Abstract
Classical Hodgkin lymphoma (CHL), a neoplasm of abnormal B lymphocytes (Hodgkin-Reed Sternberg cells), has been described to have a typical pattern of clinical presentation and dissemination often involving functionally contiguous lymph nodes. Despite the progress made in understanding CHL pathophysiology, the factors which regulate the spread of lymphoma cells in CHL are poorly understood. Sphingosine-1-Phosphate (S1P), a bioactive sphingolipid present at high concentrations in plasma and lymphatic fluid, is known to play a critical role in regulating lymphocyte trafficking mainly through S1PR1. In this study, we explore the role of the S1P-S1PR1 axis in Hodgkin lymphoma cell migration and the expression of S1PR1 in CHL cell lines and clinical cases. We found that S1PR1 is present in the KM-H2 and SUP-HD1 Hodgkin lymphoma cell lines at the mRNA and protein level. In addition, functionally, S1P potently stimulated migration of both cell lines. S1P-induced migration was inhibited by the S1PR1 antagonist, VPC44116 and the S1PR1 functional antagonist, FTY720-P, but was potentiated by the S1PR2 specific antagonist, JTE013. We also determined that S1PR1 induced migration in the KM-H2 and SUP-HD1 cells via the heterotrimeric G protein Gi and the phosphatidylinositol-3-kinase (PI3K) pathway. Immunohistochemical assessment of tissue from CHL samples revealed that a subset of cases (7/57; 12%) show strong, membranous staining for S1PR1 in Hodgkin-Reed Sternberg cells.
Altogether our data indicate that S1PR1 is a functional receptor on Hodgkin-Reed Sternberg cells which governs tumor cell migration and is expressed in a subset of CHL cases. Given the availability of S1PR1 antagonists, some of which are used clinically for modulation of the immune system, these results suggest that S1PR1 could be a future therapeutic target in the treatment of those cases of S1PR1-positive, refractory/recurrent CHL.
Keywords: Sphingosine-1-phosphate (S1P), S1PR1, classical Hodgkin lymphoma, migration, dissemination, Fingolimod
Sphingosine-1-phosphate receptor 1 (S1PR1) is a G-protein coupled receptor (GPCR) which binds the bioactive sphingolipid, sphingosine-1-phosphate (S1P) as a high affinity ligand. S1P is present at high levels (within the range of several hundred nanomolar) in blood and lymph, and plays a critical role in the homeostasis of the vascular and immune systems. S1PR1 was originally identified as an abundant transcript in endothelial cells (1) and plays a key role in regulating endothelial cell cytoskeletal structure, migration, capillary-like network formation and vascular maturation (2–7). Other GPCR in the S1PR family have also been described and characterized as binding S1P (8, 9). Interestingly, some S1PR have opposing effects on cell function; for example, while S1PR1 promotes cell migration and vascular integrity (5, 10), S1PR2 inhibits cell motility and promotes vascular permeability (11–15). More specifically, S1PR1 couples to Gi (8, 16) and activates the phosphatidylinositol-3-kinase (PI3K) pathway (17) and the small GTPase Rac (3), while S1PR2, through activation of the G12-Rho-Rho kinase (ROCK)-PTEN pathway (14, 18, 19), can antagonize S1PR1-Gi-PI3K signaling.
In the immune system, S1PR1 signaling is important in the regulation of lymphocyte migration and trafficking (20–28). Normal lymphocytes travel along a S1P gradient between lymphoid organs and lymph or blood. Inhibition of S1PR1 or disruption of the S1P gradient results in lymphocyte sequestration within lymphoid organs (29, 30). In fact, phosphorylated FTY720 (FTY720-P), the immunosuppressant and structural analog of S1P, which potently binds and activates S1PR1 signaling (10, 20, 22, 31–33), is a functional antagonist of S1PR1 since it induces irreversible internalization and degradation of S1PR1 (34, 35). Recently, FTY720 has been approved in the United States as a therapeutic option in some cases of multiple sclerosis (36–38). Additional compounds targeting S1PR have been characterized; these include JTE013, a S1PR2 specific antagonist (12, 13) and VPC44116, a S1PR1 antagonist (39). These developments highlight the fact that S1PR are becoming attractive candidates for targeted drug design in cardiovascular and immunological disorders.
Despite our understanding of the role of S1PR1 in normal lymphocyte trafficking, the expression of S1PR1 and its role in the regulation of tumor cell biology in lymphoma is just beginning to be elucidated. Classical Hodgkin lymphoma (CHL), a neoplasm of abnormal B lymphocytes (Hodgkin-Reed Sternberg cells), has a typical pattern of clinical presentation and dissemination often involving functionally contiguous lymph nodes (40, 41); however, the factors which regulate the spread of Hodgkin lymphoma cells are poorly understood. Therefore, in order to improve our understanding in this area, we set out to assess S1PR1 expression in two CHL cell lines (KM-H2 and SUP-HD1) and determine the role S1PR1 may play in CHL tumor cell migration. Herein, we report that S1PR1 is expressed in the KM-H2 and SUP-HD1 CHL cell lines and that S1P stimulates migration of both cell lines, which can be inhibited by S1PR1 antagonists (VPC44116 and FTY720-P), but not by the S1PR2 antagonist (JTE-013). Furthermore, S1P-induced migration was dependent on the Gi-PI3K pathway. Lastly, immunohistochemical assessment of tissue from CHL cases reveals that a subset show strong, membranous staining for S1PR1 in Hodgkin-Reed Sternberg cells. In summary, we demonstrate that S1PR1 can regulate the migration of HRS cells and that S1PR1 is expressed in both HRS cell lines and in a subset of CHL cases. Taken together, these data suggest that further investigation is warranted into the role of S1PR1 in CHL as well as the possibility of targeting S1PR1 in the therapy of S1PR1-positive, refractory/recurrent CHL.
MATERIALS AND METHODS
Reagents
D-erythro-S1P was purchased from Enzo Life Sciences Inc. (Farmingdale, NY). JTE013 and FTY720-P were from Cayman Chemicals (Ann Arbor, MI). VPC44116 was kindly provided by Dr. Kevin R Lynch (University of Virginia). Paraformaldehyde and fatty acid free bovine serum albumin (BSA) were purchased from Sigma (St. Louis, MI). Pertussis toxin and Y-27632 were from EMD Millipore Chemicals Inc. (San Diego, CA). LY-294002 was purchased from Cell Signaling Technology Inc. (Danvers, MA).
Cell Culture and cDNA Transfection
The KM-H2 Hodgkin lymphoma cell line was purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and were cultured in RPMI medium supplemented with 10% FBS, L-Glutamine and antibiotics. SUP-HD1 cells were kindly provided by Dr. B. Chapuy and Dr. M. Shipp (Dana Farber Cancer Institute, Boston, MA). They were cultured in McCoy’s medium, supplemented with 20% FBS, L-Glutamine and antibiotics. Human embryonic kidney cells (HEK-293T) were transfected with pcDNA 3.1 alone, S1PR1-pcDNA 3.1 or S1PR2-pcDNA 3.1 using Lipofectamine 2000 (Life Technologies Corp. Carlsbad, CA), as previously described (10, 14).
