SUMMARY
Red blood cell production is a finely tuned process that requires coordinated oxygen- and iron-dependent regulation of cell differentiation and iron metabolism. Here we show that translational regulation of HIF-2α synthesis by IRP1 is critical for controlling erythrocyte number. IRP1 null mice (Irp1−/−) display a marked transient polycythemia. HIF-2α mRNA is derepressed in kidney of Irp1−/− but not Irp2−/− mice leading to increased renal erythropoietin (Epo) mRNA and inappropriately elevated serum Epo levels. Expression of the iron transport genes DCytb, DMT1 and ferroportin as well as other HIF-2α targets is enhanced in IRP1−/− duodenum. Analysis of mRNA translation state in liver revealed IRP1-dependent dysregulation of HIF-2α mRNA translation while IRP2 deficiency derepressed translation of all other known 5′ IRE-containing mRNAs expressed in liver. These results uncover separable physiological roles of each IRP and identify IRP1 as a therapeutic target for manipulating HIF-2α action in hematologic, oncologic and other disorders.
INTRODUCTION
Erythropoiesis is a dynamic process regulated through complex interplay of cytokines, nutrient availability, and the cellular environment of erythroid progenitors (Hattangadi et al., 2011). The level of circulating erythrocytes must be tightly controlled as a deficiency leads to anemia and the consequent negative impact of hypoxic stress, while chronically high levels can lead to hyperviscosity and potentially lethal thrombotic events (Lee and Percy, 2011). Excess accumulation of erythrocytes, referred to as polycythemia or erythrocytosis, occurs physiologically, as in the adjustment to high altitude, or pathologically, due to intrinsic abnormalities in erythroid precursors or as a consequence of inappropriately high expression of Epo (Lee and Percy, 2011). Critical factors governing red cell formation include oxygen and iron sensing particularly through the transcription factor hypoxia-inducible factor 2α (HIF-2α), which regulates Epo expression as well as multiple other genes required for iron assimilation (Gruber et al., 2007; Haase, 2010; Mastrogiannaki et al., 2009; Scortegagna et al., 2005; Shah et al., 2009; Taylor et al., 2011). Elucidating the mechanisms regulating Epo production and action is relevant to understanding the etiology of, and designing treatments for, common diseases and pathological states ranging from the anemia of prematurity to the anemias associated with inflammation, cancers, aging, and renal failure (Strauss, 2010; Weiss and Goodnough, 2005).
HIFs are heterodimeric transcription factors regulated by oxygen and iron (Majmundar et al., 2010; Prabhakar and Semenza, 2012). HIFs contain a subunit from a family of inducibly degraded α proteins that have overlapping but also unique physiological and pathophysiological roles. The key role of HIF-2α in erythropoiesis has been demonstrated in part through the discovery of human HIF-2α gain-of-function mutations that cause polycythemia (Lee and Percy, 2011). Dysregulation of HIF-2α is also implicated in human cancers (Franovic et al., 2009; Keith et al., 2012; Tong et al., 2011). While much work has focused on the regulation of HIF action through oxygen- and iron-mediated protein degradation, recent work using cultured cells has revealed new mechanisms for HIF regulation including programmed changes in their synthesis (Sanchez et al., 2007; Tong et al., 2011; Young et al., 2008; Zimmer et al., 2010). Thus, physiological control of HIF-2α function and its dysregulation in disease may arise due to adaptive or maladaptive changes in HIF-2α synthesis.
Iron regulatory protein 1 (IRP1) and IRP2 are central regulators of cellular iron metabolism in metazoans (Anderson et al., 2012; Hentze et al., 2010). IRPs control the fate of mRNAs encoding proteins, including HIF-2α, involved in iron metabolism or the adaptive responses to iron deficiency. The finding that deficiency of IRP2, but apparently not IRP1, leads to dysregulated iron metabolism (Cooperman et al., 2005; Galy et al., 2005b; Meyron-Holz et al., 2004) has led to the view that IRP1 lacks a unique role in iron metabolism. However, the fact that the Fe-S switch mechanism controlling IRP1 allows for inactivation of its RNA binding in hypoxia, in contrast to IRP2 which is activated, suggests otherwise (Anderson et al., 2012). Thus, as is the case for HIF-2α mRNA in cultured cells, translation of mRNAs preferentially repressed by IRP1 would be enhanced in hypoxia (Zimmer et al., 2008). These findings suggest that an IRP1-HIF-2α regulatory axis may be a critical component of systemic mechanisms controlling erythrocyte number as well as tissue responses to iron deficiency and hypoxic stress.
Here we studied the impact of IRP1 deficiency on the function of HIF-2α in erythropoiesis and duodenal iron absorption. Our study reveals that, through its regulation of HIF-2α expression, IRP1 has a key role linking erythropoiesis and dietary iron absorption with iron and oxygen sensing, a function not attributed to IRP2. The IRP1-HIF-2α axis provides new therapeutic targets for modulating the response of cells to altered iron or oxygen levels in a wide array of hematologic and oncologic diseases.
