Abstract
For most living organisms, iron is both essential and potentially toxic, making the precise maintenance of iron homeostasis necessary for survival. To manage this paradox, bacteria regulate the acquisition, utilization, and storage of iron in response to its availability. The iron-dependent ferric uptake repressor (Fur) often mediates this iron-responsive regulation, by both direct and indirect mechanisms. In 2002, Masse and Gottesman identified a novel target of Fur-mediated regulation in Escherichia coli: a gene encoding a small regulatory RNA (sRNA) termed RyhB. Under conditions of iron-limitation, RyhB is produced and functions to regulate the expression of several target genes encoding iron-utilizing enzymes, iron acquisition systems, and iron storage factors. This pivotal finding provided the missing link between environmental iron-limitation and previously observed decreases in certain iron-dependent metabolic pathways, a phenomenon now referred to as an “iron-sparing” response. The discovery of RyhB opened the door to the rapidly expanding field of bacterial iron-regulated sRNAs, which continue to be identified and described in numerous bacterial species. Most striking are findings that the impact of iron-responsive sRNA regulation often extends beyond iron homeostasis, particularly with regard to production of virulence-associated factors by pathogenic bacteria. This review discusses trends in the collective body of work on iron-regulated sRNAs, highlighting both the regulatory mechanisms they utilize to control target gene expression and the impact of this regulation on basic processes controlling bacterial physiology and virulence.
1. Introduction
Due to its abundance and broad potential for mediating redox chemistry, iron participates in an array of biological functions and is essential for the survival of most organisms. In anaerobic environments, reduced and soluble ferrous iron is easily attainable by bacteria. In contrast, the insolubility of ferric iron found in aerobic environments limits its bioavailability, presenting a substantial barrier to aerobically growing bacteria that require this element for survival. Pathogenic bacteria face an additional challenge in acquiring iron during the course of an infection. The level of available iron within an animal host is maintained at exceedingly low concentrations by sequestration of the element within iron-binding, host-associated proteins, such as hemoglobin, myoglobin, transferrins and ferritin.1 To overcome this significant nutritional barrier, microbes have evolved to produce a variety of high-affinity iron uptake systems that are essential for survival and pathogenesis of a broad range of bacterial species.2
Despite its essentiality, iron can promote the formation of reactive oxygen species during aerobic metabolism, threatening the survival of the bacterium. To manage their dichotomous relationship with iron, bacterial cells tightly regulate the production and activity of iron uptake and storage systems in response to intra-cellular iron concentrations.2b Iron-responsive regulation of bacterial gene regulation is often mediated by Fur (ferric uptake repressor), an iron-dependent transcriptional repressor. Specifically, under iron-replete conditions, ferrated Fur binds in a sequence-specific manner within the promoter region of target genes, efficiently preventing their expression.3 Under iron-poor conditions, Fur-mediated repression is relieved, thus allowing for the expression of Fur-repressed genes.4 Fur directly influences the expression of a variety of genes, including those for iron-acquisition and virulence.5
Several groups have also observed that Fur mediates an iron-dependent increase in the expression of certain E. coli genes, yet the regulatory mechanism underlying this apparent activation remained elusive for several years.6 In 2002, Masse and Gottesman identified a Fur-regulated sRNA, termed RyhB, and demonstrated that this sRNA was responsible for many of these effects. This seminal finding paved the way for the identification of additionally iron-regulated sRNAs, and also expanded our appreciation for the significant contributions of sRNAs to bacterial physiology and virulence. While E. coli RyhB remains the most thoroughly studied example of this growing family of sRNAs, researchers have made large strides in understanding how similar sRNAs in other bacteria contribute to gene regulation and iron homeostasis. These studies have revealed considerable differences in sequence, structure, regulatory mechanisms and targets of these sRNAs, yet some common themes have emerged. A recent review by Salvail and Masse focused on the integral role of RyhB and its homologs in iron homeostasis.7 Here, we sought to highlight how the variations in function of these RNAs are intimately linked to the lifestyle of the bacteria that encode them. In spite of their diverse roles, each of these sRNAs function to redirect the cell’s resources during iron-depleted growth and affect a remarkably broad range of cellular processes.
2. The RyhB Paradigm
Appreciation of the significant contribution that iron-responsive sRNAs make to multiple complex regulatory networks began with the characterization of E. coli RyhB.8 Initial characterization of this conserved 90-nucleotide (nt) long sRNA revealed that RyhB functions to coordinate the expression of several genes in response to iron availability. Specifically, under iron-rich conditions, ferric-Fur efficiently represses RyhB production, allowing for increased expression of RyhB-repressed genes (Fig. 1). Under iron-poor conditions, Fur-mediated repression of RyhB is relieved and the sRNA functions to repress the expression of several gene targets. E. coli genes subjected to RyhB-dependent repression include those encoding iron storage proteins (ftnA and bfr), enzymes of the tricarboxylic acid cycle (acnA and fumA), succinate dehydrogenase (sdh operon) and superoxide dismutase (sodB).8 Such regulation ensures the production of iron-storage proteins and iron-containing enzymes under iron-replete conditions, when their function is most advantageous to the survival of the bacterium. Conversely, RyhB prevents the production of such factors under iron-poor conditions, when their function is not essential (Fig. 1). By inhibiting the production of iron-binding proteins during growth under iron-limited conditions, RyhB regulation ensures iron is “spared” for use in essential pathways.
Fig. 1.
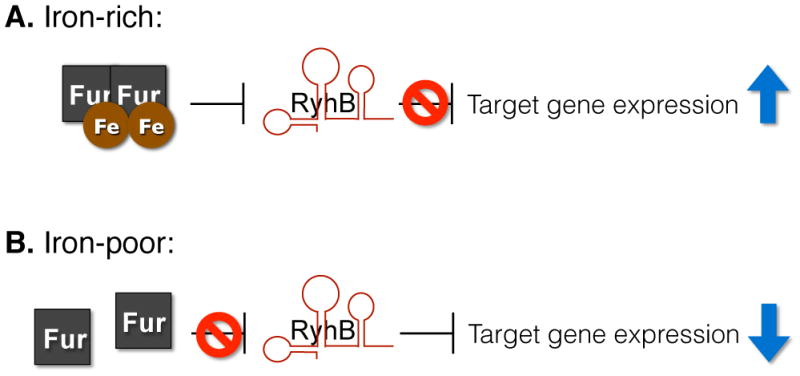
Schematic of RyhB-mediated repression of gene expression. A. In iron-rich environments, RyhB expression is repressed by the iron-bound Fur protein, resulting in derepression of target genes. B. In iron-poor conditions, Fur repression is relieved and RyhB is expressed, leading to decreased expression of specific target genes.
