostics is the detection of tuberculosis (TB) where production of a cytokine—interferon (IFN)-γ—is currently used as a diagnostic of latent TB disease.7, 8, 9 The so called IFN-γ release assays (IGRAs) are based on detecting cytokine production from TB-specific T-cells and are rapidly supplanting tuberculin skin tests. IGRAs are immunoassays that either quantify levels of IFN-γ production by enzyme linked immunosorbent assay (ELISA) or count cytokine-producing cells.7, 8, 9
The sensitivity of traditional immunoassays for cytokine detection may be improved by redesigning the assays so as to place cytokine sensors closer to cells and decreasing the volume into which cytokines are secreted. Several groups including our own have been using microfabrication platforms for detecting protein release from immune cells. These have included microwells or microengravings,10, 11, 12 microring resonators,13 aptasensors14, 15 and antibody arrays.16, 17, 18 Of particular note is the recent report by Heath lab describing the method of loading T-cells into microfluidic devices and then segmenting microfluidic channels to create pL chambers for detecting cytokine release from single cells.19
Previously, our laboratory has employed microarrays of cell- and cytokine-specific Ab spots to capture desired leukocyte subsets in the proximity of cytokine sensors.17, 18 In the present study, we wanted to further enhance detected signal by developing reconfigurable microfluidics to minimize the volume around Ab microarrays. Unlike a recent report on a similar topic by Heath et al. who loaded purified T-cells into the microfluidic device,19 we wanted to capture the desired T-cell subset from heterogeneous blood cell suspension and then deploy reconfigurable microfluidics for cytokine signal enhancement. The volume decrease within polydimethylsiloxane (PDMS) microfluidic devices may be achieved when microchannels collapse. Roof collapse in PDMS was first seen as an undesired outcome; however, more recently this effect is being harnessed for designing nanofluidic devices,20 controlling cellular communications,21, 22, 23 single-molecule detection,24 polymerase chain reaction (PCR),25 DNA sequencing,26 and ELISA-like immunoassays.27, 28
In this paper, we focus on applying reconfigurable microfluidics with a reversibly collapsible membrane to capture T-cells cells on Ab arrays and then detect important T-cell cytokines IFN-γ, tissue necrosis factor (TNF)-α, and interleukin (IL)-2. Our device enables capture of specific leukocyte subsets from heterogeneous cell suspension such as blood followed by on-chip functional analysis of captured cells. This microsystem requires minimal blood volumes for analysis and offers enhanced detection sensitivity due to reconfigurable microfluidics. Clinical applications where such devices could be of benefit include TB detection, pediatric immunology, and small animal research. Our reconfigurable microfluidic device, referred to as collapsible roof device (CRD), is fabricated using a method called fit-to-flow (F2F) assembly, which utilize 3D network of vacuum to provide reversible packaging of interchangeable microfluidic modules.29 This method of fabrication and assembly can easily incorporate a free standing, dynamically controllable membrane into the microdevice. Specifically, CRD operates in two modes: cell seeding and cytokine detection (Fig. 1). In the first mode, the channel is open and may be used to infuse blood and capture the desired T-cells. Once the cells are captured, the microfluidic configuration is switched into second mode by withdrawing the sample volume causing collapse of the roof over the Ab arrays. Because the roof is microstructured to contain chambers of 1 mm by 1 mm by 30 μm in dimensions, ∼16 spots (150 μm diameter each) become captured inside the 30 nl volume. The decrease in volume leads to 3 fold signal enhancement for TNF-α and 2 fold enhancement for both IFN-γ and IL-2. In addition to enhancing cell-secreted signal, this reconfigurable microfluidic device allows for capturing pure T-cells. An integration of reconfigurable microfluidics with Ab microarrays represents an important step towards a microdevice for capturing multiple pure leukocyte subsets from blood and establishing cytokine profiles of each subset.
Figure 1.
