Abstract
The non-structural protein 13 (nsp13) of Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) is a helicase that separates double-stranded RNA or DNA with a 5’-3’ polarity, using the energy of nucleotide hydrolysis. We have previously determined the minimal mechanism of helicase function by nsp13 where we demonstrated that the enzyme unwinds nucleic acid in discrete steps of 9.3 base-pairs each with a catalytic rate of 30 steps per second. In that study we used different constructs of nsp13 (GST and H6 constructs). GST-nsp13 showed much more efficient nucleic acid unwinding than the H6-tagged counterpart. At 0.1 second, more than 50% of the ATP is hydrolyzed hydrolysed by GST-nsp13 compared to less than 5% ATP hydrolysis by H6-nsp13. Interestingly, the two constructs have the same binding affinity for nucleic acids. We, therefore propose that the difference in the catalytic efficiency of these two constructs is due to the interference of ATP binding by the histidine tag at the amino-terminus of nsp13.
Keywords: SARS-CoV, Helicase, nucleic acids, antivirals, molecular modeling, ATP hydrolysis
INTRODUCTION
Severe acute respiratory syndrome (SARS), emerged in the Guangdong province of southern China in 2002 and it was mainly characterized by flu-like symptoms, including high fevers surpassing 38°C or 100.4°F, myalgia, dry non-productive dyspnea, lymphopaenia and infiltrate on chest radiography. The resulting pneumonia resulted in acute breathing problems requiring artificial respirators in 38% of all the cases (37). The overall mortality rate was about 10%, but varied profoundly with age: although SARS affected relatively few children and generally appeared to be milder in the pediatric age group, the mortality rate in the elderly was as high as 50% (10, 18, 36). The causative agent of SARS is SARS coronavirus (SARS-CoV). Coronaviruses are named after their corona crown-like appearance in electron micrographs, which is caused by the club-shaped peplomers that radiate outwards from the viral envelope. The positive-strand RNA genome of about 30 kb is covered by a helical capsid. (29, 31). There are 16 SARS-CoV non-structural proteins (nsps) that are expected to contribute to the formation of the viral replication complex (RC), which is also expected to consist of several unknown host factors. The composition of the complex is likely to change during the course of viral replication.
The first coronavirus proteins that have been studied in detail include viral proteinases, namely the papain-like cysteine proteinase (Plpro, nsp3) and the 3C-like cysteine proteinase (3Clpro, nsp5), which cleave the polyprotein into individual polypeptides that are required for replication and transcription (44, 45). Following the translation of the messenger RNA to yield the polyprotein, the 3C-like proteinase, also known as the main protease (Mpro), is first auto-cleaved from the polyproteins to become a mature enzyme. The Mpro further cleaves all the 11 remaining downstream non-structural proteins while nsp3 cleaves protease recognition sites between nsp1/2, nsp2/3 and nsp3/4 (17). Hence, this auto-proteolysis leads to formation of 16 non-structural proteins, including the RNA-dependent RNA polymerase (RdRp) and an NTPase/helicase that are known as non-structural proteins 12 and 13, respectively (nsp12 and nsp13). They are likely to form the core of membrane-bound replication-transcription complexes in double-membrane vesicles at perinuclear regions (47).
Helicases are motor proteins that utilize the energy derived from nucleotide hydrolysisto unwind double-stranded nucleic acids into two single-stranded nucleic acids (7, 15, 25–27, 30, 34, 35). Initially, helicases were only thought to be molecular engines that unwind nucleic acids during replication, recombination, and DNA repair (1–4, 11, 15, 21, 40, 52–54). Recent studies have shown that they are also involved in other biological processes, including displacement of proteins from nucleic acid, movement of Holliday junctions, chromatin remodeling, catalysis of nucleic acid conformational changes (9, 14, 20, 24, 48, 49), several aspects of RNA metabolism, including transcription, mRNA splicing, mRNA export, translation, RNA stability and mitochondrial gene expression (47). Some human diseases, including Bloom’s syndrome, Werner’s syndrome, and Xeroderma Pigmentosum have been associated with defects in helicase function (12, 16, 41, 55).
The specific role of nsp13 in SARS-CoV replication has yet to be established, although, many studies have shown the interaction of nsp13 with other SARS-CoV non-structural proteins, such as nsp7, nsp8 and nsp12 (32, 38, 50), and the effect of these interactions on nsp13 unwinding activity is unknown. Previously, a basic biochemical characterization of nsp13 demonstrated that it can unwind both doublestranded DNA and RNA in a 5’-3’ direction, and it can hydrolyze all deoxyribonucleotide and ribonucleotide triphosphates (19, 43). More recently, it was reported that the amplitude of nsp13 increases with longer 5’-overhang containing DNA substrates (23). These studies were conducted using construct of nsp13 that was fused with hexahistidine (H6-nsp13). H6-nsp13 was enzymatically active, but appeared to be considerably less active than other helicases (13, 28). We recently prepared GST-nsp13 using a baculovirus expression system and compared it to the MBP-nsp13 and H6-nsp13 variants of the helicase prepared in bacterial systems. We found that GST-nsp13 was dramatically more active than the other variants and had activity comparable to other helicases (5, 6) (42). We suggested that the enhanced activity of GST-nsp13 was as a result of better folding of the protein in the eukaryotic expression system. In this present study, we use both biochemical and structural studies to show that the hexahistidine tag at the N-terminus of nsp13 reduced the ability of the H6-nsp13 to bind and hydrolyze ATP, hence, the reason for the slow unwinding activity of H6-nsp13 as compared to the GST construct, as there was no difference in the DNA binding affinity of the two constructs.
