Abstract
We report that proteins labeled with fluorescein-doped silica nanoparticles (FSNPs) showed drastically different fouling behavior than those labeled with the fluorescein dye. Arrays of polymer films were covalently immobilized on silicon wafers, and were treated with protein conjugated on FSNPs. Fluorescence imaging showed that the protein-FSNP conjugate adsorbed strongly on hydrophilic polymers such as poly(ethylene oxide) (PEO) and weakly on hydrophobic polymers such as polystyrene (PS), and the extent of adsorption decreased with increasing hydrophobicity of the polymer film. Thus, carbohydrate microarrays probed with FSNP-labeled lectin showed significantly enhanced signals when PS was used as the antifouling coating than PEO as well as when bovine serum albumin was used as the blocking agent.
Keywords: anti-fouling, nanoparticle-resistant surface, dye-doped silica nanoparticles, carbohydrate microarray
Microarrays, which allow for high-density ligands to be fabricated on a single slide, have drastically increased the throughput and speed of bioanalysis, and have been frequently used in bioassays and clinical diagnosis.1 A major challenge in bioanalysis is the assay sensitivity, i.e., the signal-to-noise ratio. Generally, two strategies can be applied to increase the assay sensitivity: amplification of the specific binding interactions and reduction of the non-specific background adsorption. A high background noise can obscure the specific binding signal and thus compromise the assay result. Reduction of background noises relies on minimizing the non-specific adsorption of target molecules, such as proteins, to the substrate. Various approaches have been developed to reduce the non-specific adsorption of proteins. A common and simple method is to treat the microarray slide with a blocking agent, such as BSA (bovine serum albumin), casein or milk, before it is assayed with the target protein.2–4 The blocking agent, having moderate non-specific interactions with the substrate, would mostly remain on the background area of the slide preventing the adsorption of the target protein. In the regions where the arrayed ligands are present, the blocking agent would be displaced by the target protein that has much stronger specific binding interactions with the ligands. Besides protein-based blocking agents, anti-fouling polymers5–10 and self-assemble monolayers (SAMs)11,12 are also frequently used to effectively prevent the non-specific adsorption of target proteins.
In microarray analysis, the target molecules are often labeled, for example, with a fluorescent tag, and the ligand-target interactions are recorded by fluorescence imaging. Organic fluorophores are the most frequently used tags due to the availability of a wide range of fluorescent dyes with diverse structure, functionality, solubility, and spectral property. A major drawback of organic dyes, however, is their relatively poor photostability. Alternative labeling agents have emerged, of which luminescent nanoparticles have gained increasing popularity due to their significantly enhanced properties. For example, dye-doped silica nanoparticles,13 silica particles embedded with fluorescent dyes, offer high fluorescence intensity and excellent photostability.14–17 On the other hand, compared with organic fluorophores that are of molecular size, nanoparticles are relatively large entities. The size of nanoparticles, ranging from several to one hundred nanometers, is comparable or larger than many proteins. When nanoparticles are used as the tag to label the target protein, the resulting nanoparticle-protein conjugate may have different non-specific adsorption behavior than the protein labeled with an organic fluorophore.
In this study, we investigate the non-specific adsorption of protein labeled with fluorescein-doped silica nanoparticles (FSNPs), and compare the adsorption behavior with protein labeled with fluorescein (FITC). Arrays of polymer films with varying hydrophobicity are used in the investigation. The best antifouling polymer for FSNP-labeled protein is then applied as the background coating in the fabrication of carbohydrate microarrays. The performance of the microarrays is compared with those fabricated with poly(ethylene oxide) (PEO), a protein resistant polymer, as the background coating as well as using BSA as the blocking agent.
EXPERIMENTAL SECTION
Fabrication of Polymer Arrays and Evaluation of Adsorption of FSNP-labeled Concanavalin A (FSNP-Con A) or FSNPs by Fluorescence Imaging
PFPA-silane (Scheme 1) was synthesized according to a previously published procedure.18,19 The piranha-cleaned wafers were soaked in a solution of PFPA-silane in toluene (12.6 mM) for 4 hours. Polymer arrays were generated by manually spotting solutions of polymers (10 mg/mL) onto the PFPA-functionalized silicon wafers using a pipette tip followed by irradiating with a medium-pressure Hg lamp (450 W, Hanovia Ltd.) for 9 min (Scheme 1). A 280-nm long-path optical filter was placed on the film surface during irradiation to avoid crosslinking of the polymer films.20 The wafers were then sonicated in chloroform followed by Milli-Q water for 5 min each, and finally dried under nitrogen.