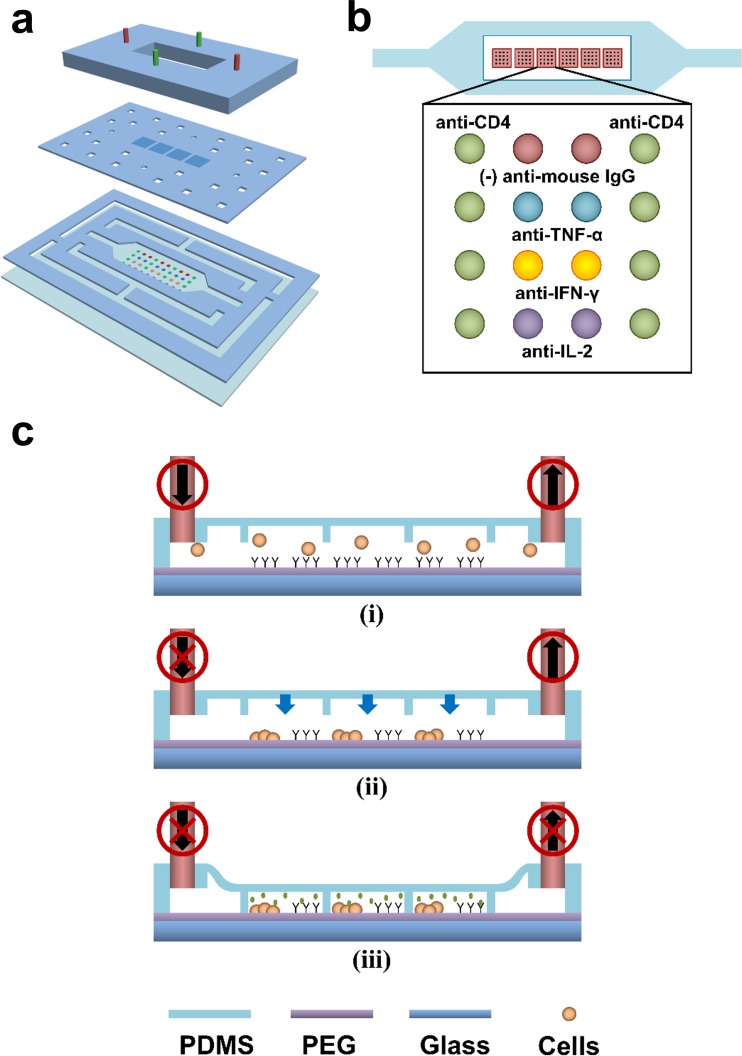
Schematic shows the principles of a collapsible roof device. (a) Fabrication of CRD is achieved through reversible assembly of PDMS layers held to the substrate by vacuum. The top layer with tubing connections, the middle membrane layer with an array of individual microchambers as the collapsible roof of the microfluidic channel, and the bottom layer with channels for sample liquid and vacuum network. (b) A diagram of the CRD on an Ab printed hydrogel substrate with its microchambers aligned to the Ab microarrays. Each microchamber has 2 columns of cell capturing Ab spots and 2 columns of anti-cytokine capturing Ab spots. (c) CRD operation process begins with (i) cell seeding and stimulation, (ii) inlet of microfluidic channel is clamped while the sample continues to be drawn out from the outlet, (iii) the roof of the channel is brought down and the outlet is clamped.
MATERIALS AND METHODS
Materials
Phosphate-buffered saline (PBS) without calcium and magnesium, paraformaldehyde (PFA) surfactant TWEEN 20, poly(ethylene glycol) diacrylate (PEG-DA, MW 575), anhydrous toluene (99.9%), and bovine serum albumin (BSA) were purchased from Sigma-Aldrich. Silane adhesion promoter (3-acryloxypropyl trichlorosilane) was purchased from Gelest, Inc. Monoclonal antibodies used for capturing T lymphocytes and cytokines consisted of the following: purified mouse antihuman Abs CD4 (clone 13b8.2) and CD8 (clone B9.11) from Beckman-Coulter. Purified mouse antihuman Abs (TNF-α (clone 28401), IFN-γ (clone K3.53), and IL-2 (clone 5355)) and biotinylated goat antihuman Abs (TNF-α, IFN-γ, and IL-2) were purchased from R&D Systems. Mouse IgG2a (OX34) was purchased from Serotec Antibodies. Antibodies used for immunostaining of surface bound cells were anti-CD8-PE (RPA-T8) and anti-CD3-FITC (UCHT1) purchased from BD Pharminogen. Streptavidin with Alexa Fluorophore 546 Conjugate and Penicillin-Streptomycin (PS) were purchased from Invitrogen. Mitogenic activation reagents: Phorbol 12-myristate 13-acetate (PMA) and ionomycin were purchased from Sigma-Aldrich. Cell culture medium RPMI (Roswell Park Memorial Institute medium) 1640 1× with L-glutamine and no phenol red was purchased from VWR. PDMS was purchased from Dow Corning. Lymphoprep was purchased from Fisher Scientific.