MATERIALS AND METHODS
Cloning, Expression and Purification of GST-nsp13 and H6-nsp13 Helicase
The cloning, expression and purification of the two constructs of nsp13 was as previously described (53).
Nucleic Acid Substrates
Synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Sequences of the DNA substrates is same as used in Adedeji et al. (5). Concentrations were determined spectrophotometrically using absorption at 260 nm and their extinction coefficients. Unlabeled oligonucleotides were annealed to corresponding 5’-Cy3 labeled 18-mer, in 50 mM Tris pH 8.0, 50 mM NaCl at a ratio of 1:1.2 by heating at 95 °C for 5 min and cooling slowly to room temperature. Unlabeled 18-mer was used as traps in all helicase assay.
Pre-Steady State Kinetics Assays
The reactions were carried out in 20 mM HEPES (pH 7.5), 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 5% glycerol at 30 °C using a Rapid Quench Flow instrument (KinTek Corp.) (39). GST-nsp13 (100 nM) and DNA substrates (5 nM) were loaded into one of the sample loops (15 µl), whereas ATP (2 mM), and unlabeled DNA substrate (1 µM) were loaded into the other sample loop (15 µl). Samples were rapidly mixed and the reaction was quenched with 100 mM EDTA, 0.2% SDS, and 20% glycerol after desired time intervals. Experiments were performed at least three times. The released single-stranded DNA (ssDNA) product and unwound double-stranded DNA (dsDNA) were resolved on a 6% non-denaturing-PAGE (polyacrylamide gel electrophoresis) using a running buffer containing 89 mM Tris-Borate pH 8.2, and run for 2 hours at 4°C and 150 V. The controls for measuring maximum unwinding were dsDNA denatured by heating for 5 min at 95 °C and loading immediately on the gel as suggested by Ahnert et al. (39). In this and subsequent assays the gels were scanned with a phosphorimager (FLA 5000, FujiFilm). The band intensities representing ssDNA and dsDNA were quantitated using the ImageQuant software (Pharmacia). The fraction of unwound DNA was plotted against time and the kinetic parameters were determined by non-linear regression using the Graphpad Prism (GraphPad Inc.).
Analyses of DNA Unwinding
To obtain the kinetic parameters associated with DNA unwinding by nsp13, the fraction of unwound DNA was plotted against time. Data fitting was carried out by non-linear regression to single-exponential equation using Prism 5.0 (GraphPad Inc.), as described below.
Fraction (ssDNA) = A.(1 − e−k.t) + n
where A is the amplitude that corresponds to the maximum fraction of ssDNA that can be generated enzymatically from the substrates. k is the pseudo first-order rate constant of DNA unwinding; t is the reaction time, and n is an additive constant representing the amount of ssDNA present before the reaction started.
Determination of KD·DNA
To determine the binding affinity of GST-nsp13 and H6-nsp13 for nucleic acid weperformed gel mobility shift assays. We measured the binding of 60/40-mer (20ss:40ds) DNA substrates at various concentrations of GST-nsp13 or H6-nsp13in a reaction mixture containing 20 mM HEPES pH 7.5, 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 5% glycerol at 30°C for 20 minutes. The concentration for each of the 5’-Cy3-labeled dsDNA substrates was 5 nM. Samples were electrophoresed at 100 V for 1.5 h at 4°C on a 6% non-denaturing polyacrylamide gel, using 89 mM Tris-Borate pH 8.2. Gels were scanned in a PhosphorImager and quantitated by ImageQuant (Amersham) (two independent experiments). The percent enzyme-dsDNA binding was calculated by quantifying the amount of uncomplexed dsDNA in each lane (experiments performed twice). Percent binding values were used to determine the KD.DNA by interpolation, using non-linear regression for one-site binding (hyperbola) using the Prism software.
UV-Mediated DNA Cross-linking with nsp13
To assess if GST-nsp13 and H6-nsp13 binds DNA as a monomer or oligomer we used UV-mediated photo cross-linking of 60/40-mer (20ss:40ds) DNA to nsp13. Varying concentrations of nsp13 (0.25, 0.50, 1 and 2 µM) were incubated with 100 nM of 5’-Cy3-labeled 60/40-mer (20ss:40ds) DNA in a buffer containing 20 mM HEPES, pH 7.5, 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA, and 5% glycerol, at 30°C, for 20 minutes. The samples were exposed to UV light (254 nm) at a dose rate of 125 mJ/cm2 for 3 min using a BioRad GS Gene Linker UV chamber (BioRad Laboratories, CA), as described previously (46, 51). Measurement of covalent attachment of labeled 60/40-mer DNA to enzyme (two independent experiments) was assessed by 8% SDS-PAGE. Bands corresponding to nsp13 cross-linked to 60/40-mer were visualized by a FLA-5000 phosphoimager.