RNA Isolation, Reverse Transcription (RT) and Quantitative PCR Analysis
Total RNA was prepared using RNeasy Mini Kit (Qiagen, Valencia, CA) with RNase-free DNase treatment (Qiagen) to remove the genomic DNA. To generate cDNA, 100 ng of RNA was reverse transcribed using random primers and SuperScript II RT-polymerase (Invitrogen, Carlsbad, CA). Primers were designed using the Primer Express oligo design program software (Applied Biosystems, Foster City, CA). All primer sets were subjected to rigorous database searches to identify potential conflicting transcript matches to pseudogenes or homologous domains within related genes. PCR primer sequences for target molecules are shown in Table 1. Amplicons generated from the primer set were analyzed for melting point temperatures using the first derivative primer melting curve software supplied by Applied BioSystems. Real-time quantitative PCR was performed using the SYBR Green I assay on the ABI 7500 Sequence Detection System (Applied Biosystems) at the Multi-Gene Transcriptional Profiling Core in the Center for Vascular Biology Research. PCR reactions for each cDNA sample were performed in duplicate and copy numbers were calculated using standard curves generated from a master template as previously described (42).
Table 1.
Primer Sequences for Quantitative RT-PCR Analysis
| Target Genes | Sequence of Primers |
|---|---|
|
| |
| Human S1PR1 | F: GCTGCTCAAGACCGTAATTATCG |
| R: ACCAGGAAGTACTCCGCTCTGA | |
| Human S1PR2 | F: TGGCCGCCTCCGATCT |
| R: GAGAGCAAGGTATTGGCTACGAA | |
| Human S1PR3 | F: GCCATCGAGCGGCACTT |
| R: GCCTCTTGTTGGCGTCGTAA | |
| Human S1PR4 | F: CCTTTGCTGGGCTGGAACT |
| R: AGAGGATGTAGCGCTTGGAGTAGA | |
| Human S1PR5 | F: GAGGTGGGAGCCATAGAAGCTT |
| R: ACACCTGTGATCACCTGAATTAGG | |
Cell Migration Assay
KM-H2 and SUP-HD1 cell migration was assayed in a modified 96 well-Boyden Chamber (Neuro Probe Inc., Gaithersburg, MD) using polycarbonate filters of 3uM pore size, as previously described (10, 14, 43). Prior to chamber assembly, cells were serum starved for one hour in plain media. Wells in the bottom half of the chamber were filled with 85uL of 0.1% BSA RPMI or 0.1% BSA McCoy’s plus vehicle or S1P in the presence or absence of the S1PR antagonists, pertussis toxin, LY-294002 or Y-27632. A suspension of 0.5×106 –1×106 cells were added to the top chamber in the presence or absence of the S1PR antagonists.
For FTY720-P pre-incubation studies, cells were incubated for 30 minutes at 37°C with vehicle or FTY720-P, they were washed and counted before adding to the top chamber. Fully assembled chamber was left to incubate at 37°C for 5–6 hours. After chamber disassembly, non-migrating cells on the surface of filter were wiped away from filter with H2O soaked gauze, and the filter was fixed in 4% paraformaldehyde and stained with 0.1% crystal violet. For quantification, crystal violet stained cells were eluted in 10% acetic acid and absorbance at 575 nm was measured using a Spectramax M5 plate reader (Molecular Devices, Sunnyvale, CA).
Immunohistochemistry
Patient samples with a diagnosis of classical Hodgkin lymphoma were identified through the Brigham and Women’s Hospital (BWH) pathology database. Immunohistochemical studies were approved by BWH Institutional Review Board (IRB # 2010P002736).
4 um thick sections of formalin fixed paraffin embedded (FFPE) cell pellets (KM-H2 and SUP-HD1) or whole tissue sections (57 clinical cases) were used for immunohistochemistry (IHC) on the Ventana Discovery XT autostainer (Ventana Medical Systems, Tucson, AZ) with heat induced antigen retrieval using a Tris-based buffer with slightly basic pH (CC1 buffer) and anti-S1PR1 antibody (catalogue # sc-25489; Santa Cruz Biotechnology, Inc., Santa Cruz, CA. 95060) at a final concentration of approximately 0.9 ug/ml followed by incubation with standard secondary antibody and diaminobenzidine-based detection reagents (Ventana Medical Systems, Tucson, AZ).
Statistical Analyses
For in vitro experiments, one-way ANOVA followed by Tukey-Kramer test was conducted using Graph Pad Prism. All experiments were conducted 3–5 times, and a representative experiment is shown. The percentage of inhibition of migration by the S1PR pharmacological modulators was calculated in each individual experiment and the average ± SEM of all experiments is reported in the text.
The Fisher’s exact test (two-sided p value) was used to compare the relative percentages of recurrence among the S1PR1-positive and S1PR1-negative cases.
RESULTS
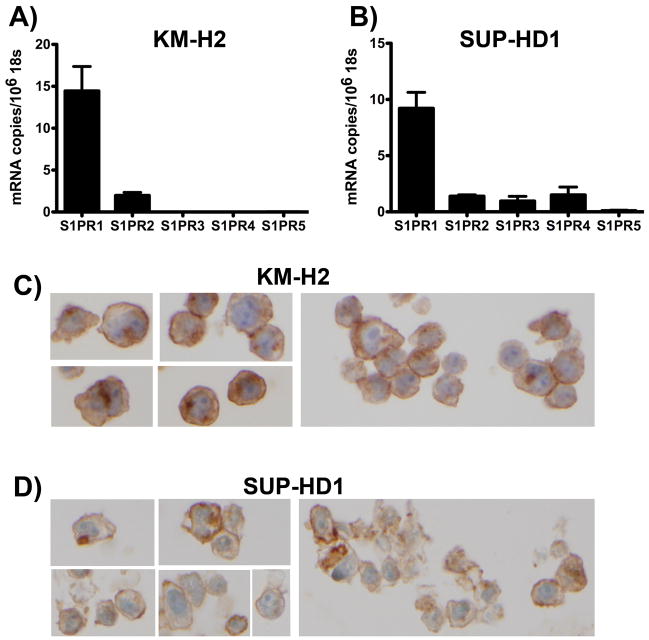
We tested two CHL cell lines for S1PR1 expression by quantitative RT-PCR analysis and found that S1PR1 was robustly expressed in both KM-H2 cells [14.4 ± 2.7 copies per 106 18S, which is equivalent to approximately 14.4 ± 2.7 copies per cell(42)] and SUP-HD1 cells (9.2 ± 1.4 copies per 106 18S) (Figure 1A, 1B). In addition, in KM-H2 cells (Figure 1A), low levels of S1PR2 transcript were detected (2.0 ± 0.4 copies per 106 18S) while the other three S1PR isoforms were not present at significant levels (<0.1 copy per cell). In SUP-HD1 (Figure 1B), low levels of S1PR2 (1.4 ± 0.1 copies per 106 18S), S1PR3 (1.0 ± 0.4 copies per 106 18S) and S1PR4 (1.5 ± 0.7 copies per 106 18S) were detected, while S1PR5 was not present at significant levels (<0.1 copy per cell).
Figure 1. Expression of S1PR1 in KM-H2 and SUP-HD1 Hodgkin lymphoma cell lines.
Quantitative RT-PCR demonstrates detectable S1PR1 expression in KM-H2 (A) and SUP-HD1 (B) cells; note that the other S1PR isoforms demonstrate much lower levels of expression than S1PR1. Data are mean ± SEM, n=4. Immunohistochemistry for S1PR1 demonstrates membranous S1PR1 protein expression in KM-H2 (C) and SUP-HD1 (D) cells. Images are shown at 60x magnification.
