Sir,
Timely diagnosis and prompt treatment of infectious tuberculosis (TB) cases is pivotal towards TB control. Multi-drug resistant tuberculosis (MDR-TB) defined as the resistance to at least rifampicin (RIF) and isoniazid (INH) poses grave challenge because of prolonged, limited and expensive treatment options with 10 to 30 per cent of cases resulting in failure of treatment and death1,2.
Conventional drug susceptibility using solid medium such as Lowenstein - Jensen (L-J) is time consuming whereas liquid medium based methods such as Mycobacterial Growth Indicator Tube (MGIT) System 960 (BD Diagnostics, USA) are sensitive and faster but involve prohibitive expenditure3. Commercial line probe assays (LPA) such as INNO-LIPA Rif TB (Innogenetics, Ghent, Belgium) and Genotype MTBDR plus assay (Hain Life Sciences, GmbH, Germany) based on reverse hybridization of amplicons to immobilized membrane based probes covering wild type and common mutation sequences of rpoB and rpoB, katG, inhA respectively, have rapid turn-around time of 48 to 72 h4.
In this study, the performance of the Genotype MTBDR plus assay was evaluated using MGIT 960 as gold standard. The study was conducted in Department of Microbiology, Lala Ram Sarup Institute of Tuberculosis and Respiratory Diseases, New Delhi. The Institute is a referral tuberculosis hospital and National Reference Laboratory (NRL) for Revised National Tuberculosis Control Programme (RNTCP). Suspected MDR-TB patients attending the out-patient unit are referred for rapid culture and drug susceptibility testing. The MGIT 960 liquid culture system in the Institute is accredited for susceptibility testing for first line and second line drugs by Supra National Reference Laboratory (SNRL), Antewerp, Belgium.
A total of 120 smear positive sputum specimens received from 120 patients during a period of five months (October 2010 - February 2011) were processed in BSL-3 laboratory by standard N-acetyl-L-cysteine-sodium hydroxide (NALC-NaOH) method5. The concentrated samples were subjected to culture and drug susceptibility of Mycobacterium tuberculosis by MGIT 9605. Final critical concentration of anti-TB drugs used was 0.1 μg/ml of INH and 1.0 μg/ml of RIF6. Following inoculation, samples were aliquoted in duplicate for DNA extraction. DNA extraction, amplification and hybridization were carried out as per manufacturer's instructions in different pre-designated, thoroughly cleaned rooms. DNA from standard strain H37Rv and molecular grade water were used as positive and negative control, respectively with every run.
For any invalid results, test was repeated on DNA extracts stored at -20° C. Each sample was run in duplicate in different batches and results matched for concordance. The result of DNA strips was interpreted as resistant for RIF and INH based on absence of hybridization to any of wild type probes and/or positive hybridization signal of common mutant probes7.
Twenty per cent of randomly selected DNA were sent for external proficiency testing to National JALMA Institute of Leprosy and Other Mycobacterial Diseases, Agra, which is an accredited National Reference Laboratory for LPA. In the internal quality control, 100 per cent concordance was obtained in the duplicate testing of Genotype MTBDR plus with all 120 samples yielding same result. These included 118 valid and two invalid results. The results of external proficiency testing were in 100 per cent concordance with our results.
Overall concordance of RIF and INH result between Genotype MTBDR plus and MGIT 960 was found to be 96.6 and 84.7 per cent, respectively. Other studies have reported good concordance of 84.4-98.1 per cent for RIF and 87.3-90.14 per cent for INH respectively between Genotype MTBDR plus and conventional drug susceptibility tests8–10.
The sensitivity, specificity, positive and negative predictive values for detection of RIF resistance by Genotype MTBDR plus was found to be 97.6, 94.4, 97.6 and 94.4 per cent respectively (Table I). High sensitivity for RIF has also been found in other studies; 98.1 per cent, (95% CI 95.9-99.1) in the meta-analysis for comparison of Genotype MTBDR and Genotype MTBDR plus assays with conventional susceptibility testing4 and 100 per cent in studies from Uganda and France7,9. Specificity for RIF resistance in various studies has been found to be 98.7 per cent (pooled specificity with 95% CI 97.3-99.4%), 96.1 and 100 per cent4,7,11. In one study, specificity of detection of RIF resistance by using INNO-LiPA Rif TB compared to conventional methods was found to be 66.7 per cent12. Such variation could be due to the intrinsic limitation of the kit mentioned above.
Table I.