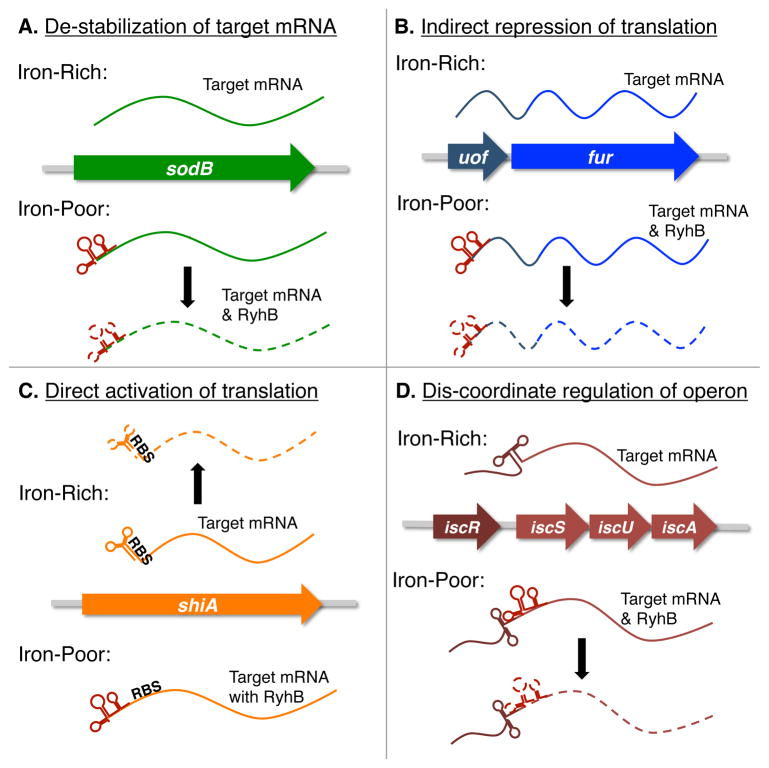
Initial investigations into the molecular mechanism underlying RyhB-mediated regulation focused primarily on the regulation of sodB, a gene encoding the superoxide dismutase enzyme. These studies demonstrated that efficient RyhB-dependent repression of sodB expression requires all of the following: nucleic acid complementarity between RyhB and the sodB transcript, the RNA-binding protein Hfq, and the RNA degrading enzymes RNaseE and RNase III.8–9 Specifically, nucleic acid complementarity and the activity of Hfq mediate the direct binding of RyhB to the sodB transcript.8–9 Following this specific interaction, the sodB mRNA molecule and RyhB are rapidly degraded in a process mediated by RNaseE and RNase III (Fig. 2A).9a, 9c Together these studies provided the first detailed evaluation of the molecular mechanism underlying RyhB-dependent repression of a specific gene target.
Fig. 2.
E. coli RyhB controls the expression of target genes via multiple regulatory mechanisms. Presented are simplified schematics of the molecular mechanisms governing RyhB-dependent regulation of target gene expression in E. coli. Only the relevant RNA molecules are depicted in each panel, and each are presented as a dashed line under conditions in which they are destabilized. RyhB is red in each panel.
Subsequent studies of E. coli RyhB have revealed additional regulatory targets of the sRNA, many of which appear to be controlled by the regulatory mechanism detailed for the control of sodB expression.10 However, other targets of RyhB-dependent regulation are controlled by unique regulatory mechanisms, summarized in Fig. 2. The impact of RyhB on E. coli physiology, as well as the variety of molecular mechanisms by which this regulation is achieved, is described below with the discussion of select targets of RyhB-dependent regulation.
Indirect regulation of fur translation
RyhB-dependent regulation of fur expression is mediated by a fascinating, novel molecular mechanism. Weak auto-regulation of fur expression allows for a basal level of Fur synthesis during growth of E. coli under iron-rich conditions.11 Under low iron conditions, auto-regulation by the Fur protein is relieved, yet fur transcript levels remain relatively unchanged.9b This paradoxical observation is explained by the fact that fur is co-transcribed with a small open reading frame termed uof, and that RyhB binds within this region of the transcript and negatively impacts the stability of this polycistronic message (Fig. 2B).9b Such regulation ensures that E. coli produces a relatively constant level of Fur during growth under both high and low iron conditions. In addition, these studies provide the first example of indirect translational regulation mediated by a trans-acting bacterial sRNA molecule.
Activation of shiA expression
Studies by Prevost, et al, implicate RyhB in directly activating the expression of shiA, a gene encoding a permease for the uptake of shikimate.12 Shikimate is utilized by E. coli for the synthesis siderophores, secreted iron scavenging molecules that facilitate bacterial survival in low iron environments.13 Specifically, RyhB binds to nucleic acid sequences within the 5′ untranslated region of shiA, and by doing so prevents the formation of an inhibitory structure that otherwise occludes the ribosomal binding site within the shiA transcript (Fig. 2C).12 Such regulation implicates RyhB in controlling the production of siderophores that promote growth under iron-limiting conditions. Additionally, this study was the first to show direct activation of target gene expression by E. coli RyhB.
Dis-coordinate regulation of genes within the polycistronic iscRSUA transcript
Two different studies have shown that RyhB mediates differential control over the expression of genes within the polycistronic icsRSUA transcript, encoding machinery required for the synthesis of Fe-S clusters.10, 14 This unique regulatory mechanism is governed by binding of RyhB to complementary sequences over-lapping the ribosomal binding site of iscS, an interaction that results in degradation of the portion of the message encoding the Fe-S cluster synthesis machinery (iscSUA, Fig. 2D). The icsR portion of the message, encoding a Fe-S responsive transcriptional regulator, is protected from RyhB-dependent degradation by the presence of a strong RNA structure within the transcript. Such regulation prevents the production of Fe-S cluster synthesis machinery in the absence of available iron, while maintaining production of the transcriptional regulator that facilitates survival under conditions of iron-limitation. In addition to expanding the reach of RyhB into regulatory pathways directly controlling the biosynthesis of Fe-S clusters, these studies were the first to document differential degradation of a polycistronic transcript mediated by a bacterial sRNA molecule.14
Collectively, these studies highlight RyhB’s influence over several key aspects of E. coli physiology by a variety of mechanisms (Fig. 2). Through these diverse mechanisms, RyhB exerts control over every facet of iron homeostasis in E. coli. Moving forward with an appreciation for the breadth of RyhB regulatory mechanisms (and the potential of others yet to be characterized) may well facilitate the identification of additional regulatory targets of RyhB, as well as similar iron-regulated sRNAs encoded by a broad range of bacterial species.