Scheme 1.
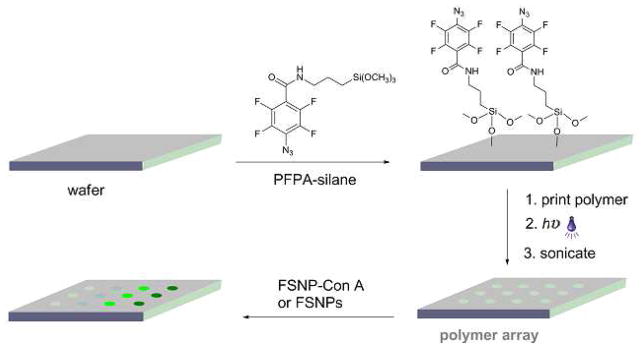
Fabrication of polymer arrays followed by treating with FSNP-labeled Con A or FSNPs.
FSNPs were prepared following a slightly modified Stöber method.17,21 Concanavalin A (Con A, a plant lectin that is known to bind α-mannoside structures22,23) was labeled with FSNPs following a previously developed procedure.24 Briefly, FSNPs were first functionalized with epoxy-silane. Con A (3.3 mg/mL) was then added to epoxy-FSNPs (9.9 mg/mL) to afford FSNP-Con A (see Supplementary Information for experimental details). The density of Con A on FSNP-Con A was determined by conjugating FITC-labeled Con A (FITC-Con A) on epoxy-functionalized silica nanoparticles (SNPs), and comparing the fluorescence intensity of the sample with a calibration curve (Figure S-1) constructed by mixing known concentration of FITC-Con A with SNPs (see Supplementary Information for experimental details). The wafers containing the polymer arrays were incubated in a solution of FSNP-Con A in phosphate buffered saline buffer (PBS, pH 7.4, 0.01 M) (1.6 mg/mL) for 1 hour, and were washed with PBS buffer followed by Milli-Q water. The wafers were then imaged with a fluorescence array scanner (GenePix 4100A, Axon Instruments Inc., Foster City, CA) at 635 nm excitation and 532 nm emission.
Adsorption of FSNPs on Polymer Arrays by Surface Plasmon Resonance Imaging (SPRi)
PFPA-disulfide was synthesized according to previously published procedures.25 The piranha-cleaned SPR chips were immediately soaked in a solution of PFPA-disulfide in chloroform (10 mM) for 24 hours to form a self-assemble monolayer on the surface. Polymer arrays were fabricated in the same manner as on silicon wafers by spotting polymer solutions on the PFPA-functionalized SPR chip followed by UV irradiation. The SPR sample was washed thoroughly in chloroform for 4 hours and dried under nitrogen.
SPRi experiments were conducted at room temperature using a SPRimager® II system (GWC Technologies, Inc.). The polymer array was primed in pH 7.4 PBS buffer until a stable baseline was reached, and then the solution of FSNP in PBS (~10 μg/mL) was injected. The SPR responses from each polymer spot were recorded simultaneously and analyzed using the Digital Optics V++ Version 4 image analysis software.
Fabrication and Evaluation of Carbohydrate Microarrays
Piranha-cleaned silicon wafers were functionalized with poly(allylamine hydrochloride) (PAAm)-PFPA following a previously reported procedure.24,26 Carbohydrate microarray was fabricated by printing aqueous solutions of carbohydrate (10 mg/mL) on PAAm-PFPA-functionalized wafers using a robotic printer (BioOdyssey Calligrapher miniarrayer, Bio-Rad Laboratories, Inc.). The wafers were spin-coated with a solution of PS 280,000 or PEO 1,000,000 in chloroform (5 mg/mL) and then UV-irradiated for 9 min in the presence of a 280-nm optical filter. Then the wafer was sonicated in chloroform and water to remove the un-attached PS and carbohydrates. The control sample was prepared following a similar procedure without the polymer coating, where the wafer was incubated in a 3% BSA solution in pH 7.4 PBS buffer at room temperature for 2 h before treating with FSNP-Con A.
