Abstract
Human bone marrow-derived mesenchymal stem cell (hMSCs) function depends on chemical factors and also on the physical cues of the microenvironmental niche. Here, this physical microenvironment is recapitulated with controlled modes of mechanical strain applied to substrata containing three-dimensional features in order to analyze the effects on cell morphology, focal adhesion distribution, and gene expression. Ten percentage of strain at 1 Hz is delivered for 48 h to hMSCs cultured on flat surfaces, or on substrata with 15 μm-high microtopographic posts spaced 75 μm apart. Adding strain to microtopography produced stable semicircular focal adhesions, and actin spanning from post to post. Strain dominated over microtopography for expression of genes for the cytoskeleton (caldesmon-1 and calponin 3), cell adhesion (integrin-α2, vinculin, and paxillin), and extracellular matrix remodeling (MMP13) (p < 0.05). Overall, attention to external mechanical stimuli is necessary for optimizing the stem cell niche for regenerative medicine.
Keywords: Micromechanics, Microarray, Adhesion, Migration
1. Introduction
Stem cells are a major focus of regenerative medicine. Among the different types of stem cells, human bone-marrow-derived mesenchymal stem cells (hMSCs) are an attractive cell source since they differentiate into a variety of cell types, such as osteoblasts, adipocytes, chondrocytes, ligament, and smooth muscle cells. This regulation depends not only on chemical factors but also on the physical cues of the microenvironmental niche, which is defined as a combination of structural and cellular components that vary from one tissue to another to control proliferation and differentiation [1]. Nonetheless, how mechanical parameters in the physical microenvironment affect cellular function is not yet well understood.
Cells remodel in response to changing physical stresses and mechanical loads of the external environment that influence gene expression patterns during development [2–4]. Furthermore, tissues present a three-dimensional (3D) environment to cells. Therefore, in addition to mechanical stimuli to cells, it is desirable to mimic the 3D of the tissue by microtopography. Cellular processes affected by engineered microtopography in culture include cell adhesion, the subcellular cytoskeleton, and differentiation [5]. External perturbation of force occurs in vivo with shear stress, extension, or compression [6–8]. Forces generated by motor proteins can reorganize the cytoskeleton in response to external stiffness, surface topography, or ligand density [9,10]. In this study, the effects of the combination of both external cyclic strain and the impact of 3D microtopography on cells are studied on cytoskeletal organization, focal adhesions, and gene expression.
2. Materials and methods
2.1. Fabrication of microtopographic substrata
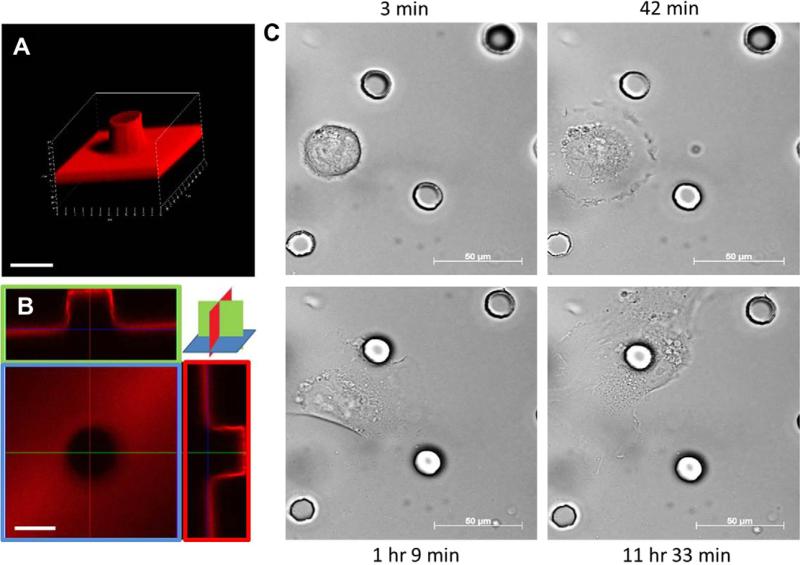
The ratio of curing agent to silicon elastomer base was 1:10, resulting in a Young's modulus of approximately 1.7 MPa. Liquid polydimethyl-siloxane (PDMS) was spread over the BioFlex plates, and the parylene mold was placed on top of the PDMS layer, which was about 1 mm thick, cured, and gently removed, resulting in flat or textured elastomeric membranes in the flex dishes. The fabrication of the parylene microtopography molds has been previously reported [11]. Flat two-dimensional sheets of PDMS were used as a control for the same surface properties. As a result, PDMS 15-μm-high and 15-μm diameter circular posts were created on top of the BioFlex plates in tetragonal array spaced 75-μm center to center [12], then coated with laminin at a concentration of 10 μg/mL for 1 h. In order to assess the uniformity of laminin distribution on the flat and microtopographic substrata, anti-laminin antibody produced in rabbit (Sigma) was incubated over night at a dilution of 1:25, and followed by the secondary antibody Alexa Fluor 568 conjugated goat-anti-rabbit antibody (Invitrogen) at a dilution of 1:200 for 30 min. All flat textured surfaces were uniformly coated with laminin as seen by confocal microscopy (Fig. 1A and B).