3. RyhB homologs in pathogenic bacteria
The first publication characterizing E. coli RyhB noted that the gene was conserved in several bacterial genera, including Salmonella, Klebsiella, Yersinia and Vibrio8. Subsequent studies have identified RyhB in these and other species of bacteria, shown in Table 1. In this section we present the major findings of the past decade of RyhB research, which highlight the role of this conserved sRNA in multiple pathogenic relatives of E. coli. Combined, these studies demonstrate how RyhB affects a wide variety of important, and often virulence-associated, processes within a collection of closely related pathogens, each employing unique life-styles and virulence strategies. For the purpose of this review, an sRNA will be considered to be a RyhB homolog if (1) the molecule contains no less than 55% sequence identify to E. coli RyhB and (2) production of the molecule is regulated in response to iron availability. If the RyhB homolog is referred to by more than one name within published literature, both names are noted.
Table 1.
Iron-responsive sRNA regulation in bacteria
| Bacterial Species | RNAReference | Affected Functions |
|---|---|---|
| Escherichia coli | RyhB8 | Iron storage; oxidative stress protection; metabolism; siderophore biosynthesis; Fur |
| Vibrio cholerae | RyhB19 | Iron storage; biofilm formation; chemotaxis; OmpT |
| Shigella flexneri | RyhB15 | Iron storage; oxidative stress protection; acid resistance |
| Shigella dysenteriae | RyhB16 | Iron storage; oxidative stress protection; virulence |
| Salmonella enterica serovar Typhimurium | RfrA/RyhB1, RfrB/RyhB220–21 | Oxidative stress protection; virulence |
| Klebsiella pneumonia | RyhB1, RyhB224–25 | None identified |
| Yersinia pestis biovar microtus | RyhB1, RyhB226 | None identified |
| Pseudomonas aeruginosa | PrrF1, PrrF227–28 | Iron storage; oxidative stress protection; metabolism; quorum sensing (PQS) |
| PrrH32a | Heme biosynthesis | |
| Neisseria meningitidis | NrrF38 | Succinate dehydrogenase |
| Neisseria gonorrhea | NrrF39 | None identified |
| Azotobacter vinelandii | ArrF33–34 | Oxidative stress protection; carbon storage; nitrogen fixation |
| Bacillus subtilis | FsrA41–42 | Metabolism; amino acid biosynthesis |
| Vibrio anguillarum | RNAα50 | Iron acquisition |
| Synechocystis | IsrR54 | Photosynthesis |
| Anabaena | α-furA58 | Photosynthesis; iron uptake |
Shigella species RyhB regulates virulence-associated traits
Not long after the discovery of RyhB, homologs were identified in several pathogens, including the closely related species of Shigella flexneri15 and Shigella dysenteriae,16 both of which cause bacillary dysentery. Given that RyhB of Shigella and E. coli are identical in nucleic acid sequence, it is not surprising that the regulatory targets of each overlap extensively.15–16 There are however, two notable exceptions to this rule, and each highlights the impact that RyhB has on virulence of Shigella species. Studies in S. flexneri have demonstrated that RyhB represses the expression of ydeP, a gene encoding a conserved, putative oxidoreductase required for the virulence-associated phenotype of extreme acid resistance. This regulation appears to occur via decreased expression of evgA, the upstream transcriptional regulator of ydeP and other genes required for extreme acid resistance.15 As a result of this regulation, the molecular machinery required to survive extreme acid stress is optimally produced by S. flexneri during growth under iron-rich conditions, when Fur actively inhibits the synthesis of RyhB. Such regulation should be advantageous for Shigella within the extremely low pH environment of the stomach, where ferric iron is soluble and therefore more bio-available. These findings indicate that RyhB promotes S. flexneri virulence by facilitating survival of the pathogen during transit through the stomach, en route to the colon where infection becomes established.
Studies in S. dysenteriae have demonstrated that RyhB represses the expression of virB, a gene encoding the transcriptional activator of several virulence-associated genes.16 RyhB-mediated repression of virB expression results in the inhibition of virulence-associated phenotypes due to decreased expression of genes encoding the type three secretion system that is required for eukaryotic cell invasion, and IcsP, a protease required for actin-based motility.16–17 Although the precise regulatory mechanism remains elusive, it is clear that RyhB represses the transcription of virB and that this regulation is independent of the upstream transcriptional activator VirF.18 Importantly, RyhB-mediated regulation of virB expression provides an example of a conserved, chromosomally-encoded sRNA molecule controlling the production of a genus specific, horizontally acquired virulence-determinant.
Acid resistance, eukaryotic cell invasion and cell-to-cell spread are all important virulence-associated processes that contribute to the success of Shigella species as primary pathogens and, remarkably, are all influenced by the activity of RyhB.
Vibrio cholerae RyhB regulates motility, chemotaxis, and bio-film formation
RyhB is less well conserved between E. coli and Vibrio cholerae, an environmental microbe and the causative agent of cholera.8 Two separate studies have demonstrated that in addition to a relatively conserved core sequence, V. cholerae RyhB possesses an additional 69 and 54 nucleotides at the 5′ and 3′ ends of the RNA, respectively.19 Despite these significant differences, V. cholerae RyhB represses expression of many of the same genes as E. coli RyhB, including sodB (superoxide dismutase), sdhCDAB (succinate dehydrogenase), fum (fumarase), and acnA (aconitase).19 In addition to these conserved targets, V. cholerae RyhB modulates the expression of several genes that facilitate the unique life-style of this pathogen, including those controlling motility, chemotaxis and biofilm formation.19 For these reasons, while RyhB is not required by V. cholerae for colonization in an infant mouse model of infection,19 it is reasonable to conclude that RyhB is important for environmental survival of this pathogen.
Salmonella enterica encodes two differentially-regulated homologs of RyhB
As predicted by Masse and Gottesman.8 Salmonella encodes two RyhB homologs, originally termed RfrA and RfrB.20 RfrA shares 82% sequence identity with E. coli RyhB and is encoded in the same genetic context as RyhB, while RfrB shares only 53% sequence identity with RfrA and is encoded within a Salmonella specific genetic locus.20–21 While the production of both sRNAs is influenced in response to iron-availability by the activity of Fur, production of RfrB is primarily regulated by the activity of the stationary phase sigma factor RpoS.20, 22 Recent analysis of the regulatory targets of RfrA and RfrB, in this study termed RyhB-1 and RyhB-2, reveals that they each influence the expression of a set of genes that is similar, but not identical, to that regulated by E. coli RyhB.22 Moreover, RfrA and RfrB play overlapping yet distinct roles in protecting against oxidative stress, bactericidal antibiotics and acid resistance, as inactivation of each has varying effects on these virulence-associated phenotypes.22 Additionally, both RfrA and RfrB appear to influence the survival of Salmonella within the unique environments of a eukaryotic epithelial cells and macrophages.20, 23 Combined, these studies suggest that Salmonella RfrA and RfrB are produced during growth under unique environmental conditions and have overlapping yet distinct roles in regulating Salmonella physiology and virulence-associated phenotypes.