Next, using an anti-S1PR1 antibody that has been used in prior publications (44) and the specificity of which we confirmed by transient transfection experiments (Supplementary Figure I), we assessed the expression of S1PR1 in the KM-H2 and SUP-HD1 cell lines at the protein level by immunohistochemistry (IHC). We found membranous staining for S1PR1 in both of the Hodgkin lymphoma cell lines tested (Figure 1C, 1D).
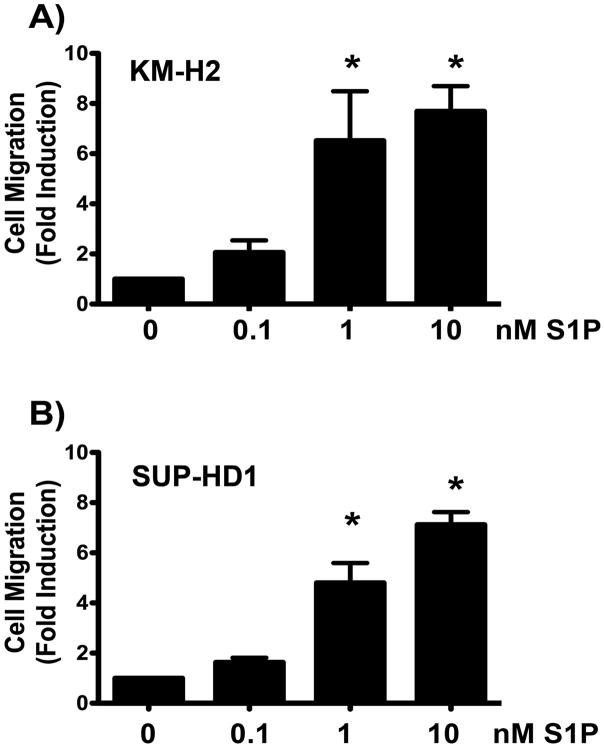
Having demonstrated expression of S1PR1 in both Hodgkin lymphoma cell lines by two different methods, we turned our attention to assessment of the functional (i.e., migratory) responses of these cell lines to the ligand for S1PR (i.e., S1P). Using a standard, in vitro, modified Boyden chamber migration assay, we found that S1P potently induced both KM-H2 and SUP-HD1 cell migration (Figure 2). The maximal migratory responses occurred at 1–10nM S1P, which are physiologically relevant concentrations consistent with the known dissociation constant of S1P for S1PR1 (8.1 nM) (2) and known concentration gradient of S1P between tissues and plasma or lymphatic fluids. In KM-H2 cells, S1P induced a 6.5 ± 2.0 and 7.7 ± 1.0 fold induction of cell migration at 1 and 10 nM, respectively (Figure 2A). S1P induced SUP-HD1 cell migration to a similar extent (4.8 ± 0.8 and a 7.1 ± 0.5 fold induction) at 1 and 10 nmM, respectively (Figure 2B).
Figure 2. S1P induces Hodgkin lymphoma cell migration in a dose-dependent way.
S1P treatment of the Hodgkin Lymphoma cell lines KM-H2 (A) and SUP-HD1 (B) lead to a dose dependent increase in cell migration at physiologically relevant doses of S1P, with maximal responses occurring at 1–10nM. The Hodgkin lymphoma cell lines, KM-H2 (A) and SUP-HD1 (B) were serum starved for 1 hour and migration experiments were conducted using S1P as chemoattractant in the bottom chamber at the doses indicated. Fold induction versus vehicle is shown. Data are mean ± SEM of triplicates. A representative experiment of n=5 is shown. *, p<0.05, S1P vs vehicle.
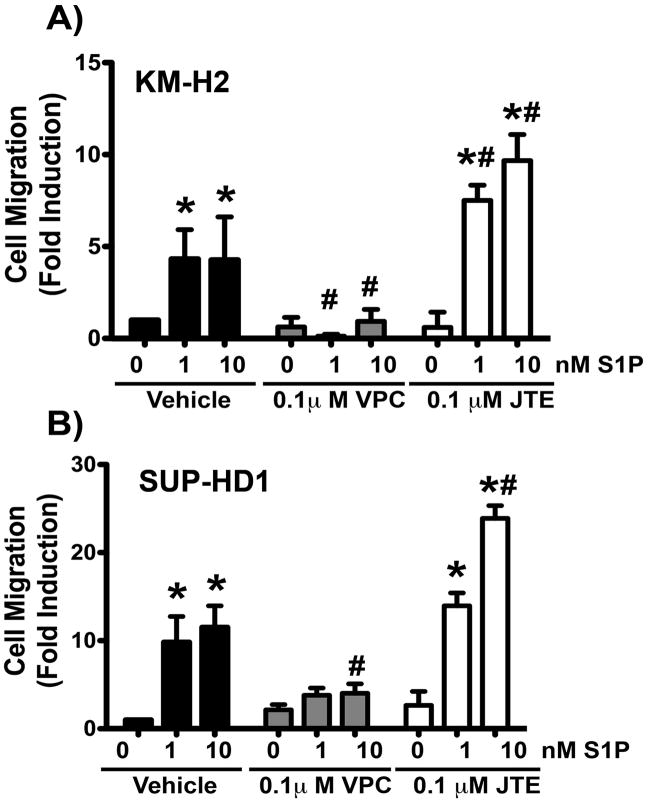
In order to further study the role of S1PR1 in Hodgkin lymphoma cell migration, we tested the effects of S1PR antagonists on the migratory responses to S1P. We found that the known S1PR1 antagonist, VPC44116, inhibited the migratory responses to 1nM and 10 nM S1P in KMH2 cells (average % inhibition over all experiments: 84.04 ± 11.9 and 46.23 ± 13.54 %, respectively, Figure 3A) and SUPHD1 cells (average % inhibition over all experiments: 68.5 ± 7.1 and 71.2 ± 6.3%, respectively, Figure 3B). On the other hand, the S1PR2 specific antagonist, JTE-013, did not impair S1P-induced migration and, in fact, potentiated migration toward S1P (Figure 3A, 3B); this potentiation of S1P-induced migration in the presence of the S1PR2 antagonist is consistent with the fact that S1PR2 is a known negative regulator of cell migration (14, 19, 45).
Figure 3. Blockade of S1PR1 signaling inhibits S1P-induced Hodgkin lymphoma cell migration.
The Hodgkin lymphoma cell lines, KM-H2 (A) and SUP-HD1 (B) were serum starved for 1 hour before conducting migration assays towards S1P at the doses indicated. Cells were pre-treated with vehicle, the S1PR1 specific antagonist VPC44116 (VPC) or the S1PR2 specific antagonist JTE013 (JTE) for 30 minutes at the doses indicated. These treatments were also present in the top and bottom chamber during migration. Fold induction versus vehicle-pre-treatment (no S1P) is shown. Data are mean ± SEM of triplicates. A representative experiment of n=3 is shown. The percentage of inhibition of migration by VPC 44116 from each experiment was calculated and the average ± SEM of all three experiments is reported in the text. *, P<0.05 S1P vs no S1P. #, P<0.05 S1PR antagonist-treated vs vehicle-treated.
