Abstract
The mechanisms that maintain sterility in the urinary tract are incompletely understood; however, recent studies stress the importance of antimicrobial peptides in protecting the urinary tract from infection. Ribonuclease 7 (RNase 7), a potent antimicrobial peptide contributing to urinary tract sterility, is expressed by intercalated cells in the renal collecting tubules and is present in the urine at levels sufficient to kill bacteria at baseline. Here, we characterize the expression and function of RNase 7 in the human urinary tract during infection. Both quantitative real-time PCR and ELISA assays demonstrated increases in RNASE7 expression in the kidney along with kidney and urinary RNase 7 peptide concentrations with infection. While immunostaining localized RNase 7 production to the intercalated cells of the collecting tubule during sterility, its expression during pyelonephritis was found to increase throughout the nephron but not in glomeruli or the interstitium. Recombinant RNase 7 exhibited antimicrobial activity against uropathogens at low micromolar concentrations by disrupting the microbial membrane as determined by atomic force microscopy. Thus, RNase 7 expression is increased in the urinary tract with infection, and has antibacterial activity against uropathogens at micromolar concentrations.
Keywords: Ribonuclease 7, Antimicrobial Peptide, Pyelonephritis, Urinary Tract Infection, Intercalated Cells, Innate Immunity, Immunology
Introduction
The urinary tract is usually sterile despite its proximity with fecal flora. The precise mechanisms by which the urinary tract maintains sterility are not well understood. Proposed mechanisms contributing to urinary tract sterility include urine flow, alterations in urine osmolality and pH, regular bladder emptying, epithelial shedding, chemical-defense components of the uroepithelium, and the influx of effector immune cells with bacterial stimulation.1 Recently, antimicrobial peptides (AMP) have been shown to play an important role in maintaining urinary tract sterility.2
AMPs, a fundamental component of the innate immune system, are small cationic molecules expressed by phagocytic white cells and epithelial cells. AMPs typically have broad-spectrum antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria, enveloped viruses, fungi, and some protozoa.3, 4 In the urinary tract, human α-defensin 5 (HD5), human β-defensin 1 (hBD-1), human β-defensin 2 (hBD-2), cathelicidin (LL-37), and hepcidin have been shown to be important epithelial derived AMPs that contribute to maintaining sterility.5–12 Uroepithelial cells constitutively produce some of these AMPs, while others are induced upon contact with microorganisms or microbial products.2, 3
Recently, we have shown that Ribonuclease 7 (RNase 7) is a potent AMP that contributes to maintaining sterility of the urinary tract.7 The uroepithelium of the lower urinary tract and the intercalated cells of the renal collecting tubules constitutively produce RNase 7. RNase 7 is present in the urine at levels sufficient to kill bacteria.7 In other organ systems, RNase 7 has potent, broad-spectrum antimicrobial activity against Gram-negative and Gram-positive bacteria.13–15 It has been stated that, on a per molar basis, RNase 7 is the most potent human AMP.16
RNase 7’s antimicrobial activity has not been previously evaluated against common uropathogenic bacteria. Therefore, this study was designed to provide the initial characterization and quantification of RNase 7 expression in the human urinary tract during infection. Here, we begin to elucidate the antimicrobial properties of recombinant RNase 7 against uropathogens.
Results
RNASE7 mRNA expression increases with acute pyelonephritis
Quantitative real-time PCR demonstrates that RNASE7 is constitutively expressed in all tested non-infected and infected human kidney samples. Quantitative real-time PCR also demonstrates that RNASE7 expression significantly increases with acute pyelonephritis. Non-infected human kidneys (n=6) had a mean RNASE7 expression of 1 028+/− 150 transcripts per 10 ng RNA while kidneys with acute pyelonephritis (n=6) expressed 2 927 +/− 591 transcripts RNASE7 per 10 ng RNA (p=0.035) (Figure 1A). In contrast, human kidneys with histological changes consistent with chronic pyelonephritis (n=6) showed a significant decrease in RNASE7 expression, compared to healthy kidneys, with a mean RNASE7 expression of 479 +/− 79 transcripts per 10 ng RNA (p=0.017) (Figure 1A).
Figure 1. RNase 7 mRNA and peptide expression increases with pyelonephritis.
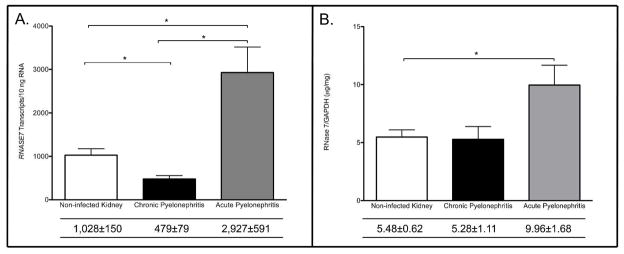
(A) RNASE7 mRNA transcript levels were quantified by real-time PCR in non-infected kidney tissue and in kidney tissue with pyelonephritis. Shown are the results for three independent samples. In the table below, the mean transcript levels are shown with the SEM. RNASE7 expression was significantly greater with acute pyelonephritis (p=0.035). (B) To confirm that the increase in RNASE7 message is accompanied by an increase in RNase 7 protein production, ELISA assays were performed on non-infected kidney tissues and kidney tissues with pyelonephritis. RNase 7 peptide production increased with acute pyelonephritis (p=0.04).
Kidney RNase 7 peptide production increases with acute pyelonephritis
To assess for concurrent increases in kidney RNase 7 peptide production with pyelonephritis, we performed enzyme linked immunoabsorbant assays (ELISA) on the same kidney tissues used for quantitative real-time PCR analysis. Results were standardized to renal GAPDH expression. Our results demonstrate that non-infected kidney tissues (n=4) expressed a mean concentration of 5.48+/−0.62 μg RNase 7/mg GAPDH. Kidney tissues with the histological diagnosis of acute pyelonephritis (n=4) expressed a mean concentration of 9.96 +/− 1.7 μg RNase 7/mg GAPDH, while kidney tissue with chronic pyelonephritis (n=4) expressed a mean concentration of 5.28 +/− 1.11 μg RNase 7/mg GAPDH (Figure 1B).
RNase 7 expression increases in the proximal nephron with pyelonephritis
To localize the increase in RNase 7 peptide production during acute pyelonephritis, we performed immunostaining using polyclonal antibodies directed against RNase 7. Immunohistochemistry (IHC) showed that RNase 7 was expressed in the tubules of the renal cortex and renal medulla in non-infected kidney tissue (n=4) and kidney tissue with both acute and chronic pyelonephritis (n=4). In both states of sterility and infection, RNase 7 was not detected in the glomeruli or interstitium of all tested specimens (Figure 2). Negative controls demonstrated no RNase 7 immunoreactivity (not shown).