Molecular Modeling of SARS-CoV Helicase
To better understand the location of conserved motifs and residues that are involved in ATP hydrolysis (Walker motifs A and B), we generated a homology-based molecular model of SARS-CoV helicase using the crystal structures of human Upf1 (8) (PDB file 2XZL) and E.coli Rep helicase (22) (PDB file 1UAA) as template-structures. The sequence alignment was generated by Multiple Sequence Viewer tool of Schrodinger Suit (Schrodinger LLC, NY). Initial homology model was predicted by Prime (Schrodinger LLC, NY) using conserved blocks of sequences. Prime was also used to predict the structure of gaps in multiple sequence alignment. Two metals (Zn2+) in nsp13 model were docked using Induced Fit Docking program of Schrodinger Suit. The side chains of Cys and His residues were manually adjusted to form metal coordination with zinc ions. The structure was further minimized for 1000 iteration using OPLS_2005 force field of Schrodinger Suit.
RESULTS AND DISCUSSION
Comparison of GST-nsp13and H6-nsp13 Nucleic Acid Unwinding Activities
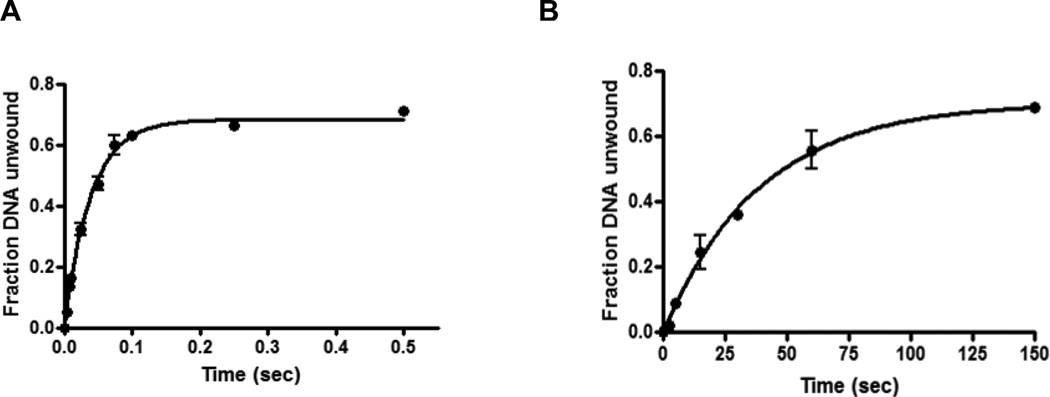
To investigate whether the GST- or H6-tags at the N-terminus have any effect on the nucleic acid unwinding activities of nsp13, we compared the rate of nucleic acid unwinding by GST-nsp13 and H6-nsp13, by performing pre-steady state kinetics to monitor the unwinding of 31/18-mer (13ss:18ds) DNA substrate in the presence of GST-nsp13 or H6-nsp13. Although, nsp13 is an RNA helicase, we used DNA substrate since it has been reported that nsp13 unwinds DNA and RNA in a similar manner (my paper). These experiments were carried out using a rapid chemical quench flow instrument, which allows for the rapid mixing and quenching of reactions on the millisecond timescale. The DNA substrate (5 nM final concentration) was preincubated with GST-nsp13 or H6-nsp13 in one syringe, with Mg2+ (5 mM) and ATP (2 mM final concentration) in the other syringe. After mixing and incubating for times ranging from 0 to 0.5 second for GST-nsp13 or 0–150 seconds for H6-nsp13 , the reactions were quenched, and samples were run on a native gel to separate the Cy3-labeled 18-mer product from the unwound 31/18-mer (13ss:18ds) DNA substrate. The fraction of 18-mer product formed was graphed as a function of time and the data were fit by nonlinear regression to the exponential equation with a rate constant of 24 ± 2.1 s−1 for GST-nsp13 and 0.10 ± 0.01 s−1 for H6-nsp13, indicating that GST-nsp13 is faster than H6-nsp13 by more than 200 folds (Fig. 1).
Figure 1.
Comparison of unwinding activity of GST- and H6- nsp13 variants. Comparison of the helicase activity at varying time points for GST-nsp13 (Glutathione Sepharose Transferase-tagged nsp13, Panel A), and H6-nsp13 (hexahistidine-tagged nsp13, Panel B) using 100 nM of each enzyme and 5 nM 31/18-mer (13ss:18ds) as substrate at 30°C. The products were separated and analyzed by 6% non-denaturing PAGE. The fraction of unwound DNA was plotted against time and the kinetic parameters were determined by non-linear regression to the exponential equation using the Graphpad Prism.
