Abstract
We report that developmental competition between sympathetic neurons for survival is critically dependent on a sensitization process initiated by target innervation and mediated by a series of feedback loops. Target-derived nerve growth factor (NGF) promoted expression of its own receptor TrkA in mouse and rat neurons and prolonged TrkA-mediated signals. NGF also controlled expression of brain-derived neurotrophic factor and neurotrophin-4, which, through the receptor p75, can kill neighboring neurons with low retrograde NGF-TrkA signaling whereas neurons with high NGF-TrkA signaling are protected. Perturbation of any of these feedback loops disrupts the dynamics of competition. We suggest that three target-initiated events are essential for rapid and robust competition between neurons: sensitization, paracrine apoptotic signaling, and protection from such effects.
In the developing peripheral nervous system, target tissues specify the amount of innervation they receive by secreting limiting amounts of neurotrophic factors, which are required for neuronal survival (1, 2). The prototypic target cell–derived neurotrophic factor, NGF, supports survival of developing sympathetic and cutaneous sensory neurons (3, 4). NGF engages its receptor tyrosine kinase TrkA on the distal axon, and the NGF-TrkA complexes travel retrogradely from the periphery to the cell body, where they induce prosurvival signaling events and NGF-dependent transcriptional programs (5–10).
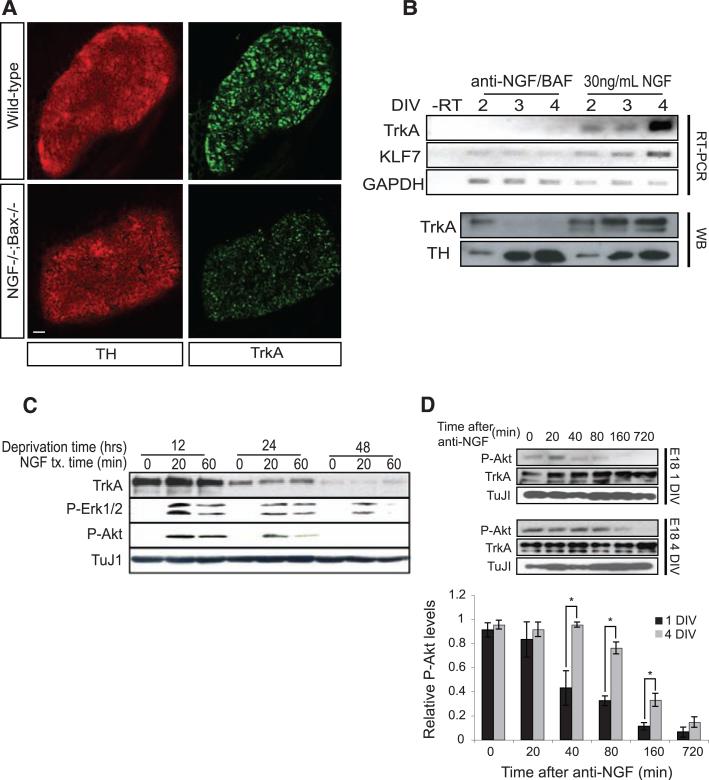
To identify NGF-dependent transcriptional events that may enable one neuron to gain competitive advantage over its neighbor with similar potential, we performed a comprehensive in vivo comparison of gene expression profiles of sympathetic neurons isolated from newly born mice with an intact or ablated NGF locus (11). To circumvent the requirement of NGF for survival, we used Bax–/– animals, which are deficient in sympathetic neuron apoptosis during development (12–14). Expression of brain-derived neurotrophic factor (BDNF), TrkA, and another neurotrophin receptor, p75, was decreased in the absence of NGF (table S1 and fig. S1A). NGF dependence of TrkA expression was confirmed by in situ hybridization, immunohistochemistry, reverse transcriptase polymerase chain reaction (RT-PCR), and immunoblot analysis (Fig. 1, A and B, and fig. S1B). NGF also regulated expression of krupple-like factor 7, a transcription factor that regulates TrkA expression (15) (Fig. 1B). Moreover, TrkA expression and downstream signaling were greatly reduced when cultured sympathetic neurons were deprived of NGF for 24 or 48 hours (Fig. 1C). We also compared the abundance of TrkA, activated phosphoTrkA (p-TrkA), and p-Akt in neurons with different maturities and found that the more mature neurons exhibited dramatic increases in all three markers of NGF prosurvival signaling (fig. S1C). In addition to the difference in NGF signaling magnitude, the duration of prosurvival signaling increased as a function of NGF exposure and neuron maturity (Fig. 1, C and D). Prosurvival signaling remained robust for at least 60 min after treatment with NGF in neurons deprived of NGF for 12 hours before treatment; neurons deprived of NGF for longer periods of time lost their ability to sustain prosurvival signaling. Taken together, these observations suggest the possibility that NGF induces feedback loops, in part through regulation of TrkA expression, that control magnitude and duration of NGF signaling.
