Abstract
Many carcinogens in tobacco smoke cause DNA damage, and some of that damage can be mitigated by the actions of DNA repair enzymes. In a case-control study nested within the Beta-Carotene and Retinol Efficacy Trial, a randomized chemoprevention trial in current and former heavy smokers, we examined whether lung cancer risk was associated with variation in 26 base excision repair, mismatch repair, and homologous recombination repair genes. Analyses were limited to Caucasians (744 cases, 1477 controls), and logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) for individual SNPs and common haplotypes, with adjustment for matching factors. Lung cancer associations were observed (p<0.05) with SNPs in MSH5 (rs3131379, rs707938), MSH2 (rs2303428), UNG (rs246079), and PCNA (rs25406). MSH5 rs3131379 is a documented lung cancer susceptibility locus in complete linkage disequilibrium with rs3117582 in BAT3, and we observed associations similar in magnitude to those in prior studies (per A allele OR 1.37, 95% CI 1.13-1.65). UNG was associated with lung cancer risk at the gene level (p=0.02), and the A allele of rs246079 was associated with an increased risk (per A allele OR 1.15, 95% CI1.01-1.31). We observed stronger associations with UNG rs246079 among individuals who carried the risk genotypes (AG/AA) for MSH5 rs3131379 (pinteraction= 0.038). Our results provide additional evidence to suggest that the MSH5/BAT3 locus is associated with increased lung cancer risk among smokers, and that associations with other SNPs may vary depending upon MSH5/BAT3 genotype. Future studies to examine this possibility are warranted.
Keywords: Lung cancer, base excision repair, mismatch repair, homologous recombination repair, DNA repair, genetic polymorphism
Introduction
Lung cancer is the leading cause of cancer death worldwide, with over a million deaths annually [1]. The large majority (80-90%) of lung cancers develop in individuals who are either current or former cigarette smokers [2]. Tobacco smoke exposure can result in various types of damage to DNA, either directly by forming DNA adducts, or through the production of reactive oxygen or nitrogen species. These lesions are repaired by a wide variety of DNA repair mechanisms, including (but not limited to) base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), and double-strand break repair (DSB) which includes homologous recombination (HR) and non-homologous end-joining [3]. Therefore, it is plausible that genetic variation in these important pathways might influence lung cancer risk. Indeed, two of the five validated lung cancer susceptibility loci to date map to regions that include genes related to DNA repair. The 6p21.33 locus in the HLA region contains the genes BAT3 and MSH5, and MSH5 is a member of the mutS homolog gene family, involved in MMR. The association with the 12p13 locus is specific to squamous cell lung cancer, and this locus contains the RAD52 homolog gene which is involved in DSB and HR [4]. A recent meta-analysis of 16 GWAS studies with 14,900 cases and 29,485 controls of European descent confirmed these associations as well as those with 5p15 (TERT/CLPTM1L), and 15q25.1 (CHRNA5/CHRNA3/CHRNB4), and reported an additional association for squamous cell carcinoma at 9p21 (CDKN2A/p16INK4A/p14ARF/CDKN2B/p15INK4B/ANRIL) [5]. An additional locus at 6p21.31, containing HLADQA1, was reported in a Japanese GWAS study [6].
Many candidate gene and candidate pathway studies as well as meta-analyses have investigated whether genetic variants in DNA repair pathways are associated with lung cancer risk, with mixed results for genes in MMR [7-15], BER [8,15-32], and HR [8,15,16,19-22,27,29,32]; NER will not be discussed since we have previously reported our findings from analyses of NER genes and lung cancer risk [33]. Herein we report results from our systematic evaluation of associations between 176 tag and functional SNP variants in genes involved in MMR (MLH1, MSH2, MSH4, MSH5, and MSH6), BER (APEX1, LIG3, MBD4, MPG, MUTYH, NEIL1, NEIL2, NTHL1, OGG1, PCNA, PNKP, POLB, POLI, PPP1R13L, RAD18, SMUG1, TDG, UNG, and XRCC1), and HR (XRCC2 and XRCC3) and risk of lung cancer in a nested case-control study of heavy smokers.
Materials and methods
Study population
Details of this study have been published previously [33]. In brief, this nested case-control study is comprised of participants from the multicenter β-Carotene and Retinol Efficacy Trial (CARET), which was a randomized, double-blinded, placebo-controlled chemoprevention trial to assess safety and efficacy of daily supplementation with β-carotene and retinyl palmitate among individuals at high risk of developing lung cancer [34-36]. The trial included men and women ages 50-69 years who were current or former heavy smokers (i.e., quit within six years prior to enrollment) with a cigarette smoking history of ≥20 pack-years (n=14,254). The trial also included men ages 45-69 years with a documented history of occupational asbestos exposure who were current or former heavy smokers (i.e., quit within fifteen years prior to enrollment) (n=4,060). Participants were asked to complete a questionnaire at baseline and annually thereafter, to obtain extensive information about smoking history as well as other risk factors. At baseline and every two years following, they were also asked to complete a food frequency questionnaire (FFQ) describing dietary intake in the prior year. After a mean of four years of follow up, the intervention was stopped in 1996 due to higher lung cancer incidence and overall mortality rates in the intervention versus placebo arm. CARET continued follow up for lung cancer and other outcomes until 2005. Tumor histology data were obtained from pathology reports collected as part of the CARET endpoint review process and through the California, Oregon, and Washington state cancer registries, since about 85% of all participants resided in these states at the time of CARET enrollment.
Participants were eligible for the present nested case-control study if they had provided a whole blood specimen for genetic research between February 1994 and January 1997. Cases included the 793 individuals who were diagnosed with primary lung cancer, and two lung cancer-free controls were matched to each case on age (±4 years), sex, race/ethnicity, enrollment year (two year intervals), baseline smoking status (current or former), history of occupational asbestos exposure, and length of follow-up. Controls were additionally required to have completed at least one FFQ. DNA was extracted from whole blood, and eighteen controls were excluded due to low DNA yield (≤10 μg), leaving a total of 793 cases and 1,568 controls available for genotyping. Three cases were excluded after genotyping, because their diagnoses were later learned to be benign or carcinoid lung tumors.
The Institutional Review Board of the Fred Hutchinson Cancer Research Center and the five other participating institutions approved all study protocols, and all participants provided written informed consent.
SNP selection and genotyping
Tag SNPs were selected from HapMap Phase I and II Centre d’Etude du Polymorphism Humain (CEU; NCBI build 36, dbSNP build 129) for the region spanning ±2,500 base pairs of each candidate gene using the ldSelect algorithm [37] to classify SNPs with a minor allele frequency (MAF) of >=5% into bins with a pair-wise linkage disequilibrium (LD) threshold of r2>=0.8. Additional putative functional SNPs were also selected (for more details, please see Sakoda et al. [33]). We assayed a total of 185 SNPs using three methods: 137 were genotyped in a custom 384-plex Illumina GoldenGate assay that included SNPs in DNA repair, cell cycle control and drug metabolism; 45 were genotyped using individual Applied Biosystem TaqMan assays; and three were genotyped using Sequenom at the Genome Analysis Core Facility at the University of California, San Francisco. Eleven SNPs failed assays, were monomorphic, or genotype frequencies among the non-Hispanic white controls deviated from those expected under Hardy-Weinberg equilibrium as assessed using Fisher’s exact test (p<0.001). After excluding these SNPs, the large majority of SNPs had genotype call success of greater than 99%; 8 SNPs had call success between 95.1 and 98.9%. Genotype concordance for all SNPs was 100% in a set of 82 randomly-placed blind duplicates. Data were excluded for 3 case and 6 control samples that failed the Illumina assays or were identified by Illumina to be gender-mismatched, leaving 787 cases and 1,562 controls available for analysis. A subset of the CARET samples (397 cases and 393 controls) were previously analyzed using the Illumina HumanHap300 BeadChip in an initial GWA study of lung cancer by Hung et al. [38], and these data (394 cases and 391 controls) are also included in the latest metaanalysis [5].
Genotype analysis
Due to small numbers of Hispanic and non-White individuals (43 cases, 85 controls), all analyses were restricted to non-Hispanic whites (744 cases, 1,477 controls). Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated using logistic regression (Stata® 11, StataCorp, College Station, TX) and were adjusted for the case-control matching variables (age, sex, enrollment year, baseline smoking status, and occupational asbestos exposure), using the most common homozygous genotype as the reference group. Per allele ORs and 95% CIs were calculated by coding SNP genotypes according to the number of minor alleles carried (0, 1, or 2).
We examined whether SNP associations varied by age (<70, ≥70 years), sex (male, female), smoking status at baseline (former, current), the number of pack-years smoked at baseline or time of blood draw (defined as the product of the average number of cigarette packs smoked per day and the total number of years smoked, divided into thirds of the distribution among controls), occupational asbestos exposure (yes, no), trial arm assignment (intervention, placebo), and tumor histology (non-small cell lung cancer, small cell lung cancer). Since MSH5 rs3131379/BAT3 rs3117582 and CHRNA5/CHRNA3/CHRNB4 rs16969968 are validated lung cancer susceptibility loci, we also examined associations stratified by these SNPs. Wald p-values of the cross product of SNP genotype and the categorical exposure of interest were generated to formally test for departure from multiplicative relationships. As these are exploratory analyses, the reported p-values are not adjusted for multiple comparisons.
Haplotype analysis
Pairwise linkage disequilibrium (LD) patterns were visualized for each gene region using Haploview, version 4.2 [39]. Haplotype imputation from tagSNP genotype data was conducted using the haplo.stats package (http://mayoresearch.mayo.edu/schaid_lab/software.cfm) in R, version 2.10.1. The expectation-maximization algorithm was used to calculate haplotype frequencies and global tests for each gene were used to evaluate whether there were case-control differences in haplotype frequencies. Additive model ORs and 95% CIs were calculated for each imputed haplotype with a frequency of >1% using the most common haplotype as the reference group, and adjusting for the matching variables.
In order to address issues of multiple testing, we performed gene-set analyses which take into account the number of SNPs tested and the LD between SNPs in each gene (PLINK version 1.04) [40]. Test statistics were averaged for SNPs in each gene and max(T) permutation was performed 10,000 times to calculate empirical p-values taking into account the matching factors.
Results
Baseline characteristics of this nested case-control study have been reported previously [33]. Two thirds of the participants were male, and 73% of participants were current smokers. The distribution of matching factors was broadly similar between cases and controls, though cases were slightly older than controls and were more likely to have reported a heavier smoking history. We successfully evaluated a total of 175 SNPs, with SNP coverage (the proportion of common SNPs represented by the genotyped SNPs through LD in the HapMap Phase I and II CEU populations) for all genes at >=95%, except for MLH1 (89%), MBD4 (93%) and XRCC2 (94%).