Comparison of GST-nsp13 and H6-nsp13 ATP Hydrolysis
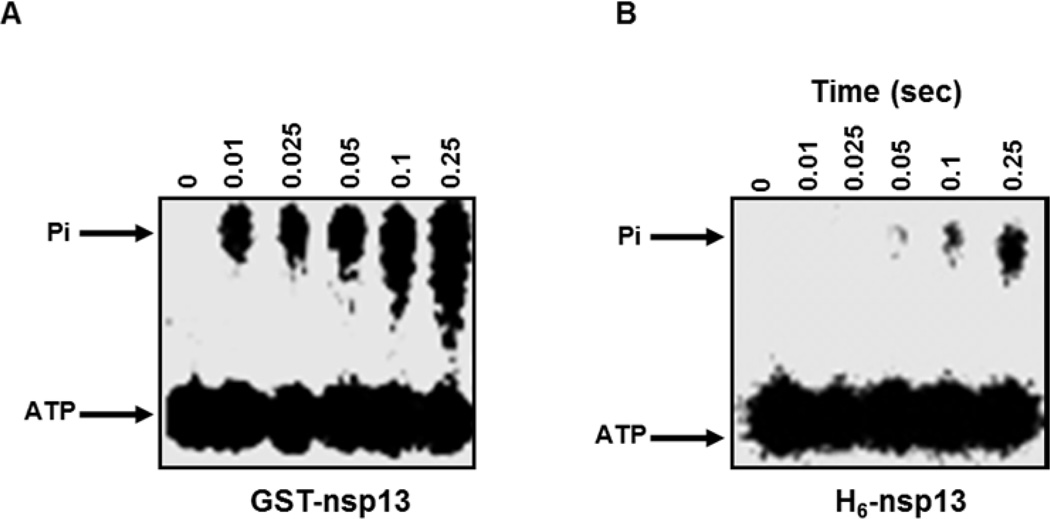
To evaluate the ATPase activity of GST-nsp13 and H6-nsp13, we investigated the amount of ATP hydrolyzed by each construct at different time points. We incubated GST-nsp13 or H6-nsp13 (100nM) with γ-32 ATP (50µM) for various time points (0.01–0.25 s) and the reactions were analyzed by thin-layer chromatography. The results presented in Fig. 2 show that more than 50% of the ATP has been hydrolyzed by GST-nsp13 at 0.25s, whereas, less than 10% of ATP has been hydrolyzed by H6-nsp13 at the same time point.
Figure 2. Hydrolysis of ATP by GST-nsp13 and H6-nsp13.
A) GST-nsp13 or B) H6-nsp13 was incubated with γ-32P-ATP (50 µM) for various reaction times. The reaction products were separated by thin-layer chromatography and visualized by autoradiography.
GSTnsp13 and H6-nsp13 have Similar Binding Affinity for Nucleic Acids
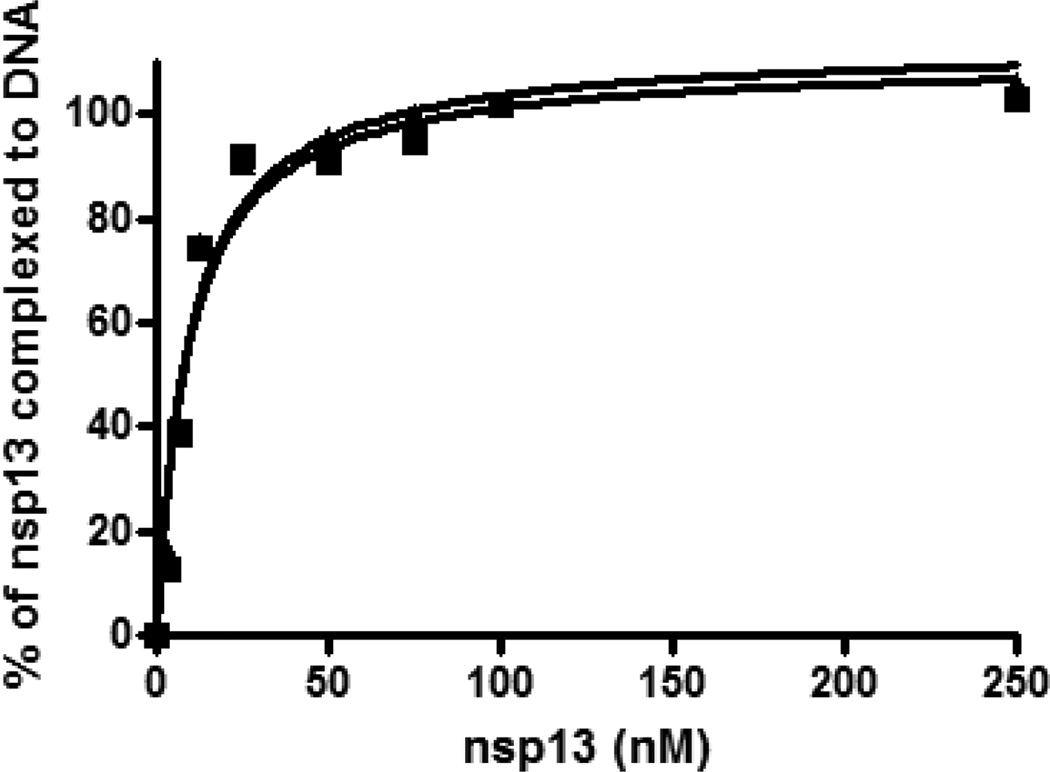
Since GSTnsp13 and H6-nsp13 exhibit significant differences in their catalytic activities, we opted to determine if they also demonstrate such differences in their binding capacity for nucleic acid. We performed gel mobility shift assays to analyze the binding of 60/40-mer (20ss:40ds) DNA substrate with various concentrations of GST-nsp13 or H6-nsp13. The concentration for the 5’-Cy3-labeled dsDNA substrate was 5 nM. As shown in Fig. 3, the two constructs of nsp13 bind DNA in a similar manner indicating that the differences in the catalytic activities of the two constructs did not result from nucleic acid binding and that the mode and affinity of nsp13 binding to nucleic acid substrate is not affected by the H6- or GST- tags. The KD.DNA values for 60/40-mer and 53/40-mer were 9.4 and 20 nM, respectively, determined by plotting the concentration of enzyme against that of complexed DNA and fitting the data to the hyperbolic curve shown in Fig. 4.
Figure 3. Binding affinity of GST-nsp13 and H6-nsp13 determined by gel shift assay.