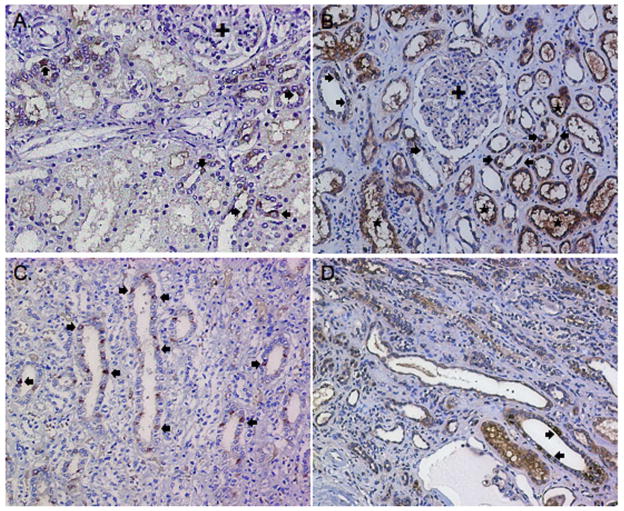
Figure 2. RNase 7 production in non-infected human kidney and human kidney with pyelonephritis.
Immunohistochemistry demonstrates RNase 7 production in isolated renal tubules (brown/arrows) in non-infected renal cortex (A) and medulla (C). With pyelonephritis, RNase 7 production increased in the renal tubules (*) of the cortex (B) and medulla (D). The glomeruli (+) show no immunostaining in non-infected and infected kidney samples. Negative controls showed no immunostaining (not shown). Magnification 20x.
Double labeling immunofluorescence (IF) allowed us to localize RNase 7 production in different segments of the nephron using segment specific markers for the proximal convoluted tubule (aquaporin-1 (AQP-1)) and loop of Henle (uromodulin). The collecting tubules were identified using aquaporin-2 (AQP-2). In all tested specimens, RNase 7 showed cell-specific expression in both the cortical and medullary collecting tubules (Figure 3). As previously shown, RNase 7 expression in the collecting tubules was limited to the intercalated cells.7 In non-infected kidney tissue, RNase 7 expression was not detected in the proximal tubules and loops of Henle. However, during both acute and chronic pyelonephritis, RNase 7 expression was detected in the proximal nephron (Figure 3 and Supplemental Figure 1). Negative controls showed no RNase 7 immunoreactivity (not shown).
Figure 3. Tubular RNase 7 expression in states of sterility and pyelonephritis.
A–B: Confocal micrographs of microdissected human collecting tubules. A: Microdissection of collecting tubules was confirmed by identifying intercalated cells with positive basolateral AE-1 staining (red/arrows) and nuclei (blue). B: Immunostaining identifying intercalated cells with basolateral AE-1 staining (red/arrows), luminal extracellular RNase 7 expression (green/(arrowheads)), and nuclei (blue). These results indicate that RNase 7 is secreted into the urinary space. Magnification 63x.
C–H: Human kidney labeled for RNase 7 (green), nuclei (blue) and nephron specific markers (red). Segment markers consisted of AQP-2 for collecting tubules (CT), uromodulin for the loop of Henle (LOH), and AQP-1 for proximal tubules (PT). C/D: RNase 7 (green/arrowheads) shows no production in the proximal tubules (red, AQP-1 staining) of non-infected kidney tissue (C) but production increases in the proximal tubules (arrows) with pyelonephritis (D). E/F: RNase 7 (green/arrowheads) was not expressed in the loop of Henle (red/arrows, uromodulin staining) in non-infected kidney tissue (E) or with pyelonephritis (F). G/H: RNase 7 (green/arrowheads) was produced by intercalated cells in the collecting tubules of non-infected kidney tissue (G) and with pyelonephritis (H). The red apical (AQP-2 staining) identifies principal cells of the collecting tubules. With pyelonephritis (H), RNase 7 expression expands outside the collecting tubules (arrows). Magnification 40X.
RNase 7 is secreted into the urine, and urinary levels of RNase 7 peptide increase with infection
Previously, we have identified measurable concentrations of RNase 7 peptide in sterile human urine samples.7 To determine if intercalated cells contribute to urinary RNase 7 production, extracellular staining was performed on microdissected human collecting tubules. Immunostaining demonstrated luminal secretion of RNase 7 into the urinary space (Figure 3). ELISA assay results demonstrate that mean urinary concentrations of RNase 7 peptide significantly increase during infection. RNase 7 was detected in non-infected urine samples (n=23) with concentrations ranging from 5.23 – 15.48 μg/mg urine creatinine (UCr), or 0.06 to 0.28 μmol/L (Figure 4). During infection, RNase 7 levels increased to 10.48 – 26.6 μg/mg UCr (n=23), or 0.43 to 0.78 μmol/L (p=0.0002). Urinary RNase 7 levels did not directly correlate with type of uropathogen, severity of infection (febrile versus non-febrile), or urine pH. In a subset of patients in which infected urine samples (E. coli) was initially obtained, follow-up sterile urine samples were collected (n=5). These results suggest that urinary RNase 7 levels increase with infection (Supplemental Figure 2). RNase 7 ELISA assays were also performed on human blood, serum, and plasma. RNase 7 was not detected in any of these tested samples (n=2).
Figure 4. Urinary RNase 7 production increases with urinary tract infection.
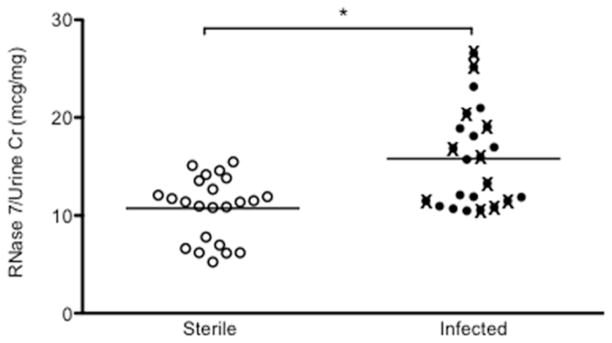
Urinary RNase 7 expression standardized to urinary creatinine in sterile and infected patient urine samples. The infected urine specimens depicted by closed circles (●) are urine isolates from patients with cystitis. The infected urine specimens depicted with (X) are urine isolates from patients with febrile infections.
RNase 7 exhibits potent antimicrobial activity against uropathogenic bacteria
To define the antimicrobial activity of recombinant RNase 7 against common uropathogens, colony count reduction assays were performed using fast performance liquid chromatography (FPLC) fractions of recombinant RNase 7 and inclusion body purified fractions of recombinant RNase 7. Uropathogenic bacteria were exposed to serial dilutions of RNase 7 (0.1–10 μM). The antimicrobial activity did not differ between FPLC or inclusion body fractions. Our results demonstrate that RNase 7 exhibits potent, broad-spectrum antimicrobial activity against Gram-positive and Gram-negative uropathogens at low micromolar concentrations (Table 1). Results are depicted as the minimal inhibitory concentration (MIC) necessary to kill 90% of bacteria (90% lethal dose [LD90] and as the minimal bactericidal concentration (MBC) necessary to kill ≥99.9% of bacteria.
Table 1. The antimicrobial activity of RNase 7 against common uropathogens.
To evaluate the antimicrobial activity of RNase 7 against uropathogenic bacteria, uropathogens were incubated with serial dilutions of recombinant RNase 7 ranging from 0.1–10 μM. Repeat testing was performed on all bacterial isolates in triplicate.
| Strain | MIC(μM)a | MBC(μM)b |
|---|---|---|
| Gram-Negative Bacteria | ||
| Escherichia coli PEDUTI-89 | 0.5 – 1.0 | 1.25 – 2.5 |
| Pseudomonas aeruginosa PEDUTI-961 | 0.6 – 0.8 | 1.25 – 2.5 |
| Klebsiella pneumoniae PEDUTI-965 | 0.2 – 0.4 | 0.8–1.25 |
| Proteus mirabilis PEDUTI-971 | 0.3–0.6 | 1.0–1.25 |
| Gram-Positive Bacteria | ||
| Enterococcus faecalis PEDUTI-983 | 0.1 – 0.2 | 0.5 – 0.75 |
| Staphylococcus saprophyticus PEDUTI-989 | 0.1 – 0.2 | 0.5 – 0.75 |
Minimal inhibitory concentration (MIC) necessary to prevent growth of 90% of the bacteria (90% lethal dose [LD90]).