Fig. 1.
Confocal images of post and cell interaction: (A) 3D image of a post 15 μm high, and 15 μm diameter with laminin coating. Scale bar, 20 μm. (B) Uniform laminin staining on flat base and sides of the post seen by confocal microscopy. Scale bar, 20 μm. (C) Time lapse images show initial migration and preferential adhesion of a cell to a post during the following 12 h. Scale bar, 50 μm.
2.2. Cell culture
Institutional approval was received to obtain and use hMSCs isolated from human bone marrow aspirates (Texas A&M Health Science Center College of Medicine Temple, TX). Microarray analyses indicate that gene expression is consistent for hMSCs from different donors, isolated and expanded as described previously [13]. Experiments were performed on passage three or lower from hMSCs obtained from 3 separate donors. hMSCs were cultured in complete culture media (CCM) consisting of MEM-α supplemented with 16.5% fetal bovine serum (FBS), 2 mM l-glutamine, 100 units/mL penicillin and 100 μg/mL streptomycin, and incubated at 37 °C.
2.3. Cyclic strain
After two days of cell culture, hMSCs were cyclically strained with 10% strain at 1 Hz for 48 h in cell culture media using the Flex-cell Strain Unit (Model FX-4000, Flexercell International, McKeesport, PA). The base plate with a diameter of 25 mm was used to produce equibiaxial strain for the majority of the area. The computer system controlled the frequency of deformation and the negative pressure applied to the culture plates. There are four conditions in this study: Flat (control group), flat-strain, post, and post-strain. All the four experiments were done under similar culture conditions.
2.4. Distribution of cells upon plating by time lapse imaging
In order to determine the initial distribution of hMSCs on the flat or microtopographic substrata, time lapse movies were recorded using the Olympus VivaView soon after plating and followed over the next 12 h. A frame is recorded every 5 min and played back 2300 times faster in the movies, see Supplement.
2.5. Actin, focal adhesion, and nuclear staining
In order to analyze subcellular features, cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 10 min at room temperature, rinsed three times with PBS and permeabilized by 0.1% Triton X-100 in PBS for 10 min, and washed 3 times with PBS. Cells were pre-incubated in blocking solution (PBS, 1% bovine serum albumin (BSA)) for 15 min and then incubated with rhoda-mine conjugated phalloidin (Molecular Probes) at a dilution of 1:400 to stain actin, or paxillin anti-rabbit antibody (Abcam) at a dilution of 1:250 for 1.5 h followed by another incubation with secondary antibody Alexa Fluor 488 conjugated goat-anti-chicken antibody (Invitrogen) at a dilution of 1:1000 for 45 min to stain the focal adhesions of the cells. DAPI (Sigma) was used for nuclear staining. Confocal images of actin and focal adhesions were obtained with Zeiss LSM 510 META and LSM 710 microscopes.
2.6. Actin and nuclear distribution from post
Actin distribution was measured morphometrically and the frequency of distribution calculated as a function of the distance from the post. The location of actin and nuclei as a function of distance was determined at 7.5 μm intervals away from a post, the presence of actin was tallied by detectable phalloidin staining at the intervals. For nuclear distribution, the region less than 37.5 μm from center of the post was considered as the “close” region, and beyond that was called the “far” region. In all, over 60 posts in 3 samples were pooled for frequency distribution and statistical analysis.