Different concentrations of GST-nsp13(▲) or H6-nsp13 (■) were incubated with 5 nM 5′ Cy3-labeled 60/40-mer (20ss:40ds) in 20 mM HEPES (pH 7.5), 20 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.1 mg/ml BSA and 5% glycerol at 30°C for 20 min. The samples were then resolved in a native 6% polyacrylamide gel. The percent of nsp13-DNA complex (two independent experiments) was determined and plotted against the concentration of unliganded GST-nsp13 and fitted to a one-site binding hyperbola using GraphPad Prism to determine KD-DNA.
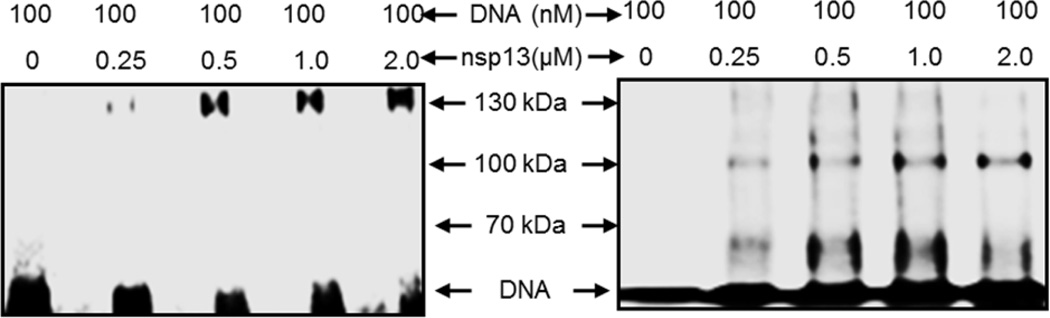
Figure 4. UV-mediated photochemical cross-linking of GST-nsp13 and H6-nsp13 to Cy3-labeled 60/40DNA/DNA.
100 nM of Cy3-labeled 60/40-mer (20ss:40ds) DNA was cross-linked to increasing amounts of GST-nsp13 (0.25, 0.5, and 1 and 2 µM) as described in the Experimental Procedures. The cross-linked species were resolved on 8% SDS-PAGE and the bands corresponding to GST-nsp13 (left panel) and H6-nsp13 (right panel) cross-linked to 60/40-mer DNA were visualized by a Fuji FLA-5000 phosphorimager. The positions of molecular weight markers in the same gel were determined by brilliant blue-coomassie staining. The shifted fluorescent bands migrated as ~125 kDa and 100 kDa complexes, in line with GST-nsp13 and 125 H6-nsp13 monomers respectively, cross-linked to the 60–40mer.
GSTnsp13 and H6-nsp13 Bind Nucleic Acid as a Monomer
Viral helicases have been known to exist and function in various oligomeric states, including monomers, dimers, trimers or hexamers (33). To ascertain the oligomeric state of nsp13 we used a photochemical cross-linking assay. UV-irradiation of a mixture of nsp13 and 60/40-mer DNA resulted in the formation of a covalent complex. The covalently linked nsp13-DNA complex migrated in an SDS-PAGE denaturing gel as a single species of ~125 kDa molecular weight for GST-nsp13 and ~100 kDa molecular weight for H6-nsp13, corresponding to a monomer of nsp13 bound to a single 60/40-mer oligonucleotide (Fig. 4). Since, we did not observe any supershifts at higher protein concentrations in the gel shift assay, it implies that nsp13 binds its substrate as a monomer regardless of the tags.