Klebsiella pneumoniae RyhB regulates capsular biosynthesis genes
Recent studies show RyhB modulates two important virulence-associated phenotypes in K. pneumoniae: biosynthesis of capsular polysaccharide and iron acquisition.24 Increased production of RyhB, as would be seen under iron-limiting conditions (Fig. 1), is associated with increased capsular polysaccharide synthesis and increased expression of multiple genes encoding components of three separate iron acquisition systems.24 Such regulation coordinates the production of capsular polysaccharide and iron acquisition systems, two factors required for survival within the human host, to growth within the iron-poor environments endured by the pathogen during the course of a natural infection. Of note, a recent study suggests that, like S. enterica and Y. pestis, K. pneumoniae encodes two RyhB homologs.25 The full extent to which each RyhB homolog influences K. pneumoniae physiology and virulence is yet to be determined.
Yersinia pestis encodes Hfq-dependent and Hfq-independent RyhB homologs
As originally predicted by Masse and Gottesman,8 Y. pestis encodes two RyhB homologs, designated RyhB1 and RyhB2.26 Studies have demonstrated that while Fur regulates the production of both RyhB1 and RyhB2 in response to iron-availability, Hfq differentially influences the stability of each.26 As observed in E. coli,9a Hfq stabilizes RyhB1, but surprisingly has no effect on the stability of RyhB2.9a, 26 The insensitivity of RyhB2 to Hfq is a feature that makes this molecule unique among RyhB homologs characterized to date. While much is left to learn about the role of RyhB1 and RyhB2 in controlling Y. pestis physiology, the inaugural investigation demonstrates that production of each sRNA is higher following growth of the pathogen in a mouse lung as compared to that measured following in vitro growth, an observation that suggests that the sRNAs may facilitate the progression of pneumonic plague in mice.26
4. Functional analogs of RyhB
Several bacteria encode for iron-responsive sRNAs that mediate a similar iron-sparing response to RyhB, yet share no significant homology with this paradigmatic sRNA. While these divergent sRNAs lack homology to RyhB, they are considered functional analogs, as (1) the production of each is repressed in response to iron via Fur, and (2) they function to regulate a similar set of genes as RyhB. As described below, these sRNAs play a significant role in modulating the iron-sparing response of the bacteria that encode them, while also controlling several other key biological processes.
The duplicated PrrF RNAs of Pseudomonas aeruginosa
Shortly after the identification of RyhB, the Fur-regulated PrrF (Pseudomonas RNA responsive to Fe) sRNAs, PrrF1 and PrrF2, were identified in the opportunistic Gram-negative pathogen Pseudomonas aeruginosa.27 While sharing no significant sequence homology with RyhB, the PrrF RNAs contribute to an analogous iron-sparing response via the repression of a similar set of gene targets.27–28 In addition to controlling the expression of regulatory targets shared with E. coli RyhB, PrrF affects the production of several factors involved in a variety of metabolic pathways, including one for the degradation of anthranilate.28 Anthranilate is a precursor for the Pseudomonas quinolone signal (PQS) quorum sensing molecule,29 and deletion of the genes encoding PrrF1 and PrrF2 results in loss of PQS production due to the constitutive degradation of anthranilate (Fig. 3).28 Thus, the PrrF RNAs spare anthranilate for PQS production during iron-restricted growth. PQS can also induce synthesis of pyoverdine, the primary siderophore of P. aeruginosa, via PQS’s ability to chelate and sequester extracellular ferric iron (Fig. 3).30 These findings have led to the proposal that PQS functions in extracellular iron entrapment to facilitate pyoverdine-mediated iron acquisition by P. aeruginosa. As such, these findings suggest a role for PrrF1 and PrrF2 in controlling iron acquisition by P. aeruginosa. Furthermore, as a quorum sensing molecule PQS is proposed to play an important role in virulence,31 suggesting these duplicated sRNAs may directly impact P. aeruginosa virulence.
Fig. 3.
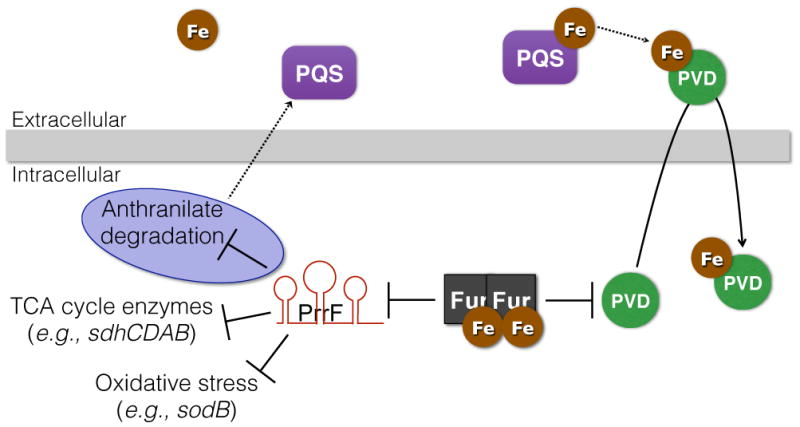
Putative role of PrrF sRNAs in pyoverdine-mediated iron uptake by P. aeruginosa. When bound to iron, the Fur protein represses expression of genes for synthesis of the pyoverdine (PVD) siderophore, which has a very high affinity for ferric iron. Fur also represses expression of the PrrF1 and PrrF2 sRNAs. The PrrF sRNAs themselves regulate a number of cell processes, most notably TCA cycle enzymes and factors that protect against oxidative stress. The PrrF sRNAs also contribute to production of the Pseudomonas quinolone signal (PQS) in low iron conditions by blocking the degradation of a vital precursor (anthranilate - regulation highlighted in purple). Additionally, PQS may enhance PVD production by chelating extracellular iron, which is subsequently scavenged by the PVD siderophore.