RESULTS AND DISCUSSION
Polycythemia and Extramedullary Erythropoiesis in IRP1-Deficient Mice
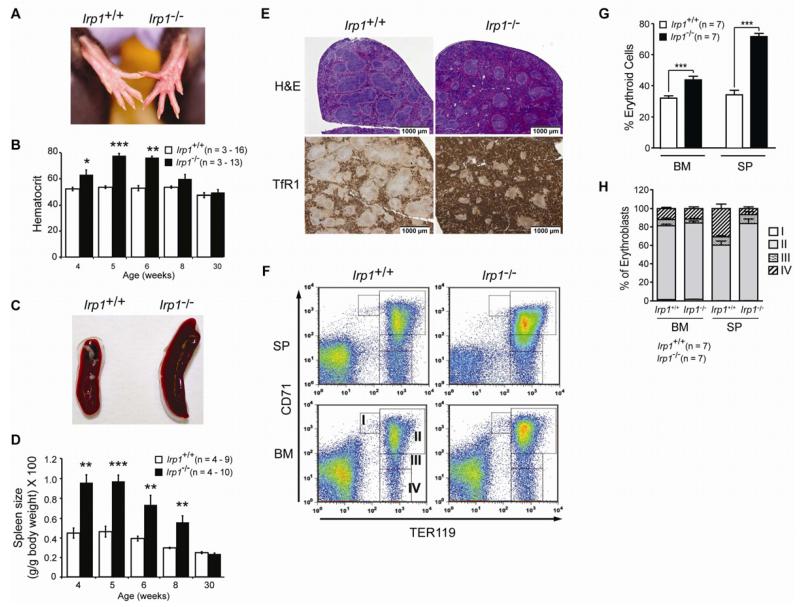
In the process of elucidating the unique physiological functions of IRP1 and IRP2, we observed that young Irp1−/− mice frequently had reddened paws around 5 weeks (wk) of age (Figure 1A) reminiscent of the “plethora” seen in human patients with polycythemias. Accordingly, we found that the hematocrit, red cell number and hemoglobin are elevated in Irp1−/− mice between 4 and 6 wk of age (Figure 1B and Table S1). The elevated hematocrit is similar to that observed in humans with HIF-2α gain-of-function mutations (Lee and Percy, 2011). Irp1−/− spleens are most enlarged at 4 and 5 wks of age but thereafter normalize (Figure 1C and Figure 1D). Serum and tissue iron parameters are indicative of iron redistribution to the erythron (Table S2). When Irp1−/− mice reach 8 wks of age, the hematocrit remains elevated although there is substantial variability from animal to animal such that only approximately one-third of the animals have elevated hematocrit. At this age the hematocrit of Irp1−/− and Irp1+/+ is not statistically different (P = 0.058), illustrating the age-dependent effect of IRP1 deficiency (Figure 1B).
Figure 1. Polycythemia and Extramedullary Erythropoiesis in IRP1-deficient Mice.
[A] Rear paws showing greater redness in Irp1−/− mice. [B] Hematocrit for Irp1+/+ and Irp1−/− mice. Samples were collected from the retroorbital sinus or by cardiac puncture and hematocrit was from CBC analysis. See Table S1. For 30 wk Irp1+/+ mice n = 2. [C] Spleen from 6 wk old WT and Irp1−/− mice. [D] Spleen weight as a percent body weight as a function of age. For 30 wk Irp1+/+ mice n = 2. [E] H & E staining (top) or immunohistochemistry for TfR1 (CD71) (bottom) in spleen. [F] FACS analysis of cells isolated from bone marrow (BM) and spleen (SP) with staining for the erythroid marker Ter119 and CD71 (TfR). The cell populations are separated into stages I – IV reflecting increasing differentiation of the erythroblasts as described (Socolovsky et al., 2001). [G] Percentage of nucleated cells that are erythroblasts in BM and SP. [H] Erythroblast differentiation state in BM and SP as determined by FACS. For panels B, D, G and H results are expressed as mean ± SEM. * = P <0.05; ** = P < 0.01; *** = P < 0.001. Unless noted 5 wk old mice were used. See Tables S1, S3 and Figure S1.
Spleens from 5-wk-old Irp1−/− mice demonstrated an altered architecture, with a decreased white pulp and a substantial increase in red pulp, signifying expanded erythropoiesis (Figure 1E). Immunohistochemistry for TfR1 (CD71) confirmed that the Irp1−/− splenic red pulp is distended by erythroblasts (Figure 1E). Maturing erythroblasts of different developmental stages were quantified by flow cytometry with the erythroid specific marker Ter119 and with CD71 (Socolovsky et al., 2001). This analysis reveals marked accumulation of erythroblasts in bone marrow and particularly in spleen of 5-wk-old Irp1−/− mice (Figures 1F and 1G). The relative abundance of cells at different stages of erythroblast maturation was not altered in BM while in the Irp1−/− spleen the distribution was skewed toward having more cells at stage II and fewer cells at stage IV (Figure 1H and Table S3). This altered pattern of maturation may reflect the response to Epo itself or changes induced by the splenic microenvironment (Socolovsky et al., 2001). Interestingly, at 6 wks of age enhanced erythropoiesis was only observed in one-quarter of the Irp1−/− mice indicating an age-dependent compensation for the loss of IRP1 (Figure S1C and S1D).