Fig. 1.
NGF promotes TrkA expression and the duration of TrkA signaling events. (A) Immunohistochemistry for TrkA and tyrosine hydroxylase (TH) on SCGs from wild-type and NGF–/–;Bax–/– animals. (B) TrkA, KLF7, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels were measured by RT-PCR and TrkA and TH by Western blot (WB) after treatment of cultured rat sympathetic neurons with NGF (30 ng/ml) for indicated times. (C) TrkA, p-ERK, p-Akt, and TuJI measured by WB in P0.5 rat sympathetic neurons that were deprived of NGF for the indicated times followed by reexposure to NGF (30 ng/ml) for 20 or 60 min. (D) Sympathetic neuron cultures from E18 rats were grown in the presence of 30 ng/ml NGF for 1 day in vitro (DIV) or 4 DIVs. NGF-specific antibody (anti-NGF) was added, and NGF signaling events were assessed at the indicated times after NGF deprivation. Graph compares relative p-Akt levels for this experiment. Scale bar in (A), 50 μm. *P < 0.05 analysis of variance (ANOVA) followed by Tukey's post hoc test.
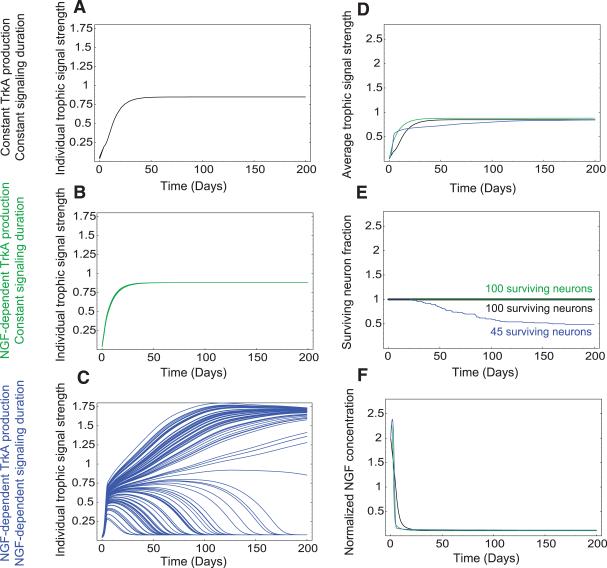
To determine whether NGF-dependent gene expression events and resultant feedback loops underlie the acquisition of competitive advantage, we turned to a computational approach to model this process. Our model assigns one differential equation per neuron, representing the relative magnitude of trophic signaling, which we define as the amount of NGF-bound TrkA within the neuron. Another differential equation was assigned for the concentration of NGF available at the target, with the assumption that the rate of NGF production is constant. The outputs of these equations changed as a function of time (figs. S2 and S3). Computer simulations were used to relate these two equations to neuronal survival; a neuron was considered dead if it reached a trophic signaling state of 10% or less of the maximum value. The results of these simulations are represented in several forms, all with respect to time: (i) trophic signaling for each modeled neuron (Fig. 2, A to C), (ii) average trophic signaling (Fig. 2D), (iii) total neuron number (Fig. 2E), and (iv) NGF concentration at the target (Fig. 2F).
Fig. 2.
Requirement for both NGF-dependent TrkA expression and signal duration during competition revealed by computer simulations. Shown are model simulation results for 100 neurons under different competitive conditions. (A to C) Simulated trophic signaling state of 100 individual neurons with indicated parameters over time. (A) TrkA production and signaling duration were held constant. (B) TrkA production was NGF-dependent but signaling duration was held constant. (C) TrkA production and signaling duration were NGF-dependent. (D to F) Comparison of various dynamic elements as a function of time in simulations from (A) to (C). (D) Average trophic signal strength; (E) cell survival; (F) relative amount of NGF at the target. The black, green, and blue lines in (D), (E), and (F) represent results of simulations described for (A), (B), and (C).