Minimal bactericidal concentration (MBC) necessary to kill ≥99.9% of the bacteria.
Recombinant RNase 7 (0.1–10 μM) did not demonstrate cytotoxic activity toward mammalian cells – including immortalized human uroepithelial cells (UROtsa cells) and human embryonic kidney cells (HEK 293 cells) (Supplemental Figure 3A). Furthermore, recombinant RNase 7 did not cause hemolysis of human red blood cells (Supplemental Figure 3B).
RNase 7 rapidly destroys uropathogens
A Live/Dead bacterial viability assay (BacLight™, Molecular Probes, Carlsbad, CA, USA) was used to confirm the bactericidal activity profiles of RNase 7 against gram-positive and gram-negative uropathogens. The Live/Dead bacterial viability assay uses a 1:1 mixture of a green-fluorescent STYO®9 dye and a red-fluorescent propidium iodide dye. SYTO®9 labels live bacteria. Propidium iodide penetrates bacteria with damaged membranes (i.e. dead/dying bacteria). The ratio of SYTO®9 to propidium iodide fluorescence provides an estimate for the percentage of viable bacteria for monitoring the kinetics of bacterial death. Changes in bacterial viability were monitored every 15 minutes by measuring the change in fluorescence of both dyes.
Serial dilutions of recombinant RNase 7 were added to 1×106 CFU/mL of stained Gram-negative uropathogenic Escherichia coli (PEDUTI-89) or Gram-positive uropathogenic Enterococcus faecalis (PEDUTI-983). Our results demonstrate that the majority of cell death in both bacterial populations occurs rapidly – within sixty minutes after exposure to recombinant RNase 7 (Figure 5A). After four hours, nearly all bacteria were killed with the addition of ≥ 1μM recombinant RNase 7. These results correlate with the MBC data depicted in Table 1.
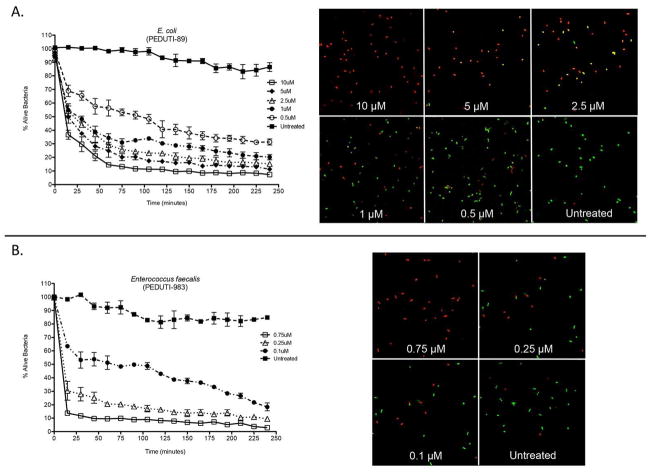
Figure 5. RNase 7 rapidly kills uropathogens at low micromolar concentrations.
(A/B) Left Panels: E. coli and E. faecalis were stained using a 1:1 mixture of SYTO®9, which labels live bacteria, and propidium iodide, which labels killed bacteria. Bacteria were incubated with increasing concentrations of recombinant RNase 7. Bacterial viability over time was analyzed integrating fluorescent changes in SYTO®9 dye and propidium iodide dye. Values are the average of three replicates and the SEM.
(A/B) Right Panels: E. coli and E. faecalis were stained using a 1:1 mixture of SYTO®9 and propidium iodide. The SYTO®9-stained cells (green) represent live cells and the propidium iodide-stained cells (red) depict killed cells. Bacterial viability was visualized after exposure to RNase 7 at 120 minutes. E. coli was incubated with 0.5–10.0 μM RNase 7 and E. faecalis was incubated with 0.1–0.75 uM RNase 7. Recombinant RNase 7 was not added to the untreated bacteria. Magnification 63x.
Next, we analyzed whether the addition of antibodies directed against RNase 7 would neutralize the antimicrobial activity of RNase 7 and enhance bacterial growth. RNase 7 antibodies inhibited the action of RNase 7 as demonstrated by increased bacterial growth of Gram-negative uropathogenic E. coli (PEDUTI-89) and Gram-positive uropathogenic E. faecalis (PEDUTI-983) (Figure 6A). Using identical techniques as previously described above, a C-terminal fragment of RNase 7 that is devoid of the initial 71 amino acids was isolated (AA 72-128). This fragment has limited antimicrobial activity against uropathogenic bacteria. To ensure the peptide purification/isolation process did not incorporate bacteriostatic/bactericidal components, this fragment was tested under identical conditions. The presence of equivalent concentrations of this mutated RNase 7 fragment had limited effects on bacterial survival (Figure 6A).
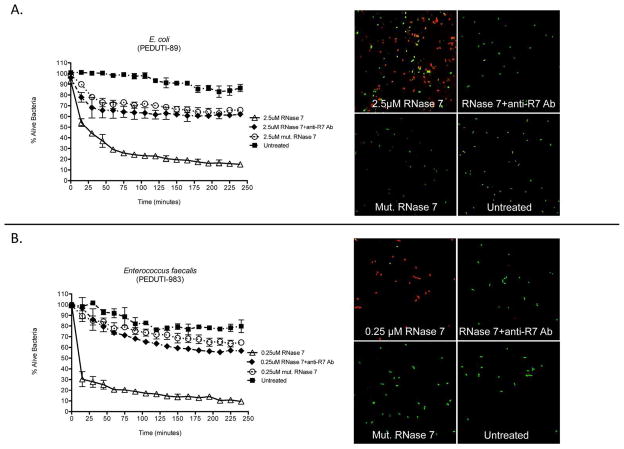
Figure 6. Neutralization of RNase 7’s antimicrobial activity.
(A/B) Left Panels: E. coli (top) and E. faecalis (bottom) were stained using a 1:1 mixture of SYTO®9 and propidium iodide. Bacteria were incubated with recombinant RNase 7, equal concentrations of recombinant RNase 7 plus neutralizing antibody directed against RNase 7 (RNase 7+anti-R7 Ab), or a low activity C-terminal fragment (AA 72-128) of RNase 7 (mut. RNase 7). Bacterial viability over time was analyzed integrating fluorescent changes in SYTO®9 dye and propidium iodide dye. Values are the average of three replicates with SEM.
(A/B) Right Panels: Bacterial viability was visualized after exposure to RNase 7 at 90 minutes. E. coli was incubated with 2.5 μM RNase 7, equal concentrations of recombinant RNase 7 plus antibody directed against RNase 7 (RNase 7+anti-R7 Ab), or 2.5 μM of a C-terminal RNase 7 fragment (mut. RNase 7). E. faecalis was incubated with 0.25 uM RNase 7, equal concentrations of recombinant RNase 7 plus antibody directed against RNase 7 (RNase 7+anti-R7 Ab), or 0.25 μM of a C-terminal RNase 7 fragment (mut. RNase 7). Recombinant RNase 7 was not added to the untreated bacteria. Magnification 63x.