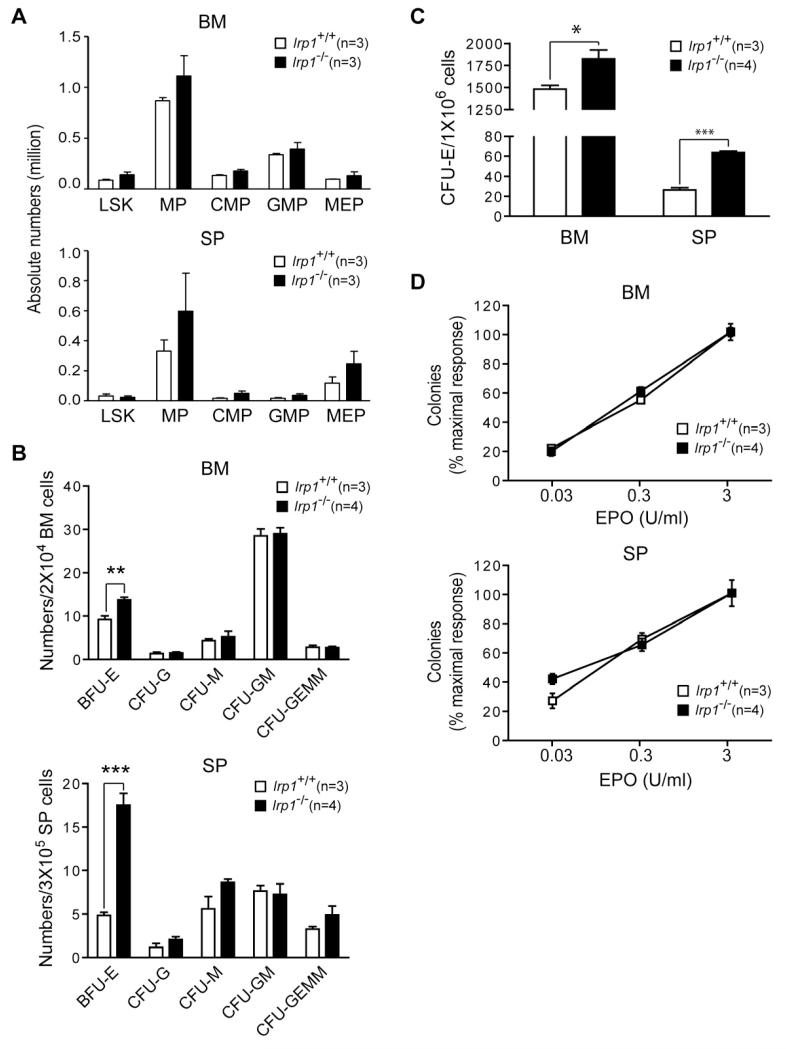
To determine if IRP1 deficiency caused a specific enhancement of erythropoiesis, the abundance of hematopoietic progenitors was determined. IRP1 deficiency does not appear to affect the abundance of primitive hematopoietic stem (LSK cells) or myeloid progenitor cells in either the bone marrow (BM) or spleen (SP) (Figure 2A). Consistent with the flow cytometric analysis, early stage myeloid progenitors formed similar numbers of colonies in vitro irrespective of genotype (Figure 2B). In contrast, the primitive erythroid BFU-E and late-stage CFU-E progenitors were elevated in BM and particularly SP of Irp1−/− mice, while progenitors for other hematopoietic lineages were not altered (Figure 2B and 2C). We then determined if IRP1 deficiency affected the sensitivity of CFU-E cells to Epo. No such effect was observed (Figure 2D). Collectively, these findings reveal that IRP1 deficiency is associated with an age-dependent increase in erythropoiesis in young mice and not a general increase in hematopoiesis. We conclude that IRP1 has a previously unrecognized role as a regulator of erythropoiesis with its dysregulation contributing to the development of polycythemia. The erythroid phenotype of IRP1 is distinct from that of IRP2 deficiency that is microcytic anemia and erythropoietic protoporphyria (Cooperman et al., 2005; Galy et al., 2005b).
Figure 2. Specific Increase in Erythroid Progenitors in Irp1−/− Mice.
[A] Abundance of myeloid progenitor cells (LSK: lineage Sca-1+, c-kit+; MP: myeloid progenitors; CMP: common myeloid progenitors; GMP: granulocyte macrophage progenitors; MEP; megakaryocyte erythroid progenitors in spleen (SP) and bone marrow (BM). For panels B, C and D, BM and SP cells were isolated from Irp1+/+ and Irp1−/− mice and plated in duplicate in M3234 medium with 3 U/ml (B and C) or varying concentrations (D) of Epo. [B] Average percentage of maximum number of colonies formed in culture. For BFU-E (burst forming unit-erythroid), CFU-G (colony forming unit (CFU)-granulocyte), CFU-M (CFU-macrophage), CFU-GM (CFU-granulocyte-macrophage) and CFU-GEMM (CFU-granulocyte, erythroid, macrophage, megakaryocytic) colony formation, BM and SP cells were plated in M3434 medium, and colonies counted at day 7 after plating. [C] CFU-E (CFU-erythroid) number in BM and SP. [D] CFU-E number as a function of Epo concentration. Error bars show SEM. * p<0.05, ** p<0.01, *** p<0.001. Mice were 5 wk old.