To assess the importance of NGF-dependent changes in both TrkA expression and signal duration during neuronal competition, we conducted a simulation in which these terms were fixed and independent of exposure to NGF. In this paradigm, all neurons rapidly reached a trophic steady state, and no competition occurred, which resulted in the survival of all neurons (Fig. 2, A and E). If either TrkA expression or signal duration was fixed and the other was allowed to change in relation to NGF exposure, competition also failed to occur (Fig. 2, B and E, and fig. S4). Both signal strength and duration had to change after neuronal exposure to NGF in order for effective competition to occur (Fig. 2, C and E). When competition occurred, some neurons obtained a high trophic signaling state and others reached a low trophic signaling state and ultimately died (Fig. 2E and fig. S4K). In this system, survival outcomes were fairly independent of starting conditions (figs. S5 and S6). However, changes in NGF production, NGF-dependent TrkA expression, and TrkA signal duration had large effects on the dynamics of neuron elimination (figs. S7 to S9). Taken together, these modeling data indicate that target innervation–dependent changes in TrkA expression and signal duration may be essential for neuronal competition.
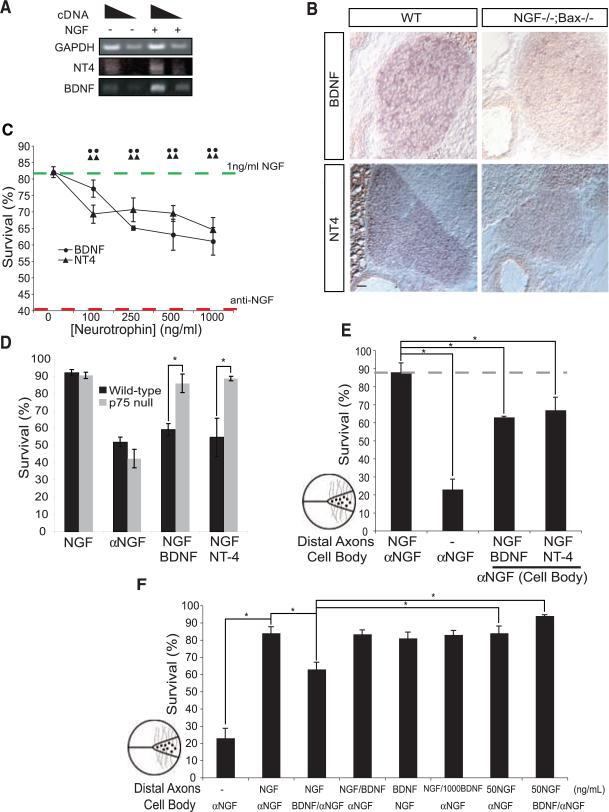
Clues from a model for synapse competition at the neuromuscular junction (16), as well as findings from our microarray analysis (fig. S1A and table S1), led us to test whether, during competition for survival, neurons with high amounts of trophic signaling actively promote apoptosis of neurons with low trophic signaling. The putative apoptotic cue should have several characteristics: (i) It should kill neurons with low trophic signaling, and (ii) it should emanate from neurons with strong trophic signaling. (iii) Neurons with strong trophic signaling should be impervious to the apoptotic cue, and (iv) apoptotic cue signaling should commence after expression of TrkA increased. BDNF and neurotrophin-4 (NT4) satisfy the criteria for the apoptotic cues: (i) BDNF and NT4 can promote apoptosis of sympathetic neurons through the receptor p75 (17, 18) (Fig. 3, C and D), and (ii) their expression is regulated by NGF in sympathetic neurons (Fig. 3, A and B, and table S1). (iii) Strong NGF-TrkA signaling blocks p75-mediated killing of sympathetic neurons (19) (Fig. 3F). (iv) Finally, p75 expression in sympathetic neurons commences in vivo 2 days after TrkA expression (20), and this expression is regulated by NGF (21, 22) (table S1). Thus, we propose that BDNF and NT4 are NGF-regulated apoptotic cues for developing sympathetic neurons and that innervation-dependent expression of p75 regulates a neuron's susceptibility to these signals.
Fig. 3.