Comparison of LPA result with MGIT 960 for RIF and INH resistance (n=118)
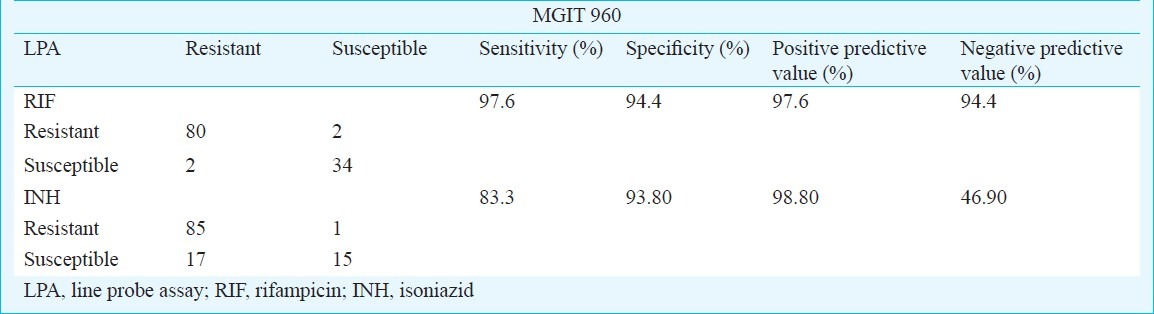
The sensitivity and specificity for INH resistance for Genotype MTBDR plus was found to be 83.3 and 93.8 per cent, respectively (Table I). Resistance in 17 isolates was not detected by Genotype MTBDR plus which could be due to inability of the test to detect mutation in other genes (ahpC-oxyR and ndh) conferring INH resistance. The low sensitivity of 83.3 per cent, obtained for INH in our study is in accordance to most other studies - 84.3 per cent (95% CI 76.6-89.8), 80.8 and 67 per cent4,7,9. However, some studies have found good sensitivity for INH (92.6, 93 and 95.3%, respectively)10,11,13.
The mutation patterns of RIF and INH produced by the Genotype MTBDR plus are displayed in Table II. Specific mutation could be detected in 58 of 82 (70.7%) RIF resistant isolates. Of these, 49 had mutation in codon S531L, 6 (7.3%) in D516V and 3 (3.7%) in H526D. Other reports from India and abroad have found S531L mutation to be commonest7,11,14–16. In 24 of 82 (29.3%) RIF resistant isolates, one or more wild type probes were missing with no gain in mutant probes. These isolates are depicted as unknown (UK). Among these maximum isolates had missing WT8-10 (41.7%). In a study from South Vietnam, 66.7 per cent isolates did not have any known mutation11.
Table II.
Mutation pattern of RIF (rpoB) and INH (katG & inhA) obtained from Genotype MTBDR plus assay
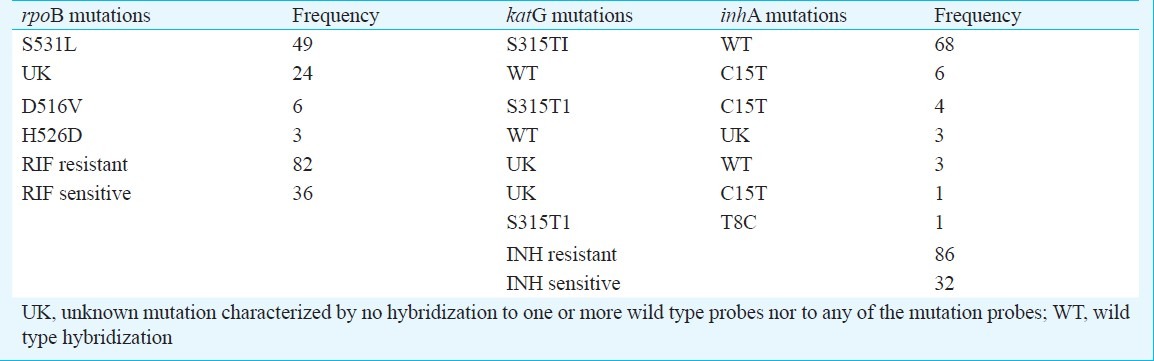
In Genotype MTBDR plus, INH resistance is detected by probes of two genes; katG and inhA Among 86 INH resistant isolates, katG mutation occurred in 77 (89.5%) of isolates. Specific mutations in codon S315T1 of katG gene was found in 73 (94.8%) isolates. Remaining four had missing wild type with no gain in mutant probes. Mutations in InhA gene occurred in 15 of 86 (17.4%) INH resistant isolates. Specific InhA mutations were found in 12 of 15 (80.0%) INH resistant isolates, of which 11 had mutation in codon C15T and one in T8C (Table II). In remaining three isolates, no specific mutation band could be detected. The mutation pattern obtained was similar to other studies in this regard7,17.
In conclusion, genotype MTBDR plus assay was found to be rapid, sensitive and specific test for routine drug susceptibility testing for diagnosis of RIF resistance which is more crucial for management of MDR-TB. All RIF resistant cases can be shifted to category IV treatment timely, thereby curtailing the spread. Larger field trials are imperative to assess the validity and usefulness of this test in field scenario if the test is deemed to play a pivotal role in tuberculosis control at national level in India.