Fabrication of collapsible roof device
CRDs were fabricated through a reversible packaging scheme previously described as fit-to-flow assembly.29 Briefly, CRD was assembled layer-by-layer with PDMS as shown in Fig. 1a. PDMS layers of varying thicknesses (50 μm to 1 mm) were prepared on a glass substrate and cured on a hotplate at 120 °C for 15 min. Laser micromachining (VersaLaser, ULS) was used to define cell capturing channel and vacuum sealing network on the cured PDMS substrates. The PDMS roof membrane was prepared on a photoresist patterned glass substrate before undergoing laser etching. The microchambers were 1 mm × 1 mm × 30 μm. The device assembly started from the bottom and worked to the top beginning with the placement of channel PDMS layer on a substrate, followed by the collapsible roof with encapsulating microchambers, and lastly, the PDMS layer with tubings on top. Alignment of each layer during the device assembly was performed by eye. The layers of the device were held together and to the substrate by vacuum suction, a physical adhesion method.
Fabrication of Ab microarray on PEG hydrogel surface
PEG hydrogel substrate was prepared as previously described17 by modifying a glass slide with acrylated silane, followed by a coating of PEG-DA pre-polymer solution, UV exposure curing, and finally, dehydration in a lyophilizer. The Ab spots were imprinted into the hydrogel surface using Spotbot 3 (Arrayit). As shown in the array layout (Fig. 1b), the Ab spots for capturing CD4 or CD8 T-cells were printed next to Ab spots for TNF-α, IFN-γ, IL-2, and mouse IgG (control). The spots printed on the surfaces were spatially separated into 6 groups, each group containing a 4 × 4 array of spots and each becoming enclosed inside one microchamber. Printed spots were ∼150 μm in diameter with 250 μm center-to-center spacing. Prior to printing, anti-CD4, anti-TNF-α, anti-IFN-γ, and anti-IL-2 were dissolved in DI water at 0.2 mg/ml and were supplemented with 0.5% v/v BSA and 0.005% v/v Tween20.
Cell capture and cytokine detection
The CRD was mounted and aligned onto an Ab printed PEG hydrogel substrate and held in position by vacuum suction. PBS with 1% BSA was flowed through and incubated in the channel for 15 min to block the surface. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood with Lymproprep and infused into the channel to permit cell seeding at 1 μl/min for 10 min as shown in Fig. 1c (i). Mitogenic solution containing PMA (50 ng/ml) and ionomycin (2 μM) diluted in RPMI 1640 with 10% fetal bovine serum (FBS) and 1% PS was flown through the channel to activate the cells.
The roof of the device was collapsed by clamping the inlet and withdrawing 40 μl from the device to pull down the 6 encapsulating microchambers (see Fig. 1c (ii)). Afterwards, the outlet was clamped and the whole apparatus was incubated at physiological conditions of 37 °C and 5% CO2 for 5 h (Fig. 1c (iii)). Following incubation, the roof was raised by unclamping both the inlet and outlet. The cell chamber was rinsed with PBS.
A cocktail of biotinylated anti-cytokine antibodies (1:10 dilution) in PBS with 1% BSA was incubated in the channel for 1 h followed by streptavidin-alexa 546 (1:100 in 1% BSA with PBS) for 30 min. PBS wash took place in between each step for a minimum of 5 min at 30 μl/min. To determine cell purity, captured T-cells were immunostained with CD3-FITC (1:10) and CD8-PE (1:10) for 2 h. Finally, cells were fixed with 4% PFA for 15 min. Brightfield and fluorescence images were captured by a Zeiss LSM 510 confocal microscope. Fluorescence of SA546 was scanned with a laser microarray scanner and quantified as described previously.17
RESULTS AND DISCUSSION
This paper describes integration of reconfigurable microfluidic device with Ab arrays for enhanced detection of T-cell secreted cytokines. The use of this reconfigurable device allowed for an input of a complex blood cell suspension, isolation of pure CD4 T-cells, and detection of TNF-α, IFN-γ and IL-2 release with a 2 to 3 fold enhancement when compared to a standard microfluidic device.