Similar to what was described for certain RyhB-encoding pathogens above, P. aeruginosa species encode for two PrrF RNAs. Pseudomonas presents an interesting twist on the duplicated RNA theme, however, in that the PrrF RNAs in P. aeruginosa are encoded in tandem by two virtually identical genes (93% identity): prrF1 and prrF2. Each PrrF RNA appears to be capable of nearly complete repression of most of its target mRNAs, as genetic deletion of both the prrF1 and prrF2 genes is required for significant derepression of target gene expression in iron-depleted growth conditions.27–28 The prrF region encodes for an additional, heme-responsive 325-nt sRNA, designated PrrH, providing a likely rationale for the duplicated prrF genes.32 Transcription of PrrH initiates at the 5′ end of prrF1, proceeds through the prrF1-prrF2 intergenic sequence (95 nt), and terminates at the 3′ end of prrF2. Thus, expression of prrH is dependent on read-through transcription at the prrF1 Rho-independent, or intrinsic, terminator. PrrH regulates genes involved in heme biosynthesis,32a suggesting this tandem prrF organization imparts unique heme-responsive regulatory activities to P. aeruginosa. The advantage that this tandem arrangement imparts to P. aeruginosa, as compared to other Pseudomonads where prrF1 and prrF2 are encoded distally, is unknown. However, it may play a role in P. aeruginosa’s unique ability to cause disease in humans, as heme is an abundant source of iron encountered during infection.
Azotobacter iron-sparing response is mediated by ArrF
Azotobacter vinelandii is a non-pathogenic, nitrogen-fixing bacteria and close relative of P. aeruginosa. Similar to P. aeruginosa, A. vinelandii requires an abundance of iron for respiratory pathways, and displays a similar iron-sparing response mediated by an iron-regulated sRNA. A. vinelandii encodes a single homolog of PrrF, termed ArrF, which shares considerable sequence homology with both PrrF1 and PrrF2 RNAs (81% and 80% respectively) of P. aeruginosa.33 Like previously described iron-regulated sRNAs, ArrF represses the expression of sodB, likely via binding of the sRNA to the target RNA molecule within a region of nucleic acid complementary.33 Unlike PrrF and RyhB however, ArrF does not appear to influence the expression of the genes encoding TCA cycle enzymes. The lack of repression of TCA cycle enzymes is likely a consequence of that fact that A. vinelandii is an obligate aerobe. As such, enzymes in the TCA cycle are essential during growth of the organism under any condition, regardless of iron availability. To maintain iron stores for aerobic respiration, ArrF represses expression of the feSII gene,33 encoding a complex that protects the iron-loaded nitrogenase enzyme required for nitrogen fixation, a non-essential function. ArrF additionally represses expression of the phbBAC operon,34 which converts acetyl-CoA to the carbon storage molecule poly-β-hydroxybutyrate. As such, ArrF spares the use of acetyl-CoA for metabolism via the TCA cycle during growth in iron-poor conditions. These variations in iron-responsive sRNA targets provide a clear example of how two closely related bacteria have evolved to utilize highly similar regulatory molecules, such as PrrF/ArrF, to uniquely promote the fitness of each organism.
Iron-sparing response of Neisseria is only partially due to NrrF
Neisseria meningitidis, a Gram-negative diplococci that causes meningitis epidemics in infants and young adults,35 also displays an iron sparing response, with Fur exerting a positive effect on the expression of several genes.36 In some cases this regulation occurs directly via binding of Fur to nucleic acid sequences within the operator regions of the activated genes.37 However, N. meningitidis also encodes a 169-nt Fur-repressed sRNA, NrrF, that is responsible for the Fur-dependent activation of the genes encoding succinate dehydrogenase.38 Despite sharing very little homology with either PrrF or RyhB, NrrF regulates succinate dehydrogenase in an Hfq-dependent manner similar, yet not identical, to that described for RyhB.38b Interestingly, Hfq is also required for the Fur-dependent activation of sodB and fumB, yet these genes are unaffected by deletion of nrrF,38b suggesting that a separate, as of yet unidentified sRNA is responsible for the repression of these regulatory targets. Sequence for a potential NrrF homolog sharing 93% identity with N. meningitidis NrrF and 195 nt in size has been identified in the related pathogen Neisseria gonorrhoeae,39 although no targets have yet been characterized for this sRNA. Also yet to be determined is the role that the NrrF sRNA plays in controlling the expression of virulence-related gene expression and/or the production of virulence factors in each of these bacterial pathogens.
Iron-responsive RNAs in Gram-positive bacteria (Bacillus subtilis FsrA and Fbp)
Fur homologs and iron sparing responses have been identified and characterized in several Gram-positive organisms, including Bacillus subtilis.40 In particular, the B. subtilis iron-sparing response is dependent on Fur-regulated expression of the 84-nt sRNA FsrA (Iron [Fe]-responsive sRNA).41 Similar to RyhB, FsrA negatively affects the expression of genes encoding iron-containing TCA cycle enzymes (succinate dehydrogenase - sdhABC; aconitase - citB), as well as related metabolic enzymes involved in amino acid biosynthesis (glutamate synthase - gltAB; isopropyl malate dehydrogenase - leuCD), a dicarboxylate permease important for succinate and fumarate utilization (dctP), and three iron-sulfur containing lactate utilization genes (lutABC). An intriguing component of the FsrA regulatory paradigm is the involvement of three small basic proteins, encoded by two distinct transcriptional units (fbpAB and fbpC), which are hypothesized to chaperone the FsrA RNA.41b The requirement of the Fbp proteins for FsrA regulation is not clearly understood, and appears to vary with FsrA regulatory target (Fig. 4). For example, FsrA repression of the lutABC genes for lactate utilization specifically requires the FbpB protein,42 while repression of leuCD is dependent on FbpC.41b As such, the Fbp proteins are proposed to function as chaperones that direct FsrA activity to mediate iron homeostasis in B. subtilis.
Fig. 4.
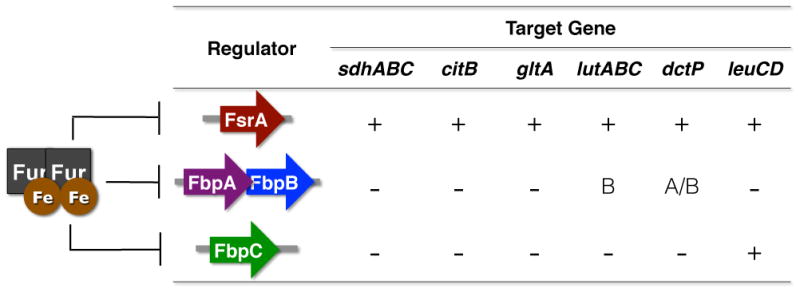
FsrA sRNA and Fbp proteins mediate iron sparing response in B. subtilis. The FsrA RNA is expressed under iron-poor conditions, when iron-dependent Fur repression is relieved. FsrA in turn represses several target genes, including the six examples shown in this figure. Fur also represses expression of three small basic proteins, FbpAB and FbpC; the fsrA, fbpAB, and fbpC transcripts are unlinked on the chromosome. The Fbp proteins make variable contributions to FsrA regulation, as indicated by the − and + signs. For one fbpAB-dependent target, lutABC, FbpB specifically has been shown to be required for FsrA-mediated regulation.