Enhanced Serum Epo and Erythroid Gene Expression in Spleen of IRP1−/− Mice
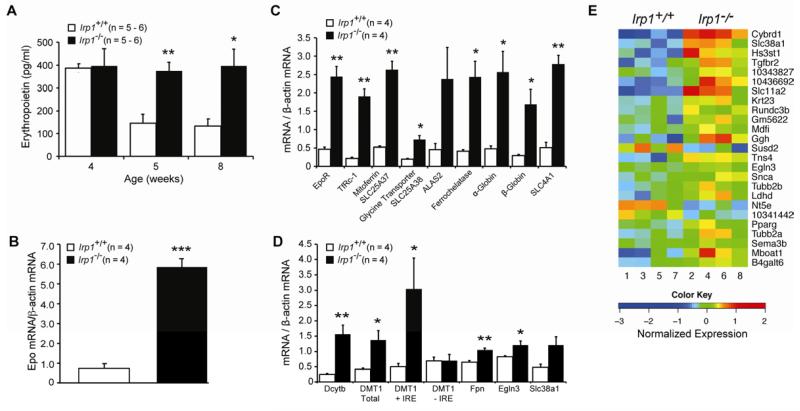
Given that HIF-2α is a major regulator of Epo gene transcription and HIF-2α mRNA translation is a direct target of IRP action (Davis et al., 2011; Sanchez et al., 2007; Zimmer et al., 2008), Epo expression was examined in rp1−/− mice. Epo expression is developmentally regulated after birth, reflecting the increased oxygen level relative to that in utero and increased demand for erythropoiesis and a relative iron deficiency as the neonate grows (Lee and Percy, 2011). The maximal changes we observed in erythropoiesis in Irp1−/− mice relative to Irp1+/+ mice occurred at 5 wks of age coincident with the slowing of growth and attendant increase in red blood cell mass as the animal reaches adult weight. To determine if the effect was Epo-dependent, we measured serum Epo concentration in 4-wk, 5-wk and 8-wk old mice. At 4 wks serum Epo concentration was high in Irp1+/+ and Irp1−/− mice, likely reflecting a physiologically appropriate erythropoietic drive in young mice (Figure 3A). However, at 5-wks, when the difference in erythropoiesis is apparent in Irp1−/− mice, serum Epo level is 2- to 3-fold higher than in Irp1+/+ mice. At this time, kidney Epo mRNA is increased by more than ~8-fold in Irp1−/− animals (Figure 3B). Furthermore, expression of multiple genes involved in erythroid development, including globins and heme biosynthetic genes was increased in Irp1−/− spleen (Figure 3C) likely reflecting increased erythroblast number. Since Epo promotes expansion of BFU-E and CFU-E cells (Peslak et al., 2012) the rise in these erythroid progenitors in Irp1−/− mice at 5 wks of age can be explained by the inability to suppress Epo expression.
Figure 3. HIF-2α Hyperactivity in IRP1-deficient Mice.
[A] Serum Epo (pg/ml) in 4 wk, 5 wk and 8 wk old mice. [B] Kidney Epo mRNA level at 5 wk. [C] Erythroid differentiation gene mRNA levels in spleen at 5 wk. [D] Duodenal mRNA level for iron transport proteins and other HIF-2α (e.g. EGLN3) targets dependent on HIF-2α for induction in iron deficiency (5 wk). [E] Microarray analysis of duodenal mucosal cells at 5 wk. Response of each of 4 mice/genotype is shown. Posterior probability of differential expression for all genes is greater the 0.95. Expression of 5 erythroid genes (globin chains α1,α2,β1,β2 and ALAS2) is not shown. Results in this figure are expressed as mean ± SEM. * = P <0.05; ** = P < 0.01; *** = P < 0.001. (see Methods and Figure S2).
Interestingly, serum Epo concentration at 8 wks of age is persistently high in IRP1−/− mice. Since erythropoiesis returns to normal by 6 wks in IRP1−/− mice (Figure S1C and S1D) it appears that erythroid differentiation has become desensitized to the elevated Epo. Epo resistance is clinically relevant in a number of chronic disease states and can be caused by iron deficiency, inflammatory cytokines or a reduction of Epo-responsive late-stage erythroid progenitors (Andrews and Bridges, 1998; Peslak et al., 2012; van der Putten et al., 2008). Because the reduction in transferrin iron in Irp1−/− mice (Table S2) does not appear to reach a level that impairs erythropoiesis, at least in vitro (Bullock et al., 2010), other factors are likely responsible. In sum, we conclude that in young Irp1−/− mice an unidentified developmental signal that normally acts through IRP1 to suppress HIF-2α synthesis is absent leading to a failure to suppress Epo expression. As Irp1−/− mice age one of many factors controlling Epo action or other aspects of erythropoiesis act to limit red cell production due to IRP1 deficiency and compensate for the inappropriately high Epo level.
HIF-2α Hyperactivity in Duodenum of IRP1−/− mice
HIF-2α induces transcription of the duodenal iron transporter genes in response to hypoxia or iron-deficiency (Taylor et al., 2011). The mRNAs encoding key components of the apical (DCytb (Cybrd1) and DMT1 (SLC11A2)) and basolateral (ferroportin (SLC40A1)) transporters are significantly increased in Irp1−/− duodenal mucosal cells (Figure 3D). The preferential impact of IRP1 deficiency on the IRE-containing form of DMT1 mRNA is consistent with this isoform being a target of HIF-2α action (Mastrogiannaki et al., 2009; Shah et al., 2009).