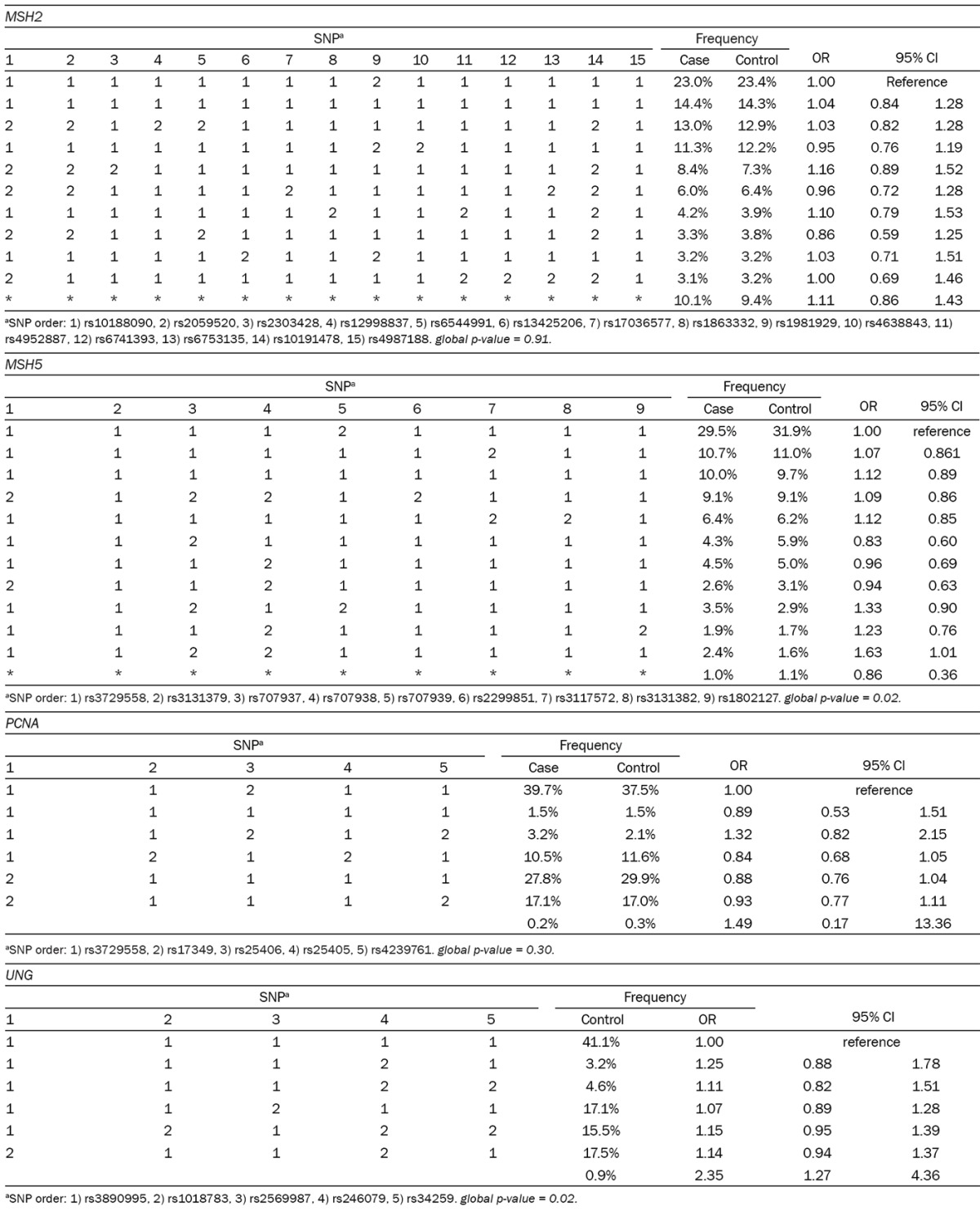
We observed associations with lung cancer for SNPs in the MMR genes MSH5 and MSH2, and in the BER genes PCNA and UNG. We observed a marginal association with a SNP in MPG. Specifically, the minor alleles of the MSH5 SNPs rs3131379 and rs707938 were associated with an increased risk of lung cancer, and there was no association with the only non-synonymous SNP in that gene (rs6905572). Perallele ORs (95% CI) for rs3131379 (A allele) and rs707938 (G allele) were, respectively 1.37 (1.13-1.65) and 1.15 (1.01-1.31) (Table 1). For rs3131379, the ORs and 95% CIs for one or two copies of the A allele (compared to none) were 1.31 (1.06-1.62) and 2.30 (1.12-4.72). The rs3131379 A allele was carried in a single haplotype that also contained the minor allele for rs707938 (although the minor allele for rs707938 was carried in several additional haplotypes), at a frequency of 11% in controls and 14% in cases. Compared to the haplotype containing no variant alleles, this haplotype was associated with an increased risk (OR 1.43, 95% CI 1.15-1.77; global p-value 0.02) (Table 3). Risk of lung cancer associated with this SNP/haplotype did not appear to vary by gender, current/former cigarette smoking, pack-years of smoking, asbestos exposure, or randomization arm, nor did it differ between small cell and non-small cell histologies (data are not shown, but are available upon request). MSH2 rs2303428 was associated with an increased risk of lung cancer (per-G-allele OR 1.24, 95% CI 1.01-1.52), and this association did not vary by any subgroup. The only haplotype that included the rs2303428 G allele was not associated with risk, nor were any other haplotypes in this gene (Table 3).
Table 1.
Per-allele ORs for BER, HR, and MMR SNPs and lung cancer risk among non-Hispanic white smokers
| Major allele | Minor allele | Genotype distributiona | All | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||||
| MAFc | Cases | Controls | (744 cases, 1,477 controls) | |||||||||||
|
| ||||||||||||||
| Pathway | Gene | SNP | (A) | (a) | (%) | AA | Aa | aa | AA | Aa | aa | OR | (95% CI) | p-value |
| MMRb | MLH1 | rs1800734 | G | A | 23.0% | 470 | 212 | 46 | 858 | 520 | 66 | 0.91 | (0.78, 1.06) | 0.215 |
| MMR | MLH1 | rs1540354 | T | A | 17.0% | 499 | 226 | 18 | 1026 | 399 | 50 | 1.05 | (0.89, 1.24) | 0.553 |
| MMR | MLH1 | rs4579 | G | A | 45.0% | 224 | 361 | 159 | 454 | 719 | 304 | 1.03 | (0.91, 1.17) | 0.652 |
| MMR | MSH2 | rs10188090 | G | A | 37.0% | 275 | 366 | 102 | 576 | 706 | 194 | 1.06 | (0.93, 1.21) | 0.394 |
| MMR | MSH2 | rs2059520 | A | G | 34.0% | 299 | 355 | 84 | 626 | 675 | 166 | 1.05 | (0.92, 1.20) | 0.442 |
| MMR | MSH2 | rs2303428 | A | G | 9.0% | 589 | 142 | 13 | 1204 | 266 | 6 | 1.24 | (1.01, 1.52) | 0.043 |
| MMR | MSH2 | rs12998837 | T | A | 13.0% | 532 | 165 | 12 | 1068 | 313 | 32 | 1.00 | (0.83, 1.21) | 0.991 |
| MMR | MSH2 | rs6544991 | A | C | 18.0% | 506 | 220 | 17 | 1008 | 413 | 52 | 0.97 | (0.82, 1.14) | 0.699 |
| MMR | MSH2 | rs13425206 | C | A | 4.0% | 690 | 53 | 1 | 1356 | 118 | 2 | 0.90 | (0.65, 1.24) | 0.509 |
| MMR | MSH2 | rs17036577 | A | G | 9.0% | 623 | 117 | 4 | 1227 | 241 | 8 | 0.96 | (0.76, 1.20) | 0.723 |
| MMR | MSH2 | rs1863332 | A | C | 8.0% | 614 | 120 | 9 | 1239 | 226 | 12 | 1.09 | (0.88, 1.36) | 0.423 |
| MMR | MSH2 | rs1981929 | A | G | 41.0% | 272 | 348 | 124 | 513 | 712 | 251 | 0.95 | (0.84, 1.08) | 0.459 |
| MMR | MSH2 | rs4638843 | G | C | 12.0% | 581 | 151 | 11 | 1135 | 313 | 27 | 0.92 | (0.76, 1.12) | 0.41 |
| MMR | MSH2 | rs4952887 | G | A | 8.0% | 627 | 108 | 9 | 1255 | 209 | 11 | 1.08 | (0.86, 1.35) | 0.506 |
| MMR | MSH2 | rs6741393 | G | A | 3.0% | 697 | 45 | 2 | 1380 | 92 | 2 | 1.00 | (0.71, 1.42) | 0.992 |
| MMR | MSH2 | rs6753135 | G | A | 12.0% | 579 | 152 | 12 | 1139 | 317 | 19 | 0.98 | (0.81, 1.19) | 0.828 |
| MMR | MSH2 | rs10191478 | G | T | 43.0% | 229 | 376 | 139 | 477 | 730 | 270 | 1.04 | (0.92, 1.18) | 0.54 |
| MMR | MSH2 | rs4987188 | G | A | 2.0% | 713 | 31 | 0 | 1428 | 47 | 1 | 1.29 | (0.82, 2.04) | 0.27 |
| MMR | MSH4 | rs5745325 | G | A | 28.0% | 384 | 306 | 54 | 773 | 576 | 128 | 0.98 | (0.86, 1.13) | 0.797 |
| MMR | MSH4 | rs5745433 | A | C | 26.0% | 424 | 247 | 69 | 823 | 547 | 106 | 1.02 | (0.89, 1.17) | 0.752 |
| MMR | MSH4 | rs3819949 | A | G | 34.0% | 339 | 297 | 91 | 633 | 614 | 188 | 0.94 | (0.82, 1.07) | 0.333 |
| MMR | MSH4 | rs2047435 | G | A | 13.0% | 552 | 173 | 17 | 1110 | 343 | 22 | 1.06 | (0.89, 1.28) | 0.507 |
| MMR | MSH4 | rs1146644 | G | A | 42.0% | 249 | 363 | 131 | 515 | 683 | 278 | 1.00 | (0.88, 1.13) | 0.995 |
| MMR | MSH4 | rs1498313 | A | G | 40.0% | 275 | 327 | 141 | 528 | 717 | 230 | 1.05 | (0.92, 1.19) | 0.477 |
| MMR | MSH4 | rs5745513 | T | A | 8.0% | 616 | 122 | 4 | 1248 | 219 | 8 | 1.10 | (0.88, 1.39) | 0.39 |
| MMR | MSH4 | rs5745549 | G | A | 3.0% | 690 | 53 | 1 | 1388 | 85 | 3 | 1.19 | (0.85, 1.67) | 0.317 |
| MMR | MSH5 | rs6905572 | G | A | 13% | 572 | 162 | 10 | 1120 | 333 | 24 | 0.95 | (0.79, 1.15) | 0.621 |
| MMR | MSH5 | rs3131379 | G | A | 11% | 548 | 180 | 16 | 1166 | 292 | 15 | 1.37 | (1.13, 1.65) | 0.001 |
| MMR | MSH5 | rs707937 | C | G | 20% | 477 | 236 | 29 | 948 | 469 | 56 | 1.01 | (0.86, 1.18) | 0.884 |
| MMR | MSH5 | rs707938 | A | G | 32% | 322 | 318 | 104 | 681 | 635 | 161 | 1.15 | (1.01, 1.31) | 0.038 |
| MMR | MSH5 | rs707939 | C | A | 35% | 326 | 343 | 73 | 620 | 673 | 176 | 0.91 | (0.80, 1.04) | 0.184 |
| MMR | MSH5 | rs2299851 | G | A | 10% | 604 | 133 | 5 | 1202 | 256 | 17 | 0.99 | (0.80, 1.22) | 0.928 |
| MMR | MSH5 | rs3117572 | G | A | 17% | 514 | 201 | 29 | 1014 | 416 | 47 | 1.00 | (0.85, 1.18) | 0.999 |
| MMR | MSH5 | rs3131382 | G | A | 6% | 645 | 80 | 6 | 1273 | 164 | 4 | 1.04 | (0.80, 1.35) | 0.76 |
| MMR | MSH5 | rs1802127 | C | T | 2% | 716 | 28 | 0 | 1427 | 49 | 1 | 1.10 | (0.69, 1.76) | 0.683 |
| MMR | MSH6 | rs1800932 | A | G | 18% | 498 | 220 | 25 | 990 | 428 | 53 | 1.00 | (0.85, 1.18) | 0.984 |
| MMR | MSH6 | rs1800937 | G | A | 11% | 599 | 136 | 9 | 1169 | 297 | 10 | 0.95 | (0.77, 1.17) | 0.615 |
| MMR | MSH6 | rs1800935 | A | G | 29% | 381 | 289 | 66 | 736 | 604 | 125 | 0.97 | (0.84, 1.11) | 0.641 |
| MMR | MSH6 | rs2710163 | A | G | 39% | 278 | 340 | 124 | 551 | 690 | 233 | 1.02 | (0.90, 1.16) | 0.748 |
| MMR | MSH6 | rs2348244 | A | G | 14% | 545 | 185 | 13 | 1084 | 358 | 33 | 0.98 | (0.82, 1.18) | 0.856 |
| MMR | MSH6 | rs3136245 | G | A | 19% | 488 | 226 | 27 | 960 | 465 | 48 | 1.00 | (0.85, 1.17) | 0.972 |
| MMR | MSH6 | rs330792 | A | C | 11% | 565 | 170 | 9 | 1156 | 307 | 14 | 1.13 | (0.93, 1.37) | 0.23 |
| MMR | MSH6 | rs1800936 | C | T | 13% | 574 | 155 | 15 | 1110 | 350 | 17 | 0.94 | (0.77, 1.13) | 0.501 |
| MMR | MSH6 | rs3136329 | A | G | 42% | 242 | 359 | 140 | 481 | 731 | 259 | 1.03 | (0.90, 1.16) | 0.701 |
| BERb | APEX1 | rs1760945 | C | T | 8% | 637 | 100 | 4 | 1249 | 213 | 10 | 0.92 | (0.73, 1.17) | 0.509 |