2.7. DNA, RNA isolation and reverse transcription
Total DNA and RNA were isolated from hMSCs from the experimental conditions of flat, flat-strain, post, and post-strain. After two days of strain, the AllPrep DNA/RNA Mini Kit (QIAGEN) was used to isolate DNA and RNA and quantified using the Qubit Quantitation Platform (Invitrogen). RNA was reverse-transcribed for 50 min at 37 °C and 15 min at 65 °C (inactivation) using M-MLV Reverse Transcriptase (Invitrogen) and a thermal cycler (BioRad iCycler, Hercules, CA).
2.8. Microarray analysis
For microarray analysis, total RNA was pooled from five independently prepared cultures of hMSCs with or without posts and with or without strain, and the control. RNA was labeled, hybridized onto 3 microarray chips per condition (Human Gene Chip ST. 1.0, Affymetrix, Santa Clara, CA) and scanned by the Genomics Core Facility at the University of Illinois at Chicago. All hybridizations passed standard quality criteria. Raw data and probe intensity levels were normalized within the DNA-Chip Analyzer (dChip) as based on the median baseline intensity of the whole array [14]. All subsequent pair-wise analysis was also performed using dChip. For pair-wise comparisons, statistically significant, differentially expressed transcripts were identified by raw local-pooled-error (LPE) test p values. For global functional clustering analysis a list of the genes was imported into the DAVID Functional Annotation Clustering tool (http://david.abcc.ncifcrf.gov). Raw LPE test p-values were then corrected for False Discovery Rate by the Benjamini–Hochberg (BH) procedure (p-value < 0.05). The “heat map” was obtained by the dChip Windows software.
2.9. Real-time polymerase chain reaction (RT-PCR)
For RT-PCR experiments, total RNA was isolated and reverse transcribed, from independently prepared flat or strained hMSCs with or without posts. Using SYBR Green PCR Master Mix and a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). Amplification was achieved by the following protocol: 1 cycle of 50 C for 2 min; 1 cycle of 95 C for 10 min; 0 cycles of 95 C for 15 s and 60 C for 1 min. To ensure specificity of PCR, melt-curve analyses were performed at the end of all PCRs. The relative amount of target cDNA was determined from the appropriate standard curve and divided by the amount of β2-microglobulin (β2M) cDNA present in each sample for normalization. Each sample was analyzed in triplicate, and results were expressed relative to control condition.
2.10. Data analysis
Data were expressed as mean ± SD (n = 3 or more experiments). Differences were analyzed by the Student's paired and unpaired t-test with significance at p < 0.05.
3. Results
In this study, hMCSs were exposed to various mechanical forces to explore the effects of physical cues while retaining chemical conditions constant. Changes in cell morphology, focal adhesions, and gene expression of hMSCs were assessed for four conditions: Flat (F), flat-strain (FS), post (P), and post-strain (PS).
3.1. Cell distribution after plating
Cell migration is random until hindered by adherence to a post as shown at different time points over the initial 12 h period (Fig. 1C and in Supplemental movies). The microtopography was 12.5% of the total surface area. Therefore, cell distribution becomes non-random because of preferential adherence.
3.2. The actin cytoskeleton
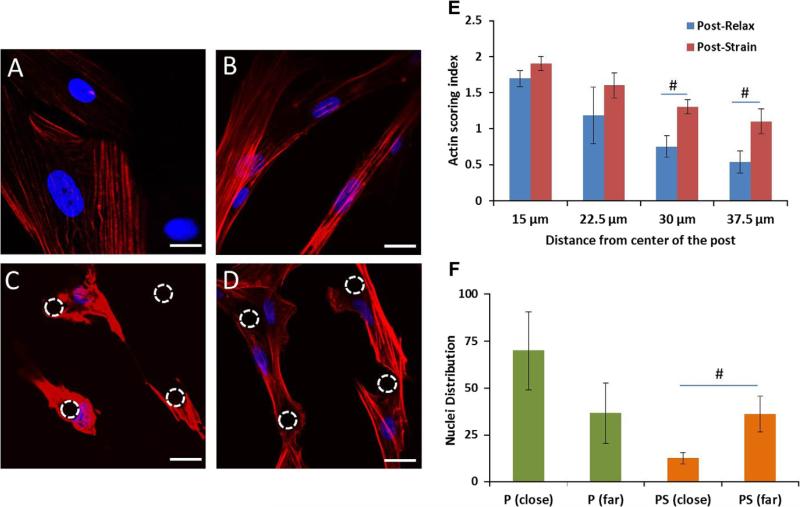
Actin distribution was changed with respect to both strain and microtopography (Fig. 2). Cells on flat surfaces contained thin layers of actin (Fig. 2A); however, by straining the cells (FS), intensely-stained bundles of actin stress fibers were observed, which elongated the cell shape (Fig. 2B). The cells in unstrained 3D microtopography had a high intensity of the actin wrapped closely around the post (Fig. 2C). Straining hMSCs on the microtopographic substrata resulted in actin stress fibers that span the 75 μm from post to post (Fig. 2D). The actin distribution was similar with or without strain nearer to the post (15 and 22.5 μm), but significant differences between the post and post-strain groups were found further away (30 and 37.5 μm) (Fig. 2E). Adding strain to microtopography shifted the nuclei midway between the adjacent posts in the cell center (Fig. 2F).