Using a transcriptomic approach, we found 25 genes that were differentially expressed between Irp1+/+ and Irp1−/− duodenum (Figure 3E and Figure S2). Amongst these genes are DCytb (Cbryd1) and DMT1 (SLC11A2). Multiple other molecular signatures emerged. First, expression of the prolyl hydroxylase EGLN3 (PHD3) that controls HIFα protein stability is increased in Irp1−/− duodenum (Figure 3D, 3E and S2). EGLN3 is part of a feedback loop for HIF-2α and the EGLN3 gene contains a bona fide hypoxia response element (Lee and Percy, 2011; Minamishima et al., 2009; Pescador et al., 2005). Second, induction of the amino acid transporter SLC38A1 and the enzyme gamma-glutamyl hydrolase (Ggh) by iron deficiency requires HIF-2α (Taylor et al., 2011). Duodenal expression of both genes is increased in Irp1−/− mice (Figure 3D and 3E). Third, in addition to DCytb, DMT1, ferroportin, SLC38A1, Ggh and EGLN3, the expression of Pparg, Nt5E, and Snca mRNAs are also altered in Irp1−/− duodenum (Figure 3E and Figure S2). Each of these genes is either a HIF-2α target or is responsive to O2 level (see Figure S2). Taken together, the polycythemia observed in Irp1−/− mice is linked with increases in HIF-2α activity and, as such, supports the notion that loss of IRP1 is similar to a HIF-2α gain of function. On this basis we conclude that IRP1 provides a critical link between fundamental pathways of cellular iron and oxygen use and systemic regulation of iron absorption and erythropoiesis.
Selective Dysregulation of HIF-2α mRNA Translation in Irp1−/− Mice
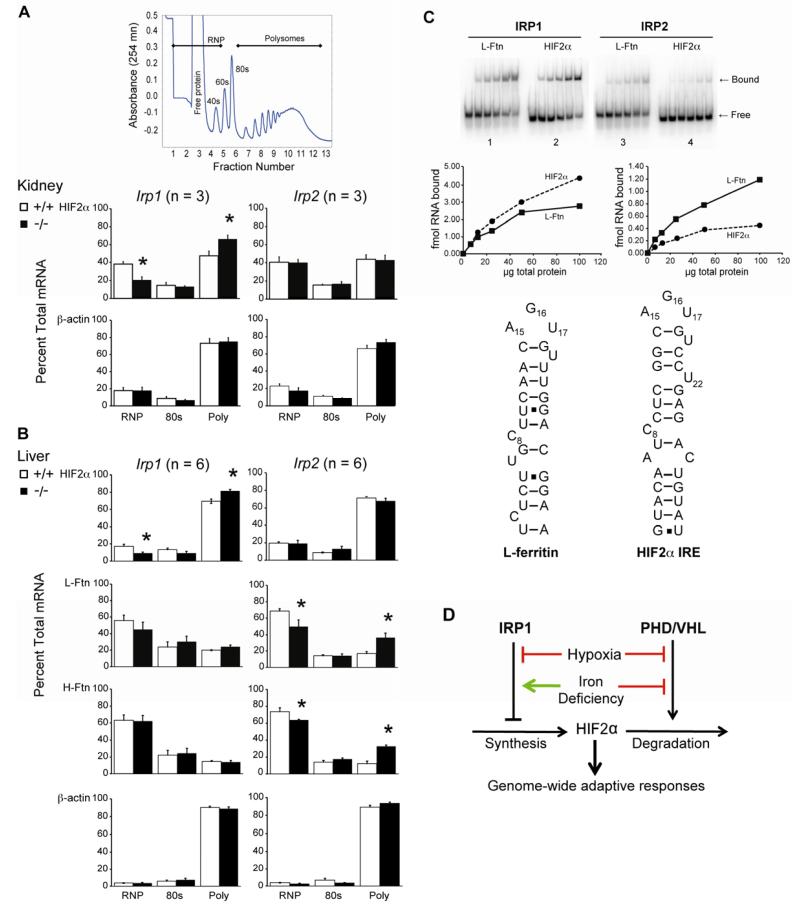
To determine the basis for development of polycythemia in Irp1−/− but not Irp2−/− mice we determined the translation state of HIF-2α and other 5′ IRE-containing mRNAs (Figure 4A and Table S4 and S5). Repressed mRNA in ribonucleoprotein particles (RNP) are separated from the 80S monosomes and the translationally active polysomes by polysome profile analysis. Given the increased expression of Epo mRNA in Irp1−/− kidney we asked whether the HIF-2α mRNA translation state was altered. In Irp1+/+ kidney nearly equal amounts of HIF-2α mRNA was in the RNP and polysome bound pools (Figure 4A). Strikingly, in Irp1−/− but not Irp2−/− kidney, HIF-2α mRNA was substantially derepressed such that the majority was polysome bound compared to Irp1+/+ mice. Thus, IRP2 does not compensate for the absence of IRP1 regarding wild type repression of HIF-2α mRNA. Deficiency of either IRP failed to affect β-actin mRNA translation (Figure 4A). Thus the polycythemia in Irp1−/− mice is associated with a selective translational derepression of HIF-2α mRNA in kidney not seen in Irp2−/− mice.
Figure 4. Selective Dysregulation of HIF-2α mRNA Translation in Irp1−/− Mice.
[A] Kidney polysome profile (PP) analysis at 8 wk. A typical PP is shown. [B] Liver PP analysis at 8 wk. [C] EMSA from Irp2−/− or Irp1−/− kidney cytosol. Panels 1 and 2 show binding of RNAs (0.5 nM) using Irp2−/− kidney cytosol so binding is to IRP1. Panels 3 and 4 show binding of RNAs (0.5 nM) using Irp1−/− kidney cytosol so binding is to IRP2. Result is representative of n = 2 experiments. Proposed secondary structure of L-ferritin and HIF-2α IRE. [D] Model of the impact of IRP1 on HIF-2α regulation. Results in panels A and B are expressed as mean ± SEM * = P <0.05; ** = P < 0.01; *** = P < 0.001. See Tables S4 and S5.