Several exceptions to this chaperone model exist, however, including the repression of sdhCDAB, citB, and gltAB by FsrA, which is not dependent on either of the Fbp proteins.41b Neither does Hfq appear to have a role in FsrA repression of sdhCDAB,41b presenting the possibility that FsrA regulation of some targets is independent of chaperone activity. The authors postulate that extensive sequence complementarity between sdhC and fsrA allows this regulation to occur in the absence of chaperone. Alternatively, a constitutively expressed chaperone, similar to the Fbp’s, may function as a FsrA chaperone for this message. Further, the Fbp proteins appear to regulate gene expression independent of FsrA, a trait reminiscent of the Hfq protein in Gram-negative bacteria. Studies into the structural basis for FsrA, FbpAB and FbpC regulation will undoubtedly be required to understand the contribution of these different RNA chaperones in the iron sparing response. The genes encoding the FsrA RNA and Fbp proteins have been identified in two other Bacillus species – Bacillus amyloquefaciens and Bacillus licheniformis, however, neither their expression nor their activity have been validated. It will also be of interest to determine if these or analogous RNAs are present in other Gram-positive bacteria that exhibit the typical Fur-dependent iron-sparing response, including Staphylococcus aureus43 and Listeria monocytogenes.44 In particular, it will be interesting to determine if the iron-sparing responses regulate genes that contribute to virulence, as has been demonstrated for many Gram-negatives that encode for iron-regulated sRNAs.
While continued research will undoubtedly add to the growing number of iron-regulated sRNAs, it is important to point out that some bacteria appear to carry out iron-sparing responses independent of sRNAs. In Helicobacter pylori, for example, Fur directly activates sodB expression.45 Although not required for iron-dependent regulation of sodB expression, numerous sRNAs have been identified in H. pylori,46 and the role these regulators play in iron-responsive gene regulation remains to be seen. Additionally, it is not yet known if bacteria utilize sRNAs to respond to other biologically important metals, such as manganese. This is a particularly interesting question with regard to the α-proteobacteria, which respond to manganese instead of iron via a Fur homolog (Mur).47
5. cis-antisense RNAs regulating iron homeostasis
Even before the identification of RyhB, the iron-responsive cis-encoded antisense RNAα was identified in the fish pathogen Vibrio anguillarum.48 RNAα is transcribed from the noncoding strand of the fatABCD operon, which encodes a transport system for the siderophore anguibactin.49 As such, the 650-nucleotide RNAα is complementary to almost two-thirds of the fatB gene.48a While binding of RNAα to the fatABCD operon has not yet been experimentally demonstrated, expression of RNAα correlates closely with loss of FatA translation.48a, 50 In contrast to RyhB and its homologs, however, the Fur protein promotes transcription initiation of RNAα in an iron-independent manner, and RNAα is posttranscriptionally stabilized under iron-replete conditions.50 The mechanism of this regulation is unknown, and is likely distinct from earlier mechanisms described in this review; yet the outcome is reminiscent of the effect of E. coli RyhB on siderophore biosynthesis,12 and provides yet another example of how an iron-responsive RNA can mediate gene expression and iron homeostasis.
The Fur proteins of cyanobacteria control the expression of genes for iron uptake systems and oxidative stress, as well as photosynthesis.51 Photosynthesis, in particular, places a particularly high iron demand on the bacterium. To compensate for loss of light-harvesting systems under low iron conditions, cyanobacteria express isiA (iron stress-induced protein A), encoding a Fur-repressed photosystem (PS) I accessory protein.52 In Synechocystis, the IsiA protein forms an additional ring around the photosystem I complex, which enhances light absorption during periods of iron depletion to compensate for the reduction in the number of PSI complexes.53 The 177-nt IsiR RNA is transcribed constitutively from the non-coding strand of isiA and appears to bind to complementary portions of the mRNA, causing coupled degradation of the RNA hybrid.54 Thus, transcription of the isiA mRNA must outcompete that of IsiR in order to function as template for IsiA protein synthesis. This competitively-based RNA regulatory network is thought to ensure quick turnover of residual isiA mRNA when intracellular iron levels are restored.
Another intriguing cis-antisense regulatory scheme exists in the cyanobacterium Anabaena, which encodes three putative Fur homologs.55 Expression of the Anabaena FurA protein is repressed by the antisense α-FurA RNA under high iron conditions.56 Mutants defective for α-FurA RNA display increased levels of the FurA protein, and are disrupted for iron homeostasis, growth, and photosynthetic efficiency.57 Thus, the paradigmatic roles of Fur and regulatory RNA appear to be reversed in Anabaena, yet work to similarly affect the overall physiology of this cyanobacterium.
6. Closing remarks
The studies presented in this review have made significant contributions to the ever-expanding appreciation for the role that iron-regulated sRNAs play in influencing overall cellular physiology and virulence in a broad range of bacterial species. Although it is obvious that iron-regulated sRNAs are important regulators, continued investigation into the regulatory targets and molecular mechanisms of iron-regulated sRNAs will be needed to reveal the full impact of these regulators on bacterial physiology and virulence. These studies also demonstrate the diversity of regulatory mechanisms and mRNA targets by which iron-responsive sRNAs function, variations that are often linked to the unique lifestyles of the organisms that encode them. Thus, it is important that research in this area move forward with awareness for the possible exceptions to the RyhB paradigm. Future studies in this field will undoubtedly yield a more comprehensive understanding of the complex role of iron in bacterial physiology.
Acknowledgments
Research in the Oglesby-Sherrouse lab is funded by the University of Maryland School of Pharmacy and National Institutes of Health grant NIAID-1K22AI089776 - 01A1. Research in the Murphy lab is funded by Ohio University College of Osteopathic Medicine as well as the Ohio University Research Committee.
Contributor Information
Amanda G. Oglesby-Sherrouse, Email: aoglesby@rx.umaryland.edu.
Erin R. Murphy, Email: murphy@oucom.ohiou.edu.