Antagonism of retrograde NGF survival signaling by NGF induced BDNF and NT4 expression in sympathetic neurons. (A) RT-PCR for BDNF, NT-4, and GAPDH with RNA from NGF-treated (24 hours) or NGF-deprived sympathetic neurons from P0 mice grown 3 DIVs. (B) In situ hybridization for BDNF and NT-4 in wild-type and NGF–/–;Bax–/– animals at P0. Scale bar, 50 μm. (C to F) Neurotrophin promoted p75-dependent cell death of cultured sympathetic neurons. Indicated neurotrophins were applied for 36 hours, cell survival was determined by Hoechst staining, and results are means ± SEM (n = 4 experiments). (C) Survival of P0 to P2 rat sympathetic neurons maintained in medium containing NGF (1 ng/ml). Green dashed line represents maximum survival, and red dashed line indicates maximum death, both conditions were assessed after 36 hours with either 1 ng/ml of NGF or NGF-specific antibody (anti-NGF), respectively. (D) Survival of P0 to P2 p75–/– or wild-type mouse sympathetic neurons treated with NGF (1 ng/ml)or NGF-specific antibody (αNGF). (E and F) P0 sympathetic neurons grown in compartmentalized chambers (represented by illustration left of graphs) for 5 to 7 days before medium was changed to contain NGF (5 ng/ml) on the distal axons. The indicated neurotrophins or NGF-specific antibody (αNGF) was applied to the cell bodies for 36 hours before assessing survival. Unless otherwise indicated, BDNF and NT-4 were applied at a concentration of 250 ng/ml *P < 0.01 ANOVA followed by Tukey's post hoc test.
We next sought to define other features of p75-mediated cell death. BDNF and NT4 caused dose-dependent killing of sympathetic neurons in the presence of concentrations of NGF insufficient to promote maximal activation of TrkA (Fig. 3C). In p75–/– neurons, these neurotrophins had no effect on survival (18) (Fig. 3D). Next, the spatial properties of these apoptotic signals were characterized in a compartmentalized culture system (23). If a low concentration of NGF was applied exclusively to distal axons and either BDNF or NT4 was applied to cell bodies, NGF-dependent survival was reduced to a similar extent as that observed in mass cultures (Fig. 3, C and E). In contrast, if BDNF was applied exclusively to distal axons, cell death was not observed, which suggested that apoptotic signals must be produced and act at the cell body to affect cell survival (Fig. 3F). It is noteworthy that induction of cell death by BDNF-p75 signaling on the cell body was suppressed when larger amounts of NGF were added to distal axons (Fig. 3F).
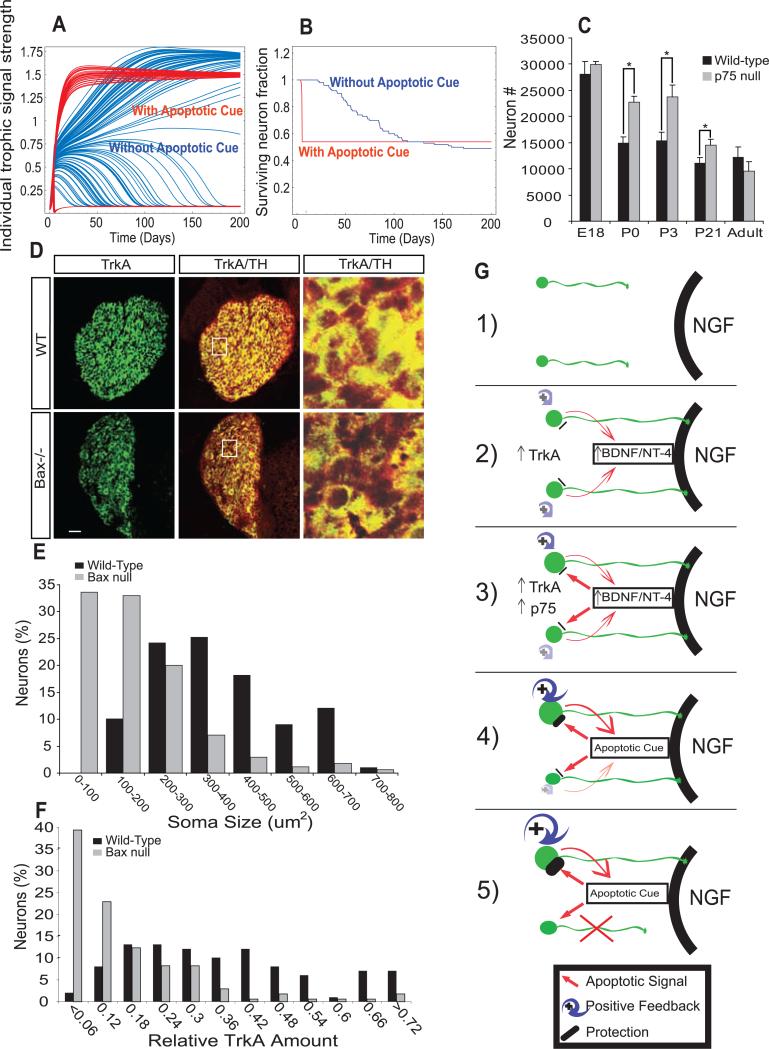
To determine the potential benefits of NGF-dependent expression of signals that promote death of neurons with low trophic signaling, we added several semiquantitative equations to the computational model presented in Fig. 2C to characterize the apoptotic signals and their receptors. As determined experimentally for BDNF and NT4, the variable representing the amount of apoptotic signal increases with neuronal exposure to NGF and has no effect on neurons with high retrograde NGF-TrkA signaling. Computer simulations revealed that the addition of this parameter hastened, by more than 10-fold, the killing of those neurons that did not gain competitive advantage through the sensitization process (Fig. 4, A and B). The extent to which the apoptotic signals hastened neuronal death was dependent on how much NGF-TrkA signaling was required for protection and the rate of production of BDNF and NT4 in response to NGF (figs. S10 and S11).