| BER | APEX1 | rs1760944 | C | A | 39% | 270 | 352 | 119 | 547 | 694 | 231 | 1.03 | (0.91, 1.18) | 0.607 |
| BER | APEX1 | rs3136817 | T | C | 25% | 411 | 276 | 57 | 840 | 541 | 96 | 1.08 | (0.93, 1.24) | 0.313 |
| BER | APEX1 | rs1130409 | A | C | 47% | 192 | 361 | 190 | 413 | 723 | 338 | 1.10 | (0.98, 1.25) | 0.116 |
| BER | LIG3 | rs3135962 | A | C | 7% | 647 | 95 | 2 | 1264 | 207 | 6 | 0.91 | (0.71, 1.16) | 0.443 |
| BER | LIG3 | rs3135989 | A | C | 6% | 644 | 97 | 2 | 1301 | 174 | 1 | 1.17 | (0.90, 1.51) | 0.249 |
| BER | LIG3 | rs2074516 | G | C | 10% | 598 | 135 | 8 | 1184 | 276 | 17 | 0.96 | (0.78, 1.18) | 0.696 |
| BER | LIG3 | rs4796030 | C | A | 42% | 242 | 377 | 123 | 486 | 730 | 261 | 0.99 | (0.87, 1.13) | 0.904 |
| BER | LIG3 | rs1052536 | G | A | 47% | 189 | 396 | 156 | 415 | 742 | 320 | 1.04 | (0.91, 1.18) | 0.584 |
| BER | MBD4 | rs3138360 | G | A | 6% | 666 | 77 | 1 | 1292 | 174 | 3 | 0.85 | (0.64, 1.12) | 0.253 |
| BER | MBD4 | rs140696 | G | A | 9% | 604 | 137 | 3 | 1210 | 257 | 10 | 1.04 | (0.84, 1.29) | 0.727 |
| BER | MBD4 | rs9821282 | G | A | 16% | 537 | 185 | 20 | 1035 | 408 | 34 | 0.94 | (0.79, 1.11) | 0.453 |
| BER | MPG | rs1013358 | T | C | 14% | 577 | 153 | 14 | 1093 | 354 | 30 | 0.86 | (0.71, 1.04) | 0.110 |
| BER | MPG | rs2562182 | A | G | 16% | 559 | 164 | 18 | 1044 | 400 | 32 | 0.84 | (0.70, 1.00) | 0.050 |
| BER | MPG | rs743725 | C | T | 19% | 518 | 203 | 23 | 978 | 450 | 49 | 0.88 | (0.75, 1.04) | 0.140 |
| BER | MUTYH | rs3219489 | G | C | 25% | 417 | 279 | 42 | 825 | 562 | 79 | 1.00 | (0.86, 1.15) | 0.948 |
| BER | MUTYH | rs3219487 | G | A | 8% | 628 | 112 | 3 | 1240 | 225 | 9 | 0.96 | (0.76, 1.21) | 0.728 |
| BER | MUTYH | rs3219484 | G | A | 7% | 638 | 106 | 0 | 1273 | 198 | 6 | 1.01 | (0.79, 1.30) | 0.930 |
| BER | MUTYH | rs3219474 | A | G | 8% | 631 | 109 | 4 | 1255 | 212 | 6 | 1.04 | (0.82, 1.32) | 0.747 |
| BER | NEIL1 | rs7182283 | G | T | 50% | 185 | 359 | 192 | 349 | 777 | 339 | 1.04 | (0.91, 1.18) | 0.578 |
| BER | NEIL1 | rs4462560 | C | G | 26% | 428 | 274 | 41 | 813 | 565 | 99 | 0.91 | (0.78, 1.05) | 0.194 |
| BER | NEIL2 | rs4841593 | C | G | 8% | 621 | 120 | 2 | 1256 | 209 | 11 | 1.08 | (0.86, 1.36) | 0.510 |
| BER | NEIL2 | rs904009 | A | C | 24% | 434 | 252 | 57 | 851 | 535 | 88 | 1.02 | (0.88, 1.18) | 0.804 |
| BER | NEIL2 | rs2010628 | G | T | 23% | 445 | 248 | 51 | 881 | 521 | 75 | 1.03 | (0.89, 1.20) | 0.664 |
| BER | NEIL2 | rs8191529 | G | C | 9% | 634 | 105 | 5 | 1229 | 240 | 8 | 0.87 | (0.69, 1.10) | 0.238 |
| BER | NEIL2 | rs804267 | A | G | 33% | 333 | 319 | 92 | 669 | 651 | 155 | 1.05 | (0.92, 1.20) | 0.456 |
| BER | NEIL2 | rs8191534 | T | A | 23% | 441 | 250 | 53 | 865 | 528 | 82 | 1.01 | (0.88, 1.17) | 0.858 |
| BER | NEIL2 | rs8191542 | G | C | 22% | 437 | 260 | 38 | 889 | 503 | 70 | 1.06 | (0.91, 1.24) | 0.422 |
| BER | NEIL2 | rs8191589 | T | A | 22% | 443 | 263 | 37 | 889 | 516 | 72 | 1.04 | (0.89, 1.20) | 0.645 |
| BER | NEIL2 | rs4840581 | G | A | 45% | 232 | 359 | 153 | 442 | 724 | 309 | 0.96 | (0.85, 1.09) | 0.563 |
| BER | NEIL2 | rs4840583 | C | T | 45% | 219 | 367 | 158 | 425 | 771 | 281 | 1.03 | (0.90, 1.17) | 0.687 |
| BER | NEIL2 | rs804256 | T | C | 36% | 304 | 338 | 102 | 597 | 695 | 185 | 1.01 | (0.88, 1.15) | 0.914 |
| BER | NEIL2 | rs8191604 | A | C | 26% | 409 | 276 | 57 | 807 | 569 | 99 | 1.00 | (0.87, 1.15) | 0.987 |
| BER | NEIL2 | rs4840585 | A | C | 8% | 627 | 115 | 2 | 1258 | 208 | 10 | 1.05 | (0.83, 1.32) | 0.700 |
| BER | NEIL2 | rs1874546 | C | G | 24% | 456 | 254 | 31 | 859 | 526 | 83 | 0.87 | (0.75, 1.01) | 0.074 |
| BER | NEIL2 | rs8191649 | C | T | 22% | 466 | 233 | 45 | 889 | 519 | 69 | 0.96 | (0.83, 1.12) | 0.620 |
| BER | NEIL2 | rs6982453 | A | G | 49% | 200 | 389 | 154 | 374 | 755 | 345 | 0.92 | (0.81, 1.04) | 0.189 |
| BER | NEIL2 | rs1534862 | G | A | 23% | 454 | 243 | 46 | 864 | 534 | 78 | 0.95 | (0.82, 1.10) | 0.464 |
| BER | NEIL2 | rs6997097 | A | G | 7% | 654 | 84 | 4 | 1281 | 186 | 7 | 0.92 | (0.71, 1.19) | 0.514 |
| BER | NEIL2 | rs1043180 | G | A | 12% | 575 | 160 | 8 | 1127 | 333 | 17 | 0.94 | (0.77, 1.14) | 0.533 |
| BER | NEIL2 | rs2645450 | T | C | 23% | 426 | 273 | 45 | 878 | 531 | 68 | 1.10 | (0.95, 1.28) | 0.187 |
| BER | NEIL2 | rs904015 | G | A | 35% | 321 | 327 | 95 | 614 | 678 | 174 | 0.99 | (0.87, 1.13) | 0.877 |
| BER | NTHL1 | rs12447809 | G | T | 19% | 474 | 233 | 37 | 960 | 463 | 54 | 1.07 | (0.92, 1.26) | 0.361 |
| BER | NTHL1 | rs1132368 | G | A | 4% | 689 | 54 | 1 | 1348 | 128 | 0 | 0.86 | (0.62, 1.20) | 0.379 |
| BER | NTHL1 | rs2531213 | A | G | 3% | 697 | 47 | 0 | 1378 | 99 | 0 | 0.93 | (0.65, 1.33) | 0.672 |
| BER | NTHL1 | rs3211995 | G | A | 17% | 517 | 205 | 22 | 1009 | 427 | 41 | 0.95 | (0.81, 1.13) | 0.579 |
| BER | NTHL1 | rs2516740 | A | C | 23% | 452 | 255 | 37 | 880 | 524 | 73 | 0.96 | (0.83, 1.12) | 0.593 |
| BER | NTHL1 | rs2516739 | G | A | 22% | 459 | 248 | 37 | 886 | 517 | 72 | 0.95 | (0.82, 1.10) | 0.495 |
| BER | OGG1 | rs159153 | A | G | 29% | 353 | 307 | 84 | 748 | 584 | 140 | 1.11 | (0.97, 1.27) | 0.127 |
| BER | OGG1 | rs1052133 | C | G | 23% | 440 | 265 | 39 | 873 | 519 | 85 | 1.00 | (0.86, 1.16) | 0.992 |
| BER | OGG1 | rs293795 | A | G | 18% | 498 | 222 | 24 | 990 | 438 | 49 | 0.98 | (0.83, 1.15) | 0.815 |
| BER | OGG1 | rs293794 | A | G | 18% | 499 | 221 | 24 | 986 | 439 | 49 | 0.97 | (0.83, 1.14) | 0.736 |
| BER | OGG1 | rs293796 | G | A | 8% | 623 | 110 | 8 | 1260 | 207 | 8 | 1.14 | (0.91, 1.43) | 0.253 |
| BER | PCNA | rs3729558 | G | C | 47% | 223 | 373 | 148 | 422 | 719 | 335 | 0.92 | (0.81, 1.05) | 0.205 |
| BER | PCNA | rs17349 | G | A | 12% | 594 | 140 | 8 | 1147 | 309 | 19 | 0.88 | (0.72, 1.07) | 0.199 |
| BER | PCNA | rs25406 | G | A | 40% | 239 | 370 | 135 | 553 | 675 | 249 | 1.14 | (1.01, 1.29) | 0.038 |
| BER | PCNA | rs25405 | A | G | 12% | 591 | 141 | 8 | 1145 | 306 | 19 | 0.89 | (0.73, 1.09) | 0.262 |
| BER | PCNA | rs4239761 | A | G | 19% | 476 | 233 | 34 | 970 | 439 | 68 | 1.04 | (0.90, 1.22) | 0.587 |
| BER | PNKP | rs7257463 | T | A | 34% | 319 | 327 | 98 | 628 | 685 | 164 | 1.04 | (0.92, 1.19) | 0.518 |
| BER | PNKP | rs1290646 | G | A | 50% | 188 | 383 | 172 | 373 | 734 | 366 | 0.96 | (0.85, 1.09) | 0.558 |
| BER | PNKP | rs3739177 | C | T | 8% | 615 | 124 | 5 | 1259 | 211 | 7 | 1.20 | (0.96, 1.51) | 0.112 |
| BER | PNKP | rs2257103 | G | A | 39% | 265 | 362 | 115 | 546 | 699 | 224 | 1.04 | (0.92, 1.19) | 0.519 |
| BER | PNKP | rs2353005 | G | A | 16% | 543 | 187 | 14 | 1057 | 379 | 41 | 0.92 | (0.77, 1.10) | 0.356 |
| BER | POLB | rs3136711 | T | C | 8% | 627 | 111 | 6 | 1247 | 219 | 11 | 1.03 | (0.82, 1.29) | 0.829 |
| BER | POLB | rs2976244 | A | T | 7% | 645 | 95 | 2 | 1288 | 177 | 11 | 0.98 | (0.76, 1.25) | 0.851 |
| BER | POLB | rs3136790 | A | C | 11% | 585 | 154 | 5 | 1170 | 286 | 18 | 1.00 | (0.82, 1.22) | 0.991 |
