Abstract
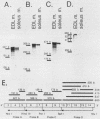
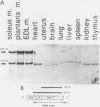
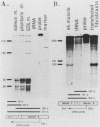
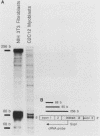
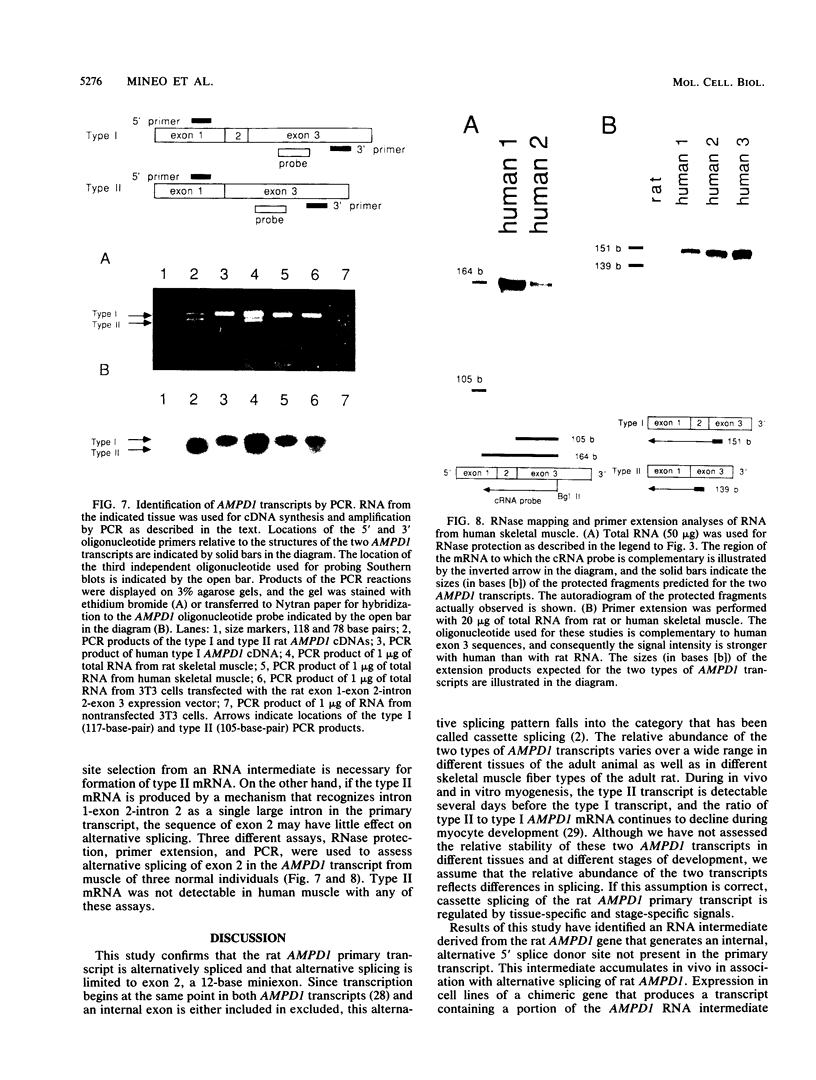
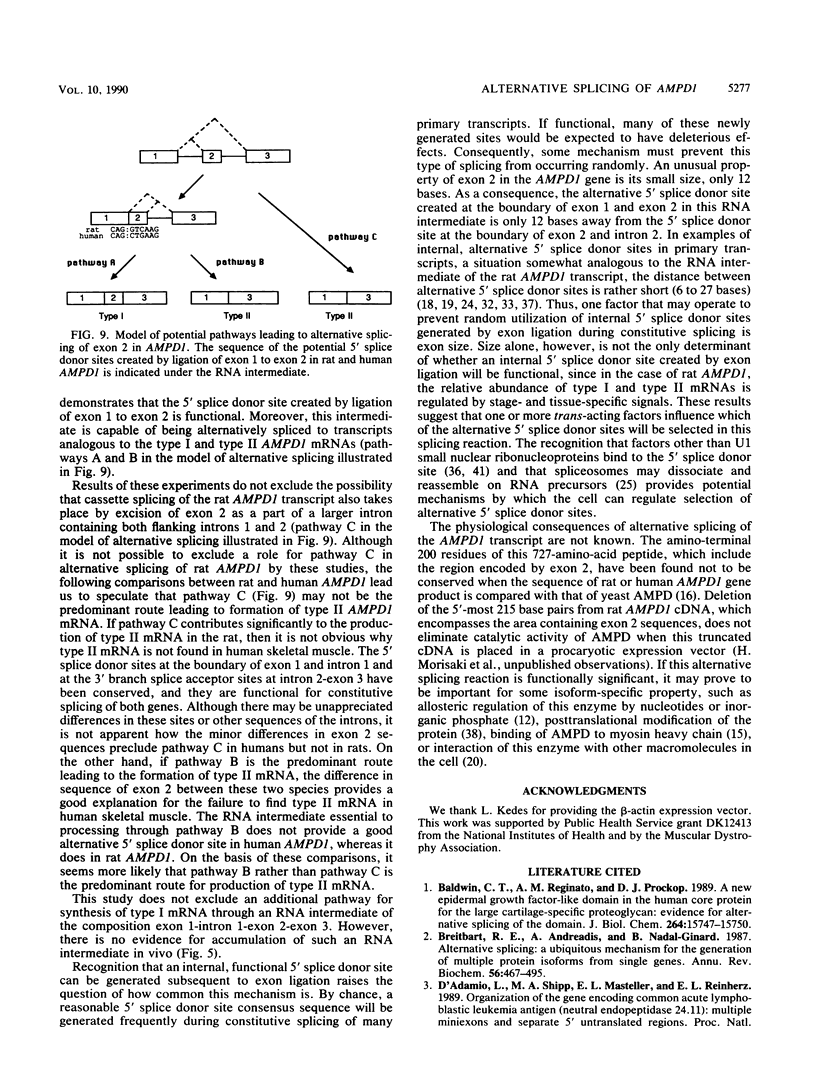
AMP deaminase (AMPD) is a central enzyme in eucaryotic energy metabolism, and tissue-specific as well as stage-specific isoforms are found in many vertebrates. This study demonstrates the AMPD1 gene product in rat is alternatively spliced. The second exon, a 12-base miniexon, was found to be excluded or included in a tissue-specific and stage-specific pattern. This example of cassette splicing utilizes a unique pathway through an RNA intermediate that generates an alternative 5' splice donor site at the point where exon 2 is ligated to exon 1. In the analogous intermediate of human AMPD1, the potential 5' splice donor site created at the boundary of exon 1 and exon 2 was a poor substrate for splicing because of differences in exon 2 sequences, and human AMPD1 was not alternatively spliced. These results demonstrate that in some cases alternative splicing may proceed through an RNA intermediate that generates an alternative splice donor site not present in the primary transcript. Discrimination between alternative 5' splice donor sites in the RNA intermediate of AMPD1 is apparently controlled by tissue-specific and stage-specific signals.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin C. T., Reginato A. M., Prockop D. J. A new epidermal growth factor-like domain in the human core protein for the large cartilage-specific proteoglycan. Evidence for alternative splicing of the domain. J Biol Chem. 1989 Sep 25;264(27):15747–15750. [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Feener C. A., Koenig M., Kunkel L. M. Alternative splicing of human dystrophin mRNA generates isoforms at the carboxy terminus. Nature. 1989 Apr 6;338(6215):509–511. doi: 10.1038/338509a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gallego M. E., Nadal-Ginard B. Myosin light-chain 1/3 gene alternative splicing: cis regulation is based upon a hierarchical compatibility between splice sites. Mol Cell Biol. 1990 May;10(5):2133–2144. doi: 10.1128/mcb.10.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gunning P., Leavitt J., Muscat G., Ng S. Y., Kedes L. A human beta-actin expression vector system directs high-level accumulation of antisense transcripts. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4831–4835. doi: 10.1073/pnas.84.14.4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig M., Monaco A. P., Kunkel L. M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988 Apr 22;53(2):219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Lipson K. E., Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9774–9777. doi: 10.1073/pnas.86.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi J. P., Holmes E. W. Control of the purine nucleotide cycle in extracts of rat skeletal muscle: effects of energy state and concentrations of cycle intermediates. Arch Biochem Biophys. 1984 Sep;233(2):515–529. doi: 10.1016/0003-9861(84)90475-2. [DOI] [PubMed] [Google Scholar]