Fig. 2.
Actin remodels with microtopography and strain. Actin cytoskeleton and nuclei of hMSCs (A) flat, (B) flat-strain, (C) post, and (D) post-strain, seen with confocal microscopy after two days of strain at 1 Hz and 10%. (C) The cells containing actin fibers are wrapped around the post. (D) The actin fibers are elongated in the cells and span from post to post. (E) Frequency histogram of actin distribution at 15, 22.5, 30 and 37.5 μm from post center. (F) Nuclear distribution shifted from close to far regions from a post with cyclic strain. The dashed circles indicate the location of post. Actin stained by rhodamine phalloidin (red); nuclei with DAPI (blue). Mean ± SE, n = 3 human samples, *All vs. flat (control), #close vs. far p < 0.05. Scale bar, 20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Focal adhesions
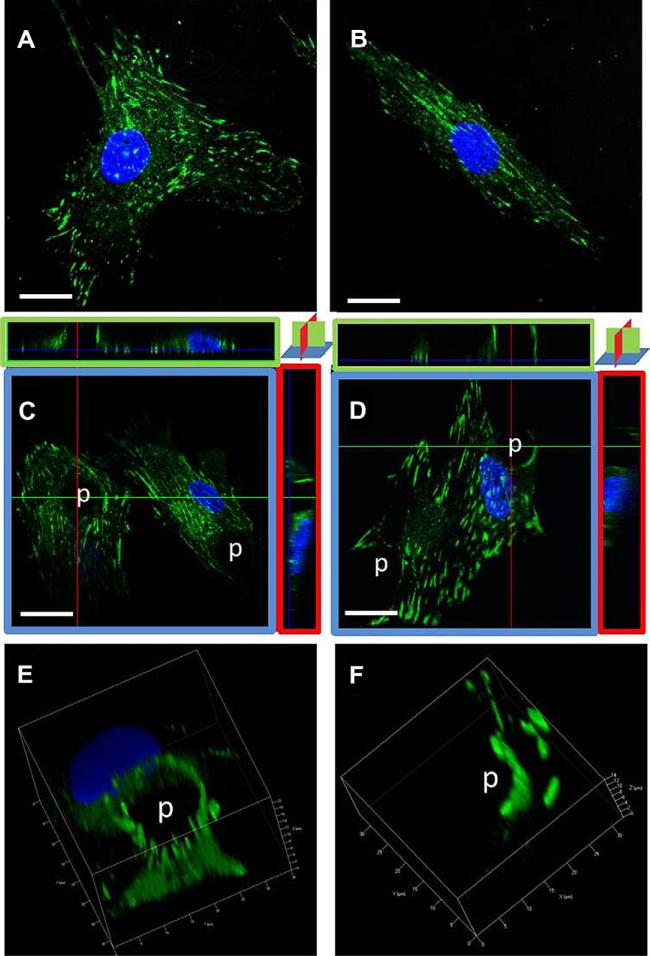
In this study, focal adhesions were identified by paxillin. Microtopography and strain both alter the distribution of paxillin. On flat, unstrained surfaces, paxillin is sparse (Fig. 3A); while with strain the paxillin is aligned with the long axis of the cell (Fig. 3B). The presence of the post causes a complete ring of paxillin to form, distributed evenly around the post (as seen in top view with Fig. 3C); when hMSCs are strained, the paxillin adopts a semi-circular appearance around the post (Fig. 3D). The cross-sectional views of confocal images (X–Y, X–Z, and Y–Z planes) show paxillin along the vertical walls of the posts in conditions with or without strain (Fig. 3C and D). Full 3D volume renders the bright, punctuate paxillin on the sides and the top edge of the posts (Fig. 3E and F).