We then investigated the role each IRP had in controlling the translation state of 5′ IRE-containing mRNAs in the liver, a tissue in which HIF-2α has many metabolic roles (Haase, 2010). Liver expresses less IRP1 RNA binding activity compared to kidney (Meyron-Holz et al., 2004). Thus, it was not surprising that a larger fraction of HIF-2α mRNA was polysome-bound compared to kidney (Figure 4B and Table S5). Similar to kidney, loss of IRP1 but not IRP2, led to translational derepression of HIF-2α mRNA. However, all other 5′ IRE-containing mRNAs in liver showed the opposite result. Translational derepression of H- and L-ferritin, ferroportin and mitochondrial aconitase mRNAs was observed in Irp2−/− but not Irp1−/− liver (Figure 4B and Table S5). Taken together, our findings illustrate unique roles for each IRP in orchestrating the fate of 5′ IRE-containing mRNAs.
We next asked if the selective dysregulation of HIF-2α in IRP1−/− mice related to the level or selectivity of IRP1 and IRP2 binding activity in kidney. Previous studies in 786-0 cells found that the level of IRP2 expressed was insufficient to bind to the HIF-2α IRE (Zimmer et al., 2008). Kidney cytosol extracts from IRP1−/− or IRP2−/− mice were the source of IRP2 or IRP1, respectively, in electrophoretic mobility shift assays (EMSA) (Figure 4C). HIF-2α IRE binds to IRP1 as well or better than the L-ferritin IRE (Figure 4C, subpanels 1 and 2). Consistent with previous studies, IRP2 binding activity was much lower than that for IRP1 in kidney (Meyron-Holz et al., 2004). However, in contrast to IRP1, IRP2 bound to the L-ferritin IRE better than it did to the HIF-2α IRE. Thus, the specific derepression of HIF-2α mRNA in Irp1−/− mice is associated with a reduced level of IRP2 RNA binding activity in kidney compared to IRP1 coupled with a greater preference of IRP2 for the L-ferritin IRE.
Our study establishes new paradigms concerning the integrated regulation of iron and oxygen metabolism in mammals. First, the polycythemic phenotype of Irp1−/− mice illustrates that control of HIF-2α synthesis is a major aspect of IRP1’s function. A key consequence of this is that IRP1 will amplify the impact of prolyl hydroxlase-mediated regulation of HIF-2α accumulation in response to oxygen while dampening the action of iron (Fig. 4D). Since the Irp1−/− mice used here were fed a normal diet under normoxic conditions altered exposure to iron or oxygen will likely exacerbate the phenotype of IRP1 deficiency. Second, IRP1 has a physiologic role separable from IRP2 that suggests IREs can be functionally distinct in vivo. The presence of a similar non-canonical 3′ unpaired nucleotide in the DMT1 IRE, as is predicted for HIF-2α (Fig. 4C), and its’ preferential recognition by IRP1 suggests a mechanism for IRP selectivity (Gunshin et al., 2001). Third, the key role of the Fe-S switch in controlling IRP1 RNA binding activity predicts unanticipated links between major pathways of iron metabolism (e.g. Fe-S cluster biogenesis), HIF-2α, and oxygen sensing that when perturbed may contribute to disease phenotype. Fourth, the finding that the oxygen-sensitivity of the Fe-S switch mechanism of IRP1 may be modulated by S138 phosphorylation suggests novel mechanisms that may contribute to pathological abnormalities in Epo expression or other aspects of HIF-2α action (Andrews and Bridges, 1998; Deck et al., 2009; van der Putten et al., 2008).
Sensing of oxygen and iron is a central aspect of the homeostatic responses in a wide spectrum of physiological scenarios and its dysregulation is a pathophysiological feature of diseases including polycythemias and other erythropoietic diseases, pulmonary syndromes, renal disease, cancers and other disorders (Prabhakar and Semenza, 2012). While HIF-2α and IRP1 respond to hypoxia, iron deficiency and other metabolic stresses the extent to which they do so in a concerted manner has only recently come into greater focus (Taylor et al., 2011; Zimmer et al., 2010). The current demonstration that IRP1 is the key IRP regulator of HIF-2α establishes a broad framework through which the regulation of iron and oxygen homeostasis is integrated and provides a new paradigm for defining how maladaptive regulation of this network contributes to human disease.
EXPERIMENTAL PROCEDURES
Mice
Generation of tissue-wide Irp1−/− and Irp2−/− mice has been described (Galy et al., 2005a). Irp1−/− and Irp2−/− mice used here were backcrossed 6 generations with C57BL/6 mice. Irp1+/− or Irp2+/− were bred to generate Irp1−/− and Irp2−/− and their wildtype littermates. Animal use met UW-Madison IACUC requirements.
Hematologic and Iron Parameters
Blood was drawn from the retroorbital sinus or heart for complete blood count analysis (CBC), calculated or manual hematocrit analysis or for serum for the Epo quantikine kit ELISA MEP00B (R&D Systems). For CBC and hematocrit, blood was collected into heparin- or EDTA-treated tubes. CBC analysis was by the UW School of Vet. Med. Blood and tissue were analyzed for serum iron or tissue non-heme iron as described (Schmidt et al., 2010).
Flow cytometry
For hematopoetic cell lineage analysis flow cytometric analyses were as described (Socolovsky et al., 2001; Zhang et al., 2009). LSK cells and myeloid progenitors were also determined by flow cytometry (Wang et al., 2011). Stained cells were analyzed on a FACSCalibur (BD Biosciences) or MACSQuant Analyzer (Miltenyi Biotec). Colony forming assays for quantization of BFU-E and various CFU progenitors and Epo-sensitivity of CFU-E used growth in methylcellulose with defined media (StemCell Technologies). Antibodies to surface antigens and other details are described in the supplement.