To evaluate the morphological changes in the bacterial cell populations after incubation with increasing concentrations of RNase 7, RNase 7 antibodies, and the RNase 7 C-terminal fragment, uropathogenic E. coli and E. faecalis were labeled with STYO®9 and propidium iodide and visualized using confocal microscopy. These results confirm that micromolar concentrations rapidly kill uropathogenic bacteria (Figure 5B and 6B). The addition of antibodies directed against RNase 7 decreased bacterial death while the addition of the C-terminal RNase 7 fragment caused minimal cell death. With cell death, bacterial aggregation, agglutination, or clumping was not observed.
RNase 7 disrupts the microbial cell membrane of uropathogenic bacteria
To visualize the effects of RNase 7 on uropathogenic bacterial membranes, atomic force microscopy (AFM) was utilized. Recombinant RNase 7 (2.5 μM) was added to 1×106 CFU/mL of uropathogenic E. coli (PEDUTI-89), P. aeruginosa (PEDUTI-961), or E. faecalis (PEDUTI-983). After 90-minute incubation, AFM imaging was performed and demonstrated that RNase 7 has a dramatic impact on the structural integrity of bacteria when compared to untreated controls. Untreated bacteria revealed a relatively smooth surface with no ruptures or large pores, while uropathogens treated with recombinant RNase 7 showed a loss of cellular integrity with membrane splitting and bleb formation (Figure 7).
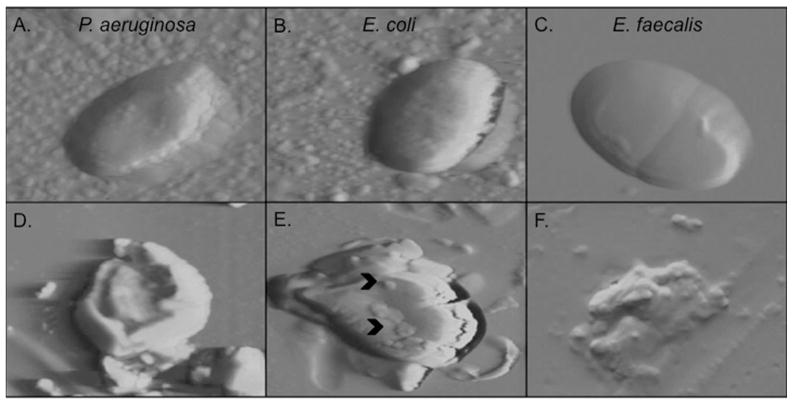
Figure 7. RNase 7 disrupts the microbial membrane at low micromolar concentrations.
Atomic force microscopy demonstrates that 2.5 μM of RNase 7 disrupts the structural integrity of P. aeruginosa, E. coli, and E. faecalis after 45 minutes of exposure. A–C: Images of untreated uropathogens revealed intact cell morphology with a relatively smooth surface. D–F: uropathogens treated with RNase 7 showed loss of cellular integrity with membrane splitting and bleb formation (^). Each panel is 5 micron.
Discussion
RNase 7 is a novel AMP expressed in the human urinary tract that has an important antimicrobial role in maintaining urinary tract sterility.7 In this study, we demonstrate that RNase 7 expression increases with infection and that RNase 7 rapidly exhibits potent, broad-spectrum antimicrobial activity against common uropathogenic bacteria.
Previously, we demonstrated that RNASE7 is constitutively expressed at high levels in the urinary tract.7 In this study, quantitative real-time PCR analysis demonstrates that RNASE7 expression increases with acute pyelonephritis. These results suggest that RNase 7 may be an important AMP for the defense and prevention of early infection in the upper urinary tract. These results also support prior studies suggesting that RNase 7 is induced upon bacterial stimulation. RNASE7 expression in keratinocytes, oral epithelial cells, and human hair follicles has been shown to acutely increase with bacterial exposure.13, 17–19,20 In contrast, prior studies also demonstrate that RNASE7 expression is not induced by human papillomavirus associated infections.21, 22 These results suggest that the regulation of RNASE7 expression may differ between viral and bacterial infections.
In the urinary tract, other epithelial-derived AMPs including DEFA5, DEFB4, and CAMP (the genes encoding HD5, hBD-2, and cathelicidin, respectively) also demonstrate an inducible pattern of mRNA expression with bacterial infection.5, 6, 9 These AMPs may work synergistically to produce a more robust antimicrobial effect or to facilitate a local inflammatory response by attracting immunonocompetent cells.2 At this time, the interactions of RNase 7 with other AMPs in the urinary tract remain to be determined.
Our quantitative real-time PCR results also demonstrate a lack of RNASE7 induction with chronic pyelonephritis. An analogous pattern of RNASE7 expression has been demonstrated in patients with aging oral biofilms and with chronic bacterial skin infections. Eberhard et al demonstrate that RNASE7 expression is up-regulated in early oral biofilm stages and then decreases over time.17 Similarly, Hofmann et al demonstrate that RNASE7 expression decreases with chronic hydradenitis suppurativa, irrespective of disease severity.23 These findings support the notion that RNase 7 may be critical to mounting an innate immune response that favors rapid microbial clearance. Thus, RNase 7 may be more important during early states of infection than in later stages of disease. The observed decrease in RNASE7 expression with chronic infection may represent a pathogenic mechanism aimed at overwhelming the innate immune response, inhibiting RNase 7 synthesis, and allowing pathogens to evade innate defenses to cause disease.
ELISA assays complement our real-time PCR data by demonstrating that RNase 7 kidney and urinary peptide production increases with infection. In the kidney, we also demonstrate that the intercalated cells of the collecting tubules produce RNase 7 and secrete it into the tubular lumen. Chassin et al demonstrate that pyelonephritis-associated E. coli isolates specifically target and attach to the apical membrane of murine intercalated cells.24 Therefore, RNase 7 is ideally positioned to defend the kidney from ascending UTIs as intercalated cells are one of the initial cell types targeted by ascending microbes before they infiltrate the renal parenchyma.
During acute pyelonephritis, the increased concentrations of RNase 7 in the kidney and urinary stream may originate from increased production by the intercalated cells or from increased expression in the proximal nephron. Alternatively, because urinary levels of RNase 7 do not significantly vary between pyelonephritis and cystitis, the increased concentrations of urinary RNase 7 may also result from increased RNase 7 production from the urothepithelium of the lower urinary tract. Previously, we have demonstrated that the uroepithelium expresses high titers of RNase 7.7 Alternatively, we cannot exclude the possibility that neuro-hormonal “crosstalk” between the bladder and kidney leads to increased production in the kidney during uncomplicated cystitis.25
During sterility and/or infection, urinary levels of RNase 7 are significantly greater than previously described epithelial-derived urinary tract AMPs such as HD5, hBD-1, and cathelicidin (Table 2). At these concentrations, RNase 7 has been shown to have antimicrobial activity against pathogens in other organ systems.13, 19, 26 Our results demonstrate that RNase 7 has potent, broad-spectrum antimicrobial activity against Gram-negative and Gram-positive uropathogenic bacteria at these concentrations – with the MIC or LD90 ranging from 0.1–1.0 μM. At these micromolar concentrations, RNase 7 is more potent than the previously described urinary tract AMPs.5, 8, 9, 27 Live/Dead bacterial viability assays demonstrate that RNase 7 kills uropathogens within minutes – faster than conventional antibiotics. These results, when used in conjunction with our previously published data showing that microbial growth increases in human urine when urinary RNase 7’s antibacterial activity is neutralized, demonstrates that RNase 7 has an important role in protecting the urinary tract against infection.7
Table 2. Urinary antimicrobial peptide concentrations during sterility and infection.