Fig. 3.
Focal adhesions redistributed with microtopography and strain. Paxillin staining of hMSCs after two days with or without straining at 1 Hz and 10% strain seen in 3D with confocal microscopy. (A) flat, (B) flat-strain, (C) post confocal orthogonal views, (D) post-strain orthogonal views. Volumetric renditions of 3D for (E) post and (F) post-strain. (C) Paxillin surrounds the post and goes up its full height. (D) Paxillin forms a semicircular ring up the side of the post, and extends from post to post on the lower surface of the cell. (E, F) 3D reconstructions show punctate paxillin on the sides of the post. Paxillin (green) and DAPI (blue). Scale bar, 20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Gene expression
Microarray results of 31,000 genes reveal many transcript-level differences between hMSCs cultured in the four experimental conditions; Differentially expressed transcripts were grouped as a “heat map” by hierarchal clustering into enriched functional groups including matrix and focal adhesions, muscle proteins, proliferation, and differentiation. Higher relative expression (red) under flat-strain, and post-strain groups from the “heat map” confirm the dominant effect of strain over microtopography (Fig. S3 and Table S1). The RNA/DNA ratios did not change between the four conditions (data not shown).
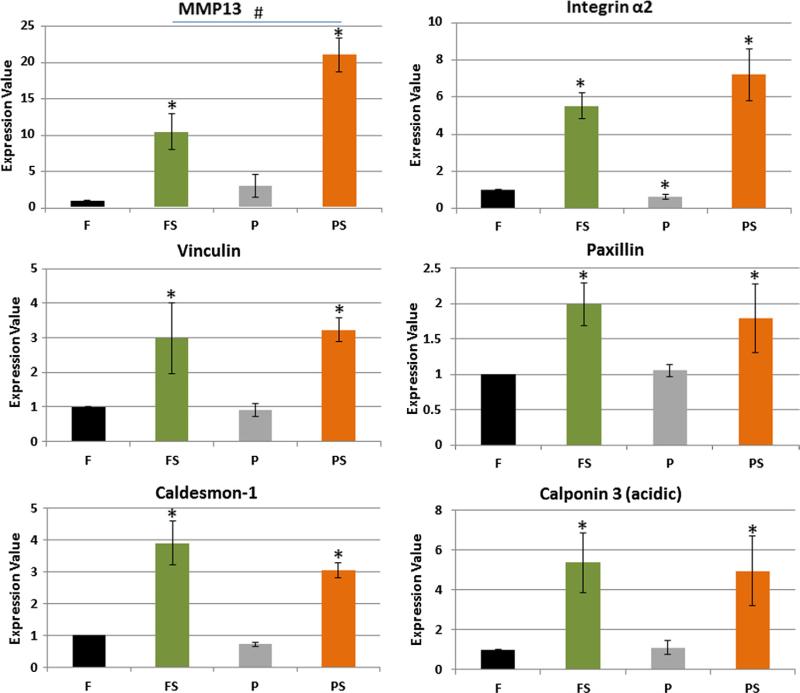
In total 6 genes were analyzed with RT-PCR (Fig. 4). The genes confirmed for three functional groups, namely matrix (matrix metalloproteinase 13, MMP13); focal adhesions (paxillin, integrin-α2, and vinculin); and actin binding proteins (caldesmon-1 and calponin-3). Among these genes, MMP13 showed the most difference between flat-strain and post-strain with an almost twofold increase. However, the most significant differences for these quantified genes came from straining rather than from 3D microtopography (Fig. 4).
Fig. 4.
Strain is dominant over microtopography in regulating gene expression. RT-PCR of paxillin, vinculin, and integrin-α2 from focal adhesions; caldesmon-1 and calponin3 from actin proteins; and MMP13 from matrix; MMP13 had the highest fold change with post-strain, but the flat-strain and post-strain affected rest of the genes similarly. However, integrin-α2 was slightly decreased with microtopography. Mean ± SE, n = 4. *All vs. flat, #FS vs. PS, p < 0.05.