Acknowledgment
Authors acknowledge the technical support of Foundation of Innovative Diagnostics (FIND), India in the study.
References
- 1.CDC. Emergence of Mycobacterium tuberculosis with extensive resistance to second line drugs-world-wide, 2000-2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–5. [PubMed] [Google Scholar]
- 2.Banerjee S, Siddiqui N, Hasnain SE. Drug resistant tuberculosis. In: Sharma SK, Alladi M, editors. Tuberculosis. 2nd ed. New Delhi: Jaypee; 2009. pp. 682–713. [Google Scholar]
- 3.Gupta A, Sharma SK, Pande JN. Diagnostic methods for tuberculosis. Indian J Chest Dis Allied Sci. 1993;35:63–84. [PubMed] [Google Scholar]
- 4.Ling DI, Zwerling AA, Pai M. GenoType MTBDR assays for the diagnosis of multi-drug resistant tuberculosis. Eur Respir J. 2008;32:1165–74. doi: 10.1183/09031936.00061808. [DOI] [PubMed] [Google Scholar]
- 5.Kent PT, Kubica GP. Public health mycobacteriology, A guide for level III laboratory. Atlanta, GA: Center for Disease Control, Division of Laboratory Training and Consultation. US Department of Health and Human Services; 1985. [Google Scholar]
- 6.Berner P, Palicova F, Rusch-Gerdes S, Drugeon HB, Pfyffer GE. Multi-center evaluation of fully automated BACTEC mycobacterial growth indicator tube system MGIT 960 for susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:150–4. doi: 10.1128/JCM.40.1.150-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert H, Bwanga F, Mukkada S, Nyesiga B, Ademun JP, Lukyamuzi G, et al. Rapid screening of MDR-TB using molecular line probe assay is feasible in Uganda. BMC Infect Dis. 2010;10:41. doi: 10.1186/1471-2334-10-41. (1-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makinen J, Marttila HJ, Marjamaki M, Viljanen MK, Soini H. Comparison of two commercially available DNA line probe assays for detection of multi-drug resistant Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:350–2. doi: 10.1128/JCM.44.2.350-352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brossier F, Veziris N, Pernot CT, Jarlier V, Sougakoff W. Performance of the GenoType MTBDR line probe assay for detection of resistance to rifampicin and isoniazid in strains of Mycobacterium tuberculosis with low and high level resistance. J Clin Microbiol. 2006;44:3569–4. doi: 10.1128/JCM.01054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balabanova Y, Drobniewski F, Nikolayevskyy V, Kruuner A, Malomanova N, Simak T, et al. An integrated approach to rapid diagnosis of tuberculosis and multi-drug resistance using liquid culture and rapid methods in Russia. PLoS One. 2009;4(9):e7129. doi: 10.1371/journal.pone.0007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huyen MNT, Tiemersma EW, Nguyen TNL, Cobelens FGJ, Nguyen HD, Sy DH, et al. Validation of the GenoType MTBDRplus assay for diagnosis of multi-drug resistant tuberculosis in South Vietnam. BMC Infect Dis. 2010;10:149. doi: 10.1186/1471-2334-10-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viveiros M, Leandro C, Rodrigues L, Almeida J, Bettencourt R, Couto I, et al. Direct application of the INNO-LiPA Rif TB Line probe assay for rapid identification of Mycobacterium tuberculosis complex strains and detection of rifampicin resistance in 360 smear positive respiratory specimens from an area of high incidence of multi-drug resistant tuberculosis. J Clin Microbiol. 2005;43:4880–4. doi: 10.1128/JCM.43.9.4880-4884.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anek-Vorapong R, Sinthuwattanawibool C, Podewils LJ, McCarthy K, Ngamlert K, Promsarin B, et al. Validation of the GenoType MTBDRplus assay for detection of MDR-TB in a public health laboratory in Thailand. BMC Infect Dis. 2010;20:123. doi: 10.1186/1471-2334-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suresh N, Singh UB, Arora J, Pant H, Seth P, Sola C, et al. RpoB gene sequencing and spoligotyping of multi-drug resistant Mycobacterium tuberculosis isolates from India. Infect Genet Evol. 2006;6:474–83. doi: 10.1016/j.meegid.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Mani C, Selvakumar N, Narayanan S, Narayanan PR. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J Clin Microbiol. 2001;39:2987–90. doi: 10.1128/JCM.39.8.2987-2990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddiqi N, Shamim M, Hussain S, Choudhary RK, Ahmed N, Prachee, et al. Molecular characterization of multidrug-resistant isolates of Mycobacterium tuberculosis from patients in North India. Antimicrob Agents Chemother. 2002;46:443–50. doi: 10.1128/AAC.46.2.443-450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazbón MH, Brimacombe M, Valle MB, Cavatore M, Guerrero MI, Varma-Basil M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–9. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