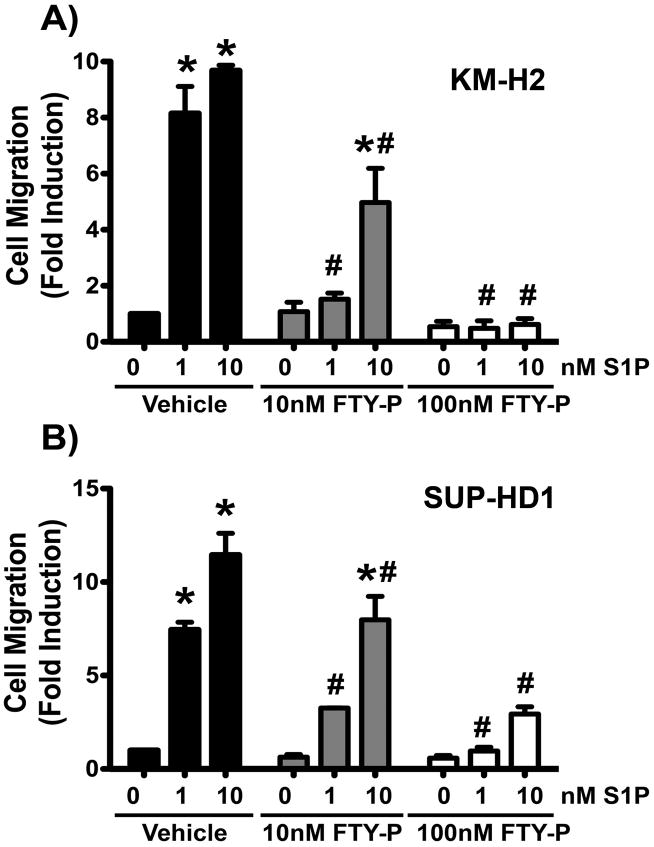
Next, we tested the effects of FTY720-P, the active form of FTY720, a clinically approved functional antagonist of S1PR1. We found that pre-incubation with FTY720-P lead to a dose dependent inhibition of S1P-induced migration in both cell lines (Figure 4). In KM-H2 cells, 10 nM FTY720-P pre-incubation resulted in impaired S1P-induced migration (average % inhibition over all experiments: 67.25 ± 12.8% towards 1nM S1P and 63 ± 12.6% inhibition towards 10nM S1P, Figure 4A). Pre-treatment with 100nM FTY720-P resulted in further inhibition of S1P-induced cell migration (average % inhibition over all experiments: 90.8 ± 4.6% and 90.25 ± 3.3% towards 1 and 10nM S1P, respectively, Figure 4A). Similarly, in SUP-HD1 cells, pre-treatment with 10nM FTY720-P inhibited S1P-induced motility (average % inhibition over all experiments: 57.6 ± 1.8% for 1nM S1P and 31.6 ± 1.9% for 10 nM S1P, Figure 4B) and pretreatment with 100nM FTY720-P resulted in further inhibition of S1P-induced migration (average % inhibition over all experiments: 88.6 ± 2.1% and 69.5 ± 6.8%, respectively, Figure 4B). These findings indicate that pre-incubation with FTY720-P desensitizes S1PR1 and renders the cells unresponsive to S1P gradient (34, 35). Taken together, these migration data demonstrate that the S1PR1 expressed in KM-H2 and SUP-HD1 cell lines is capable of actively regulating migratory cell function in responses to S1P stimulation and that this effect can be effectively altered by treatment with known S1PR antagonists.
Figure 4. FTY7720-P pre-incubation inhibits Hodgkin lymphoma cell migration towards S1P gradient in a dose-dependent way.
The Hodgkin lymphoma cell lines, KM-H2 (A) and SUP-HD1 (B) were serum starved for 1 hour in the presence or absence of FTY720-P at the doses indicated. Then, cells were washed and counted and migration experiments towards S1P gradient were conducted as described. Fold induction compared to vehicle-pre-treatment (no S1P) is shown. Data are mean ± SEM of triplicates. A representative experiment of n=3 is shown. The percentage of inhibition of migration by FTY720-P from each experiment was calculated and the average ± SEM of all three experiments is reported in the text. *, P<0.05 S1P vs no S1P. #, P<0.05 FTY720-P-pre-treated vs vehicle-pre-treated.
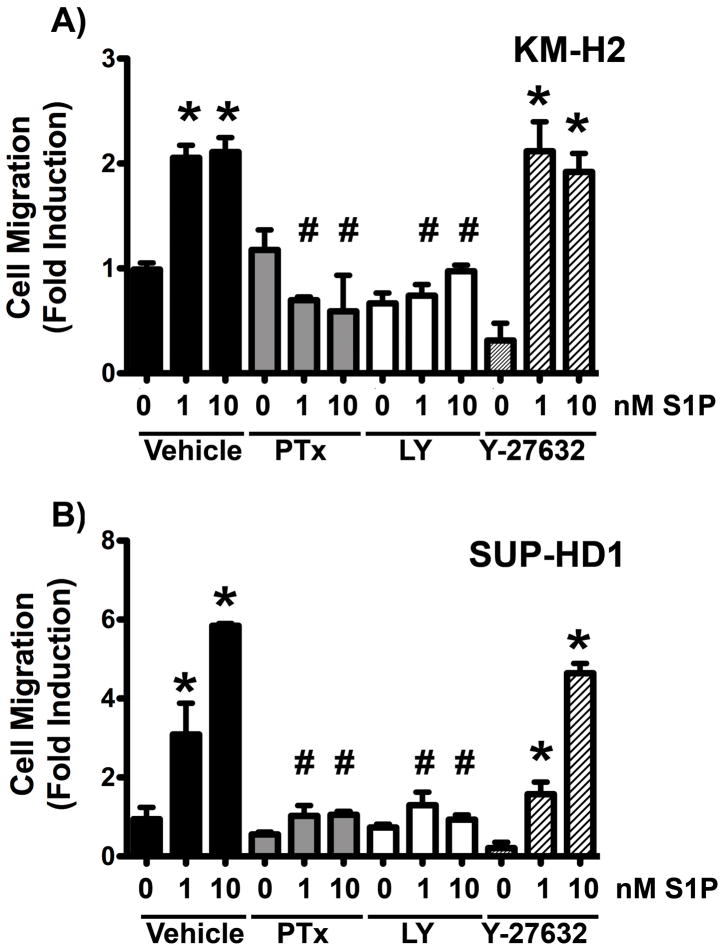
In order to gain mechanistic insights into the regulation of Hodgkin lymphoma cell migration by S1PR1 we tested the role of the Gi-PI3K pathway, which has previously shown to be activated by S1PR1 in a pertussis toxin sensitive manner (16, 17). We used pertussis toxin to inhibit Gi proteins and LY-294002 to inhibit PI3K activity. As shown in Figure 5A, both pertussis toxin (200 ng/mL) and LY-294002 (50 μM) potently inhibited the ability of S1P to induce KM-H2 cell migration. Inactivation of Gi proteins resulted in 67±4% inhibition of migration to 1nM S1P and 69±7.5% inhibition towards 10 nM S1P. LY-294002 treatment resulted in 60.3±9% and 57.7±3% inhibition of migration to 1 and 10 nM S1P, respectively (Figure 5A). On the contrary, S1P-induced migration was not dependent on Rho kinases, since the ROCK inhibitor, Y-27632, did not affect S1P-induced migration. Similarly, S1P induced SUP-HD1 cell migration was dependent on the Gi-PI3K pathway (Figure 5B) (75±1% and 89±0.9% inhibition by pertussis toxin and 61±2.4% and 72.5+11% by LY-294002) and was not impaired by the ROCK inhibitor. Altogether these data indicate that S1P-induced Hodgkin lymphoma cell migration is mediated by the S1PR1-Gi-PI3K pathway but not the Rho-ROCK pathway.