References
- 1.(a) Otto BR, Verweijvan Vught AM, MacLaren DM. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Critical reviews in microbiology. 1992;18:217–33. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]; (b) Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron - a metal at the host-pathogen interface. Cellular microbiology. 2010;12:1691–702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- 2.(a) Payne SM. Iron acquisition in microbial pathogenesis. Trends in microbiology. 1993;1:66–9. doi: 10.1016/0966-842x(93)90036-q. [DOI] [PubMed] [Google Scholar]; (b) Braun V. Iron uptake mechanisms and their regulation in pathogenic bacteria. International journal of medical microbiology. IJMM. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]; (c) Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infection and immunity. 2010;78:4977–89. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands JB. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–7. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 4.Hantke K. Iron and metal regulation in bacteria. Current opinion in microbiology. 2001;4:172–7. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter BM, Whitmire JM, Merrell DS. This is not your mother’s repressor: the complex role of fur in pathogenesis. Infection and immunity. 2009;77:2590–601. doi: 10.1128/IAI.00116-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Molecular & general genetics : MGG. 1987;210:135–9. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]; (b) Dubrac S, Touati D. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. Journal of bacteriology. 2000;182:3802–8. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. Journal of bacteriology. 1995;177:2305–14. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Horsburgh MJ, Ingham E, Foster SJ. Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. Journal of bacteriology. 2001;183:468–75. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salvail H, Masse E. Regulating iron storage and metabolism with RNA: an overview of posttranscriptional controls of intracellular iron homeostasis. Wiley interdisciplinary reviews. RNA. 2012;3:26–36. doi: 10.1002/wrna.102. [DOI] [PubMed] [Google Scholar]
- 8.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4620–5. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets inEscherichia coli. Genes & development. 2003;17:2374–83. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vecerek B, Moll I, Blasi U. Control of Fur synthesis by the non-coding RNA RyhB and iron-responsive decoding. The EMBO journal. 2007;26:965–75. doi: 10.1038/sj.emboj.7601553. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic acids research. 2005;33:1678–89. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masse E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. Journal of bacteriology. 2005;187:6962–71. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Lorenzo V, Herrero M, Giovannini F, Neilands JB. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. European journal of biochemistry/FEBS. 1988;173:537–46. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 12.Prevost K, Salvail H, Desnoyers G, Jacques JF, Phaneuf E, Masse E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Molecular microbiology. 2007;64:1260–73. doi: 10.1111/j.1365-2958.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann KM, Weaver LM. The Shikimate Pathway. Annual review of plant physiology and plant molecular biology. 1999;50:473–503. doi: 10.1146/annurev.arplant.50.1.473. [DOI] [PubMed] [Google Scholar]
- 14.Desnoyers G, Morissette A, Prevost K, Masse E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. The EMBO journal. 2009;28:1551–61. doi: 10.1038/emboj.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oglesby AG, Murphy ER, Iyer VR, Payne SM. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Molecular microbiology. 2005;58:1354–67. doi: 10.1111/j.1365-2958.2005.04920.x. [DOI] [PubMed] [Google Scholar]
- 16.Murphy ER, Payne SM. RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infection and immunity. 2007;75:3470–7. doi: 10.1128/IAI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Africa LA, Murphy ER, Egan NR, Wigley AF, Wing HJ. The iron-responsive Fur/RyhB regulatory cascade modulates the Shigella outer membrane protease IcsP. Infection and immunity. 2011;79:4543–9. doi: 10.1128/IAI.05340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broach WH, Egan N, Wing HJ, Payne SM, Murphy ER. VirF-independent regulation of Shigella virB transcription is mediated by the small RNA RyhB. PLoS ONE. 2012;7:e38592. doi: 10.1371/journal.pone.0038592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infection and immunity. 2005;73:5706–19. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. Journal of bacteriology. 2005;187:4005–14. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic acids research. 2008;36:1913–27. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellermeier JR, Slauch JM. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. Journal of bacteriology. 2008;190:476–86. doi: 10.1128/JB.00926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JN, Kwon YM. Genetic and phenotypic characterization of the RyhB regulon in Salmonella typhimurium. Microbiological research. 2012 doi: 10.1016/j.micres.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Ortega A, Gonzalo-Asensio J, Garcia-Del Portillo F. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA biology. 2012;9:469–88. doi: 10.4161/rna.19317. [DOI] [PubMed] [Google Scholar]
- 24.Huang SH, Wang CK, Peng HL, Wu CC, Chen YT, Hong YM, Lin CT. Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC microbiology. 2012;12:148. doi: 10.1186/1471-2180-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Hong JS, Qiu Y, Nagarajan H, Seo JH, Cho BK, Tsai SF, Palsson BO. Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS genetics. 2012;8:e1002867. doi: 10.1371/journal.pgen.1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z, Meng X, Su S, Liu Z, Ji X, Zhang Y, Zhao X, Wang X, Yang R, Han Y. Two sRNA RyhB homologs from Yersinia pestis biovar microtus expressed in vivo have differential Hfq-dependent stability. Research in microbiology. 2012;163:413–8. doi: 10.1016/j.resmic.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9792–7. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oglesby AG, Farrow JM, 3rd, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. The Journal of biological chemistry. 2008;283:15558–67. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrow JM, 3rd, Pesci EC. Two distinct pathways supply anthranilate as a precursor of the Pseudomonas quinolone signal. Journal of bacteriology. 2007;189:3425–33. doi: 10.1128/JB.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chemistry & biology. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]; (b) Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environmental microbiology. 2006;8:1318–29. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 31.(a) Calfee MW, Coleman JP, Pesci EC. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11633–7. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, Schwab U, Gilligan P, Pesci EC. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS microbiology letters. 2002;215:41–6. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- 32.(a) Oglesby-Sherrouse AG, Vasil ML. Characterization of a heme-regulated non-coding RNA encoded by the prrF locus of Pseudomonas aeruginosa. PLoS ONE. 2010;5:e9930. doi: 10.1371/journal.pone.0009930. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt 1):185–98. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 33.Jung YS, Kwon YM. Small RNA ArrF regulates the expression of sodB and feSII genes in Azotobacter vinelandii. Current microbiology. 2008;57:593–7. doi: 10.1007/s00284-008-9248-z. [DOI] [PubMed] [Google Scholar]