Reversible F2F microfluidic assembly
Microfluidic device packaging plays a critical role in Lab-on-a-Chip microfluidics as biological components such as cells, proteins, and biomarkers need to retain functionalities throughout the device fabrication process. Using a reversible packaging method, the device can be disassembled as intended to safely extract the sample of interest without risk of damage or contamination. F2F is a reversible microfluidic device packaging scheme that utilizes vacuum suction to hold the microfluidic device to the biofunctionalized substrate.29 This allows functionalization of the substrate with fragile biomolecules or cells and then assembles the microfluidic device without exposing biological components to harsh chemical environment. After completion of the experiment, the substrate with cells or biomolecules may be easily separated from the microfluidic channels and then used for further analysis.
Fig. 1a shows assembly of the device from three modular PDMS layers. The first layer containing the microfluidic channels and the vacuum network is placed onto gel-coated glass substrates. Above this first layer is the second layer—a 200 μm thick membrane with an array of microchambers that serves as the roof of the microfluidic channel. During roof collapse, the membrane with microchambers descends onto the Ab spots to minimize volume during cytokine release. The second layer also has the through-holes located above the vacuum network channels to expose the above layer to the suction force thereby holding all PDMS layers together. Lastly, the third, topmost layer provides structural integrity for the entire microfluidic device and allows tubing access to the cell chamber and vacuum network. An opening is made in the top layer that is directly above cell chamber such that the chamber roof membrane can deflect to collapsed and raised configurations. Each layer of the device is interchangeable permitting versatility and reusability of modular components. The F2F assembled microfluidic device enables on-chip processes from cell seeding on biofunctionalized substrate, device reconfiguration to minimizing working volume, and performing immunofluorescent staining. Once on-chip processes are completed, the device can be detached for further analysis of cytokine captured substrate.
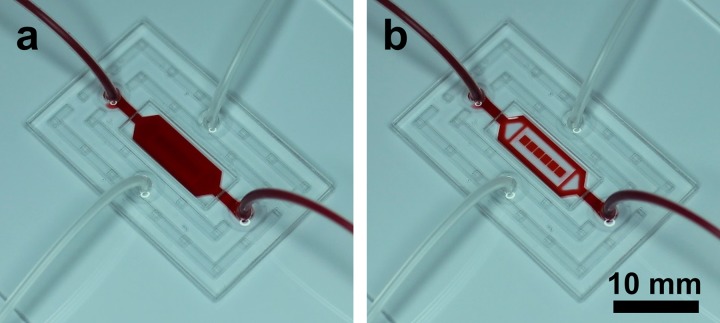
Infusion of food dye was used to show principle of the device operation. Fig. 2a shows solution with food dye uniformly distributed throughout the channel—this configuration of the microfluidics was used during cell infusion and capture. Once the microfluidic channel was reconfigured and flushed with PBS (Fig. 2b), the food dye was only trapped inside microchambers, each with a volume of 30 nl. This second configuration was used to confine cells and cytokine sensors inside a smaller volume to enhance cytokine signal.
Figure 2.
The photographs show the CRD loaded with red dye in the (a) open and (b) collapsed configurations. A clear volume change between the two states illustrates isolation and localization of working samples within individual microchambers.