Figure 5. S1PR1-induced Hodgkin lymphoma cell migration is mediated by the Gi-PI3K pathway.
The Hodgkin lymphoma cell lines, KM-H2 (A) and SUP-HD1 (B) were serum starved for 1 hour before conducting migration assays towards S1P at the doses indicated. Cells were pre-treated with vehicle, 200 ng/mL pertussis toxin (PTx), 50μM LY-294002 (LY) or Y-27632 (10μM) during the serum starvation period. These treatments were also present in the top and bottom chamber during migration. Fold induction versus vehicle-pre-treated (no S1P) is shown. Data are mean ± SEM of triplicates. A representative experiment of n=3–4 is shown. The percentage of inhibition of migration from each experiment was calculated and the average ± SEM of all three experiments is reported in the text. *, P<0.05 S1P vs no S1P. #, P<0.05 inhibitor-treated vs vehicle-treated.
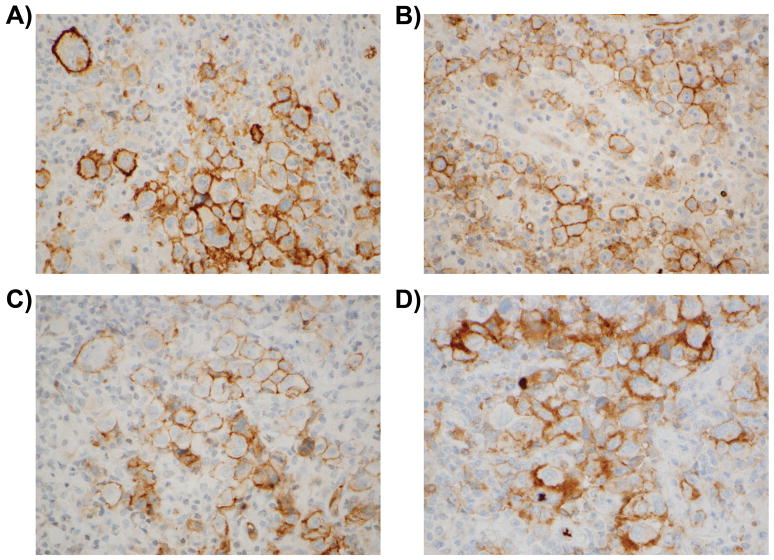
Lastly, in order to further assess the potential clinical relevance of these findings, we performed immunohistochemical analysis to assess S1PR1 expression in tissue from clinical cases of CHL. We found membranous S1PR1 staining in 25–90% of Hodgkin-Reed Sternberg (HRS) cells in 7 of 57 cases tested (12%, Figure 6). Clinical data was available for 6 of these 7 patients (Table 2). Three patients (3/6) died of their disease after multiple (two or more) recurrences. Two patients (2/6) remain alive after multiple recurrences that required multiple therapeutic approaches including allogeneic bone marrow stem cell transplantation. One patient (1/6) entered complete remission after induction chemotherapy, indicating that S1PR1 positivity is not uniformly associated with disease recurrence. Follow up data was available for 45 out of the 50 patients whose HRS cells were negative for S1PR1 expression. Four of these patients died of their disease (4/45) after multiple recurrences. Sixteen patients remain alive (16/45) but had recurrent disease, 12 of which have had multiple recurrences (12/45). The remaining 25 patients entered complete remission with appropriate, standard chemotherapy (25/45). Interestingly, although the overall prevalence of S1PR1 positivity in all cases studies was 12% (7/57), the prevalence of S1PR1 positivity in the subgroup comprised of all of the CHL cases that had multiple recurrences was 24% (5/21). Nevertheless, the relative proportion of cases with any recurrence among the S1PR1-positive (5/6) and S1PR1-negative (20/45) groups was not statistically different (p=0.099, Fisher’s exact test).
Figure 6. S1PR1 is expressed in Hodgkin-Reed Sternberg cells in a subset of cHL cases.
Representative images of S1PR1 immunohistochemistry in cases of classical Hodgkin lymphoma. Formalin fixed paraffin embedded tissue sections were used for immunohistochemical staining of S1PR1 as described in the methods section. The images demonstrate membranous staining for S1PR1 in Hodgkin-Reed Sternberg cells in Case 1 (A), Case 2 (B), Case 3 (C) and Case 7 (D). Images are shown at 60x magnification.
Table 2.
Available Clinical and Pathological Features of S1PR1-Positive CHL Cases.
| At Initial Diagnosis | ||||||||
|---|---|---|---|---|---|---|---|---|
| Case # | M/F | Age | Stage | Initial Therapy | Initial CR | Follow Up Therapy* | Alive/Dead* | Specimen Available for S1PR1 IHC |
| 1 | M | 28 | IV B | ABVD x 6 cycles | No | ICE therapy. No CR. Auto-SCT. RadRx for multiple recurrences (at 15 mo.). Non-myeloablative Allo-SCT (at 18 mo.). Recurrence (at 23 mo.). DLI. Second Non-myeloablative Allo-SCT (at 41 mo.). Recurrence (at 62 mo.). RadRx. | Dead (at 67 mo.) | Recurrence (at 23 mo.): Sheets of HRS cells. CD30+, CD15+, CD20 (subset)+. EBER: Negative. |
| 2 | M | 52 | III B | ABVD x 3 cycles plus (Doxil, Bleomycin, Vinblastine and Dacarbazine) x 3 cycles | No | COPP x 3 cycles. 1st recurrence (at 10 mo.): ICE therapy. CR. Auto-SCT. 2nd recurrence (at 26 mo.): GND x 5 cycles. | Dead (at 36 mo.) | 2nd recurrence (at 26 mo.): Sheets of HRS cells. CD30+, CD15(focal)+, CD20(subset)+, BSAP+. EBER:Negative. |
| 3 | M | NA | NA | NA | NA | NA | NA | Focal Clusters of HRS cells. CD30+ CD15+ BSAP+ CD20−, LCA−. EBER:Negative |
| 4 | F | 27 | II A | ABVD x 4 cycles | No | Refractory/recurrent disease (at 7 mo.). ICE therapy. GND therapy. Rad Rx. Auto-SCT (at 12 mo.). Recurrence (at 17 mo.). Bendamustine. Neulasta. | Dead (at 20 mo.) | Recurrence (at 17 mo.): Focal clusters of HRS cells. |
| 5 | M | 62 | II A | ABVD x 6 cycles | Yes | Not necessary. | Alive | Initial Sample. Focal clusters of HRS cells. CD30+, CD15−, CD20−. |
| 6 | M | 52 | IV A | ABVD x 6 cycles | Yes | 1st Recurrence (at 18 mo.). ICE therapy. Auto-SCT (at 20 mo.). 2nd recurrence (at 26 mo). Rad Rx. 3rd recurrence (at 39 mo.). Allo-SCT (at 48 mo.). 4th recurrence (at 51 mo.). DLI. | Alive | 3rd recurrence (at 39 mo.) Frequent HRS cells. CD30+, CD15+, BSAP+, CD20−. EBER:Negative. |
| 7 | M | 19 | II A | ABVD x 4 cycles. Rad Rx. | Yes | 1st recurrence(at 12 mo.). GDP therapy. CR. Auto-SCT. 2nd recurrence (at 21 mo.). Rad Rx. CR. 3rd recurrence (at 61 mo.). GND+Rituxan therapy. Non- myeloablative Allo-SCT (at 65 mo.). 4th recurrence (CHL plus EBV+DLBCL;at 70 mo.). DLI. Brentuximab. | Alive | 3rd recurrence (at 61 mo.) HRS cells. CD30+, CD15+, BSAP+, Fascin+, CD20−, CD45−. EBER:Negative. Concurrent EBV+ DLBCL: CD20+, CD45+, CD30+(subset), Fascin−, CD15−. |
Abbreviations: ABVD: Adriamycin, Bleomycin, Vinblastine, Dacarbazine; Allo-SCT: Allogeneic Stem Cell Transplant, Auto-SCT: Autologous Stem Cell Transplant; CHL: Classical Hodgkin Lymphoma; COPP: Cyclophosphamide, Oncovin, Procarbazine, Prednisone; CR: Complete Remission; DLBCL: Diffuse Large B Cell Lymphoma; DLI: Donor Lymphocyte Infusion; EBER: Epstein-Barr Virus Encoded RNA; GDP: Gemcitabine, Dexamethasone, Cisplatin; GND: Gemcitabine, Vinorelbine, Doxorubicin; HRS: Hodgkin-Reed Sternberg cells; ICE: Ifosfamide, Carboplatin, Etoposide; IHC: Immunohistochemistry; M: Male; F: Female; RadRx: Radiation Therapy.