- Marquetant R., Desai N. M., Sabina R. L., Holmes E. W. Evidence for sequential expression of multiple AMP deaminase isoforms during skeletal muscle development. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2345–2349. doi: 10.1073/pnas.84.8.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquetant R., Sabina R. L., Holmes E. W. Identification of a noncatalytic domain in AMP deaminase that influences binding to myosin. Biochemistry. 1989 Oct 31;28(22):8744–8749. doi: 10.1021/bi00448a010. [DOI] [PubMed] [Google Scholar]
- Meyer S. L., Kvalnes-Krick K. L., Schramm V. L. Characterization of AMD, the AMP deaminase gene in yeast. Production of amd strain, cloning, nucleotide sequence, and properties of the protein. Biochemistry. 1989 Oct 31;28(22):8734–8743. doi: 10.1021/bi00448a009. [DOI] [PubMed] [Google Scholar]
- Morisaki T., Sabina R. L., Holmes E. W. Adenylate deaminase. A multigene family in humans and rats. J Biol Chem. 1990 Jul 15;265(20):11482–11486. [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine Y., Pearson D., Grattan M. Exon-intron boundary sliding in the generation of two mRNAs coding for porcine urokinase-like plasminogen activator. Biochem Biophys Res Commun. 1985 Oct 30;132(2):563–569. doi: 10.1016/0006-291x(85)91170-2. [DOI] [PubMed] [Google Scholar]
- Nagata S., Tsuchiya M., Asano S., Yamamoto O., Hirata Y., Kubota N., Oheda M., Nomura H., Yamazaki T. The chromosomal gene structure and two mRNAs for human granulocyte colony-stimulating factor. EMBO J. 1986 Mar;5(3):575–581. doi: 10.1002/j.1460-2075.1986.tb04249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley K. L., Anhalt M. J., Martin B. M., Kelsoe J. R., Winfield S. L., Ginns E. I. Isolation and characterization of the human tyrosine hydroxylase gene: identification of 5' alternative splice sites responsible for multiple mRNAs. Biochemistry. 1987 Nov 3;26(22):6910–6914. doi: 10.1021/bi00396a007. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y. Interaction of AMP deaminase with RNA. Biochim Biophys Acta. 1981 Sep 15;661(1):164–169. doi: 10.1016/0005-2744(81)90096-6. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Nishigaki I., Itoh T., Hasegawa I., Park K. S. Deficiency of AMP deaminase in erythrocytes. Hum Genet. 1987 Jan;75(1):15–18. doi: 10.1007/BF00273831. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Watanabe T., Asano T. AMP deaminase isozymes in human tissues. Biochim Biophys Acta. 1982 Feb 2;714(2):298–306. doi: 10.1016/0304-4165(82)90337-3. [DOI] [PubMed] [Google Scholar]