Capturing pure T-cell subsets in the reconfigurable microfluidic devices
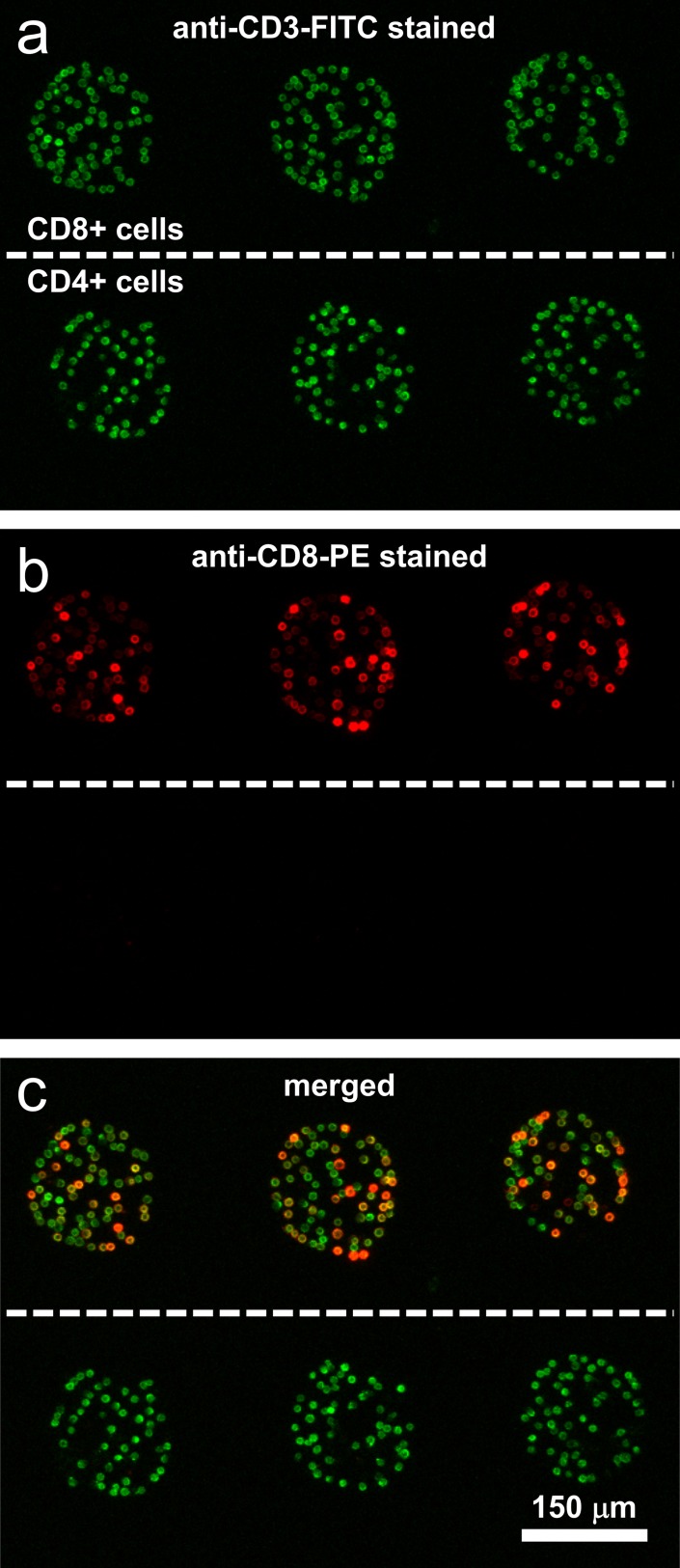
Cytokines such as TNF-α or IFN-γ may be produced by several leukocyte types; therefore, one needs to isolate/capture the cell type of interest to accurately attribute cytokine production. Preciously, our lab proposed to capture T-cells on Ab microarray and then detect secreted cytokines on anti-cytokine spots in the same array.17 In these previous studies, the cell purity was demonstrated to be ∼95% for CD4 and CD8 T-cells.17 In this paper, we performed cell capture and purity assessment experiments to verify that similar results may be obtained in reconfigurable microfluidic channels. The results of a representative experiment are summarized in Fig. 3. In this experiment, cells from PBMCs isolated human blood were infused into the channel containing Abs against CD4 and CD8 surface markers present on T-cells. After capture, cells were stained with anti-CD3-FITC (green) and anti-CD8-PE (red). As shown in Fig. 3, cells on both CD4 and CD8 Ab spots were stained green—an expected outcome since CD3 antigen is present on all T-cells. The majority of cells on anti-CD8 spots were also stained red, pointing to the presence of CD8 T-cells which are generally described as CD3+CD8+ cells. The CD4 T-cells on the other hand are not expected to express CD8 surface markers so that the cells on anti-CD4 spots were only stained with green fluorescence. Merging brightfield and fluorescence images and analyzing cell labeling, we determined the purity of T-cells on the Ab spots to be ∼90%. It is worth noting that T-cells only represent 20% to 30% of the cell suspension infused into the channel.
Figure 3.