Structure of SARS-CoV Helicase
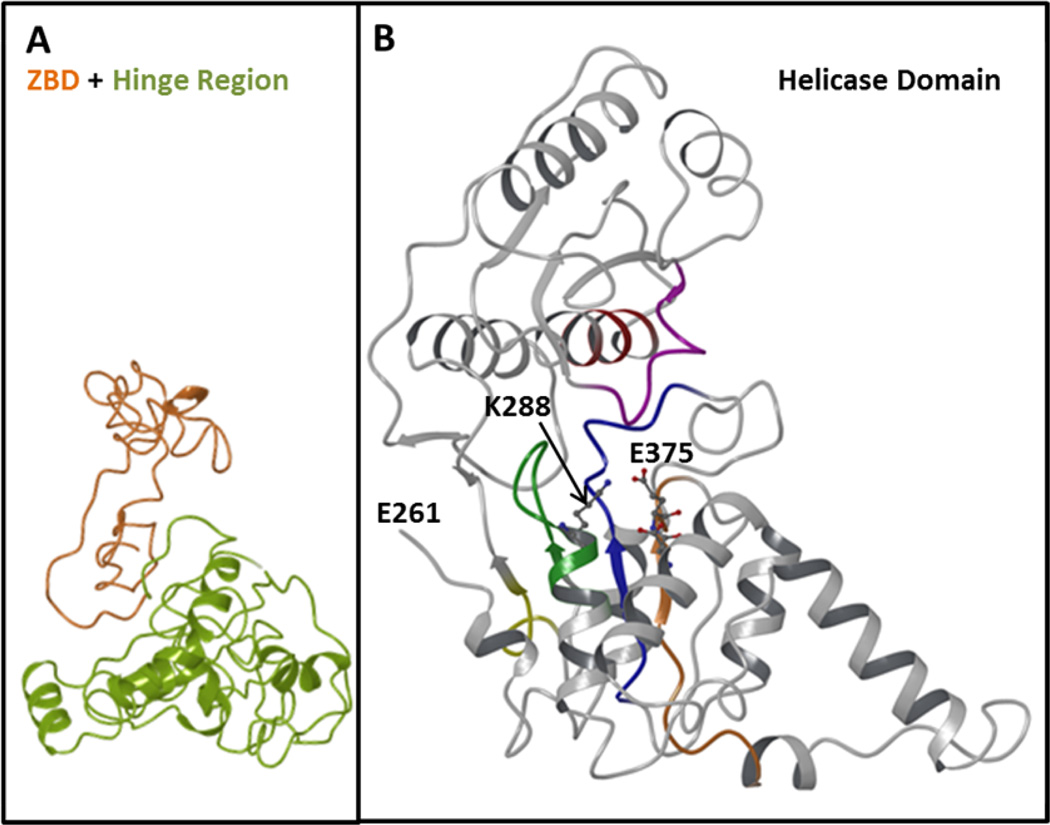
In the absence of the crystal structure of SARS-CoV nsp13, we constructed the molecular model of this enzyme using the crystal structures of human Upf1 (8) and E.coli Rep helicase (22). Human Upf1 (up-frameshift 1) is a nucleic acid-dependent ATPase and possesses a 5'-to-3' helicase activity. It is known to play central role in cytoplasmic RNA quality control. The E.coli Rep helicase is a ATP-dependent 3’-to-5’ helicase. The sequence alignment showed that nsp13 has 30 and 26 % sequence identity with Upf1 and Rep helicase, respectively. sequence annotation by Ivanov et al., (19) have shown that SARS-CoV nsp13 can be divided into three domains: (i) N-terminal Zn2+ binding domain and (ii) hinge domain and (iii) helicase domain. The Zn2+ binding domain has 12 Cys/His conserved residues, which can bind at least three Zn2+ ions. Figure 5 shows the overall all fold of nsp13. Zn2+ (orange) and hinge domain (lime) are shown in Figure 5A. The helicase domain is shown in Figure 5B. The helicase domain has six conserved motifs. Two of these motifs are called Walker A motif (GXXXXGK(T/S) (green), which contains a conserved Lys residue. The tentative Walker B motif (R/K)XXXXGXXXXLhhhhDE) blue containing a Asp and a Glu residue is shown in blue. Both Lys in Walker A and Asp/Glu in motif II are known to participate in ATP-hydrolysis by DNA/RNA helicases. The conserved motifs are and appear to be located in a channel like structure.
Fig. 5. Predicted Structure of nsp13.
This figure shows overall fold of nsp13 helicase structure. Panel A shows the backbone of Zn2+ binding region (orange) and hinge region (lime). Six conserved motifs of helicase domain are shown in different colors: motif I (Walker A), green; motif II, orange; motif III, blue; motif IV, yellow; motif V, magenta and motif V, red. The residues most likely to involve in ATP hydrolysis (K288, D374 and D375) are rendered as ball-and-stick.