Previously defined urinary AMP concentrations for human alpha defensin 5 (HD5), human beta defensin 1 (hBD-1), cathelicidin (LL-37), and RNase 7. HD5 is not detectable (N.D.) in sterile urine.
In this study, immunostaining suggests that RNase 7 production is induced in the proximal nephron during acute and chronic pyelonephritis. Given the size of RNase 7 (14.5 kDa), it is possible that the increased expression observed in the proximal nephron originates, at least in part, from plasma filtration and subsequent proximal tubule endocytosis. However, our ELISA assays do not detect RNase 7 in human blood, serum, or plasma, which is supported by previously published reports that do not detect RNase 7 in human plasma.15 To determine if RNase 7 expression increased in the proximal nephron at the RNA level during pyelonephritis, we attempted to perform in situ hybridization. However, due to limitations of obtaining architecturally preserved human tissue with no RNA degradation, these experiments are ongoing. Moreover, at this time, a lower order vertebrate orthologue of RNase 7 has not been identified. Future studies will involve humanizing the murine kidney and urinary tract to further gain insight into its function. We believe the information gained from these experiments will provide great insight and value as AMPs, like RNase 7, are developed as novel therapeutics.
Currently, the mechanisms for RNase 7’s antimicrobial properties are not completely understood. Its bactericidal activity has been linked to its capacity to permeate and disrupt the bacterial cell membrane, independent of its ribonuclease activity.18, 19 RNase 7, like many human AMPs, is highly cationic protein (with a pI of approximately 10.7). Using NMR spectroscopy and mutagenesis studies, Huang et al have identified clusters of flexible cationic lysine residues on the surface of RNase 7 that are critical for its antimicrobial activity.28 The authors postulate that RNase 7 binds to negatively-charged bacterial membranes through these cationic residues. Upon binding to the bacterial membrane, RNase 7 incorporates itself within the membrane, disrupting its physical and functional characteristics, killing the microbe. The toxicity of RNase 7 is directed mainly towards bacteria, as our results suggest that RNase 7 is not cytotoxic to human uroepithelial cells or red blood cells. Similarly, Harder et al have demonstrated that RNase 7 is not cytotoxic toward human keratinocytes.29 The bactericidal mechanism(s) of RNase 7 warrant in depth studies, and these experiments are ongoing in our laboratory.
To evaluate the effects of RNase 7 on the microbial membranes of common uropathogens, we performed AFM. Our results demonstrate that RNase 7 causes membrane splitting, bleb formation, and significant cell death. These antimicrobial mechanisms of physical bacterial membrane disruption markedly differ from the mechanisms of conventional antibiotics, which typically exhibit antimicrobial effects by inhibiting cell wall synthesis, DNA replication, RNA transcription, or protein synthesis. These unique antimicrobial mechanisms offer potential insights into the development of AMPs, like RNase 7, as a new class of therapeutic agents against bacterial infections like UTIs.
In conclusion, this is the first study to identify and quantitate the expression and production of RNase 7 in the urinary tract during infection. Our results suggest that RNase 7 is a potent epithelial-derived AMP that plays an important role in the innate immunity of the human uroepithelium. Elucidation of the factors that regulate RNase 7 production and its antimicrobial mechanisms may lend novel insight into the pathogenesis of UTIs and help develop RNase 7 as a new therapeutic agent.
Materials and Methods
Study approval
Informed written consent was obtained from all patients participating in this study. For subjects less than 18 year of age, written parental/guardian consent was obtained. The Nationwide Children’s Hospital (NCH) Institutional Review Board approved this study along with the consent process and documents (IRB07-00383). Uropathogenic bacterial isolates were obtained from patients seeking treatment for UTIs at NCH as approved by the Institutional Review Board (IRB-06-00603).
Human Samples
Non-infected kidney samples (n=6) were obtained from patients undergoing nephrectomy for renal tumors. Tissue samples were free of microscopic signs of disease or inflammation. Pyelonephritis kidney samples were obtained from patients undergoing nephrectomy with the clinical diagnosis of chronic pyelonephritis (n=12). Two independent pathologists confirmed the histopathologic diagnosis of pyelonephritis. The pyelonephritis kidney samples showed no evidence of eosinophilic infiltration, granulomas, or interstitial nephritis. The kidney samples with acute plus chronic pyelonephritis were identified by the presence of neutrophils. Kidney tissue was provided by the Cooperative Human Tissue Network, which is funded by the National Cancer Institute.30 Other investigators may have received specimens from the same subjects. Tissue samples were snap-frozen or preserved as neutral formalin-fixed paraffin-embedded sections.
Non-infected and infected urines (n=23) samples were obtained from children presenting to the NCH emergency department or the nephrology clinic. The diagnosis of a UTI was made by a positive urine culture according to the American Academy of Pediatrics Guidelines.31 All infected urine samples had >105 CFU/mL of bacteria and evidence of pyuria. E. coli was detected in 20/23 of the urine samples. The remaining samples were infected with S. saprophyticus (n=1) or E. faecalis (n=2). Urine samples were centrifuged to remove urine sediment and protease inhibitor cocktail was added (Thermo Scientific, Rockford, IL, USA).
Human blood, plasma, and serum samples (n=2) were provided by the Cooperative Human Tissue Network.
Ribonucleic Acid Isolation and Reverse Transcription
Total RNA was isolated from frozen tissue using the Promega Total RNA Isolation System (Promega, Madison, WI, USA). For cDNA synthesis, 4–8 μg of total RNA was reverse transcribed with Superscript III reverse transcriptase using an oligo-(dT)12–18 primer according to the supplier’s protocol (Invitrogen, Carlsbad, CA, USA). A single cDNA preparation from each specimen was used for the assay of all antimicrobial products tested.
Cloning of Gene Specific Plasmids for Standard Curves
cDNA encoding RNASE7 and GAPDH were cloned into a 4-Topo plasmid vector (Invitrogen) according to the manufacturer’s instructions. Plasmids were sequenced to confirm that the correct constructs were obtained. Serial dilutions of gene specific plasmids were quantitated (by both spectrophotometric absorbance at 260 nm and ethidium bromide staining agarose gel electrophoresis with DNA standards) and used in real-time PCR experiments to generate standard curves for each reaction.
Quantitative Real-time PCR
Real-time PCR was performed using single-stranded cDNA from human kidney tissue with specific oligonucleotide primer pairs using the 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) as previously described.7 PCR primers were selected using previously published standards and sequences were confirmed using DNAstar® Laser Gene SeqBuilder (RNase 7 forward primer: 5′-GGA GTC ACA GCA CGA AGA CCA-3′ and RNase 7 reverse primer 5′-CAT GGC TGA GTT GCA TGC TTG A-3′).26 Gene specific plasmid standards were included with every set of reactions. Absolute transcript levels are shown per 10 ng total RNA.