Immunofluorescent staining of cells captured on adjacent anti-CD4 and anti-CD8 microarrays. The top row cells are CD8+ cells and the bottom row cells are CD4+ cells. (a) Fluorescent image of CD4+ and CD8+ cells stained with anti-CD3-FITC (green). (b) Fluorescent image of CD4+ and CD8+ cells stained with anti-CD8-PE (red). (c) Merged fluorescent image of the two images showing both CD4 and CD8 cells were stained for anti-CD3-FITC (green) while only CD8 cells were stained for anti-CD8-PE (red) thus indicating a high purity and specificity of cell capture.
Enhanced detection of cytokines in reconfigurable microfluidic devices
To determine the signal enhancement derived from reconfiguring the microfluidic channel, two devices were set-up for parallel analysis. Upon capturing CD4 T-cells in both devices, one device was reconfigured to decrease the volume while the other device was not. In both open and reconfigured scenarios, the T-cells were mitogenically activated for 5 h and the arrays were consequently stained for capture of TNF-α, IFN-γ, and IL-2. As shown schematically in Fig. 1b, each microchamber covered a 4 × 4 array of 150 μm diameter Ab spots of which 2 columns of Ab spots were designated for cell capture and 2 other columns for cytokine detection. The design of the array places small groups of T-cells (∼40 cells per spot) 100 μm away from cytokine sensing spots, thus satisfying cell-to-sensor proximity requirement. In the collapsed configuration, each microchamber sequesters the cell/sensing Ab spots within a 30 nl volume, compared to a 3 μl volume of an open chamber.
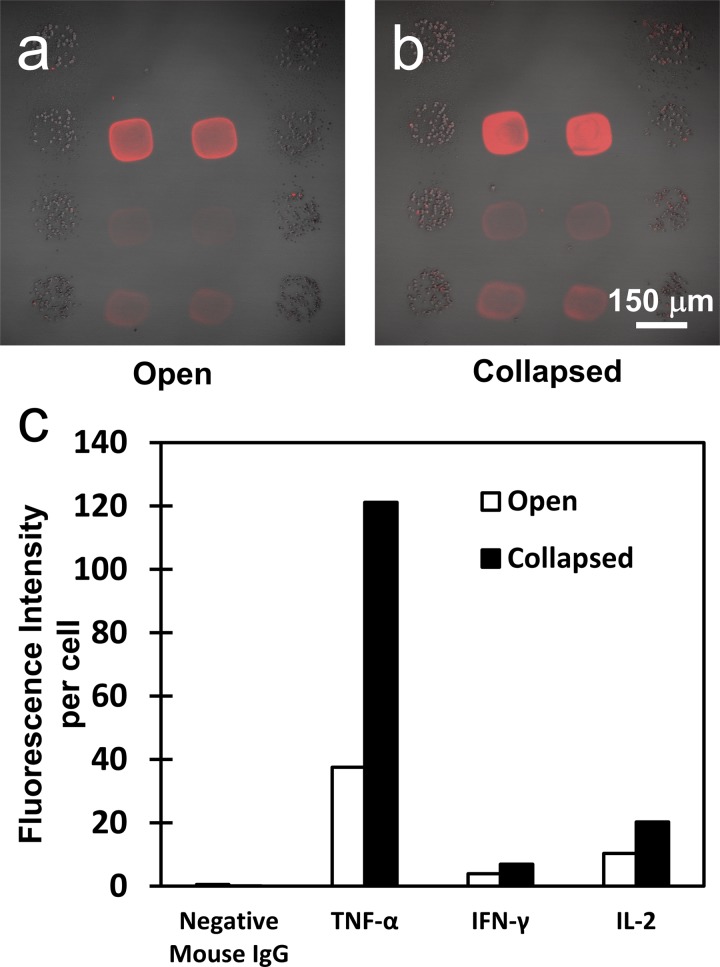
Fig. 4 summarizes results of cytokine detection experiments in open and reconfigured microfluidic channels. Immunofluorescent staining for captured cytokines shows much stronger signal for TNF-α, IFN-γ, and IL-2 in a collapsed vs. open device (Figs. 4a, 4b). Quantifying results of an experiment using microarray scanner showed that fluorescence intensity was 3 fold higher for TNF-α and 2 fold higher for IFN-γ and IL-2 in a reconfigured vs. open device (Fig. 4c). Relative fluorescent intensity values were normalized based on a per cell basis. Thus reconfigurable microfluidics offer significant enhancement of the signal that may be leveraged in decreasing the experiment time or decreasing the number of cells used in an experiment.