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants AI076119, AI094715, AI079801, and AI074389. We also acknowledge support by a grant from the Ministry of Knowledge and Economy, Bilateral International Collaborative R&D Program, Republic of Korea. B.M. was the recipient of an amfAR Mathilde Krim Fellowship and a CIHR Fellowship.
Abbreviations
- bp
base-pair
- CoV
coronavirus
- ds
double stranded
- GST
glutathione S-transferase
- H6
hexahistidine
- nsp
non-structural protein
- nt
nucleotide
- RC
replication complex
- SARS
severe acute respiratory syndrome
- ss
single stranded
REFERENCES
- 1.Abdel-Monem M, Chanal MC, Hoffmann-Berling H. DNA unwinding enzyme II of Escherichia coli. 1. Purification and characterization of the ATPase activity. Eur J Biochem. 1977;79:33–38. doi: 10.1111/j.1432-1033.1977.tb11780.x. [DOI] [PubMed] [Google Scholar]
- 2.Abdel-Monem M, Durwald H, Hoffmann-Berling H. DNA unwinding enzyme II of Escherichia coli. 2. Characterization of the DNA unwinding activity. Eur J Biochem. 1977;79:39–45. doi: 10.1111/j.1432-1033.1977.tb11781.x. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Monem M, Durwald H, Hoffmann-Berling H. Enzymic unwinding of DNA. 2. Chain separation by an ATP-dependent DNA unwinding enzyme. Eur J Biochem. 1976;65:441–449. doi: 10.1111/j.1432-1033.1976.tb10359.x. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Monem M, Hoffmann-Berling H. Enzymic unwinding of DNA. 1. Purification and characterization of a DNA-dependent ATPase from Escherichia coli. Eur J Biochem. 1976;65:431–440. doi: 10.1111/j.1432-1033.1976.tb10358.x. [DOI] [PubMed] [Google Scholar]
- 5.Adedeji AO, Marchand B, Te Velthuis AJ, Snijder EJ, Weiss S, Eoff RL, Singh K, Sarafianos SG. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PloS one. 2012;7:e36521. doi: 10.1371/journal.pone.0036521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, Weiss S, Sarafianos SG. Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrobial agents and chemotherapy. 2012;56:4718–4728. doi: 10.1128/AAC.00957-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benarroch D, Selisko B, Locatelli GA, Maga G, Romette JL, Canard B. The RNA helicase, nucleotide 5'-triphosphatase, and RNA 5'-triphosphatase activities of Dengue virus protein NS3 are Mg2+-dependent and require a functional Walker B motif in the helicase catalytic core. Virology. 2004;328:208–218. doi: 10.1016/j.virol.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Donnelly CA, Ghani AC, Leung GM, Hedley AJ, Fraser C, Riley S, Abu-Raddad LJ, Ho LM, Thach TQ, Chau P, Chan KP, Lam TH, Tse LY, Tsang T, Liu SH, Kong JH, Lau EM, Ferguson NM, Anderson RM. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet. 2003;361:1761–1766. doi: 10.1016/S0140-6736(03)13410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duguet M, Yarranton G, Gefter M. The rep protein of Escherichia coli: interaction with DNA and other proteins. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):335–343. doi: 10.1101/sqb.1979.043.01.040. [DOI] [PubMed] [Google Scholar]
- 12.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 13.Eoff RL, Raney KD. Intermediates revealed in the kinetic mechanism for DNA unwinding by a monomeric helicase. Nat Struct Mol Biol. 2006;13:242–249. doi: 10.1038/nsmb1055. [DOI] [PubMed] [Google Scholar]
- 14.Flores MJ, Sanchez N, Michel B. A fork-clearing role for UvrD. Mol Microbiol. 2005;57:1664–1675. doi: 10.1111/j.1365-2958.2005.04753.x. [DOI] [PubMed] [Google Scholar]
- 15.Geider K, Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- 16.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 17.Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hon KL, Leung CW, Cheng WT, Chan PK, Chu WC, Kwan YW, Li AM, Fong NC, Ng PC, Chiu MC, Li CK, Tam JS, Fok TF. Clinical presentations and outcome of severe acute respiratory syndrome in children. Lancet. 2003;361:1701–1703. doi: 10.1016/S0140-6736(03)13364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004;78:5619–5632. doi: 10.1128/JVI.78.11.5619-5632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNAprotein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 21.Kornberg A, Scott JF, Bertsch LL. ATP utilization by rep protein in the catalytic separation of DNA strands at a replicating fork. J Biol Chem. 1978;253:3298–3304. [PubMed] [Google Scholar]
- 22.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to singlestranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 23.Lee NR, Kwon HM, Park K, Oh S, Jeong YJ, Kim DE. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res. 2010;38:7626–7636. doi: 10.1093/nar/gkq647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lestini R, Michel B. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 2007;26:3804–3814. doi: 10.1038/sj.emboj.7601804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohman TM. Escherichia coli DNA helicases: mechanisms of DNA unwinding. Mol Microbiol. 1992;6:5–14. doi: 10.1111/j.1365-2958.1992.tb00831.x. [DOI] [PubMed] [Google Scholar]