Immunostaining
IHC was performed as previously described.7 Double-labeled immunofluorescence was performed to localize RNase 7 expression in the kidney. Sections were double-labeled for principal cells with goat polyclonal anti-human AQP-2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA); loops of Henle with mouse monoclonal anti-human uromodulin antibody (Sigma Aldrich); and the proximal tubules with goat polyclonal anti-human AQP-1 antibody (Santa Cruz biotechnology). Rhodamine donkey polyclonal anti-goat (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), rhodamine goat anti-mouse (Jackson ImmunoResearch Laboratories), and FITC donkey polyclonal anti-rabbit (Santa Cruz) served as the secondary antibodies.
All sections were prepared as previously described.7 Sections were incubated with a mixture of antisera against RNase 7 (1:200) (Sigma-Aldrich) and antisera against AQP-1 (1:300), uromodulin (1:500), or AQP-2 (1:500) at room temperature for 2 hours. The secondary antibody was applied for 2 hours at room temperature and the sections were mounted using mounting media with DAPI. Non-immune serum was used as a negative control.
Extracellular labeling of microdissected human collecting tubules was performed by incubation with anti-RNase7 antibody at 1:100 dilution in PBS/1% BSA at 4°C prior to fixation with formalin. Post-fixation the tubules were double labeled for α-intercalated cells with mouse IgG1 monoclonal anti-human AE-1 antibody (gift from M. Jennings) followed by rhodamine goat anti-mouse (Jackson ImmunoResearch Laboratories).32
Enzyme Linked Immunosorbent Assay
Commercial antibodies against RNase 7 were used (Abcam, Cambridge, MA, USA; Novus Biologicals, Littleton, CO, USA; and Sigma-Aldrich, St. Louis, MO, USA) as previously described.7 A RNase 7 ELISA assay was used as previously described.7 To quantitate kidney RNase 7 production, 250 μg of kidney protein lysate, as determined by modified Bradford assay, was added per well. Samples were measured in triplicate and standardized to kidney GAPDH production. Kidney GAPDH concentrations were determined using the eBioscience InstantOne™ GAPDH ELISA (San Diego, California, USA). To determine urinary RNase 7 levels, 100 μl of human urine was added to each well. Samples were measured in duplicate. Results from the urine ELISA assay were divided by urine creatinine to establish standardized urine RNase 7-to-creatinine ratios (μg/mg) to account for urine dilution. Urine creatinine concentrations were determined using the Oxford Biomedical Research creatinine microplate assay (Rochester Hills, Michigan, USA).
Generation of Recombinant RNase 7 from Inclusion Body
Using PCR, the full-length human RNASE7 sequence (AA1-128) was generated from a kidney cDNA library and cloned into E. coli expression vector pDEST17 (Invitrogen, Carlesbad, CA, USA), which adds a six-residue histidine tag at the N-terminus. The full-length RNASE7 containing plasmids were transformed into E. coli BL21 AI (Invitrogen) to allow for L-arabinose inducible expression. Bacteria were grown in 2L cultures to mid-log phase and protein expression was induced by L-arabinose (0.2%) for 3–4 hours. The cells were harvested and recombinant RNase 7 was purified from inclusion body as previously described.33
Denatured RNase 7 was then resuspended in 9 mL of 8M guanidinium with 4mM DTT and refolded as follows: 1 mL aliquots of denatured protein was slowly added into 200 mL of cold refolding buffer (100 mM Tris-HCl pH8.0, 400 mM L-Arginine, 0.5 mM ox-glutathione, and 5 mM red-glutathione) while stirring at 4° C for 8 hours. Refolded protein was concentrated and dialyzed against DN(0) buffer [25 mM Tris (pH 7.0), 0.1 mM EDTA, 10% glycerol], and peptide concentrations were determined with a Bradford protein assay (Bio-Rad, Hercules, CA, USA). The presence of the full-length RNase 7 was subjected to SDS-PAGE and visualized by silver stain and Western blot (Supplemental Figure 4).
Generation of Recombinant RNase 7 and a C-terminal RNase 7 Fragment using Fast Protein Liquid Chromatography
An additional full-length RNase 7 peptide (AA1-128) and a C-terminal RNase 7 fragment were generated (AA 72-128) by PCR and cloned into vector pDEST17 (Invitrogen). The C-terminal fragment was generated from the full-length RNase 7 template. This C-terminal fragment has limited antimicrobial activity and was used as a control peptide for the experiments described below. The full length (AA1-128) and C-terminal fragment constructs (AA 72-128) were transformed into E. coli BL21 AI (Invitrogen). Bacteria were grown in 2 L cultures to mid-log phase and peptide expression was induced by L-arabinose for 3–4 hours. The cells were harvested, and the pellets were resuspended in 30 mL 20 mM sodium phosphate pH 7.4, 500 mM NaCl, 0.5 mM phenylmethylsulphonyl fluoride, and protease inhibitor cocktail (Sigma, St. Louis, MO, USA) and lysed by sonication. Cell lysate was pelleted at 13 000 rpm for 20 minutes. The supernatant was applied to a Ni2+ charged HiTrap Chelating HP column (General Electric, Piscataway, NJ, USA). After washing, proteins were eluted with increasing concentrations of imidazole.26 Purified peptides were dialyzed against DN (0) buffer [25 mM Tris (pH 7.0), 0.1 mM EDTA, 10% glycerol], and peptide concentrations were determined with a Bradford protein assay (Bio-Rad).
Antimicrobial Kill Assay
Antimicrobial activity of recombinant RNase 7 was estimated using a colony count reduction assay against uropathogenic E. coli (PEDUTI-89), P. aeruginosa (PEDUTI-961), E. faecalis (PEDUTI-983), Klebsiella pneumoniae (PEDUTI-965), Proteus mirabilis (PEDUTI-971), and S. saprophyticus (PEDUTI-989). These bacterial strains were isolated from positive urine cultures of patients at NCH.
Single cell colonies were grown to mid-exponential growth phase in a 37°C shaking incubator. Working dilutions of 106 bacteria/ml in 0.9% sodium chloride solution (pH 7.4) were prepared (OD670 = 0.06 ~ 2 × 108 bacteria/mL). Test uropathogens were incubated with serial dilutions of recombinant RNase 7 (0.1–10μM) for three hours at 37°C. The antimicrobial activity of RNase 7 was analyzed by plating the incubation mixtures and determining the CFUs the following day. Repeat testing was performed on all bacterial isolates in triplicate. Both the FPLC and the inclusion body fractions of recombinant RNase 7 were tested against E. coli (PEDUTI-89) and S. saprophyticus (PEDUTI-989). Because the antimicrobial profiles were similar between both protein isolates, the remaining assays were performed using recombinant RNase 7 generated from inclusion bodies.