Dates of recurrences and death are relative to date of initial diagnosis.
In order to assess for the possible modulation of S1PR1 expression during the different phases of disease (initial diagnosis, treatment, recurrence, etc), we tested additional biopsy material for 10 of the S1PR1-negative patients with recurrent disease who had additional biopsies available from other time points during their disease course (ie, biopsies either prior to or subsequent to the biopsy first tested for S1PR1 expression). We found that the additional biopsies were S1PR1 negative, similar to the original biopsies tested for each patient.
DISCUSSION
S1PR1 has been shown to be important in the physiologic trafficking of lymphocytes (20–28). However, our understanding of the role it plays in B-cell lymphomas is limited. Recent work has shown that S1PR1 is expressed in some B cell lymphomas (44). Nishimura et al. found that S1PR1 was consistently expressed in mantle cell lymphomas and was expressed in a subset of chronic lymphocytic leukemias/small lymphocytic lymphomas (CLL/SLL) as well as in a subset of diffuse large B cell lymphomas (DLBCL); however, they found that S1PR1 expression was negative in cases of follicular lymphoma and marginal zone lymphoma. These expression data suggest that S1PR1 may regulate tumor cell functions in some types of B cell lymphoma. Along these lines, the recent work by Liu et al. (46) has shed some light on the functional role that S1PR1 may play in lymphoma; more specifically, using a diffuse large B cell lymphoma xenograft tumor model, Liu et al. demonstrated that inhibition of S1PR1 expression down regulated Signal Transducer and Activator of Transcription 3 activity, leading to inhibition of lymphoma tumor cell growth in vitro and in vivo.
Given that the clinical presentation and pattern(s) of spread in CHL are somewhat different than other types of B-cell lymphomas (40, 41) and that the factors regulating the spread of Hodgkin lymphoma cells are poorly understood, we focused our studies on the expression and functional role of S1PR1 in CHL. We found that S1PR1 was the major S1PR isoform expressed in two different Hodgkin lymphoma cell lines (KM-H2 and SUP-HD1) by quantitative RT-PCR analysis. The expression of S1PR1 was confirmed at the protein level using immunohistochemistry. Furthermore, the functional relevance of the S1PR1 expressed in these cell lines was demonstrated by its ability to regulate S1P-induced migration, consistent with the key role that S1PR1 is known to play in physiologic trafficking of normal lymphocytes. Also consistent with previously published studies in other cell systems, S1PR1-mediated migration in the HRS cell lines was found to be dependent on the heterotrimeric G protein, Gi-PI3K pathway. Lastly, the S1PR1 antagonists tested, including FTY720-P, the known functional antagonist for S1PR1 which induces internalization and desensitization of S1PR1 (34, 35), were effective at impairing S1P-induced migration. Of note, FTY720, the precursor of FTY720-P, is approved as therapeutic option for some cases of multiple sclerosis (36–38).
The potential pathophysiologic relevance of our findings is supported by the known significance of S1PR1 in regulating the trafficking of normal lymphocytes, the known importance of S1P concentration gradients in vivo between various tissue compartments (29, 30) and by our finding that membranous S1PR1 is detectable by immunohistochemical analysis in HRS cells in an albeit small subset of clinical cases of CHL (7/57 cases, 12%). In addition, the recent work by Liu et al., using a xenograft model of human DLBCL tumor growth and invasion, demonstrated that downregulation of S1PR1 by shRNA inhibited tumor growth and reduced the spread of the lymphoma as evidenced by the formation of fewer lung nodules (46).
The evaluation of the potential clinical significance of S1PR1 expression in CHL is limited by the small size of our clinical case cohort and by the incomplete clinical and pathologic information available for the cases tested. Also, unlike the high prevalence of S1PR1 positivity that has been reported in mantle cell lymphoma, there is a relatively low prevalence of S1PR1 positivity among the CHL cases tested here, which may be a reflection of the fact that in some types of lymphoma, only a subset of cases will be positive for S1PR1, as has been reported for DLBCL and CLL/SLL (44). Further immunohistochemical analysis of S1PR1 on a larger series of CHL cases with more complete clinical and pathologic annotation will be required to better characterize the prevalence of S1PR1 expression in CHL. Such studies may clarify if the apparent low prevalence of S1PR1 positivity observed in CHL is due, in part, to aberrant down regulation of S1PR1 in HRS cells and/or if the prevalence of S1PR1 expression varies according to the clinical/pathologic stage of disease. Lastly, the relationship, if any, between S1PR1 expression and recurrence rate/outcome in CHL also remains to be definitively determined.
In conclusion, we demonstrate that i) S1PR1 is a functional receptor on HRS cells which governs tumor cell migration, ii) antagonists of S1PR1 effectively impair S1P-induced migration in HRS cells, iii) S1PR1 is expressed in a subset of clinical cases of CHL. These results suggest that further study into the role(s) of S1PR1 in CHL is warranted and may shed light on the potential utility of S1PR1-targeted therapies in the future treatment of S1PR1-positive cases of recurrent/refractory CHL.
Supplementary Material
Acknowledgments
This work is supported by NIH grant HL094465 (to TS).
We would like to thank Dan Li and Dr. Shou-ching Jaminet from the Multi-Gene Transcriptional Profiling Core Facility at the Center for Vascular Biology Research for the quantitative PCR analysis and Dr. Kevin Lynch (University of Virginia) for VPC44116.