The carbohydrate microarrays were incubated in a solution of FSNP-Con A in HEPES buffer pH 7.5) containing CaCl2 (1 mM) and MnCl2 (1 mM) (0.5 mg/mL) for 1 h, and were then rinsed with HEPES buffer. The wafers were then imaged using a fluorescence array scanner at 635 nm excitation and 532 nm emission.
RESULTS AND DISCUSION
To evaluate the non-specific adsorption of FSNP-labeled protein, arrays of polymer films were fabricated following a general procedure developed in our laboratory.18,19,24,27–30 The photocoupling agent, PFPA, was introduced on wafers or glass slides by treating them with PFPA-silane (Scheme 1). Polymer solutions were spotted on the PFPA-functionalized surfaces using a micropipettor tip. Upon photoactivation, PFPA is converted to a singlet perfluorophenyl nitrene which subsequently forms covalent adducts with the polymers via CH insertion reaction (Scheme S-1).29 Polymer arrays were obtained after the unattached polymers were removed by sonication in the solvent.
FSNPs were synthesized following a previously developed procedure.17 Con A was covalently immobilized on FSNPs by first functionalizing FSNPs with an epoxy silane and then treating the nanoparticles with Con A.24 The density of Con A on FSNP-Con A was determined, and results show a surface coverage of 20±5% against the theoretical maximum surface coverage (see Supplementary Information Part D and Table S-1 for detailed calculation and results). This was comparable to the result reported by Zimmt el al. where 15–30% surface coverage was obtained for FSNP-Con A prepared by NHS/EDC coupling at the Con A loading concentration of 1 mg/mg FSNPs.31
The polymer arrays were then treated with FSNP-Con A, and the fluorescence intensities were measured using a microarray scanner (Figure 1). The hydrophilic neutral polymers such as PEO and PEOX (poly(2-ethyl-2-oxazoline)) showed the highest fluorescence signals, indicating that FSNP-Con A was efficiently absorbed on these polymers. On the hydrophobic polymers such as PS (polystyrene), PMMA (poly(methyl methacrylate)) and PVA (poly(vinyl acetate)), the images were negative (Figure 1). The darker images on these spots imply that FSNP-Con A adsorbed less on these polymers than the background, which was the photoactivated PFPA.32 When the fluorescence intensity was plotted against the contact angle of the polymer film, it can be seen that the fluorescence intensity increased with the hydrophilicity of the polymer (Figure S-3). In other words, FSNP-Con A adsorbed more on hydrophilic polymers than the hydrophobic ones, meaning that hydrophobic polymers were more effective than hydrophilic polymers in resisting the non-specific adsorption of FSNP-Con A. This is in sharp contrast to organic dye-labeled or unlabeled proteins where hydrophilic polymers such as PEO and PEOX are excellent in resisting protein adsorption, whereas hydrophobic polymers such as PS exhibit high non-specific protein adsorption.5–9,33–35
Figure 1.
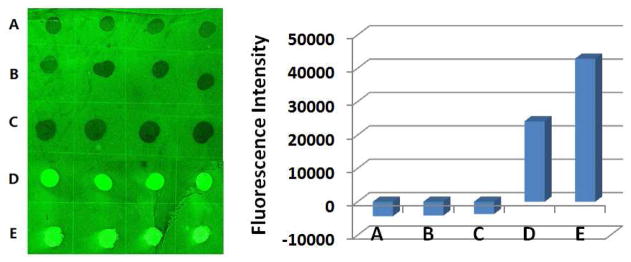
Fluorescence image (left) and intensities (right) of polymer array after treating with FSNP-Con A. The array contains immobilized PS 280,000 (A), PMMA 97,000 (B), PVA 100,000 (C), PEOX 500,000 (D), and PEO 1,000,000 (E). On the fluorescence intensity plot (right), each data was the average of the 4 spots on the array, and the error bars were omitted for clarity.