| BER | POLB | rs3136797 | C | G | 2% | 716 | 28 | 0 | 1430 | 46 | 1 | 1.15 | (0.72, 1.84) | 0.569 |
| BER | POLB | rs2073664 | G | A | 6% | 647 | 89 | 2 | 1288 | 168 | 11 | 0.96 | (0.75, 1.24) | 0.761 |
| BER | POLI | rs3730668 | C | A | 41% | 283 | 336 | 116 | 524 | 681 | 254 | 0.91 | (0.80, 1.03) | 0.135 |
| BER | POLI | rs476630 | G | A | 29% | 367 | 300 | 77 | 750 | 600 | 126 | 1.08 | (0.95, 1.24) | 0.248 |
| BER | POLI | rs686881 | A | G | 6% | 643 | 100 | 1 | 1310 | 161 | 6 | 1.20 | (0.92, 1.54) | 0.173 |
| BER | POLI | rs3730814 | C | A | 23% | 431 | 272 | 40 | 886 | 499 | 89 | 1.04 | (0.90, 1.20) | 0.593 |
| BER | POLI | rs3218786 | A | G | 3% | 701 | 40 | 2 | 1391 | 82 | 0 | 1.06 | (0.73, 1.53) | 0.771 |
| BER | POLI | rs8305 | A | G | 30% | 359 | 315 | 70 | 716 | 630 | 131 | 1.03 | (0.90, 1.18) | 0.673 |
| BER | POLI | rs596986 | G | C | 6% | 643 | 100 | 1 | 1310 | 161 | 6 | 1.20 | (0.92, 1.54) | 0.173 |
| BER | PPP1R13L | rs6966 | T | A | 16% | 539 | 185 | 20 | 1048 | 377 | 47 | 0.95 | (0.80, 1.12) | 0.524 |
| BER | PPP1R13L | rs4803817 | A | G | 23% | 451 | 255 | 36 | 874 | 510 | 88 | 0.94 | (0.81, 1.09) | 0.425 |
| BER | PPP1R13L | rs10412761 | A | G | 40% | 282 | 357 | 105 | 541 | 681 | 252 | 0.92 | (0.81, 1.05) | 0.204 |
| BER | PPP1R13L | rs1005165 | G | A | 17% | 527 | 194 | 22 | 1016 | 410 | 45 | 0.93 | (0.78, 1.10) | 0.367 |
| BER | RAD18 | rs4389469 | C | T | 40% | 282 | 349 | 113 | 547 | 691 | 239 | 0.97 | (0.85, 1.10) | 0.619 |
| BER | RAD18 | rs369032 | A | G | 38% | 285 | 357 | 102 | 579 | 674 | 224 | 0.99 | (0.87, 1.13) | 0.915 |
| BER | RAD18 | rs2035221 | G | A | 9% | 613 | 123 | 6 | 1227 | 235 | 10 | 1.05 | (0.85, 1.31) | 0.641 |
| BER | RAD18 | rs593205 | G | C | 8% | 605 | 136 | 3 | 1251 | 211 | 12 | 1.23 | (0.99, 1.53) | 0.066 |
| BER | RAD18 | rs373572 | A | G | 26% | 402 | 283 | 56 | 800 | 571 | 105 | 1.02 | (0.88, 1.17) | 0.805 |
| BER | RAD18 | rs13088787 | C | A | 13% | 569 | 165 | 10 | 1127 | 326 | 24 | 0.98 | (0.81, 1.19) | 0.868 |
| BER | RAD18 | rs615967 | T | C | 21% | 461 | 253 | 30 | 920 | 480 | 77 | 0.99 | (0.85, 1.15) | 0.850 |
| BER | RAD18 | rs604092 | A | G | 18% | 501 | 220 | 22 | 1004 | 414 | 56 | 1.00 | (0.85, 1.18) | 0.962 |
| BER | SMUG1 | rs971 | G | A | 34% | 324 | 323 | 93 | 654 | 647 | 172 | 1.03 | (0.90, 1.17) | 0.654 |
| BER | SMUG1 | rs3087404 | G | A | 46% | 226 | 358 | 157 | 437 | 714 | 325 | 0.96 | (0.85, 1.09) | 0.573 |
| BER | TDG | rs172814 | A | G | 16% | 548 | 186 | 10 | 1052 | 384 | 40 | 0.87 | (0.72, 1.03) | 0.113 |
| BER | TDG | rs4135054 | G | A | 11% | 573 | 163 | 8 | 1170 | 291 | 16 | 1.11 | (0.91, 1.35) | 0.290 |
| BER | TDG | rs4135061 | A | G | 27% | 404 | 297 | 41 | 788 | 578 | 109 | 0.93 | (0.80, 1.07) | 0.305 |
| BER | TDG | rs4135064 | G | A | 9% | 599 | 140 | 4 | 1221 | 246 | 9 | 1.13 | (0.91, 1.40) | 0.274 |
| BER | TDG | rs4135081 | A | G | 37% | 273 | 370 | 101 | 579 | 695 | 200 | 1.06 | (0.93, 1.21) | 0.388 |
| BER | TDG | rs3751206 | G | A | 7% | 644 | 96 | 2 | 1292 | 174 | 9 | 1.04 | (0.81, 1.34) | 0.735 |
| BER | TDG | rs4135087 | G | A | 10% | 610 | 130 | 3 | 1184 | 278 | 13 | 0.87 | (0.70, 1.08) | 0.197 |
| BER | TDG | rs167715 | A | G | 11% | 576 | 160 | 8 | 1164 | 294 | 19 | 1.05 | (0.87, 1.28) | 0.598 |
| BER | TDG | rs10861152 | G | A | 39% | 291 | 350 | 97 | 538 | 715 | 216 | 0.91 | (0.80, 1.03) | 0.142 |
| BER | TDG | rs1866074 | A | G | 51% | 178 | 387 | 178 | 377 | 689 | 409 | 0.96 | (0.85, 1.09) | 0.517 |
| BER | TDG | rs4135106 | A | G | 7% | 654 | 86 | 2 | 1282 | 183 | 9 | 0.88 | (0.68, 1.14) | 0.326 |
| BER | TDG | rs4135128 | G | C | 9% | 617 | 123 | 3 | 1241 | 218 | 17 | 1.02 | (0.82, 1.27) | 0.849 |
| BER | UNG | rs3890995 | A | G | 18% | 491 | 233 | 20 | 986 | 461 | 30 | 1.06 | (0.89, 1.25) | 0.529 |
| BER | UNG | rs1018783 | T | A | 16% | 492 | 232 | 20 | 1033 | 403 | 41 | 1.13 | (0.96, 1.33) | 0.152 |
| BER | UNG | rs2569987 | A | G | 17% | 501 | 223 | 19 | 1009 | 421 | 45 | 1.01 | (0.86, 1.19) | 0.893 |
| BER | UNG | rs246079 | A | G | 42% | 217 | 381 | 145 | 485 | 750 | 241 | 1.15 | (1.01, 1.31) | 0.034 |
| BER | UNG | rs34259 | C | G | 20% | 446 | 266 | 30 | 938 | 476 | 62 | 1.10 | (0.94, 1.28) | 0.245 |
| BER | XRCC1 | rs25487 | C | T | 37% | 288 | 365 | 91 | 604 | 664 | 209 | 1.00 | (0.88, 1.14) | 0.950 |
| BER | XRCC1 | rs25486 | A | G | 37% | 288 | 365 | 90 | 599 | 664 | 209 | 1.00 | (0.87, 1.13) | 0.946 |
| BER | XRCC1 | rs25489 | C | T | 4% | 685 | 57 | 2 | 1348 | 128 | 1 | 0.94 | (0.69, 1.29) | 0.701 |
| BER | XRCC1 | rs1799782 | G | A | 5% | 661 | 82 | 1 | 1320 | 153 | 4 | 1.05 | (0.80, 1.38) | 0.733 |
| BER | XRCC1 | rs3213344 | G | C | 5% | 661 | 80 | 1 | 1320 | 150 | 5 | 1.03 | (0.78, 1.35) | 0.844 |
| BER | XRCC1 | rs3213334 | G | A | 24% | 434 | 266 | 44 | 866 | 509 | 102 | 0.99 | (0.85, 1.14) | 0.851 |
| BER | XRCC1 | rs2023614 | G | C | 8% | 633 | 107 | 1 | 1249 | 220 | 6 | 0.92 | (0.72, 1.18) | 0.513 |
| BER | XRCC1 | rs2854510 | A | G | 21% | 470 | 245 | 28 | 936 | 461 | 78 | 0.96 | (0.82, 1.12) | 0.586 |
| BER | XRCC1 | rs2854509 | C | A | 22% | 456 | 242 | 42 | 914 | 480 | 80 | 1.02 | (0.88, 1.18) | 0.793 |
| BER | XRCC1 | rs3213266 | G | A | 8% | 630 | 111 | 2 | 1238 | 232 | 7 | 0.92 | (0.73, 1.16) | 0.477 |
| BER | XRCC1 | rs3213255 | A | G | 43.0% | 242 | 375 | 126 | 491 | 712 | 274 | 0.98 | (0.86, 1.11) | 0.759 |
| HRb | XRCC2 | rs3218536 | G | A | 7% | 631 | 109 | 2 | 1262 | 210 | 5 | 1.05 | (0.82, 1.33) | 0.715 |
| HR | XRCC2 | rs6964582 | G | C | 4% | 663 | 77 | 2 | 1349 | 124 | 2 | 1.26 | (0.95, 1.68) | 0.114 |
| HR | XRCC2 | rs3218438 | T | C | 9% | 598 | 138 | 8 | 1215 | 247 | 15 | 1.10 | (0.89, 1.35) | 0.378 |
| HR | XRCC2 | rs3218408 | A | C | 22% | 430 | 278 | 35 | 901 | 494 | 78 | 1.08 | (0.93, 1.25) | 0.316 |
| HR | XRCC2 | rs3218373 | C | A | 9% | 617 | 119 | 7 | 1230 | 235 | 10 | 1.03 | (0.82, 1.28) | 0.808 |
| HR | XRCC2 | rs2040639 | G | A | 48% | 212 | 366 | 166 | 387 | 767 | 322 | 0.97 | (0.85, 1.10) | 0.625 |
| HR | XRCC3 | rs861539 | G | A | 39% | 307 | 333 | 104 | 536 | 724 | 217 | 0.89 | (0.78, 1.01) | 0.067 |
| HR | XRCC3 | rs3212102 | C | T | 3% | 711 | 33 | 0 | 1402 | 75 | 0 | 0.88 | (0.58, 1.33) | 0.539 |
| HR | XRCC3 | rs3212090 | G | A | 32% | 311 | 354 | 78 | 694 | 628 | 151 | 1.12 | (0.98, 1.28) | 0.087 |
| HR | XRCC3 | rs3212079 | G | A | 7% | 644 | 99 | 1 | 1271 | 191 | 12 | 0.93 | (0.73, 1.19) | 0.585 |
| HR | XRCC3 | rs861530 | C | T | 29% | 362 | 328 | 54 | 733 | 625 | 119 | 1.01 | (0.88, 1.17) | 0.839 |
| HR | XRCC3 | rs1799794 | A | G | 18% | 477 | 245 | 22 | 980 | 442 | 48 | 1.08 | (0.92, 1.27) | 0.340 |
| HR | XRCC3 | rs861528 | G | A | 26% | 427 | 267 | 45 | 807 | 550 | 107 | 0.90 | (0.78, 1.04) | 0.157 |
Among all cases and controls; AA, homozygous major allele; Aa, heterozygous; aa, homozygous minor allele; numbers do not sum to total due to missing.