4. Discussion
The major findings of this study are that both strain and microtopography contribute to the remodeling of the actin cytoskeleton and focal adhesions in hMSCs. Focal adhesions formed on the vertical side of the post with the actin fibers wrapping around. With the addition of strain, the cytoskeleton elongated and the focal adhesion attachment on the post became asymmetric. The subcellular actin distribution became more distant from the post with both strain and microtopography.
Cells evaluate the level of external force and adjust internally. The cytoskeleton filaments and their linkages to transmembrane focal adhesions assemble, break down, and reassemble in response to local forces. As cells probe their environment, their anchorage is determined by amount and direction of micromechanical forces, which cluster integrins stabilizing new focal adhesions over time [15]. The cell tension is increased when microtopography stabilizes anchorage to the static forces of the microposts, and to an even greater extent when dynamic external strain is delivered. Thus, in individual cells, the subcellular distribution of focal adhesions and the direction of actin cytoskeleton fibers depend on local traction forces.
4.1. Microtopography
Early studies of cell adherence to microtopography was done by our group and showed focal adhesion and actin filament remodeling for myocytes [6,12] and fibroblasts [16] on substrata with 15 μm diameter posts. Microtopography affects this local traction force, with a one micron diameter micropillar being sufficient to stabilize the focal adhesions visualized by vinculin staining [17]. The focal adhesion complex is the anchor for the cytoskeletal organization of the cells plated on posts, and depends on both focal adhesion kinase and the contractile forces generated by myosin II [18]. RhoA regulates the actin polymerization in the topography-induced focal adhesion formation [19]. Here, hMSC encircles the post by forming actin bundles attached by focal. Myosin heavy chain of cardiomyocytes extended over the full height of the posts of 10 or 15 μm height [5,20]. Fibroblasts spread in between the posts with 20 μm height, 5 μm diameter, and spaced 4–12 μm apart, where they form arc-like shapes that presented an inward curvature that increased with time, and the importance of cell tension was confirmed because the wrapping process can be reversed with a myosin II inhibitor showing the importance of cell tension [18].
4.2. Strain
Focal adhesion formation can also be induced through application of external force by strain [18]. RhoA/ROCK and FAK regulate actin fiber formation and cytoskeleton reorganization of cells with mechanical strain [21]. Fifteen percentage of uniaxial strain at 1 Hz reoriented the actin fibers of hMSC [22]. Integrin is attached to the actin cytoskeleton through several actin-associated proteins such as paxillin, vinculin, talin, tensin, and α-actinin. In this study, two-day strain of hMSCs resulted in up-regulation of isoforms of integrin (integrin-α2, integrin-α5, and integrin-β6) and of increased focal adhesion molecules (paxillin, vinculin, α-actinin and talin) (see the heat map in the supplementary data). RT-PCR confirmed the enhanced expression of integrin-α2, paxillin and vinculin with strain (Fig. 4).
4.3. Combination of microtopography and strain
Adding mechanical strain to the microtopographic substrata caused the redistribution of focal adhesions for anchorage so that the hMSCs were still elongated but also preferentially anchored to the posts rather than the intervening flat spaces. The dynamic flexing of the substrata increases the internal tension above strain alone, but the focal adhesion stabilized in a semi-circle on the side of the post because of the local increase and direction of traction forces.
4.4. Matrix remodeling
hMSCs not only remodel their interior architecture in response to 3D microtopography and strain, but they also remodel the extracellular matrix in which they reside. This was achieved by regulating transcription of collagen, laminin, fibronectin, and metalloproteinases (MMPs). Strikingly, MMP13 was affected by the combination of microtopography and strain with twofold changes more than the strain alone. MMP13 (collagenase 3) has been shown to degrade the native interstitial collagens in several tissues and to participate in situations where rapid and effective remodeling of collagenous extracellular matrix (ECM) is required. Biaxial strain increased ECM degradation of osteoblastic cells by MMP13 [23]. Since the microenvironmental niche is thought to affect gene expression, microtopography and strain were used to determine whether there was any regulation of actin and focal adhesions genes. Strained hMSCs gene expression confirmed by RT-PCR showed significant difference for two actin proteins (calponin-3, acidic, and caldesmon-1). The acidic calponin 3 (CNN3) is an actin filament-associated regulatory protein, which has been found in smooth muscle and non-muscle cells [24]. CNN3 has been shown to up-regulate with combination of transforming growth factor beta and cyclic mechanical straining of cultured hMSC with microgrooves [25]. Caldesmon-1 is a calmodulin binding protein that plays an essential role in the regulation of smooth muscle and non-muscle contraction. Ten percentage of cyclic strain increased caldesmon-1 [4,26]. Thus, despite the predictions that the combination of mechanical cues from physical deformation by externally applied force and also from microtopography might both contribute significantly, the addition of microtopography to strain did not yield any notable cell regulation.