Figure 4.
Merged brightfield and fluorescent images of (a) sample incubated in the open CRD under mitogenic stimulation for 5 h and (b) sample incubated in the collapsed CRD under mitogenic stimulation for 5 h. (c) Relative fluorescent intensity values were normalized based on a per cell basis and graphed to compare the signal advantage in cytokine capture that the collapsed configuration has over the open configuration.
CONCLUSION
In this study, our goals were to enhance cytokine detection capabilities while retaining the ability to capture pure leukocyte subsets from a heterogeneous cell sample. To accomplish this, we designed a reconfigurable microfluidic device that could be combined with microarrays of cell-capture and cytokine-detection Abs. The open configuration of the device was used to capture pure T-cells from PBMCs isolated blood on Ab spots. Subsequently, the device was reconfigured, limiting the volume around cell-capture and cytokine-detection spots to 30 nl. Through this confinement, the detection signal of three important T-cells secreted cytokines TNF-α, IFN-γ, and IL-2 was enhanced by 2 to 3 fold. The use of Fit-to-Flow microfabrication concept allowed modular microfluidic devices simple to assemble or disassemble. Overall, this study explored the use of microfluidic devices that could be reconfigured once the experiment has commencement and that could serve different functions in different configurations, in our case cell seeding and cytokine detection. We envision extending the use of such devices for detecting cytokine profiles of multiple leukocyte subsets isolated in the same device and further enhancing detection limits of antibody microarrays.
ACKNOWLEDGMENTS
The authors would like to thank NSF EFRI and NSF CAREER Program (ECCS-0846502) for financial support. We thank Mr. Anil Singapuri and Dr. Chiamvimonvat for help with fluorescence microscopy. UC Davis School of Medicine Capital Equipment Grant provided funding for the fluorescence microscope instrument. Scanning of the microarrays was performed at Expression Analysis Facility of UC Davis.
References
- Douek D. C., Brenchley J. M., Betts M. R., Ambrozak D. R., Hill B. J., Okamoto Y., Casazza J. P., Kuruppu J., Kuntsman K., Wolinsky S., Grossman Z., Dybul M., Oxenius A., Price D. A., Connors M., and Koup R. A., Nature 417(6884), 95–98 (2002). 10.1038/417095a [DOI] [PubMed] [Google Scholar]
- Sherman G. G., Galpin J. S., Patel J. M., Mendelow B. V., and Glencross D. K., J. Immunol. Methods 222(1–2), 209–217 (1999). 10.1016/S0022-1759(98)00172-0 [DOI] [PubMed] [Google Scholar]
- Clerici M. and Shearer G. M., Immunol. Today 14(3), 107–110 (1993). 10.1016/0167-5699(93)90208-3 [DOI] [PubMed] [Google Scholar]
- Clerici M. and Shearer G. M., Immunol. Lett. 51(1–2), 69–73 (1996). 10.1016/0165-2478(96)02557-6 [DOI] [PubMed] [Google Scholar]
- Pantaleo G. and Koup R. A., Nat. Med. 10(8), 806–810 (2004). 10.1038/nm0804-806 [DOI] [PubMed] [Google Scholar]
- Zimmerli S. C., Harari A., Cellerai C., Vallelian F., Bart P. A., and Pantaleo G., Proc. Natl. Acad. Sci. U.S.A. 102(20), 7239–7244 (2005). 10.1073/pnas.0502393102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey R., Blumenkrantz D., Millington K., Montamat-Sicotte D., Kon O. M., Wickremasinghe M., Bremang S., Magtoto M., Sridhar S., Connell D., and Lalvani A., PLoS ONE 5(12), e15619 (2010). 10.1371/journal.pone.0015619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheda K., Smit R. V., Badri M., and Pai M., Curr. Opin. Pulm. Med. 15(3), 188–200 (2009). 10.1097/MCP.0b013e32832a0adc [DOI] [PubMed] [Google Scholar]