MMR, mismatch repair; BER, base excision repair; HR, homologous recombination.
MAF, minor allele frequency.
Table 3.
MSH2, MSH5, PCNA, and UNG haplotypes and lung cancer risk
|
For the BER genes, the A allele of PCNA rs25406 was associated with an increased risk of lung cancer (per-allele OR 1.14, 95% CI 1.01-1.29; Table 1), with an association present only among individuals ages 70 years and older (per allele OR 1.38 (1.14-1.66)); among women (1.37 (1.10-1.70)); and among participants who had not been exposed to asbestos (1.22 (1.06-1.40)) (data are not shown, but are available upon request). The G allele of UNG rs246079 was associated with lung cancer risk (per-allele OR 1.15, 95% CI 1.01-1.31; Table 1), and associations did not vary by subgroup. The p-value for gene-level significance for UNG was 0.02, and the haplotype that contained the major allele for all of the SNPs was more frequent in controls than cases (41.1% versus 37.9%, respectively). Four out of five of the other haplotypes included the minor allele of rs246079 and all had ORs that were greater than 1. Only the combined rare genotypes were strongly associated with an increased risk (OR 2.35, 95% CI 1.27-4.36; Table 3). The G allele of MPG rs2562182 was marginally associated with a decreased risk of lung cancer (per-allele OR 0.84, 95% CI 0.70, 1.00; Table 1), and this association was present only among individuals receiving placebo (per allele OR 0.68 (0.51-0.90)). While the p-value for gene-level significance for XRCC2 was 0.03, no SNPs (Table 1) or haplotypes in this gene were individually associated with risk. None of the SNPs in XRCC1 were associated with lung cancer risk overall, but the magnitude of the associations between 4 SNPs (representing 2 SNPs with r2<0.80) in XRCC1 and lung cancer risk appeared to differ between men and women, with interaction p-values less than 0.004 and 0.0001 for rs3213334 (data are not shown, but are available upon request).
In exploratory analyses stratified by the known lung cancer susceptibility loci CHRNA5 rs16969968 and MSH5 rs3131379 genotypes, we observed a departure from a multiplicative relationship (p<0.05) for SNPs in MSH2, MSH4, MSH5, LIG3, and XRCC2 by rs16969968 genotype, with generally stronger associations among individuals carrying the rs16969968 GG genotype than the AG/AA (risk) genotypes. When we stratified by MSH5 rs3131379 genotype, associations with lung cancer were generally stronger among individuals carrying at least one of the rs3131379 A (risk) alleles compared to the GG genotype (Table 2), with a departure from a multiplicative relationship for at least one SNP in each of the MMR genes studied (MLH1, MSH2, MSH4, and MSH6), as well as in the BER genes MUTYH, NTHL1, RAD18, and UNG and the HR gene XRCC2. Among SNPs for which we observed an overall association with lung cancer risk, associations varied by rs3131379 genotype only for UNG rs246079. The per G allele ORs and 95% CIs among AA/AG carriers and among GG carriers were 1.48 (1.13, 1.94) and 1.07 (0.92, 1.24), respectively (pinteraction= 0.038; Table 2).
Table 2.
Per-allele ORs for BER, HR, and MMR SNPs and lung cancer risk among non-Hispanic white smokers, stratified by MSH5 rs3131379 and CHRNA5 rs16969968 genotypes
| rs16969968 GG | rs16969968 AG/AA | rs3131379 GG | rs3131379 AG/AA | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (258 ca, 624 co) | (483 ca, 852 co) | (548 ca, 1,166 co) | (196 ca, 307 co) | |||||||||
|
|
|
|
|
|||||||||
| Pathway | Gene | SNP | OR | (95% CI) | OR | (95% CI) | pc | OR | (95% CI) | OR | (95% CI) | pc |
| MMRb | MLH1 | rs1800734 | 0.99 | (0.77, 1.28) | 0.85 | (0.70, 1.03) | 0.410 | 0.93 | (0.78, 1.11) | 0.84 | (0.61, 1.15) | 0.505 |
| MMR | MLH1 | rs1540354 | 0.99 | (0.76, 1.30) | 1.11 | (0.90, 1.37) | 0.605 | 0.95 | (0.78, 1.15) | 1.40 | (1.01, 1.95) | 0.039 |
| MMR | MLH1 | rs4579 | 1.01 | (0.82, 1.25) | 1.03 | (0.88, 1.21) | 0.905 | 1.07 | (0.92, 1.23) | 0.91 | (0.70, 1.19) | 0.344 |
| MMR | MSH2 | rs10188090 | 1.13 | (0.91, 1.41) | 1.00 | (0.85, 1.18) | 0.392 | 1.01 | (0.87, 1.17) | 1.22 | (0.93, 1.61) | 0.190 |
| MMR | MSH2 | rs2059520 | 1.12 | (0.89, 1.40) | 1.02 | (0.86, 1.21) | 0.561 | 1.00 | (0.85, 1.16) | 1.25 | (0.95, 1.65) | 0.141 |
| MMR | MSH2 | rs2303428 | 1.42 | (1.01, 2.01) | 1.13 | (0.87, 1.47) | 0.280 | 1.30 | (1.03, 1.64) | 1.05 | (0.67, 1.66) | 0.457 |
| MMR | MSH2 | rs12998837 | 1.01 | (0.75, 1.36) | 1.01 | (0.79, 1.28) | 0.992 | 0.88 | (0.71, 1.10) | 1.47 | (1.01, 2.13) | 0.025 |
| MMR | MSH2 | rs6544991 | 0.99 | (0.75, 1.30) | 0.95 | (0.77, 1.18) | 0.840 | 0.86 | (0.71, 1.04) | 1.41 | (1.00, 1.98) | 0.012 |
| MMR | MSH2 | rs13425206 | 1.00 | (0.59, 1.69) | 0.84 | (0.55, 1.28) | 0.642 | 0.89 | (0.62, 1.29) | 0.88 | (0.43, 1.81) | 0.973 |
| MMR | MSH2 | rs17036577 | 0.77 | (0.52, 1.15) | 1.08 | (0.81, 1.43) | 0.179 | 0.94 | (0.72, 1.22) | 0.98 | (0.62, 1.56) | 0.796 |
| MMR | MSH2 | rs1863332 | 1.08 | (0.74, 1.57) | 1.12 | (0.86, 1.47) | 0.865 | 1.09 | (0.86, 1.40) | 1.17 | (0.73, 1.88) | 0.816 |
| MMR | MSH2 | rs1981929 | 0.90 | (0.73, 1.11) | 0.98 | (0.84, 1.15) | 0.549 | 0.97 | (0.84, 1.12) | 0.91 | (0.70, 1.17) | 0.606 |
| MMR | MSH2 | rs4638843 | 0.98 | (0.72, 1.33) | 0.90 | (0.70, 1.15) | 0.595 | 1.07 | (0.86, 1.33) | 0.57 | (0.38, 0.86) | 0.009 |
| MMR | MSH2 | rs4952887 | 1.53 | (1.05, 2.22) | 0.88 | (0.66, 1.16) | 0.022 | 1.12 | (0.87, 1.44) | 1.00 | (0.62, 1.61) | 0.663 |
| MMR | MSH2 | rs6741393 | 1.20 | (0.66, 2.16) | 0.85 | (0.55, 1.32) | 0.342 | 1.12 | (0.76, 1.65) | 0.65 | (0.30, 1.44) | 0.260 |
| MMR | MSH2 | rs6753135 | 0.90 | (0.64, 1.26) | 1.01 | (0.79, 1.28) | 0.616 | 0.99 | (0.80, 1.24) | 0.90 | (0.60, 1.35) | 0.768 |
| MMR | MSH2 | rs10191478 | 1.16 | (0.93, 1.44) | 0.97 | (0.83, 1.14) | 0.197 | 1.01 | (0.87, 1.17) | 1.15 | (0.89, 1.50) | 0.354 |
| MMR | MSH2 | rs4987188 | 1.55 | (0.73, 3.27) | 1.09 | (0.61, 1.97) | 0.487 | 1.71 | (1.04, 2.83) | 0.38 | (0.11, 1.39) | 0.034 |
| MMR | MSH4 | rs5745325 | 1.26 | (1.01, 1.57) | 0.85 | (0.71, 1.02) | 0.009 | 0.90 | (0.77, 1.06) | 1.26 | (0.96, 1.66) | 0.035 |
| MMR | MSH4 | rs5745433 | 0.97 | (0.77, 1.23) | 1.05 | (0.88, 1.25) | 0.589 | 0.97 | (0.83, 1.14) | 1.20 | (0.91, 1.58) | 0.250 |