Fig. 4.
Mathematical modeling predicts changes in competition dynamics that are corroborated in vivo. (A and B) Simulations of neuronal competition with and without NGF-dependent production of an apoptotic cue (red and blue, respectively). (A) Trophic signaling strength of 100 individual neurons as a function of time. (B) Cell survival as a function of time. (C) Cell counts of Nissl-stained SCG sections from p75–/– or wild-type mice at indicated developmental ages. Results are means ± SEM (n = 3 for each age). (D) Immunostaining for TH and TrkA in SCGs from Bax–/– and wild-type animals. Scale bar, 50 μm. (Right) Boxed region in middle panels magnified 5×. (E and F) Quantification of soma size (E) and relative TrkA amount (F) in P5 Bax–/– versus wild-type SCGs. Results are represented as a percentage of total neurons counted. *P < 0.01 ANOVA followed by Tukey's post hoc test. (G) Model for developmental competition: 1) Before target innervation neurons are modestly responsive to NGF; 2) upon target innervation and exposure to NGF, levels of TrkA, then BDNF and NT-4 are increased; 3) induction of p75 expression, as well as differential sensitization of neurons, by modulation of NGF-TrkA signal strength and duration. 4) BDNF and NT-4 (apoptotic cues) kill neurons with low NGF-TrkA signaling; neurons with high NGF-TrkA signaling are resistant; 5) selection and neuronal death.
We thus propose a model (Fig. 4G) in which, before target innervation, sympathetic neurons are modestly responsive to NGF because of low TrkA levels. Upon target innervation, neurons acquire NGF, and subtle differences in initial amounts of NGF signaling are amplified through transcription-dependent feedback loops into large cell-autonomous differences in both strength and duration of TrkA signaling, which are the ultimate determinants for whether a neuron lives or dies. We further propose that expeditious competition requires target innervation–dependent expression of apoptotic cues (BDNF and NT-4), susceptibility to such signals (expression of p75), and protection from apoptotic signals (strong retrograde NGF-TrkA signaling) (Fig. 4G and fig. S12). These features enable competition that is rapid, robust, and stable, even in a scenario in which all neurons arrive at their target simultaneously and are virtually equivalent in their initial responsiveness to target-derived NGF.
Several predictions arise from this model. One prediction is that competing neurons distinguish themselves from one another on the basis of their amount of TrkA expression; those with abundant TrkA live, whereas those with low amounts die. Consistent with this prediction, in Bax–/– mice at postnatal day 5 (P5), over half of the neurons had extremely low levels of TrkA and small soma areas, whereas the rest of these neurons had larger amounts of TrkA and cell body areas comparable to those of wild-type controls (Fig. 4, D to F, and figs. S13 and S14). The model also predicts that atrophic neurons in superior cervical ganglia (SCG) from P5 Bax–/– mice should display persistent p75-mediated apoptotic signals given that these are neurons that would normally have died during developmental competition. Indeed, compared with neurons with high TrkA expression, atrophic neurons in Bax–/– ganglia have large amounts of p75 and p75-dependent signaling as measured by immunostaining of the phosphorylated c-Jun transcription factor (figs. S17 and S18).