- Ogasawara N., Goto H., Yamada Y., Watanabe T. Distribution of AMP-deaminase isozymes in rat tissues. Eur J Biochem. 1978 Jun 15;87(2):297–304. doi: 10.1111/j.1432-1033.1978.tb12378.x. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. An early hierarchic role of U1 small nuclear ribonucleoprotein in spliceosome assembly. Science. 1988 Nov 18;242(4881):1028–1035. doi: 10.1126/science.2973660. [DOI] [PubMed] [Google Scholar]
- Sabina R. L., Marquetant R., Desai N. M., Kaletha K., Holmes E. W. Cloning and sequence of rat myoadenylate deaminase cDNA. Evidence for tissue-specific and developmental regulation. J Biol Chem. 1987 Sep 15;262(26):12397–12400. [PubMed] [Google Scholar]
- Sabina R. L., Morisaki T., Clarke P., Eddy R., Shows T. B., Morton C. C., Holmes E. W. Characterization of the human and rat myoadenylate deaminase genes. J Biol Chem. 1990 Jun 5;265(16):9423–9433. [PubMed] [Google Scholar]
- Sabina R. L., Ogasawara N., Holmes E. W. Expression of three stage-specific transcripts of AMP deaminase during myogenesis. Mol Cell Biol. 1989 May;9(5):2244–2246. doi: 10.1128/mcb.9.5.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausville E. A., Lebacq-Verheyden A. M., Spindel E. R., Cuttitta F., Gazdar A. F., Battey J. F. Expression of the gastrin-releasing peptide gene in human small cell lung cancer. Evidence for alternative processing resulting in three distinct mRNAs. J Biol Chem. 1986 Feb 15;261(5):2451–2457. [PubMed] [Google Scholar]
- Schulz R. A., Cherbas L., Cherbas P. Alternative splicing generates two distinct Eip28/29 gene transcripts in Drosophila Kc cells. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9428–9432. doi: 10.1073/pnas.83.24.9428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B., Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989 Oct 20;59(2):349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel C. W., Rio D. C. Regulated splicing of the Drosophila P transposable element third intron in vitro: somatic repression. Science. 1990 Jun 8;248(4960):1200–1208. doi: 10.1126/science.2161558. [DOI] [PubMed] [Google Scholar]
- Stein J. P., Catterall J. F., Kristo P., Means A. R., O'Malley B. W. Ovomucoid intervening sequences specify functional domains and generate protein polymorphism. Cell. 1980 Oct;21(3):681–687. doi: 10.1016/0092-8674(80)90431-6. [DOI] [PubMed] [Google Scholar]
- Tovmasian E. K., Hairapetian R. L., Bykova E. V., Severin S. E., Jr, Haroutunian A. V. Phosphorylation of the skeletal muscle AMP-deaminase by protein kinase C. FEBS Lett. 1990 Jan 1;259(2):321–323. doi: 10.1016/0014-5793(90)80037-j. [DOI] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Zapp M. L., Berget S. M. Evidence for nuclear factors involved in recognition of 5' splice sites. Nucleic Acids Res. 1989 Apr 11;17(7):2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]