Similar results were obtained from FSNP-Con A using different loading concentration of Con A (Figure S4–S6). These data imply that FSNPs may play a key role dictating the non-specific adsorption behavior of the FSNP-Con A conjugate. The non-specific adsorption property of FSNPs was then evaluated by treating the polymer array with FSNPs. Consistent with our hypothesis, the hydrophilic polymers (PEOX, PEO) showed high fluorescence intensities whereas negative images were observed on the hydrophobic polymers (PS, PMMA, PVA) (Figure S-7).
The non-specific adsorption behavior of FSNPs was further evaluated using SPRi, a label-free and real-time sensing technique that allows simultaneous measurements of multiple interactions of the target with each ligand on a microarray. In this study, an array consisting of immobilized PEOX and PS were prepared on a PFPA-functionalized Au sensor chip (see Supplementary Information for detailed experimental procedures).30,36 The sensor containing the polymer array was then fixed in a flow cell, and FSNPs were introduced at a flow rate of 100 μL/min. The change in reflectivity (%ΔR) on each polymer spot was monitored simultaneously in real time. Upon injection of FSNPs, %ΔR increased on the PEOX spots, and the increase continued with time (Figure S-8). On PS spots, however, no obvious change was observed when FSNPs solution was injected (Figure S-8).
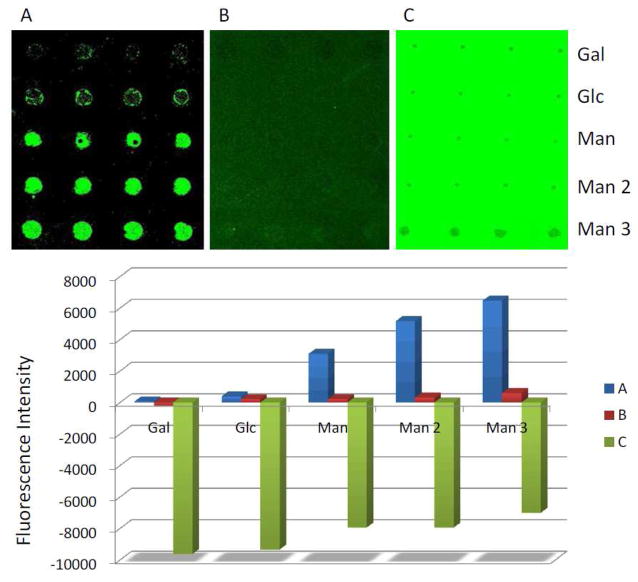
The results obtained above were subsequently applied where PS was used as the anti-fouling coating on a carbohydrate microarray probed with FSNP-labeled lectin. The carbohydrate array was prepared by printing aqueous solutions of 3,6-di-O-(α-D-mannopyranosyl)-D-mannopyranose (Man 3), 2-O-α-D-mannopyranosyl-D-mannopyranose (Man 2), D-(+)-mannose (Man), D-(+)glucose (Glc) and D-(+)-galactose (Gal) on a wafer that was functionalized with PFPA-modified polyallylamine (PAAm-PFPA) (Scheme 2). PAAm-PFPA was used as it showed significantly higher immobilization efficiency for small molecules such as mono- and oligo-saccharides.26 PS was then coated for anti-fouling purpose, and the generated microarray was treated with FSNP-Con A (Scheme 2). Results show that the microarray gave higher signals for the Con A-binding ligands of Man3, Man2, and Man (Figure 2A), and the affinity rank order was consistent with those reported in the literature.22,23 Two control experiments were carried out. First, a carbohydrate microarray without any polymer coating was pre-treated with BSA, a widely-used blocking agent for reducing non-specific protein adsorption, before subjecting to binding with FSNP-Con A. The signals (Figure 2B) were noticeably lower than those when PS was used as the background coating (Figure 2A). In the second control experiment, PEO instead of PS was used as the background coating and the microarray was treated with FSNP-Con A. The signals became negative in this case (Figure 2C), indicating that the FSNP-Con A adsorbed more on the PEO background than on the carbohydrate ligands. These results are consistent with those shown in Figure 1 where FSNP-Con A adsorbed strongly on PEO and weakly on PS. This is a clear contrast to our previous studies on FITC-Con A that the non-specific adsorption was much higher on PS than on PEO.26,37
Scheme 2.