| MMR | MSH4 | rs3819949 | 0.86 | (0.69, 1.07) | 0.97 | (0.82, 1.15) | 0.407 | 0.97 | (0.83, 1.13) | 0.84 | (0.64, 1.10) | 0.359 |
| MMR | MSH4 | rs2047435 | 0.97 | (0.71, 1.31) | 1.13 | (0.90, 1.43) | 0.464 | 1.18 | (0.96, 1.45) | 0.74 | (0.50, 1.09) | 0.053 |
| MMR | MSH4 | rs1146644 | 0.85 | (0.69, 1.05) | 1.09 | (0.93, 1.28) | 0.072 | 1.11 | (0.96, 1.28) | 0.69 | (0.53, 0.89) | 0.002 |
| MMR | MSH4 | rs1498313 | 0.95 | (0.78, 1.17) | 1.11 | (0.94, 1.30) | 0.275 | 1.05 | (0.91, 1.22) | 1.03 | (0.80, 1.33) | 0.842 |
| MMR | MSH4 | rs5745513 | 1.64 | (1.16, 2.31) | 0.87 | (0.64, 1.19) | 0.008 | 1.06 | (0.81, 1.38) | 1.23 | (0.78, 1.94) | 0.526 |
| MMR | MSH4 | rs5745549 | 0.83 | (0.48, 1.44) | 1.60 | (1.02, 2.51) | 0.067 | 1.59 | (1.08, 2.34) | 0.45 | (0.21, 0.97) | 0.005 |
| MMR | MSH5 | rs6905572 | 1.01 | (0.74, 1.38) | 0.93 | (0.73, 1.19) | 0.785 | 1.01 | (0.82, 1.25) | 0.80 | (0.48, 1.34) | 0.385 |
| MMR | MSH5 | rs3131379 | 1.12 | (0.82, 1.52) | 1.55 | (1.22, 1.97) | 0.101 | NA | NA | |||
| MMR | MSH5 | rs707937 | 0.93 | (0.72, 1.21) | 1.07 | (0.88, 1.31) | 0.361 | 1.02 | (0.86, 1.22) | 1.30 | (0.84, 2.01) | 0.361 |
| MMR | MSH5 | rs707938 | 0.96 | (0.77, 1.19) | 1.28 | (1.09, 1.51) | 0.028 | 1.03 | (0.87, 1.21) | 1.16 | (0.79, 1.71) | 0.593 |
| MMR | MSH5 | rs707939 | 1.18 | (0.95, 1.47) | 0.78 | (0.65, 0.92) | 0.003 | 0.96 | (0.83, 1.12) | 0.98 | (0.67, 1.42) | 0.975 |
| MMR | MSH5 | rs2299851 | 0.90 | (0.63, 1.28) | 1.05 | (0.80, 1.37) | 0.418 | 1.05 | (0.83, 1.32) | 0.85 | (0.48, 1.49) | 0.485 |
| MMR | MSH5 | rs3117572 | 0.90 | (0.69, 1.19) | 1.06 | (0.86, 1.30) | 0.435 | 1.11 | (0.93, 1.32) | 0.68 | (0.40, 1.13) | 0.094 |
| MMR | MSH5 | rs3131382 | 0.82 | (0.53, 1.28) | 1.19 | (0.86, 1.65) | 0.217 | 1.15 | (0.88, 1.52) | 0.60 | (0.24, 1.49) | 0.186 |
| MMR | MSH5 | rs1802127 | 0.90 | (0.37, 2.19) | 1.18 | (0.68, 2.05) | 0.596 | 1.02 | (0.61, 1.71) | 1.99 | (0.58, 6.82) | 0.342 |
| MMR | MSH6 | rs1800932 | 0.93 | (0.71, 1.22) | 1.05 | (0.86, 1.29) | 0.550 | 1.08 | (0.90, 1.31) | 0.75 | (0.54, 1.04) | 0.068 |
| MMR | MSH6 | rs1800937 | 0.79 | (0.55, 1.15) | 1.03 | (0.80, 1.33) | 0.298 | 0.81 | (0.64, 1.04) | 1.53 | (1.01, 2.33) | 0.012 |
| MMR | MSH6 | rs1800935 | 0.88 | (0.70, 1.11) | 1.02 | (0.86, 1.22) | 0.370 | 0.98 | (0.83, 1.14) | 0.91 | (0.69, 1.21) | 0.751 |
| MMR | MSH6 | rs2710163 | 1.05 | (0.85, 1.29) | 1.03 | (0.87, 1.20) | 0.914 | 1.09 | (0.94, 1.26) | 0.81 | (0.62, 1.06) | 0.063 |
| MMR | MSH6 | rs2348244 | 1.07 | (0.80, 1.42) | 0.95 | (0.75, 1.19) | 0.562 | 1.05 | (0.85, 1.28) | 0.82 | (0.56, 1.21) | 0.271 |
| MMR | MSH6 | rs3136245 | 1.12 | (0.86, 1.44) | 0.94 | (0.76, 1.16) | 0.371 | 1.03 | (0.86, 1.23) | 0.93 | (0.66, 1.32) | 0.569 |
| MMR | MSH6 | rs330792 | 1.19 | (0.87, 1.63) | 1.09 | (0.85, 1.40) | 0.675 | 1.14 | (0.91, 1.43) | 1.08 | (0.72, 1.60) | 0.801 |
| MMR | MSH6 | rs1800936 | 0.85 | (0.62, 1.19) | 0.99 | (0.78, 1.25) | 0.585 | 0.84 | (0.67, 1.05) | 1.38 | (0.93, 2.04) | 0.032 |
| MMR | MSH6 | rs3136329 | 1.04 | (0.84, 1.30) | 1.01 | (0.86, 1.18) | 0.799 | 0.96 | (0.83, 1.11) | 1.24 | (0.96, 1.61) | 0.090 |
| BERb | APEX1 | rs1760945 | 0.83 | (0.55, 1.26) | 0.96 | (0.72, 1.29) | 0.521 | 0.97 | (0.74, 1.27) | 0.81 | (0.49, 1.33) | 0.576 |
| BER | APEX1 | rs1760944 | 0.96 | (0.78, 1.18) | 1.09 | (0.92, 1.28) | 0.307 | 1.05 | (0.91, 1.21) | 0.99 | (0.76, 1.30) | 0.622 |
| BER | APEX1 | rs3136817 | 1.05 | (0.84, 1.32) | 1.10 | (0.92, 1.32) | 0.729 | 1.01 | (0.86, 1.20) | 1.21 | (0.92, 1.61) | 0.288 |
| BER | APEX1 | rs1130409 | 1.21 | (0.99, 1.49) | 1.05 | (0.90, 1.23) | 0.327 | 1.06 | (0.92, 1.22) | 1.23 | (0.96, 1.58) | 0.376 |
| BER | LIG3 | rs3135962 | 0.78 | (0.51, 1.21) | 0.97 | (0.71, 1.33) | 0.348 | 1.01 | (0.76, 1.33) | 0.63 | (0.35, 1.14) | 0.147 |
| BER | LIG3 | rs3135989 | 1.72 | (1.11, 2.66) | 0.94 | (0.68, 1.30) | 0.029 | 1.18 | (0.88, 1.60) | 1.09 | (0.63, 1.87) | 0.807 |
| BER | LIG3 | rs2074516 | 0.86 | (0.61, 1.21) | 1.01 | (0.78, 1.31) | 0.586 | 0.97 | (0.77, 1.22) | 1.01 | (0.64, 1.58) | 0.883 |
| BER | LIG3 | rs4796030 | 0.88 | (0.71, 1.09) | 1.04 | (0.88, 1.22) | 0.202 | 0.99 | (0.86, 1.15) | 0.95 | (0.73, 1.24) | 0.785 |
| BER | LIG3 | rs1052536 | 1.18 | (0.96, 1.46) | 0.98 | (0.84, 1.16) | 0.190 | 1.03 | (0.89, 1.19) | 1.07 | (0.82, 1.38) | 0.812 |
| BER | MBD4 | rs3138360 | 0.87 | (0.56, 1.35) | 0.85 | (0.59, 1.22) | 0.907 | 0.84 | (0.61, 1.17) | 0.83 | (0.48, 1.44) | 0.985 |
| BER | MBD4 | rs140696 | 1.02 | (0.72, 1.45) | 1.09 | (0.82, 1.44) | 0.702 | 1.02 | (0.79, 1.31) | 1.14 | (0.73, 1.78) | 0.756 |
| BER | MBD4 | rs9821282 | 0.94 | (0.71, 1.25) | 0.95 | (0.76, 1.19) | 0.882 | 0.93 | (0.76, 1.14) | 0.95 | (0.67, 1.33) | 0.977 |
| BER | MPG | rs1013358 | 1.04 | (0.76, 1.42) | 0.76 | (0.61, 0.97) | 0.146 | 0.84 | (0.68, 1.03) | 0.95 | (0.63, 1.42) | 0.537 |
| BER | MPG | rs2562182 | 0.91 | (0.67, 1.25) | 0.80 | (0.64, 0.99) | 0.525 | 0.82 | (0.67, 1.01) | 0.92 | (0.61, 1.39) | 0.501 |
| BER | MPG | rs743725 | 0.95 | (0.71, 1.26) | 0.86 | (0.70, 1.05) | 0.628 | 0.88 | (0.73, 1.06) | 0.92 | (0.63, 1.32) | 0.758 |
| BER | MUTYH | rs3219489 | 0.90 | (0.70, 1.16) | 1.04 | (0.86, 1.25) | 0.419 | 0.90 | (0.76, 1.07) | 1.30 | (0.97, 1.74) | 0.028 |
| BER | MUTYH | rs3219487 | 1.22 | (0.86, 1.75) | 0.85 | (0.62, 1.16) | 0.153 | 0.93 | (0.71, 1.21) | 1.06 | (0.66, 1.69) | 0.628 |
| BER | MUTYH | rs3219484 | 0.80 | (0.52, 1.23) | 1.16 | (0.85, 1.58) | 0.159 | 1.10 | (0.83, 1.45) | 0.77 | (0.44, 1.34) | 0.277 |
| BER | MUTYH | rs3219474 | 0.86 | (0.58, 1.28) | 1.20 | (0.89, 1.62) | 0.182 | 1.09 | (0.82, 1.44) | 0.91 | (0.58, 1.43) | 0.487 |
| BER | NEIL1 | rs7182283 | 1.14 | (0.92, 1.40) | 0.98 | (0.83, 1.15) | 0.267 | 1.04 | (0.90, 1.21) | 1.01 | (0.78, 1.30) | 0.780 |
| BER | NEIL1 | rs4462560 | 0.88 | (0.70, 1.12) | 0.93 | (0.77, 1.13) | 0.705 | 0.86 | (0.72, 1.01) | 1.11 | (0.82, 1.51) | 0.137 |
| BER | NEIL2 | rs4841593 | 1.01 | (0.70, 1.46) | 1.16 | (0.86, 1.56) | 0.561 | 1.14 | (0.88, 1.48) | 0.91 | (0.56, 1.51) | 0.462 |
| BER | NEIL2 | rs904009 | 1.07 | (0.84, 1.37) | 0.98 | (0.82, 1.17) | 0.569 | 0.97 | (0.82, 1.14) | 1.20 | (0.90, 1.62) | 0.221 |
| BER | NEIL2 | rs2010628 | 1.09 | (0.85, 1.40) | 1.00 | (0.83, 1.20) | 0.572 | 1.01 | (0.86, 1.20) | 1.09 | (0.81, 1.47) | 0.718 |