Finally, the notion that the NGF-dependent apoptotic signal hastens but does not change the fundamental elements of competition was tested in mice lacking p75. A developmental time course comparing numbers of SCG neurons in wild-type and p75–/– animals was performed at embryonic day 18 (E18), P0, P3, P21, and 6 months of age. As predicted by the modeling data (Fig. 4, A and B), SCGs from wild-type and p75–/– animals had a similar number of neurons at E18, whereas P0 and P3 p75–/– SCGs had ~40% excess of neurons compared with wild-type animals. At 6 months of age, p75–/– SCGs had the same number of neurons as control SCGs (Fig. 4C) [see also (18, 24)].
These genetic and computational modeling data show that two neurotrophin receptors and potentially three neurotrophin ligands orchestrate developmental competition between SCG neurons through a series of feedback loops. Although the cast of players for other populations of competing neurons is almost certain to be different from those that coordinate competition in the SCG, we suggest that the underlying principles of target initiated sensitization, paracrine apoptotic signaling, and protection from apoptotic signals are common features of neuronal competition at large.
Supplementary Material
Acknowledgments
We thank C. Jie and F. Martinez Murillo of the Johns Hopkins Medical Institute microarray core for their assistance in microarray analysis; and N. Gaiano, A. Harrington, R. Kuruvilla, A. Levchenko, W. Luo, and K. Wright for helpful comments during manuscript preparation. This work is supported by NIH fellowship NS053187 (C.D.D.), A Woodrow Wilson Undergraduate Research Fellowship (N.S.), and NIH grants NS34814 (D.D.G.) and EY016281 (E.N.). D.D.G. is an investigator of the Howard Hughes Medical Institute. The microarray analysis data have been deposited in National Center for Biotechnology Information's Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/), accession no. GSE10498.
Footnotes
Supporting Online Material
References and Notes
- 1.Hamburger V, Levi-Montalcini R. J. Exp. Zool. 1949;111:457. doi: 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- 2.Levi-Montalcini R. Science. 1987;237:1154. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 3.Levi-Montalcini R, Booker B. Proc. Natl. Acad. Sci. U.S.A. 1960;46:384. doi: 10.1073/pnas.46.3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowley C, et al. Cell. 1994;76:1001. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 5.Heerssen HM, Pazyra MF, Segal RA. Nat. Neurosci. 2004;7:596. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- 6.Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Science. 1999;286:2358. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 7.Watson FL, et al. Nat. Neurosci. 2001;4:981. doi: 10.1038/nn720. [DOI] [PubMed] [Google Scholar]
- 8.Ye H, Kuruvilla R, Zweifel LS, Ginty DD. Neuron. 2003;39:57. doi: 10.1016/s0896-6273(03)00266-6. [DOI] [PubMed] [Google Scholar]
- 9.Delcroix JD, et al. Neuron. 2003;39:69. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- 10.Howe CL, Mobley WC. Curr. Opin. Neurobiol. 2005;15:40. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Materials and methods are available as supporting material on Science Online.
- 12.Glebova NO, Ginty DD, Neurosci J. 2004;24:743. doi: 10.1523/JNEUROSCI.4523-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel TD, Jackman A, Rice FL, Kucera J, Snider WD. Neuron. 2000;25:345. doi: 10.1016/s0896-6273(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 14.Deckwerth TL, et al. Neuron. 1996;17:401. doi: 10.1016/s0896-6273(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 15.Lei L, Ma L, Nef S, Thai T, Parada LF. Development. 2001;128:1147. doi: 10.1242/dev.128.7.1147. [DOI] [PubMed] [Google Scholar]
- 16.Sanes JR, Lichtman JW. Annu. Rev. Neurosci. 1999;22:389. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- 17.Yeiser EC, Rutkoski NJ, Naito A, Inoue J, Carter BD. J. Neurosci. 2004;24:10521. doi: 10.1523/JNEUROSCI.1390-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bamji SX, et al. J. Cell Biol. 1998;140:911. doi: 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majdan M, Walsh GS, Aloyz R, Miller FD. J. Cell Biol. 2001;155:1275. doi: 10.1083/jcb.200110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wyatt S, Davies AM. J. Cell Biol. 1995;130:1435. doi: 10.1083/jcb.130.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuruvilla R, et al. Cell. 2004;118:243. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 22.Miller FD, Mathew TC, Toma JG. J. Cell Biol. 1991;112:303. doi: 10.1083/jcb.112.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campenot RB. Methods Enzymol. 1979;58:302. doi: 10.1016/s0076-6879(79)58146-4. [DOI] [PubMed] [Google Scholar]
- 24.Brennan C, Rivas-Plata K, Landis SC. Nat. Neurosci. 1999;2:699. doi: 10.1038/11158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.