Tissue staining and immunohistochemistry
Spleens were fixed in 10% buffered formalin and embedded in paraffin. Deparaffinized sections of tissue were stained with hematoxylin and eosin or CD71 antibody (TfR1) in the Children’s Hospital Boston, Dept of Pathology Histology Lab. Images were acquired using a 4×/0.13 objective lens on a B×51 microscope with a DP71 Digital Camera using Olympus MicroSuite™ FIVE Imaging Software.
qPCR
RNA was isolated using STAT-60 (Tel-Test, Inc). Reverse-transcription used total RNA, Superscript III (Invitrogen, Carlsbad, CA) and random hexamers. Real Time PCR on cDNA used SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7000.
Polysome profile analysis
Tissues were minced and homogenized in 3 volumes PB (40 mM HEPES pH 7.4, 100 mM KCl, 5 mM MgCl2, 2 mM citrate, and 1 mM DTT) in a Potter Elvehjem homogenizer. The homogenate was centrifuged at 5000 ×g at 4 °C for 20 min. The upper 2/3 of supernatant was brought to 1% Na deoxycholate, 1% Triton X-100 in PB. 500 μL sample was loaded on a linear 15% to 60% sucrose gradient in PB and centrifuged at 180,000 ×g in a Sorvall TH641 rotor for 2 h at 4°C. Gradient fractionation used an ISCO UA-6 and 254 nm detector linked to an Agilent integrator.
EMSA
RNA binding assay with [32P] IREs (~15,000 dpm/fmol) was as described (Goforth et al., 2010).
Microarray
Mice were fasted overnight and killed under isoflurane. Total RNA was isolated from frozen duodenal mucosal cells (see qPCR), labeled using the Ambion Gene Chip WT Expression Kit (Ambion) and hybridized to Affymetrix Mouse Gene 1.0 ST Arrays.
Statistical Methods
Group means were tested for differences by Student’s T-tests. Error bars are SEM. Statistical analysis of arrays is described in the Supplement.
Supplementary Material
Research Highlights.
-
■
Derepression of HIF-2α mRNA in Irp1−/− mice causes age-dependent polycythemia.
-
■
HIF-2α hyperactivity is observed in multiple tissues of Irp1−/− mice.
-
■
The mRNA regulons of IRP1 and IRP2 are separable in vivo.
-
■
The IRP1/HIF-2α axis is a therapeutic target for hematologic or oncologic disorders.
ACKNOWLEDGEMENTS
This study was supported: RE: NIH RO1DK66600, the Iron Metabolism Research Fund, the UW-Madison Graduate School and Hatch Project WIS01324; JZ: NIH R01CA152108, a Shaw Scientist Award from the Greater Milwaukee Foundation, a V Scholar Award from the V Foundation for Cancer Research, and an Investigator Initiated Grant from the UW Comprehensive Cancer Center Support (UWCCC); MDF: NIH R01 DK 080011; CK: NIH GM102756 and grant 1UL1RR025011 from the Clinical and Translational Science Award program of the National Center for Research Resources, National Institutes of Health. We thank UWCCC for use of its flow cytometry and experimental pathology units. Partial support from NIH/NCI P30 CA014520 for the UWCCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson CP, Shen M, Eisenstein RS, Leibold EA. Mammalian iron metabolism and its control by IRPs. Biochim. Biophys. Acta. 2012;1823:1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Bridges KR. Disorders of iron metabolism and sideroblastic anemia. In: Nathan DG, Oski SH, editors. Nathan and Oski’s Hematology in Infancy and Childhood. W.B. Saunders; Philadelphia, PA: 1998. pp. 423–461. [Google Scholar]
- Bullock GC, Delehanty LL, Talbot AL, Gonias SL, Tong WH, Rouault TA, Dewar B, Macdonald JM, Chruma JJ, Goldfarb AN. Iron control of erythroid development by a novel aconitase-associated regulatory pathway. Blood. 2010;116:97–108. doi: 10.1182/blood-2009-10-251496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of IRP 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MR, Shawron KM, Rendina E, Peterson SK, Lucas EA, Smith BJ, Clarke SL. Hif-2α is translationally repressed in response to dietary iron deficiency. J. Nutr. 2011;141:1590–1596. doi: 10.3945/jn.111.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deck KM, Vasanthakumar A, Anderson SA, Goforth JB, Kennedy MC, Antholine WE, Eisenstein RS. Evidence that phosphorylation of IRP 1 at S138 destabilizes the [4Fe-4S] cluster in cytosolic aconitase by enhancing 4Fe-3Fe cycling. J. Biol. Chem. 2009;284:12701–12709. doi: 10.1074/jbc.M807717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franovic A, Holterman CE, Payette J, Lee S. Human cancers converge at the HIF-2α oncogenic axis. Proc. Natl. Acad. Sci. USA. 2009;106:21306–21311. doi: 10.1073/pnas.0906432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy B, Ferring D, Hentze MW. Generation of conditional alleles of the murine IRP-1 and -2 genes. Genesis. 2005a;43:181–188. doi: 10.1002/gene.20169. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring D, Minana B, Bell O, Janser HG, Muckenthaler M, Schumann K, Hentze MW. Altered body iron distribution and microcytosis in mice deficient in IRP2. Blood. 2005b;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- Goforth JB, Anderson SA, Nizzi CP, Eisenstein RS. Multiple determinants within IREs dictate IRP binding and regulatory hierarchy. RNA. 2010;16:154–169. doi: 10.1261/rna.1857210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. Acute postnatal ablation of HIF-2α results in anemia. Proc. Natl. Acad. Sci. USA. 2007;104:2301–2306. doi: 10.1073/pnas.0608382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA. Iron-dependent regulation of DMT1. FEBS Letts. 