- Lalvani A. and Pareek M., Enferm. Infecc. Microbiol. Clin. 28(4), 245–252 (2010). 10.1016/j.eimc.2009.05.012 [DOI] [PubMed] [Google Scholar]
- Jin A., Ozawa T., Tajiri K., Obata T., Kondo S., Kinoshita K., Kadowaki S., Takahashi K., Sugiyama T., Kishi H., and Muraguchi A., Nat. Med. 15(9), 1088–U1146 (2009). 10.1038/nm.1966 [DOI] [PubMed] [Google Scholar]
- Han Q., Bradshaw E. M., Nilsson B., Hafler D. A., and Love J. C., Lab Chip 10(11), 1391–1400 (2010). 10.1039/b926849a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J. C., Ronan J. L., Grotenbreg G. M., van der Veen A. G., and Ploegh H. L., Nat. Biotechnol. 24(6), 703–707 (2006). 10.1038/nbt1210 [DOI] [PubMed] [Google Scholar]
- Luchansky M. S. and Bailey R. C., Anal. Chem. 82(5), 1975–1981 (2010). 10.1021/ac902725q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kwa T., and Revzin A., Biomaterials 33(30), 7347–7355 (2012). 10.1016/j.biomaterials.2012.06.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yan J., Howland M. C., Kwa T., and Revzin A., Anal. Chem. 83(21), 8286–8292 (2011). 10.1021/ac202117g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey R. C., Kwong G. A., Radu C. G., Witte O. N., and Heath J. R., J. Am. Chem. Soc. 129(7), 1959–1967 (2007). 10.1021/ja065930i [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Stybayeva G., Macal M., Ramanculov E., George M. D., Dandekar S., and Revzin A., Lab Chip 8(12), 2197–2205 (2008). 10.1039/b810244a [DOI] [PubMed] [Google Scholar]
- Stybayeva G., Mudanyali O., Seo S., Silangcruz J., Macal M., Ramanculov E., Dandekar S., Erlinger A., Ozcan A., and Revzin A., Anal. Chem. 82(9), 3736–3744 (2010). 10.1021/ac100142a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Fan R., Ahmad H., Shi Q. H., Comin-Anduix B., Chodon T., Koya R. C., Liu C. C., Kwong G. A., Radu C. G., Ribas A., and Heath J. R., Nat. Med. 17(6), 738–U133 (2011). 10.1038/nm.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. M., Huh Y. S., Craighead H. G., and Erickson D., Proc. Natl. Acad. Sci. U.S.A. 106(37), 15549–15554 (2009). 10.1073/pnas.0904004106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Hegde M., and Jayaraman A., Lab Chip 10(1), 43–50 (2010). 10.1039/b911367c [DOI] [PubMed] [Google Scholar]
- Gao Y. D., Majumdar D., Jovanovic B., Shaifer C., Lin P. C., Zijlstra A., Webb D. J., and Li D. Y., Biomed. Microdevices 13(3), 539–548 (2011). 10.1007/s10544-011-9523-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar D., Gao Y. D., Li D. Y., and Webb D. J., J. Neurosci. Methods 196(1), 38–44 (2011). 10.1016/j.jneumeth.2010.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shon M. J. and Cohen A. E., J. Am. Chem. Soc. 134(35), 14618–14623 (2012). 10.1021/ja3062425 [DOI] [PubMed] [Google Scholar]
- Men Y. F., Fu Y. S., Chen Z. T., Sims P. A., Greenleaf W. J., and Huang Y. Y., Anal. Chem. 84(10), 4262–4266 (2012). 10.1021/ac300761n [DOI] [PubMed] [Google Scholar]
- Sims P. A., Greenleaf W. J., Duan H. F., and Xie S., Nat. Methods 8(7), 575–U584 (2011). 10.1038/nmeth.1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cordero J. L. and Maerkl S. J., Chem. Commun. 49(13), 1264–1266 (2013). 10.1039/c2cc37740c [DOI] [PubMed] [Google Scholar]
- Zheng C. H., Wang J. W., Pang Y. H., Wang J. B., Li W. B., Ge Z. G., and Huang Y. Y., Lab Chip 12(14), 2487–2490 (2012). 10.1039/c2lc40145b [DOI] [PubMed] [Google Scholar]
- Chen A. N. and Pan T. R., Biomicrofluidics 5(4), 046505 (2011). 10.1063/1.3670368 [DOI] [PMC free article] [PubMed] [Google Scholar]