| BER | NEIL2 | rs8191529 | 0.81 | (0.56, 1.18) | 0.92 | (0.68, 1.24) | 0.666 | 0.91 | (0.70, 1.18) | 0.75 | (0.44, 1.25) | 0.512 |
| BER | NEIL2 | rs804267 | 1.09 | (0.87, 1.36) | 1.03 | (0.87, 1.21) | 0.716 | 1.03 | (0.89, 1.20) | 1.11 | (0.84, 1.46) | 0.675 |
| BER | NEIL2 | rs8191534 | 1.05 | (0.82, 1.34) | 0.99 | (0.82, 1.18) | 0.699 | 0.97 | (0.82, 1.15) | 1.16 | (0.86, 1.55) | 0.333 |
| BER | NEIL2 | rs8191542 | 1.20 | (0.94, 1.55) | 0.98 | (0.80, 1.18) | 0.198 | 1.04 | (0.88, 1.24) | 1.13 | (0.83, 1.54) | 0.662 |
| BER | NEIL2 | rs8191589 | 1.15 | (0.90, 1.48) | 0.96 | (0.80, 1.17) | 0.292 | 1.02 | (0.85, 1.21) | 1.09 | (0.80, 1.48) | 0.724 |
| BER | NEIL2 | rs4840581 | 0.89 | (0.73, 1.09) | 1.02 | (0.87, 1.20) | 0.308 | 0.99 | (0.86, 1.15) | 0.87 | (0.68, 1.12) | 0.372 |
| BER | NEIL2 | rs4840583 | 1.07 | (0.86, 1.33) | 1.00 | (0.85, 1.17) | 0.593 | 1.00 | (0.86, 1.16) | 1.12 | (0.86, 1.46) | 0.436 |
| BER | NEIL2 | rs804256 | 0.98 | (0.80, 1.21) | 1.04 | (0.88, 1.23) | 0.781 | 1.03 | (0.89, 1.20) | 0.94 | (0.72, 1.22) | 0.584 |
| BER | NEIL2 | rs8191604 | 0.98 | (0.78, 1.25) | 1.00 | (0.84, 1.20) | 0.949 | 0.97 | (0.82, 1.14) | 1.11 | (0.84, 1.48) | 0.395 |
| BER | NEIL2 | rs4840585 | 0.99 | (0.68, 1.45) | 1.11 | (0.82, 1.49) | 0.640 | 1.12 | (0.86, 1.46) | 0.87 | (0.52, 1.44) | 0.417 |
| BER | NEIL2 | rs1874546 | 0.77 | (0.59, 0.99) | 0.94 | (0.78, 1.14) | 0.207 | 0.87 | (0.73, 1.04) | 0.85 | (0.63, 1.14) | 0.869 |
| BER | NEIL2 | rs8191649 | 1.03 | (0.80, 1.33) | 0.91 | (0.75, 1.10) | 0.447 | 0.93 | (0.78, 1.11) | 1.07 | (0.79, 1.44) | 0.448 |
| BER | NEIL2 | rs6982453 | 0.90 | (0.72, 1.11) | 0.92 | (0.79, 1.09) | 0.834 | 0.92 | (0.80, 1.07) | 0.89 | (0.69, 1.16) | 0.787 |
| BER | NEIL2 | rs1534862 | 1.02 | (0.79, 1.30) | 0.89 | (0.74, 1.08) | 0.432 | 0.92 | (0.78, 1.10) | 1.02 | (0.76, 1.37) | 0.562 |
| BER | NEIL2 | rs6997097 | 0.85 | (0.55, 1.30) | 0.94 | (0.68, 1.30) | 0.728 | 0.84 | (0.62, 1.13) | 1.18 | (0.71, 1.96) | 0.237 |
| BER | NEIL2 | rs1043180 | 0.94 | (0.69, 1.29) | 0.96 | (0.74, 1.23) | 0.984 | 0.94 | (0.75, 1.17) | 1.03 | (0.67, 1.58) | 0.688 |
| BER | NEIL2 | rs2645450 | 1.05 | (0.82, 1.33) | 1.16 | (0.96, 1.40) | 0.550 | 1.09 | (0.92, 1.29) | 1.14 | (0.84, 1.55) | 0.757 |
| BER | NEIL2 | rs904015 | 1.10 | (0.88, 1.36) | 0.92 | (0.78, 1.09) | 0.225 | 0.96 | (0.82, 1.12) | 1.08 | (0.83, 1.40) | 0.466 |
| BER | NTHL1 | rs12447809 | 0.95 | (0.72, 1.24) | 1.13 | (0.93, 1.37) | 0.336 | 1.00 | (0.83, 1.20) | 1.30 | (0.95, 1.79) | 0.145 |
| BER | NTHL1 | rs1132368 | 1.04 | (0.62, 1.75) | 0.73 | (0.48, 1.12) | 0.329 | 0.65 | (0.43, 0.96) | 1.90 | (1.01, 3.58) | 0.007 |
| BER | NTHL1 | rs2531213 | 0.83 | (0.47, 1.46) | 1.09 | (0.68, 1.77) | 0.466 | 1.02 | (0.68, 1.54) | 0.66 | (0.32, 1.39) | 0.316 |
| BER | NTHL1 | rs3211995 | 0.89 | (0.67, 1.20) | 0.97 | (0.79, 1.20) | 0.753 | 0.91 | (0.75, 1.11) | 1.03 | (0.73, 1.44) | 0.501 |
| BER | NTHL1 | rs2516740 | 1.01 | (0.78, 1.30) | 0.92 | (0.76, 1.11) | 0.465 | 0.88 | (0.74, 1.05) | 1.18 | (0.87, 1.60) | 0.097 |
| BER | NTHL1 | rs2516739 | 0.97 | (0.75, 1.25) | 0.92 | (0.76, 1.11) | 0.650 | 0.88 | (0.73, 1.05) | 1.17 | (0.86, 1.58) | 0.107 |
| BER | OGG1 | rs159153 | 1.01 | (0.80, 1.26) | 1.18 | (1.00, 1.40) | 0.256 | 1.07 | (0.92, 1.25) | 1.24 | (0.95, 1.62) | 0.332 |
| BER | OGG1 | rs1052133 | 1.02 | (0.80, 1.29) | 1.01 | (0.83, 1.22) | 0.997 | 1.01 | (0.85, 1.19) | 0.97 | (0.71, 1.32) | 0.804 |
| BER | OGG1 | rs293795 | 0.79 | (0.60, 1.05) | 1.11 | (0.90, 1.35) | 0.066 | 0.95 | (0.79, 1.15) | 1.08 | (0.76, 1.54) | 0.424 |
| BER | OGG1 | rs293794 | 0.81 | (0.61, 1.07) | 1.08 | (0.88, 1.32) | 0.114 | 0.95 | (0.79, 1.14) | 1.06 | (0.75, 1.50) | 0.484 |
| BER | OGG1 | rs293796 | 0.96 | (0.65, 1.41) | 1.26 | (0.95, 1.67) | 0.228 | 1.15 | (0.88, 1.49) | 1.13 | (0.71, 1.81) | 0.918 |
| BER | PCNA | rs3729558 | 0.93 | (0.76, 1.14) | 0.91 | (0.78, 1.07) | 0.977 | 0.94 | (0.81, 1.08) | 0.87 | (0.67, 1.13) | 0.607 |
| BER | PCNA | rs17349 | 1.07 | (0.78, 1.47) | 0.78 | (0.60, 1.02) | 0.165 | 0.81 | (0.64, 1.02) | 1.11 | (0.74, 1.69) | 0.204 |
| BER | PCNA | rs25406 | 1.07 | (0.87, 1.32) | 1.19 | (1.02, 1.40) | 0.522 | 1.16 | (1.01, 1.34) | 1.08 | (0.82, 1.40) | 0.636 |
| BER | PCNA | rs25405 | 1.07 | (0.77, 1.47) | 0.81 | (0.62, 1.05) | 0.212 | 0.83 | (0.66, 1.05) | 1.11 | (0.73, 1.68) | 0.263 |
| BER | PCNA | rs4239761 | 1.10 | (0.86, 1.41) | 1.03 | (0.85, 1.26) | 0.751 | 1.02 | (0.85, 1.22) | 1.10 | (0.81, 1.51) | 0.670 |
| BER | PNKP | rs7257463 | 1.02 | (0.82, 1.27) | 1.05 | (0.89, 1.24) | 0.821 | 1.08 | (0.93, 1.26) | 0.96 | (0.73, 1.26) | 0.472 |
| BER | PNKP | rs1290646 | 0.97 | (0.78, 1.21) | 0.96 | (0.82, 1.12) | 0.859 | 0.92 | (0.80, 1.07) | 1.09 | (0.84, 1.41) | 0.297 |
| BER | PNKP | rs3739177 | 1.40 | (0.97, 2.02) | 1.09 | (0.81, 1.45) | 0.309 | 1.24 | (0.96, 1.59) | 1.11 | (0.66, 1.87) | 0.775 |
| BER | PNKP | rs2257103 | 1.00 | (0.81, 1.24) | 1.07 | (0.91, 1.26) | 0.553 | 1.06 | (0.92, 1.23) | 0.99 | (0.76, 1.30) | 0.688 |
| BER | PNKP | rs2353005 | 0.76 | (0.57, 1.02) | 1.04 | (0.83, 1.30) | 0.106 | 0.98 | (0.81, 1.20) | 0.76 | (0.53, 1.10) | 0.221 |
| BER | POLB | rs3136711 | 0.99 | (0.67, 1.45) | 1.05 | (0.79, 1.39) | 0.853 | 1.02 | (0.79, 1.33) | 1.02 | (0.64, 1.63) | 0.952 |
| BER | POLB | rs2976244 | 1.14 | (0.76, 1.71) | 0.88 | (0.64, 1.21) | 0.324 | 0.95 | (0.71, 1.26) | 1.05 | (0.64, 1.73) | 0.706 |
| BER | POLB | rs3136790 | 1.11 | (0.79, 1.54) | 0.94 | (0.73, 1.22) | 0.468 | 0.96 | (0.76, 1.22) | 1.12 | (0.74, 1.67) | 0.562 |
| BER | POLB | rs3136797 | 1.79 | (0.93, 3.44) | 0.73 | (0.35, 1.49) | 0.076 | 0.88 | (0.48, 1.63) | 1.67 | (0.76, 3.67) | 0.211 |
| BER | POLB | rs2073664 | 1.14 | (0.75, 1.72) | 0.88 | (0.64, 1.22) | 0.330 | 0.94 | (0.70, 1.27) | 1.00 | (0.60, 1.66) | 0.834 |
| BER | POLI | rs3730668 | 0.89 | (0.72, 1.10) | 0.89 | (0.76, 1.05) | 0.892 | 0.94 | (0.81, 1.08) | 0.83 | (0.64, 1.08) | 0.423 |
| BER | POLI | rs476630 | 1.02 | (0.82, 1.27) | 1.15 | (0.97, 1.37) | 0.362 | 1.14 | (0.98, 1.34) | 0.94 | (0.70, 1.25) | 0.210 |
| BER | POLI | rs686881 | 1.14 | (0.74, 1.77) | 1.20 | (0.87, 1.65) | 0.888 | 1.17 | (0.87, 1.56) | 1.37 | (0.78, 2.42) | 0.581 |