2001;509:309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- Haase VH. Hypoxic regulation of erythropoiesis and iron metabolism. Amer. J. Physiol. 2010;299:F1–13. doi: 10.1152/ajprenal.00174.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nature Rev. Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Ann. Rev. Pathol. 2011;6:165–192. doi: 10.1146/annurev-pathol-011110-130321. [DOI] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. HIFs and the response to hypoxic stress. Mol. Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2α, but not HIF-1α, promotes iron absorption in mice. J. Clin. Invest. 2009;119:1159–1166. doi: 10.1172/JCI38499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyron-Holz EG, Ghosh MC, Iwai K, LaVaute T, Brazzolotto X, Berger UV, Land W, Olivarerre-Wilson H, Grinberg A, Love P, et al. Genetic ablations of IRP 1 and 2 reveal why IRP 2 dominates iron homeostasis. EMBO J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamishima YA, Moslehi J, Padera RF, Bronson RT, Liao R, Kaelin WG., Jr. A feedback loop involving the PHD3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol. Cell. Biol. 2009;29:5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landazuri MO, Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the EGLN3/PHD3 gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peslak SA, Wenger J, Bemis JC, Kingsley PD, Koniski AD, McGrath KE, Palis J. Epo-mediated expansion of late-stage erythroid progenitors in the bone marrow initiates recovery from sublethal radiation stress. Blood. 2012;120:2501–2511. doi: 10.1182/blood-2011-11-394304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by HIFs 1 and 2. Physiol Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Galy B, Muckenthaler MU, Hentze MW. IRPs limit HIF-2α expression in iron deficiency. Nat. Struct. Mol. Biol. 2007;14:420–426. doi: 10.1038/nsmb1222. [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Andrews NC, Fleming MD. Hepcidin induction by transgenic overexpression of Hfe does not require the Hfe cytoplasmic tail, but does require hemojuvelin. Blood. 2010;116:5679–5687. doi: 10.1182/blood-2010-04-277954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortegagna M, Ding K, Zhang Q, Oktay Y, Bennett MJ, Bennett M, Shelton JM, Richardson JA, Moe O, Garcia JA. HIF-2α regulates murine hematopoietic development in an Epo-dependent manner. Blood. 2005;105:3133–3140. doi: 10.1182/blood-2004-05-1695. [DOI] [PubMed] [Google Scholar]
- Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal HIFs are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–164. doi: 10.1016/j.cmet.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Nam H, Fleming MD, Haase VH, Brugnara C, Lodish HF. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Strauss RG. Anaemia of prematurity: pathophysiology and treatment. Blood Rev. 2010;24:221–225. doi: 10.1016/j.blre.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Qu A, Anderson ER, Matsubara T, Martin A, Gonzalez FJ, Shah YM. HIF-2α mediates the adaptive increase of intestinal ferroportin during iron deficiency in mice. Gastroenterology. 2011;140:2044–2055. doi: 10.1053/j.gastro.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong WH, Sourbier C, Kovtunovych G, Jeong SY, Vira M, Ghosh M, Romero VV, Sougrat R, Vaulont S, Viollet B, et al. The glycolytic shift in fumarate-hydratase-deficient kidney cancer lowers AMPK levels, increases anabolic propensities and lowers cellular iron levels. Cancer cell. 2011;20:315–327. doi: 10.1016/j.ccr.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Putten K, Braam B, Jie KE, Gaillard CA. Mechanisms of disease: Epo resistance in patients with both heart and kidney failure. Nature Clin. Pract. Nephrol. 2008;4:47–57. doi: 10.1038/ncpneph0655. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu Y, Li Z, Wang Z, Tan LX, Ryu MJ, Meline B, Du J, Young KH, Ranheim E, et al. Endogenous oncogenic Nras mutation initiates hematopoietic malignancies in a dose- and cell type-dependent manner. Blood. 2011;118:368–379. doi: 10.1182/blood-2010-12-326058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G, Goodnough LT. Anemia of chronic disease. N. Engl. J. Med. 2005;352:1011–1023. doi: 10.1056/NEJMra041809. [DOI] [PubMed] [Google Scholar]
- Young RM, Wang SJ, Gordan JD, Ji X, Liebhaber SA, Simon MC. Hypoxia-mediated selective mRNA translation by an internal ribosome entry site-independent mechanism. J. Biol. Chem. 2008;283:16309–16319. doi: 10.1074/jbc.M710079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang J, Liu Y, Sidik H, Young KH, Lodish HF, Fleming MD. Oncogenic Kras-induced leukemogeneis: hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113:1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Ebert BL, Neil C, Brenner K, Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T, et al. Small-molecule inhibitors of HIF-2α translation link its 5′IRE to O2 sensing. Mol. Cell. 2008;32:838–848. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Lamb J, Ebert BL, Lynch M, Neil C, Schmidt E, Golub TR, Iliopoulos O. The connectivity map links IRP-1-mediated inhibition of HIF-2α translation to the anti-inflammatory 15-deoxy-delta12,14-prostaglandin J2. Cancer Res. 2010;70:3071–3079. doi: 10.1158/0008-5472.CAN-09-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.