5. Conclusion and summary
The mechanical cues of microtopography and strain altered local forces, which in turn affected focal adhesion formation and cytoskeletal reorganization. Gene expression in hMSCs was induced more readily by strain than microtopography, which reinforces the expected dominance of external strain stimuli over the static influence of microtopography. However, with the combination of strain and microtopography, local remodeling of cell shape and morphology was accomplished by the focal adhesion formation on the vertical post that stabilizes the cytoskeleton over time so that there is preferential elongation from post to post.
Clearly more work is needed to understand how a cell type with potential clinical use responds to mechanical strain in the presence of tightly controlled 3D geometry. Exactly how the mechanical forces link to the cellular activities remains elusive but actin fiber reorganization and focal adhesion redistribution appear to be the first steps for regulation of specific cellular functions.
Supplementary Material
Acknowledgments
This investigation was funded in part by the National Institutes of Health through grants HL090523 (BR) and T32 HL07692 (GD). Human MSCs were supplied under the auspices of an NIH grant ‘Preparation and Distribution of Adult Stem Cells’ (P 40 RR017447) to Darwin J. Prockop, M.D., Ph.D., from Texas A&M Health Science Center College of Medicine, Temple, TX.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2012.11.120.
References
- 1.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414(6859):98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat. Rev. Mol. Cell. Biol. 2009;10(1):34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gwak SJ, Bhang SH, Kim IK, Kim S, Cho SW, Jeon O, Yoo KJ, Putnam AJ, Kim BS. The effect of cyclic strain on embryonic stem cell-derived cardiomyocytes. Biomaterials. 2008;29:844–856. doi: 10.1016/j.biomaterials.2007.10.050. [DOI] [PubMed] [Google Scholar]
- 4.Jang JY, Lee SW, Park SH, Shin JW, Mun C, Kim SH, Kim DH, Shin JW. Combined effects of surface morphology and mechanical straining magnitudes on the differentiation of mesenchymal stem cells without using biochemical reagents. J. Biomed. Biotechnol. 2011;2011:860652. doi: 10.1155/2011/860652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Motlagh D, Senyo SE, Desai TA, Russell B. Microtextured substrata alter gene expression, protein localization and the shape of cardiac myocytes. Biomaterials. 2003;24:2436–2476. doi: 10.1016/s0142-9612(02)00644-0. [DOI] [PubMed] [Google Scholar]
- 6.Chen KD, Li YS, Kim M, Li S, Yuan S, Chien S, Shyy JY. Mechanotransduction in response to shear stress: roles of receptor tyrosine kinases, integrins, and Shc. J. Biol. Chem. 1999;274:18393–18400. doi: 10.1074/jbc.274.26.18393. [DOI] [PubMed] [Google Scholar]
- 7.Ingber DE. Tensegrity: the architectural basis of cellular mechanotransduction. Annu. Rev. Physiol. 1997;59:575–599. doi: 10.1146/annurev.physiol.59.1.575. [DOI] [PubMed] [Google Scholar]
- 8.Szafranski JD, Grodzinsky AJ, Burger E, Gaschen V, Hung HH, Hunziker EB. Chondrocyte mechanotransduction: effects of compression on deformation of intracellular organelles and relevance to cellular biosynthesis. Osteoarthritis Cartilag. 2004;12:937–946. doi: 10.1016/j.joca.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Pelham RJ, Wang YL. Cell locomotion and focal adhesions are regulated by the mechanical properties of the substrate. Biol. Bull. 1998;194:348–349. doi: 10.2307/1543109. [DOI] [PubMed] [Google Scholar]