- 34.(a) Pyla R, Kim TJ, Silva JL, Jung YS. Proteome analysis of Azotobacter vinelandii arrF mutant that overproduces poly-beta-hydroxybutyrate polymer. Applied microbiology and biotechnology. 2010;88:1343–54. doi: 10.1007/s00253-010-2852-4. [DOI] [PubMed] [Google Scholar]; (b) Pyla R, Kim TJ, Silva JL, Jung YS. Overproduction of poly-beta-hydroxybutyrate in the Azotobacter vinelandii mutant that does not express small RNA ArrF. Applied microbiology and biotechnology. 2009;84:717–24. doi: 10.1007/s00253-009-2002-z. [DOI] [PubMed] [Google Scholar]
- 35.Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27(Suppl 2):B51–63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 36.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Molecular microbiology. 2004;52:1081–90. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 37.(a) Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, Rappuoli R, Grandi G, Genco CA. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9542–7. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shaik YB, Grogan S, Davey M, Sebastian S, Goswami S, Szmigielski B, Genco CA. Expression of the iron-activated nspA and secY genes in Neisseria meningitidis group B by Fur-dependent and -independent mechanisms. Journal of bacteriology. 2007;189:663–9. doi: 10.1128/JB.01638-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(a) Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. Journal of bacteriology. 2007;189:3686–94. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Metruccio MM, Fantappie L, Serruto D, Muzzi A, Roncarati D, Donati C, Scarlato V, Delany I. The Hfq-dependent small noncoding RNA NrrF directly mediates Fur-dependent positive regulation of succinate dehydrogenase in Neisseria meningitidis. Journal of bacteriology. 2009;191:1330–42. doi: 10.1128/JB.00849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ducey TF, Jackson L, Orvis J, Dyer DW. Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microbial pathogenesis. 2009;46:166–70. doi: 10.1016/j.micpath.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.(a) Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. Journal of bacteriology. 2006;188:3664–73. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fuangthong M, Helmann JD. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. Journal of bacteriology. 2003;185:6348–57. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.(a) Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, Helmann JD. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. Journal of bacteriology. 2012;194:2594–605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11927–32. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. Journal of bacteriology. 2012;194:2586–93. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedman DB, Stauff DL, Pishchany G, Whitwell CW, Torres VJ, Skaar EP. Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS pathogens. 2006;2:e87. doi: 10.1371/journal.ppat.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Applied and environmental microbiology. 2010;76:406–16. doi: 10.1128/AEM.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst FD, Homuth G, Stoof J, Mader U, Waidner B, Kuipers EJ, Kist M, Kusters JG, Bereswill S, van Vliet AH. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. Journal of bacteriology. 2005;187:3687–92. doi: 10.1128/JB.187.11.3687-3692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao B, Li W, Guo G, Li B, Liu Z, Jia K, Guo Y, Mao X, Zou Q. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Current microbiology. 2009;58:258–63. doi: 10.1007/s00284-008-9318-2. [DOI] [PubMed] [Google Scholar]
- 47.Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2007;20:501–11. doi: 10.1007/s10534-007-9085-8. [DOI] [PubMed] [Google Scholar]
- 48.(a) Salinas PC, Waldbeser LS, Crosa JH. Regulation of the expression of bacterial iron transport genes: possible role of an anti-sense RNA as a repressor. Gene. 1993;123:33–8. doi: 10.1016/0378-1119(93)90535-b. [DOI] [PubMed] [Google Scholar]; (b) Waldbeser LS, Chen Q, Crosa JH. Antisense RNA regulation of the fatB iron transport protein gene in Vibrio anguillarum. Molecular microbiology. 1995;17:747–56. doi: 10.1111/j.1365-2958.1995.mmi_17040747.x. [DOI] [PubMed] [Google Scholar]
- 49.Actis LA, Fish W, Crosa JH, Kellerman K, Ellenberger SR, Hauser FM, Sanders-Loehr J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1) Journal of bacteriology. 1986;167:57–65. doi: 10.1128/jb.167.1.57-65.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Q, Crosa JH. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. The Journal of biological chemistry. 1996;271:18885–91. doi: 10.1074/jbc.271.31.18885. [DOI] [PubMed] [Google Scholar]
- 51.Su Z, Olman V, Mao F, Xu Y. Comparative genomics analysis of NtcA regulons in cyanobacteria: regulation of nitrogen assimilation and its coupling to photosynthesis. Nucleic acids research. 2005;33:5156–71. doi: 10.1093/nar/gki817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.(a) Laudenbach DE, Straus NA. Characterization of a cyanobacterial iron stress-induced gene similar to psbC. Journal of bacteriology. 1988;170:5018–26. doi: 10.1128/jb.170.11.5018-5026.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Vinnemeier J, Kunert A, Hagemann M. Transcriptional analysis of the isiAB operon in salt-stressed cells of the cyanobacterium Synechocystis sp. PCC 6803. FEMS microbiology letters. 1998;169:323–30. doi: 10.1111/j.1574-6968.1998.tb13336.x. [DOI] [PubMed] [Google Scholar]
- 53.(a) Bibby TS, Nield J, Barber J. Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyano-bacteria. Nature. 2001;412:743–5. doi: 10.1038/35089098. [DOI] [PubMed] [Google Scholar]; (b) Boekema EJ, Hifney A, Yakushevska AE, Piotrowski M, Keegstra W, Berry S, Michel KP, Pistorius EK, Kruip J. A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature. 2001;412:745–8. doi: 10.1038/35089104. [DOI] [PubMed] [Google Scholar]
- 54.Duhring U, Axmann IM, Hess WR, Wilde A. An internal anti-sense RNA regulates expression of the photosynthesis gene isiA. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7054–8. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hernandez JA, Lopez-Gomollon S, Bes MT, Fillat MF, Peleato ML. Three fur homologues from Anabaena sp. PCC7120: exploring reciprocal protein-promoter recognition. FEMS microbiology letters. 2004;236:275–82. doi: 10.1016/j.femsle.2004.05.050. [DOI] [PubMed] [Google Scholar]
- 56.Hernandez JA, Pellicer S, Huang L, Peleato ML, Fillat MF. FurA modulates gene expression of alr3808, a DpsA homologue in Nostoc (Anabaena) sp. PCC7120. FEBS letters. 2007;581:1351–6. doi: 10.1016/j.febslet.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez JA, Alonso I, Pellicer S, Luisa Peleato M, Cases R, Strasser RJ, Barja F, Fillat MF. Mutants of Anabaena sp. PCC 7120 lacking alr1690 and alpha-furA antisense RNA show a pleiotropic phenotype and altered photosynthetic machinery. Journal of plant physiology. 2010;167:430–7. doi: 10.1016/j.jplph.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Hernandez JA, Muro-Pastor AM, Flores E, Bes MT, Peleato ML, Fillat MF. Identification of a furA cis antisense RNA in the cyanobacterium Anabaena sp. PCC 7120. Journal of molecular biology. 2006;355:325–34. doi: 10.1016/j.jmb.2005.10.079. [DOI] [PubMed] [Google Scholar]