- 26.Lohman TM. Helicase-catalyzed DNA unwinding. J Biol Chem. 1993;268:2269–2272. [PubMed] [Google Scholar]
- 27.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 28.Lucius AL, Vindigni A, Gregorian R, Ali JA, Taylor AF, Smith GR, Lohman TM. DNA unwinding step-size of E. coli RecBCD helicase determined from single turnover chemical quenched-flow kinetic studies. J Mol Biol. 2002;324:409–428. doi: 10.1016/s0022-2836(02)01067-7. [DOI] [PubMed] [Google Scholar]
- 29.Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, Vogrig R, Ward D, Watson B, Brunham RC, Krajden M, Petric M, Skowronski DM, Upton C, Roper RL. The Genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 30.Matson SW, Kaiser-Rogers KA. DNA helicases. Annu Rev Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 31.Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, Chu CM, Hui PK, Mak KL, Lim W, Yan KW, Chan KH, Tsang NC, Guan Y, Yuen KY, Peiris JS. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan J, Peng X, Gao Y, Li Z, Lu X, Chen Y, Ishaq M, Liu D, Dediego ML, Enjuanes L, Guo D. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PloS one. 2008;3:e3299. doi: 10.1371/journal.pone.0003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papageorgiou N, Coutard B, Lantez V, Gautron E, Chauvet O, Baronti C, Norder H, de Lamballerie X, Heresanu V, Ferte N, Veesler S, Gorbalenya AE, Canard B. The 2C putative helicase of echovirus 30 adopts a hexameric ring-shaped structure. Acta Crystallogr D Biol Crystallogr. 2010;66:1116–1120. doi: 10.1107/S090744491002809X. [DOI] [PubMed] [Google Scholar]
- 34.Patel SS, Donmez I. Mechanisms of helicases. J Biol Chem. 2006;281:18265–18268. doi: 10.1074/jbc.R600008200. [DOI] [PubMed] [Google Scholar]
- 35.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 36.Peiris JS, Chu CM, Cheng VC, Chan KS, Hung IF, Poon LL, Law KI, Tang BS, Hon TY, Chan CS, Chan KH, Ng JS, Zheng BJ, Ng WL, Lai RW, Guan Y, Yuen KY. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng TK, Yuen KY. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schrock RD, Alberts B. Processivity of the gene 41 DNA helicase at the bacteriophage T4 DNA replication fork. J Biol Chem. 1996;271:16678–16682. doi: 10.1074/jbc.271.28.16678. [DOI] [PubMed] [Google Scholar]
- 40.Scott JF, Eisenberg S, Bertsch LL, Kornberg A. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc Natl Acad Sci U S A. 1977;74:193–197. doi: 10.1073/pnas.74.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol Biol Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sims AC, Burkett SE, Yount B, Pickles RJ. SARS-CoV replication and pathogenesis in an in vitro model of the human conducting airway epithelium. Virus Res. 2008;133:33–44. doi: 10.1016/j.virusres.2007.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanner JA, Watt RM, Chai YB, Lu LY, Lin MC, Peiris JS, Poon LL, Kung HF, Huang JD. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J Biol Chem. 2003;278:39578–39582. doi: 10.1074/jbc.C300328200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thiel V, Herold J, Schelle B, Siddell SG. Viral replicase gene products suffice for coronavirus discontinuous transcription. J Virol. 2001;75:6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 46.Tumino G, Casazza AP, Engelmann E, Garlaschi FM, Zucchelli G, Jennings RC. Fluorescence lifetime spectrum of the plant photosystem II core complex: photochemistry does not induce specific reaction center quenching. Biochemistry. 2008;47:10449–10457. doi: 10.1021/bi800831j. [DOI] [PubMed] [Google Scholar]
- 47.van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4:e1000054. doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005;24:180–189. doi: 10.1038/sj.emboj.7600485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 50.von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R, Roberts R, Baric R, Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS one. 2007;2:e459. doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their"coupling" within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 52.Wilcox KW, Smith HO. Mechanism of DNA degradation by the ATP-dependent DNase from Hemophilus influenzae Rd. J Biol Chem. 1976;251:6127–6134. [PubMed] [Google Scholar]
- 53.Yarranton GT, Das RH, Gefter ML. Enzyme-catalyzed DNA unwinding. A DNAdependent ATPase from E. coli. J Biol Chem. 1979;254:11997–12001. [PubMed] [Google Scholar]
- 54.Yarranton GT, Das RH, Gefter ML. Enzyme-catalyzed DNA unwinding. Mechanism of action of helicase III. J Biol Chem. 1979;254:12002–12006. [PubMed] [Google Scholar]
- 55.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]