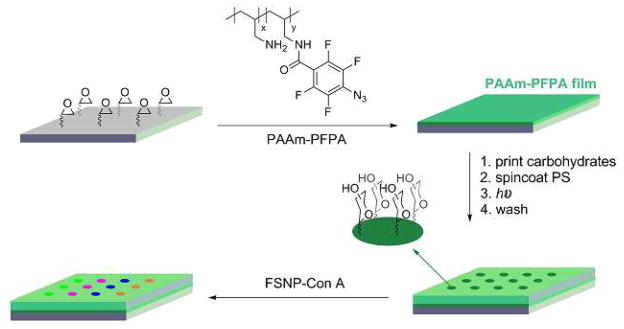
Fabrication of carbohydrate microarrays followed by treating with FSNP-Con A.
Figure 2.
Fluorescence images (top) and fluorescence intensities (bottom) after carbohydrate microarrays were treated with FSNP-Con A. Microarrays were fabricated using PS (A) or PEO (C) as the background coating. In B, no polymer coating was used, and instead, the microarray was incubated with BSA before treating with FSNP-Con A.
CONCLUSIONS
In summary, we found that FSNP-labeled Con A adsorbed strongly on hydrophilic polymers and weakly on hydrophobic ones. This is in sharp contrast with FITC-labeled Con A that has the completely opposite non-specific adsorption behavior. The data demonstrate that when using nanomaterials as labels in bioanalysis, one must take into consideration the adsorption property of the nanomaterials themselves which could drastically alter the assay outcome. Results from these studies were successfully applied to microarray analysis where PS was used as the anti-fouling coating for FSNP-Con A on a carbohydrate microarray. Low non-specific adsorption led to significantly reduced background noise and thus markedly increased assay sensitivity. In this microarray configuration, both the carbohydrate ligands and the anti-fouling polymer were immobilized simultaneously in a single step by a fast light activation. Because the photocoupling reaction applies to a wide range of molecules and materials including small molecules, polymers, nanoparticles and carbon materials,29,38 tailor-made microarrays can be readily fabricated where efficient anti-fouling coating can be incorporated to enhance assay sensitivity.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of General Medical Science (NIGMS) under NIH Award Numbers 1R01GM080295 and 2R15GM066279.
Footnotes
Experimental details, static water contact angles of immobilized polymer films, dynamic light scattering characterization of FSNPs, surface coverage of FITC-Con A on SNPs, fluorescence images of polymer arrays after treating with FSNP, and SPR sensorgram monitoring the adsorption of FSNPs on a polymer array. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Matson R. Microarray Methods and Protocols. CRC press; Boca Raton: 2009. [Google Scholar]
- 2.Alegria-Schaffer A, Lodge A, Vattem K. Methods Enzymol. 2009;463:573–599. doi: 10.1016/S0076-6879(09)63033-0. [DOI] [PubMed] [Google Scholar]
- 3.Nemzek J, Call D, Ebong S, Newcomb D, Bolgos G, Remick D. Am J Physiol Lung Cell Mol Physiol. 2000;278:L512–520. doi: 10.1152/ajplung.2000.278.3.L512. [DOI] [PubMed] [Google Scholar]
- 4.Wolfman J, Palmby T, Der C, Wolfman A. Mol Cell Biol. 2002;22:1589–1606. doi: 10.1128/mcb.22.5.1589-1606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrade JD, Hlady V. Adv Polym Sci. 1986;79:1–63. [Google Scholar]
- 6.Gombotz WR, Guanghui W, Horbett TA, Hoffman AS. J Biomed Mater Res. 1991;25:1547–1562. doi: 10.1002/jbm.820251211. [DOI] [PubMed] [Google Scholar]
- 7.Alcantar NA, Aydil ES, Israelachvili JN. J Biomed Mater Res. 2000;51:343–351. doi: 10.1002/1097-4636(20000905)51:3<343::aid-jbm7>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 8.Adams N, Schubert US. Adv Drug Deliv Rev. 2007;59:1504–1520. doi: 10.1016/j.addr.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 9.Hoogenboom R. Angew Chem, Int Ed. 2009;48:7978–7994. doi: 10.1002/anie.200901607. [DOI] [PubMed] [Google Scholar]