- 10.Xiao Y, Truskey GA. Effect of receptor-ligand affinity on the strength of endothelial cell adhesion. Biophys J. 1996;71:2869–2884. doi: 10.1016/S0006-3495(96)79484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch J, Motlagh D, Russell B, Desai TA. Fabrication of microtextured membranes for cardiac myocyte attachment and orientation. J. Biomed. Mater. Res. Appl. Biomater. 2000;53:267–275. doi: 10.1002/(sici)1097-4636(2000)53:3<267::aid-jbm12>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Biehl JK, Yamanaka S, Desai TA, Boheler KR, Russell B. Proliferation of mouse embryonic stem cell progeny and the spontaneous contractile activity of cardiomyocytes are affected by microtopography. Dev. Dyn. 2009;238(8):1964–1973. doi: 10.1002/dvdy.22030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins JM, Ayala P, Desai TA, Russell B. Three-dimensional culture with stiff microstructures increases proliferation and slows osteogenic differentiation of human mesenchymal stem cells. Small. 2009;6(3):355–360. doi: 10.1002/smll.200901757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li C, Wong WH. Model based analysis of oligonucleotide arrays expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheehy SP, Parker KK. The role of mechanical forces in guiding tissue differentiation. In: Bernstein H, editor. Tissue Engineering in Regenerative Medicine. Springer; 2011. pp. 77–97. [Google Scholar]
- 16.Boateng SY, Hartman TJ, Ahluwalia N, Vidula H, Desai TA, Russell B. Inhibition of fibroblast proliferation in cardiac myocyte cultures by surface microtopography. Am. J. Physiol: Cell. 2003;285:C171–C182. doi: 10.1152/ajpcell.00013.2003. [DOI] [PubMed] [Google Scholar]
- 17.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK independent mechanism. J. Cell. Biol. 2001;153(6):1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghibaudo M, Di Meglio JM, Hersen P, Ladoux B. Mechanics of cell spreading within 3D-micropatterned environments. Lab. Chip. 2011;11:805–812. doi: 10.1039/c0lc00221f. [DOI] [PubMed] [Google Scholar]
- 19.Seo CH, Furukawa K, Montagne K, Jeong H, Ushida T. The effect of substrate microtopography on focal adhesion maturation and actin organization via the RhoA/ROCK pathway. Biomaterials. 2011;32(36):9568–9575. doi: 10.1016/j.biomaterials.2011.08.077. [DOI] [PubMed] [Google Scholar]
- 20.Patel AA, Desai TA, Kumar S. Microtopographical assembly of cardiomyocytes. Integr. Biol. (Camb.) 2011;3:1011–1019. doi: 10.1039/c1ib00024a. [DOI] [PubMed] [Google Scholar]
- 21.Xu B, Song G, Ju Y, Li X, Song Y, Watanabe S. RhoA/ROCK, cytoskeletal dynamics, and focal adhesion kinase are required for mechanical stretch induced tenogenic differentiation of human mesenchymal stem cells. J. Cell. Physiol. 2012;227(6):2722–2729. doi: 10.1002/jcp.23016. [DOI] [PubMed] [Google Scholar]
- 22.Ghazanfari S, Shadpour MT, Shokrgozar MA. Effects of cyclic stretch on proliferation of mesenchymal stem cells and their differentiation to smooth muscle cells. Biochem. Biophys. Res. Commun. 2009;388(3):601–605. doi: 10.1016/j.bbrc.2009.08.072. [DOI] [PubMed] [Google Scholar]
- 23.Yang CM, Chien CS, Yao CC, Hsiao LD, Huang YC, Wu CB. Mechanical strain induces collagenase-3 (MMP-13) expression in MC3T3-E1 osteoblastic cells. J. Biol. Chem. 2004;279(21):22158–22165. doi: 10.1074/jbc.M401343200. [DOI] [PubMed] [Google Scholar]
- 24.Applegate D, Feng W, Green RS, Taubman MB. Cloning and expression of a novel acidic calponin isoform from aortic vascular smooth muscle. J. Biol. Chem. 1994;269:10683–10690. [PubMed] [Google Scholar]
- 25.Kurpinski K, Chu J, Wang D, Li S. Proteomic profiling of mesenchymal stem cell responses to mechanical strain and TGF-beta1. Cell. Mol. Bioeng. 2009;2(4):606–614. doi: 10.1007/s12195-009-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cevallos M, Riha GM, Wang X, Yang H, Yan S, Li M, Chai H, Yao Q, Chen C. Cyclic strain induces expression of specific smooth muscle cell markers in human endothelial cells. Differentiation. 2006;74(9–10):552–556. doi: 10.1111/j.1432-0436.2006.00089.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.