Analysis of Cytotoxic Activity
To determine if recombinant RNase 7 has cytotoxic activity, immortalized human uroepithelial cells (UROtsa cells) (kindly provided by D. Sens) and human embryonic kidney cells (HEK 293 cells) were seeded into a 96-well tissue culture pate (105 cells/well) and incubated with increasing concentrations of recombinant RNase 7 (0.1–10μM) for three hours at 37°C.34 After incubation, cell viability was determined using a Live/Dead mammalian cell viability/cytotoxicity kit in accordance with the manufacturer’s instructions (Molecular Probes, Carlsbad, CA, USA). Cells were stained with calcein AM that is retained in live cells and ethidium heterodimer-1 that is detected in dead cells. Addition of 70% ethanol served as a positive control and untreated cells served as the negative control. The fluorescent intensity was measured using the Spectramax M2 multi-mode microplate reader (Molecular Devices, Sunnyvale, California, USA). The percentage of live bacteria was determined by comparing the calcein AM signal/live bacteria (530 nm) in the RNase 7 treated cells to the control live cells (not exposed to RNase 7). The percentage of dead bacteria was determined by comparing the ethidium homodimer-1 signal/dead bacteria (645 nm) in the RNase 7 treated cells to the positive control cells (exposed to 70% ethanol).
To assess if recombinant RNase 7 induces hemolysis of human red blood cells, human red blood cells were obtained from the Nationwide Children’s Hospital blood bank. 1 mL of human red blood cells was washed with phosphate buffered saline (PBS) and resuspended in 10 mL PBS. The red blood cells were then incubated with increasing concentrations of RNase 7. Addition of 70% ethanol served as a positive control and untreated cells served as the negative control. After three hours of incubation, the cells were centrifuged at 150g for ten minutes. The supernatant was collected and the absorbance was measured at 405nm. The percentage of hemolysis was calculated as: 100 × [(absorbance of sample – absorbance of blank)/(highest absorbance of positive control-absorbance of blank].35
Bacterial Viability Assay
Bacterial viability assays were performed using a Live/Dead BacLight™ bacterial viability kit in accordance with the manufacturer’s instructions. Uropathogenic E. coli (UTI-89) and E. faecalis (PEDUTI-983) were grown at 37°C to the mid-exponential phase, centrifuged at 2500×g for 15 min, and resuspended in a 0.9% sodium chloride solution. Bacteria were diluted to a working concentration of 106 bacteria/mL and stained using a 1:1 mixture of a green-fluorescent STYO®9 dye and a red-fluorescent propidium iodide dye as provided in the Live/Dead BacLight™ bacterial viability kit. Serial dilutions of RNase 7 were added to 200 μL of stained E. coli or E. faecalis. E. coli was incubated with 0.25–10.0 μM recombinant RNase 7 and E. faecalis was incubated with 0.1–1.0 μM recombinant RNase 7.
To determine if the addition of antibodies directed against RNase 7 lead to enhanced bacterial growth, equal concentrations of recombinant RNase 7 and antibodies directed against RNase 7 (Novus Biologicals, Littleton, CO) were incubated together at 37°C for one hour and then added to stained E. coli or E. faecalis. To serve as a control, equal concentrations of the C-terminal RNase 7 fragment, which has minimal activity, were also added to stained bacteria.
The fluorescent intensity was measured using the Spectramax M2 multi-mode microplate reader (Molecular Devices, Sunnyvale, California, USA). The excitation wavelength was centered at 485 nm. The percentage of live bacteria was determined every 15 minutes by comparing the STYO®9 signal/live bacteria (510–540 nm) to the propidium iodide signal/dead bacteria (610–640 nm). These results were compared to a standard curve that was generated using increasing mixtures of live:dead bacteria. Killed bacteria were generated by incubating the test uropathogen in 70% isopropyl alcohol for 1 hour. Samples were performed in triplicate.
Confocal microscopy
Bacterial isolates were prepared as outline above. 50 μL of stained uropathogenic E. coli (PEDUTI-89) or E. faecalis (PEDUTI-983) were incubated with recombinant RNase 7, recombinant RNase 7 plus antibodies directed against RNase 7, or the C-terminal RNase 7 fragment. 10 μL of this mixture was added to poly-l-lysine coated microscope slides (Polysciences, Inc, Warrington, PA, USA) and confocal images of the bacteria were obtained using a Zeiss LSM 710 confocal laser-scanning microscope (Carl Zeiss LLC, Thornwood, NY, USA). SYTO®9 was excited using an argon laser (448–500 nm emission collected) and propidium iodide was excited using an orange diode (555–600nm emission collected). Samples were imaged in triplicate.
Atomic Force Microscopy
Uropathogenic E. coli (UTI-89), P. aeruginosa (UTI-949), and E. faecalis (PEDUTI-983) were prepared as described above for the antimicrobial kill assays. 2.5 μM of recombinant RNase 7 was added to ~106/mL of bacteria. Control samples were not treated with RNase 7. For AFM analysis, the RNase 7 treated samples were imaged after 45-minute incubation at 37°C. AFM imaging was performed using the Bioscope II microscope (Digital Instruments, Piscataway, NJ) in contact mode. All images were obtained with a scan speed of 1.0 Hz. A silicon noncontact low-resonance-frequency cantilever (Nanosensors, Nauchatel, Switzerland) with a resonance frequency of 160 kHz and a spring constant of ~50 N/m was used.
To determine the effect of RNase 7 on the cell membrane, an average of four individual bacterial cells was imaged per slide. Analysis was done using duplicate cultures for each pathogen. Each scan resulted in a topography image and a phase image, which were acquired simultaneously. Images were processed using software provided by Brucker AXS (Madison, WI).
Acknowledgments
We would like to acknowledge the Human Cooperative Human Tissue Network for providing the human samples. We would also like to acknowledge Dr. Amy Leber and Dr. Sheryl Justice for providing the bacterial isolates and Dr. Dan Cohen and the NCH Emergency Department staff for the infected urine samples. We thank Dr. Gunjan Agarwal at The Ohio State University for her assistance with AFM. JDS is supported by the National Institute of Health Grant K08 DK094970-01. ALS and DSH are supported by the National Institute of Health Grant 1RC4DK090937-01.
Abbreviations
- AMP
Antimicrobial Peptide
- RNase 7
Ribonuclease 7
- UTI
Urinary tract Infection
- HD5
Human alpha defensin 5
- hBD-1
Human beta defensin 1
- hBD-2
Human beta defensin 2
- LL-37
Cathelicidin
- ELISA
Enzyme linked immunoabsorbant assay
- AQP-1
Aquaporin -1
- AQP-2
Aquaporin-2
- IHC
Immunohistochemistry
- IF
Immunofluorescence
- MIC
Minimal inhibitory concentration
- MBC
Minimal bactericidal concentration
- FPLC
Fast performance liquid chromatography
- NCH
Nationwide Children’s Hospital
Footnotes
Financial Disclosure and Conflict of Interest: The authors have no conflicts of interest or financial relationships relevant to this article to disclose.
Disclosure:
All the authors in this manuscript declared no competing interests.