- 10.Dalsin J, Messersmith P. Mater Today. 2005;8:38–46. [Google Scholar]
- 11.Ostuni E, Chapman RG, Holmlin RE, Takayama S, Whitesides GM. Langmuir. 2001;17:5605–5620. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 12.Holmlin RE, Chen X, Chapman RG, Takayama S, Whitesides GM. Langmuir. 2001;17:2841–2850. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 13.Van Blaaderen A, Vrij A. Langmuir. 1992;8:2921–2931. [Google Scholar]
- 14.Wang L, Wang KM, Santra S, Zhao XJ, Hilliard LR, Smith JE, Wu JR, Tan WH. Anal Chem. 2006;78:646–654. [Google Scholar]
- 15.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 16.De M, Ghosh PS, Rotello VM. Adv Mater. 2008;20:4225–4241. [Google Scholar]
- 17.Wang X, Ramstrom O, Yan MD. Chem Commun. 2011;47:4261–4263. doi: 10.1039/c0cc05299j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu L, Engelhard MH, Yan M. J Am Chem Soc. 2006;128:14067–14072. doi: 10.1021/ja062802l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett MA, Yan M. Adv Mater. 2001;13:1449–1451. [Google Scholar]
- 20.Yan M, Harnish B. Adv Mater. 2003;15:244–248. [Google Scholar]
- 21.Ow H, Larson DR, Srivastava M, Baird BA, Webb WW, Wiesner U. Nano Lett. 2005;5:113–117. doi: 10.1021/nl0482478. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz FP, Puri KD, Bhat RG, Surolia A. J Biol Chem. 1993;268:7668–7677. [PubMed] [Google Scholar]
- 23.Mandal DK, Kishore N, Brewer CF. Biochemistry. 1994;33:1149–1156. doi: 10.1021/bi00171a014. [DOI] [PubMed] [Google Scholar]
- 24.Tong Q, Wang X, Wang H, Kubo T, Yan M. Anal Chem. 2012;84:3049–3052. doi: 10.1021/ac203455b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Ramstrom O, Yan M. J Mater Chem. 2009;19:8944–8949. doi: 10.1039/B917900C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo TWX, Tong Q, Yan M. Langmuir. 2011;27:9372–9378. doi: 10.1021/la201324h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Yan M. Angew Chem, Int Ed. 2006;45:6207–6210. doi: 10.1002/anie.200602097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan M. Chem— Eur J. 2007;13:4138–4144. doi: 10.1002/chem.200700317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu LH, Yan M. Acc Chem Res. 2010;43:1434–1443. doi: 10.1021/ar100066t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Li L, Tong Q, Yan M. ACS Appl Mater Interfaces. 2011;3:3463–3471. doi: 10.1021/am200690s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang YQ, Gildersleeve JC, Basu A, Zimmt MB. J Phys Chem B. 2010;114:14487–14494. doi: 10.1021/jp101854m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu LH, Zorn G, Castner D, Solanki R, Lerner M, Yan M. J Mater Chem. 2010;20:5041–5046. doi: 10.1039/C0JM00509F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham NB. In: Poly(ethylene Glycol) Chemistry. Harris JM, editor. Plenum; New York: 1992. pp. 263–282. [Google Scholar]
- 34.Prime KL, Whitesides GM. Science. 1991;252:1164–1167. doi: 10.1126/science.252.5009.1164. [DOI] [PubMed] [Google Scholar]
- 35.Prime KL, Whitesides GM. J Am Chem Soc. 1993;115:10714–10721. [Google Scholar]
- 36.Wang H, Ren J, Hlaing A, Yan M. J Colloid Interface Sci. 2011;354:160–167. doi: 10.1016/j.jcis.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang H, Zhang Y, Yuan X, Chen Y, Yan M. Bioconjugate Chem. 2011;22:26–32. doi: 10.1021/bc100251f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J, Yan M. Acc Chem Res. 2013 doi: 10.1021/ar300172h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.