ABBREVIATIONS
- BSA
Bovine Serum Albumin
- CHL
Classical Hodgkin Lymphoma
- CLL/SLL
Chronic Lymphocytic Leukemia/Small Lymphocytic Lymphoma
- DLBCL
Diffuse Large B Cell Lymphoma
- FFPE
Formalin Fixed Paraffin Embedded
- GPCR
G Protein Coupled Receptor
- HRS
Hodgkin-Reed Sternberg
- PI3K
Phosphatidylinositol-3-Kinase
- PTEN
Phosphatase and Tensin Homolog Deleted on Chromosome 10
- qPCR
quantitative PCR
- ROCK
Rho kinase
- RT
Reverse Transcription
- S1P
Sphingosine-1-Phosphate
- S1PR
Sphingosine-1-Phosphate Receptors
Footnotes
DISCLOSURE:
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Laboratory Investigation website (http://www.nature.com/labinvest).
References
- 1.Hla T, Maciag T. An abundant transcript induced in differentiating human endothelial cells encodes a polypeptide with structural similarities to G-protein-coupled receptors. J Biol Chem. 1990 Jun 5;265(16):9308–13. [PubMed] [Google Scholar]
- 2.Lee MJ, Van Brocklyn JR, Thangada S, Liu CH, Hand AR, Menzeleev R, et al. Sphingosine-1-phosphate as a ligand for the G protein-coupled receptor EDG-1. Science. 1998 Mar 6;279(5356):1552–5. doi: 10.1126/science.279.5356.1552. [DOI] [PubMed] [Google Scholar]
- 3.Lee MJ, Thangada S, Claffey KP, Ancellin N, Liu CH, Kluk M, et al. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell. 1999 Oct 29;99(3):301–12. doi: 10.1016/s0092-8674(00)81661-x. [DOI] [PubMed] [Google Scholar]
- 4.Liu CH, Thangada S, Lee MJ, Van Brocklyn JR, Spiegel S, Hla T. Ligand-induced trafficking of the sphingosine-1-phosphate receptor EDG-1. Mol Biol Cell. 1999 Apr;10(4):1179–90. doi: 10.1091/mbc.10.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000 Oct;106(8):951–61. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik JH, Chae S, Lee MJ, Thangada S, Hla T. Sphingosine 1-phosphate-induced endothelial cell migration requires the expression of EDG-1 and EDG-3 receptors and Rho-dependent activation of alpha vbeta3- and beta1-containing integrins. J Biol Chem. 2001 Apr 13;276(15):11830–7. doi: 10.1074/jbc.M009422200. [DOI] [PubMed] [Google Scholar]
- 7.Wang F, Van Brocklyn JR, Hobson JP, Movafagh S, Zukowska-Grojec Z, Milstien S, et al. Sphingosine 1-phosphate stimulates cell migration through a G(i)-coupled cell surface receptor. Potential involvement in angiogenesis. J Biol Chem. 1999 Dec 10;274(50):35343–50. doi: 10.1074/jbc.274.50.35343. [DOI] [PubMed] [Google Scholar]
- 8.Ancellin N, Hla T. Differential pharmacological properties and signal transduction of the sphingosine 1-phosphate receptors EDG-1, EDG-3, and EDG-5. J Biol Chem. 1999 Jul 2;274(27):18997–9002. doi: 10.1074/jbc.274.27.18997. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez T, Hla T. Structural and functional characteristics of S1P receptors. J Cell Biochem. 2004 Aug 1;92(5):913–22. doi: 10.1002/jcb.20127. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez T, Estrada-Hernandez T, Paik JH, Wu MT, Venkataraman K, Brinkmann V, et al. Phosphorylation and action of the immunomodulator FTY720 inhibits vascular endothelial cell growth factor-induced vascular permeability. J Biol Chem. 2003 Nov 21;278(47):47281–90. doi: 10.1074/jbc.M306896200. [DOI] [PubMed] [Google Scholar]
- 11.Kon J, Sato K, Watanabe T, Tomura H, Kuwabara A, Kimura T, et al. Comparison of intrinsic activities of the putative sphingosine 1-phosphate receptor subtypes to regulate several signaling pathways in their cDNA-transfected Chinese hamster ovary cells. J Biol Chem. 1999 Aug 20;274(34):23940–7. doi: 10.1074/jbc.274.34.23940. [DOI] [PubMed] [Google Scholar]
- 12.Osada M, Yatomi Y, Ohmori T, Ikeda H, Ozaki Y. Enhancement of sphingosine 1-phosphate-induced migration of vascular endothelial cells and smooth muscle cells by an EDG-5 antagonist. Biochem Biophys Res Commun. 2002 Dec 6;299(3):483–7. doi: 10.1016/s0006-291x(02)02671-2. [DOI] [PubMed] [Google Scholar]
- 13.Arikawa K, Takuwa N, Yamaguchi H, Sugimoto N, Kitayama J, Nagawa H, et al. Ligand-dependent inhibition of B16 melanoma cell migration and invasion via endogenous S1P2 G protein-coupled receptor. Requirement of inhibition of cellular RAC activity. J Biol Chem. 2003 Aug 29;278(35):32841–51. doi: 10.1074/jbc.M305024200. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez T, Thangada S, Wu MT, Kontos CD, Wu D, Wu H, et al. PTEN as an effector in the signaling of antimigratory G protein-coupled receptor. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4312–7. doi: 10.1073/pnas.0409784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanchez T, Skoura A, Wu MT, Casserly B, Harrington EO, Hla T. Induction of vascular permeability by the sphingosine-1-phosphate receptor-2 (S1P2R) and its downstream effectors ROCK and PTEN. Arterioscler Thromb Vasc Biol. 2007 Jun;27(6):1312–8. doi: 10.1161/ATVBAHA.107.143735. [DOI] [PubMed] [Google Scholar]
- 16.Lee MJ, Evans M, Hla T. The inducible G protein-coupled receptor edg-1 signals via the G(i)/mitogen-activated protein kinase pathway. J Biol Chem. 1996 May 10;271(19):11272–9. doi: 10.1074/jbc.271.19.11272. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi J, Michel T. Sphingosine 1-phosphate and isoform-specific activation of phosphoinositide 3-kinase beta. Evidence for divergence and convergence of receptor-regulated endothelial nitric-oxide synthase signaling pathways. J Biol Chem. 2001 Sep 28;276(39):36281–8. doi: 10.1074/jbc.M105628200. [DOI] [PubMed] [Google Scholar]
- 18.Ishii I, Ye X, Friedman B, Kawamura S, Contos JJ, Kingsbury MA, et al. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate (S1P) receptors, S1P(2)/LP(B2)/EDG-5 and S1P(3)/LP(B3)/EDG-3. J Biol Chem. 2002 Jul 12;277(28):25152–9. doi: 10.1074/jbc.M200137200. [DOI] [PubMed] [Google Scholar]
- 19.Sugimoto N, Takuwa N, Okamoto H, Sakurada S, Takuwa Y. Inhibitory and stimulatory regulation of Rac and cell motility by the G12/13-Rho and Gi pathways integrated downstream of a single G protein-coupled sphingosine-1-phosphate receptor isoform. Mol Cell Biol. 2003 Mar;23(5):1534–45. doi: 10.1128/MCB.23.5.1534-1545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandala S, Hajdu R, Bergstrom J, Quackenbush E, Xie J, Milligan J, et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 2002 Apr 12;296(5566):346–9. doi: 10.1126/science.1070238. [DOI] [PubMed] [Google Scholar]
- 21.Graeler M, Goetzl EJ. Activation-regulated expression and chemotactic function of sphingosine 1-phosphate receptors in mouse splenic T cells. Faseb J. 2002 Dec;16(14):1874–8. doi: 10.1096/fj.02-0548com. [DOI] [PubMed] [Google Scholar]