References
- 1.Weichhart T, Haidinger M, Horl WH, et al. Current concepts of molecular defence mechanisms operative during urinary tract infection. Eur J Clin Invest. 2008;38 (Suppl 2):29–38. doi: 10.1111/j.1365-2362.2008.02006.x. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Antimicrobial peptides, innate immunity, and the normally sterile urinary tract. J Am Soc Nephrol. 2007;18:2810–2816. doi: 10.1681/ASN.2007050611. [DOI] [PubMed] [Google Scholar]
- 3.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 4.Ali AS, Townes CL, Hall J, et al. Maintaining a sterile urinary tract: the role of antimicrobial peptides. J Urol. 2009;182:21–28. doi: 10.1016/j.juro.2009.02.124. [DOI] [PubMed] [Google Scholar]
- 5.Chromek M, Slamova Z, Bergman P, et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 6.Lehmann J, Retz M, Harder J, et al. Expression of human beta-defensins 1 and 2 in kidneys with chronic bacterial infection. BMC Infect Dis. 2002;2:20. doi: 10.1186/1471-2334-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spencer JD, Schwaderer AL, Dirosario JD, et al. Ribonuclease 7 is a potent antimicrobial peptide within the human urinary tract. Kidney Int. 2011;80:174–180. doi: 10.1038/ki.2011.109. [DOI] [PubMed] [Google Scholar]
- 8.Valore EV, Park CH, Quayle AJ, et al. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spencer JD, Hains DS, Porter E, et al. Human alpha defensin 5 expression in the human kidney and urinary tract. PloS one. 2012;7:e31712. doi: 10.1371/journal.pone.0031712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park CH, Valore EV, Waring AJ, et al. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. The Journal of biological chemistry. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 11.Nitschke M, Wiehl S, Baer PC, et al. Bactericidal activity of renal tubular cells: the putative role of human beta-defensins. Exp Nephrol. 2002;10:332–337. doi: 10.1159/000065296. [DOI] [PubMed] [Google Scholar]
- 12.Townes CL, Ali A, Robson W, et al. Tolerance of bacteriuria after urinary diversion is linked to antimicrobial peptide activity. Urology. 2011;77:509 e501–508. doi: 10.1016/j.urology.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 13.Jaremko JL, Poncet P, Ronsky J, et al. Comparison of Cobb angles measured manually, calculated from 3-D spinal reconstruction, and estimated from torso asymmetry. Comput Methods Biomech Biomed Engin. 2002;5:277–281. doi: 10.1080/10255840290032649. [DOI] [PubMed] [Google Scholar]
- 14.Torrent M, Badia M, Moussaoui M, et al. Comparison of human RNase 3 and RNase 7 bactericidal action at the Gram-negative and Gram-positive bacterial cell wall. FEBS J. 2010;277:1713–1725. doi: 10.1111/j.1742-4658.2010.07595.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Dyer KD, Rosenberg HF. Human RNase 7: a new cationic ribonuclease of the RNase A superfamily. Nucleic acids research. 2003;31:602–607. doi: 10.1093/nar/gkg157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boix E, Nogues MV. Mammalian antimicrobial proteins and peptides: overview on the RNase A superfamily members involved in innate host defence. Mol Biosyst. 2007;3:317–335. doi: 10.1039/b617527a. [DOI] [PubMed] [Google Scholar]
- 17.Eberhard J, Menzel N, Dommisch H, et al. The stage of native biofilm formation determines the gene expression of human beta-defensin-2, psoriasin, ribonuclease 7 and inflammatory mediators: a novel approach for stimulation of keratinocytes with in situ formed biofilms. Oral Microbiol Immunol. 2008;23:21–28. doi: 10.1111/j.1399-302X.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- 18.Harder J, Dressel S, Wittersheim M, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. The Journal of investigative dermatology. 2010;130:1355–1364. doi: 10.1038/jid.2009.432. [DOI] [PubMed] [Google Scholar]
- 19.Simanski M, Dressel S, Glaser R, et al. RNase 7 protects healthy skin from Staphylococcus aureus colonization. The Journal of investigative dermatology. 2010;130:2836–2838. doi: 10.1038/jid.2010.217. [DOI] [PubMed] [Google Scholar]
- 20.Reithmayer K, Meyer KC, Kleditzsch P, et al. Human hair follicle epithelium has an antimicrobial defence system that includes the inducible antimicrobial peptide psoriasin (S100A7) and RNase 7. The British journal of dermatology. 2009;161:78–89. doi: 10.1111/j.1365-2133.2009.09154.x. [DOI] [PubMed] [Google Scholar]
- 21.Erhart W, Alkasi O, Brunke G, et al. Induction of human beta-defensins and psoriasin in vulvovaginal human papillomavirus-associated lesions. The Journal of infectious diseases. 2011;204:391–399. doi: 10.1093/infdis/jir079. [DOI] [PubMed] [Google Scholar]
- 22.Gambichler T, Skrygan M, Tigges C, et al. Significant upregulation of antimicrobial peptides and proteins in lichen sclerosus. Br J Dermatol. 2009;161:1136–1142. doi: 10.1111/j.1365-2133.2009.09273.x. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann SC, Saborowski V, Lange S, et al. Expression of innate defense antimicrobial peptides in hidradenitis suppurativa. J Am Acad Dermatol. 2011 doi: 10.1016/j.jaad.2011.07.020. [DOI] [PubMed] [Google Scholar]
- 24.Chassin C, Goujon JM, Darche S, et al. Renal collecting duct epithelial cells react to pyelonephritis-associated Escherichia coli by activating distinct TLR4-dependent and -independent inflammatory pathways. J Immunol. 2006;177:4773–4784. doi: 10.4049/jimmunol.177.7.4773. [DOI] [PubMed] [Google Scholar]
- 25.Taub DD. Neuroendocrine interactions in the immune system. Cellular immunology. 2008;252:1–6. doi: 10.1016/j.cellimm.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koten B, Simanski M, Glaser R, et al. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PloS one. 2009;4:e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiratsuka T, Nakazato M, Ihi T, et al. Structural analysis of human beta-defensin-1 and its significance in urinary tract infection. Nephron. 2000;85:34–40. doi: 10.1159/000045627. [DOI] [PubMed] [Google Scholar]
- 28.Huang YC, Lin YM, Chang TW, et al. The flexible and clustered lysine residues of human ribonuclease 7 are critical for membrane permeability and antimicrobial activity. The Journal of biological chemistry. 2007;282:4626–4633. doi: 10.1074/jbc.M607321200. [DOI] [PubMed] [Google Scholar]
- 29.Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. The Journal of biological chemistry. 2002;277:46779–46784. doi: 10.1074/jbc.M207587200. [DOI] [PubMed] [Google Scholar]
- 30.LiVolsi VA, Clausen KP, Grizzle W, et al. The Cooperative Human Tissue Network. An update Cancer. 1993;71:1391–1394. doi: 10.1002/1097-0142(19930215)71:4<1391::aid-cncr2820710434>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 31.Roberts KB. Urinary tract infection: clinical practice guideline for the diagnosis and management of the initial UTI in febrile infants and children 2 to 24 months. Pediatrics. 2011;128:595–610. doi: 10.1542/peds.2011-1330. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe S, Tsuruoka S, Vijayakumar S, et al. Cyclosporin A produces distal renal tubular acidosis by blocking peptidyl prolyl cis-trans isomerase activity of cyclophilin. Am J Physiol Renal Physiol. 2005;288:F40–47. doi: 10.1152/ajprenal.00218.2004. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg HF, Dyer KD. Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. The Journal of biological chemistry. 1995;270:30234. [PubMed] [Google Scholar]
- 34.Rossi MR, Masters JR, Park S, et al. The immortalized UROtsa cell line as a potential cell culture model of human urothelium. Environmental health perspectives. 2001;109:801–808. doi: 10.1289/ehp.01109801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naxin J, Tan N, Ho B, et al. Measurement of the red blood cell lysis by bacterial hemolysin. Protocol Exchange. 2007 [Google Scholar]