- 22.Brinkmann V, Davis MD, Heise CE, Albert R, Cottens S, Hof R, et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J Biol Chem. 2002;277(24):21453–7. doi: 10.1074/jbc.C200176200. [DOI] [PubMed] [Google Scholar]
- 23.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004 Jan 22;427(6972):355–60. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 24.Allende ML, Dreier JL, Mandala S, Proia RL. Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J Biol Chem. 2004 Apr 9;279(15):15396–401. doi: 10.1074/jbc.M314291200. [DOI] [PubMed] [Google Scholar]
- 25.Allende ML, Tuymetova G, Lee BG, Bonifacino E, Wu YP, Proia RL. S1P1 receptor directs the release of immature B cells from bone marrow into blood. J Exp Med. 2010 May 10;207(5):1113–24. doi: 10.1084/jem.20092210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira JP, Xu Y, Cyster JG. A role for S1P and S1P1 in immature-B cell egress from mouse bone marrow. PLoS One. 2010;5(2):e9277. doi: 10.1371/journal.pone.0009277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thangada S, Khanna KM, Blaho VA, Oo ML, Im DS, Guo C, et al. Cell-surface residence of sphingosine 1-phosphate receptor 1 on lymphocytes determines lymphocyte egress kinetics. J Exp Med. 2010 Jul 5;207(7):1475–83. doi: 10.1084/jem.20091343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnon TI, Xu Y, Lo C, Pham T, An J, Coughlin S, et al. GRK2-dependent S1PR1 desensitization is required for lymphocytes to overcome their attraction to blood. Science. 2011 Sep 30;333(6051):1898–903. doi: 10.1126/science.1208248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwab SR, Pereira JP, Matloubian M, Xu Y, Huang Y, Cyster JG. Lymphocyte sequestration through S1P lyase inhibition and disruption of S1P gradients. Science. 2005 Sep 9;309(5741):1735–9. doi: 10.1126/science.1113640. [DOI] [PubMed] [Google Scholar]
- 30.Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol. 2007 Dec;8(12):1295–301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- 31.Halin C, Scimone ML, Bonasio R, Gauguet JM, Mempel TR, Quackenbush E, et al. The S1P-analog FTY720 differentially modulates T-cell homing via HEV: T-cell-expressed S1P1 amplifies integrin activation in peripheral lymph nodes but not in Peyer patches. Blood. 2005 Aug 15;106(4):1314–22. doi: 10.1182/blood-2004-09-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kharel Y, Lee S, Snyder AH, Sheasley-O’neill SL, Morris MA, Setiady Y, et al. Sphingosine kinase 2 is required for modulation of lymphocyte traffic by FTY720. J Biol Chem. 2005 Nov 4;280(44):36865–72. doi: 10.1074/jbc.M506293200. [DOI] [PubMed] [Google Scholar]
- 33.Zemann B, Kinzel B, Muller M, Reuschel R, Mechtcheriakova D, Urtz N, et al. Sphingosine kinase type 2 is essential for lymphopenia induced by the immunomodulatory drug FTY720. Blood. 2006 Feb 15;107(4):1454–8. doi: 10.1182/blood-2005-07-2628. [DOI] [PubMed] [Google Scholar]
- 34.LaMontagne K, Littlewood-Evans A, Schnell C, O’Reilly T, Wyder L, Sanchez T, et al. Antagonism of sphingosine-1-phosphate receptors by FTY720 inhibits angiogenesis and tumor vascularization. Cancer Res. 2006 Jan 1;66(1):221–31. doi: 10.1158/0008-5472.CAN-05-2001. [DOI] [PubMed] [Google Scholar]
- 35.Oo ML, Thangada S, Wu MT, Liu CH, Macdonald TL, Lynch KR, et al. Immunosuppressive and anti-angiogenic sphingosine 1-phosphate receptor-1 agonists induce ubiquitinylation and proteasomal degradation of the receptor. J Biol Chem. 2007 Mar 23;282(12):9082–9. doi: 10.1074/jbc.M610318200. [DOI] [PubMed] [Google Scholar]
- 36.Brinkmann V, Billich A, Baumruker T, Heining P, Schmouder R, Francis G, et al. Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat Rev Drug Discov. 2010 Nov;9(11):883–97. doi: 10.1038/nrd3248. [DOI] [PubMed] [Google Scholar]
- 37.Mehling M, Johnson TA, Antel J, Kappos L, Bar-Or A. Clinical immunology of the sphingosine 1-phosphate receptor modulator fingolimod (FTY720) in multiple sclerosis. Neurology. 2011 Feb 22;76(8 Suppl 3):S20–7. doi: 10.1212/WNL.0b013e31820db341. [DOI] [PubMed] [Google Scholar]
- 38.Scott LJ. Fingolimod: a review of its use in the management of relapsing-remitting multiple sclerosis. CNS Drugs. 2011 Aug 1;25(8):673–98. doi: 10.2165/11207350-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 39.Foss FW, Jr, Snyder AH, Davis MD, Rouse M, Okusa MD, Lynch KR, et al. Synthesis and biological evaluation of gamma-aminophosphonates as potent, subtype-selective sphingosine 1-phosphate receptor agonists and antagonists. Bioorg Med Chem. 2007 Jan 15;15(2):663–77. doi: 10.1016/j.bmc.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg SA, Kaplan HS. Evidence for an orderly progression in the spread of Hodgkin’s disease. Cancer Res. 1966 Jun;26(6):1225–31. [PubMed] [Google Scholar]
- 41.Roth SL, Sack H, Havemann K, Willers R, Kocsis B, Schumacher V. Contiguous pattern spreading in patients with Hodgkin’s disease. Radiother Oncol. 1998 Apr;47(1):7–16. doi: 10.1016/s0167-8140(97)00208-9. [DOI] [PubMed] [Google Scholar]
- 42.Shih SC, Smith LE. Quantitative multi-gene transcriptional profiling using real-time PCR with a master template. Exp Mol Pathol. 2005 Aug;79(1):14–22. doi: 10.1016/j.yexmp.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Kluk MJ, Hla T. Role of the sphingosine 1-phosphate receptor EDG-1 in vascular smooth muscle cell proliferation and migration. Circ Res. 2001 Sep 14;89(6):496–502. doi: 10.1161/hh1801.096338. [DOI] [PubMed] [Google Scholar]
- 44.Nishimura H, Akiyama T, Monobe Y, Matsubara K, Igarashi Y, Abe M, et al. Expression of sphingosine-1-phosphate receptor 1 in mantle cell lymphoma. Mod Pathol. 2010 Mar;23(3):439–49. doi: 10.1038/modpathol.2009.194. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto H, Takuwa N, Yokomizo T, Sugimoto N, Sakurada S, Shigematsu H, et al. Inhibitory regulation of Rac activation, membrane ruffling, and cell migration by the G protein-coupled sphingosine-1-phosphate receptor EDG5 but not EDG1 or EDG3. Mol Cell Biol. 2000 Dec;20(24):9247–61. doi: 10.1128/mcb.20.24.9247-9261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Deng J, Wang L, Lee H, Armstrong B, Scuto A, et al. S1PR1 is an effective target to block STAT3 signaling in activated B cell-like diffuse large B-cell lymphoma. Blood. 2012 Aug 16;120(7):1458–65. doi: 10.1182/blood-2011-12-399030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.