| BER | POLI | rs3730814 | 0.99 | (0.78, 1.25) | 1.11 | (0.92, 1.34) | 0.389 | 1.12 | (0.95, 1.32) | 0.83 | (0.61, 1.15) | 0.093 |
| BER | POLI | rs3218786 | 1.62 | (0.85, 3.08) | 0.84 | (0.53, 1.33) | 0.087 | 1.07 | (0.71, 1.63) | 1.16 | (0.48, 2.81) | 0.924 |
| BER | POLI | rs8305 | 1.12 | (0.89, 1.41) | 1.00 | (0.84, 1.18) | 0.471 | 0.94 | (0.81, 1.11) | 1.26 | (0.96, 1.66) | 0.059 |
| BER | POLI | rs596986 | 1.14 | (0.74, 1.77) | 1.20 | (0.87, 1.65) | 0.888 | 1.17 | (0.87, 1.56) | 1.37 | (0.78, 2.42) | 0.581 |
| BER | PPP1R13L | rs6966 | 0.99 | (0.74, 1.33) | 0.91 | (0.74, 1.13) | 0.735 | 0.95 | (0.78, 1.16) | 0.94 | (0.67, 1.33) | 0.933 |
| BER | PPP1R13L | rs4803817 | 0.96 | (0.75, 1.22) | 0.94 | (0.78, 1.14) | 0.996 | 0.99 | (0.83, 1.17) | 0.80 | (0.58, 1.09) | 0.252 |
| BER | PPP1R13L | rs10412761 | 0.92 | (0.75, 1.14) | 0.91 | (0.78, 1.07) | 0.936 | 0.94 | (0.81, 1.09) | 0.87 | (0.67, 1.12) | 0.657 |
| BER | PPP1R13L | rs1005165 | 0.91 | (0.69, 1.20) | 0.91 | (0.73, 1.12) | 0.840 | 0.90 | (0.74, 1.10) | 1.01 | (0.71, 1.44) | 0.563 |
| BER | RAD18 | rs4389469 | 0.93 | (0.75, 1.15) | 0.99 | (0.84, 1.16) | 0.623 | 1.00 | (0.87, 1.16) | 0.87 | (0.67, 1.13) | 0.307 |
| BER | RAD18 | rs369032 | 0.93 | (0.75, 1.15) | 1.04 | (0.88, 1.22) | 0.414 | 1.00 | (0.87, 1.16) | 0.96 | (0.73, 1.25) | 0.724 |
| BER | RAD18 | rs2035221 | 1.02 | (0.70, 1.49) | 1.05 | (0.80, 1.38) | 0.922 | 1.23 | (0.95, 1.58) | 0.67 | (0.42, 1.08) | 0.029 |
| BER | RAD18 | rs593205 | 1.37 | (0.96, 1.97) | 1.15 | (0.87, 1.53) | 0.453 | 1.28 | (1.00, 1.66) | 1.06 | (0.67, 1.66) | 0.439 |
| BER | RAD18 | rs373572 | 1.01 | (0.80, 1.28) | 1.01 | (0.84, 1.21) | 0.970 | 1.04 | (0.88, 1.22) | 0.95 | (0.71, 1.26) | 0.533 |
| BER | RAD18 | rs13088787 | 0.94 | (0.69, 1.29) | 1.01 | (0.80, 1.29) | 0.666 | 1.09 | (0.88, 1.35) | 0.72 | (0.48, 1.09) | 0.072 |
| BER | RAD18 | rs615967 | 0.95 | (0.74, 1.21) | 1.02 | (0.84, 1.25) | 0.584 | 0.94 | (0.78, 1.12) | 1.13 | (0.83, 1.53) | 0.356 |
| BER | RAD18 | rs604092 | 0.99 | (0.76, 1.29) | 1.01 | (0.82, 1.24) | 0.927 | 0.95 | (0.79, 1.15) | 1.14 | (0.83, 1.57) | 0.383 |
| BER | SMUG1 | rs971 | 0.92 | (0.73, 1.14) | 1.10 | (0.94, 1.30) | 0.181 | 0.95 | (0.82, 1.11) | 1.28 | (0.98, 1.67) | 0.064 |
| BER | SMUG1 | rs3087404 | 0.92 | (0.75, 1.13) | 0.99 | (0.85, 1.16) | 0.601 | 0.92 | (0.80, 1.06) | 1.10 | (0.85, 1.43) | 0.200 |
| BER | TDG | rs172814 | 1.05 | (0.79, 1.41) | 0.77 | (0.61, 0.97) | 0.107 | 0.88 | (0.72, 1.08) | 0.81 | (0.56, 1.18) | 0.711 |
| BER | TDG | rs4135054 | 1.03 | (0.74, 1.44) | 1.17 | (0.92, 1.50) | 0.578 | 1.13 | (0.90, 1.42) | 1.05 | (0.70, 1.57) | 0.817 |
| BER | TDG | rs4135061 | 0.98 | (0.77, 1.25) | 0.90 | (0.75, 1.07) | 0.518 | 0.90 | (0.76, 1.06) | 1.04 | (0.77, 1.41) | 0.409 |
| BER | TDG | rs4135064 | 0.95 | (0.66, 1.37) | 1.25 | (0.95, 1.64) | 0.264 | 1.14 | (0.89, 1.47) | 1.08 | (0.70, 1.66) | 0.848 |
| BER | TDG | rs4135081 | 1.10 | (0.89, 1.37) | 1.03 | (0.87, 1.21) | 0.620 | 1.04 | (0.89, 1.21) | 1.12 | (0.85, 1.47) | 0.651 |
| BER | TDG | rs3751206 | 1.06 | (0.68, 1.66) | 1.03 | (0.76, 1.39) | 0.891 | 1.10 | (0.83, 1.46) | 0.91 | (0.52, 1.57) | 0.483 |
| BER | TDG | rs4135087 | 0.83 | (0.59, 1.19) | 0.92 | (0.70, 1.22) | 0.598 | 0.85 | (0.66, 1.10) | 0.86 | (0.57, 1.29) | 0.968 |
| BER | TDG | rs167715 | 0.91 | (0.64, 1.27) | 1.13 | (0.89, 1.44) | 0.348 | 0.97 | (0.77, 1.21) | 1.43 | (0.95, 2.16) | 0.096 |
| BER | TDG | rs10861152 | 0.95 | (0.77, 1.18) | 0.89 | (0.75, 1.05) | 0.637 | 0.88 | (0.75, 1.02) | 0.99 | (0.76, 1.30) | 0.408 |
| BER | TDG | rs1866074 | 1.09 | (0.89, 1.33) | 0.88 | (0.75, 1.03) | 0.115 | 0.96 | (0.84, 1.11) | 0.95 | (0.73, 1.24) | 0.940 |
| BER | TDG | rs4135106 | 0.88 | (0.57, 1.37) | 0.87 | (0.63, 1.19) | 0.912 | 0.87 | (0.65, 1.17) | 0.94 | (0.54, 1.61) | 0.789 |
| BER | TDG | rs4135128 | 0.96 | (0.65, 1.42) | 1.03 | (0.79, 1.36) | 0.796 | 1.09 | (0.85, 1.39) | 0.86 | (0.52, 1.42) | 0.360 |
| BER | UNG | rs3890995 | 0.95 | (0.72, 1.27) | 1.12 | (0.91, 1.39) | 0.375 | 1.03 | (0.85, 1.25) | 1.16 | (0.82, 1.65) | 0.601 |
| BER | UNG | rs1018783 | 1.05 | (0.80, 1.39) | 1.16 | (0.95, 1.43) | 0.576 | 1.03 | (0.85, 1.25) | 1.45 | (1.04, 2.03) | 0.085 |
| BER | UNG | rs2569987 | 1.01 | (0.77, 1.34) | 1.00 | (0.81, 1.23) | 0.848 | 1.08 | (0.89, 1.30) | 0.82 | (0.58, 1.16) | 0.175 |
| BER | UNG | rs246079 | 1.06 | (0.85, 1.31) | 1.21 | (1.03, 1.43) | 0.326 | 1.07 | (0.92, 1.24) | 1.48 | (1.13, 1.94) | 0.038 |
| BER | UNG | rs34259 | 1.13 | (0.87, 1.45) | 1.07 | (0.88, 1.29) | 0.723 | 1.02 | (0.86, 1.22) | 1.34 | (0.98, 1.83) | 0.128 |
| BER | XRCC1 | rs25487 | 0.91 | (0.73, 1.13) | 1.05 | (0.90, 1.24) | 0.309 | 0.98 | (0.84, 1.14) | 1.09 | (0.83, 1.42) | 0.566 |
| BER | XRCC1 | rs25486 | 0.90 | (0.73, 1.13) | 1.04 | (0.88, 1.23) | 0.343 | 0.97 | (0.84, 1.13) | 1.08 | (0.83, 1.41) | 0.548 |
| BER | XRCC1 | rs25489 | 1.06 | (0.64, 1.74) | 0.87 | (0.58, 1.31) | 0.604 | 0.87 | (0.61, 1.24) | 1.28 | (0.64, 2.58) | 0.323 |
| BER | XRCC1 | rs1799782 | 1.07 | (0.68, 1.68) | 1.05 | (0.74, 1.49) | 0.997 | 1.05 | (0.77, 1.42) | 1.17 | (0.60, 2.27) | 0.784 |
| BER | XRCC1 | rs3213344 | 1.09 | (0.69, 1.71) | 1.00 | (0.70, 1.42) | 0.799 | 1.02 | (0.75, 1.39) | 1.17 | (0.60, 2.27) | 0.732 |
| BER | XRCC1 | rs3213334 | 1.09 | (0.86, 1.38) | 0.95 | (0.79, 1.14) | 0.400 | 0.98 | (0.83, 1.16) | 0.99 | (0.74, 1.32) | 0.990 |
| BER | XRCC1 | rs2023614 | 0.87 | (0.59, 1.30) | 0.96 | (0.70, 1.31) | 0.705 | 0.86 | (0.65, 1.14) | 1.18 | (0.71, 1.97) | 0.269 |
| BER | XRCC1 | rs2854510 | 1.01 | (0.78, 1.31) | 0.93 | (0.76, 1.12) | 0.574 | 1.01 | (0.84, 1.20) | 0.82 | (0.60, 1.12) | 0.299 |
| BER | XRCC1 | rs2854509 | 1.17 | (0.92, 1.50) | 0.96 | (0.79, 1.15) | 0.203 | 1.02 | (0.86, 1.21) | 1.01 | (0.75, 1.35) | 0.952 |
| BER | XRCC1 | rs3213266 | 0.88 | (0.60, 1.30) | 0.94 | (0.70, 1.27) | 0.792 | 0.85 | (0.65, 1.12) | 1.20 | (0.73, 1.96) | 0.226 |
| BER | XRCC1 | rs3213255 | 1.12 | (0.91, 1.39) | 0.91 | (0.78, 1.07) | 0.124 | 1.01 | (0.87, 1.17) | 0.88 | (0.68, 1.14) | 0.387 |
| HRb | XRCC2 | rs3218536 | 0.90 | (0.60, 1.36) | 1.18 | (0.87, 1.60) | 0.227 | 1.08 | (0.82, 1.42) | 0.95 | (0.57, 1.58) | 0.644 |
| HR | XRCC2 | rs6964582 | 1.43 | (0.90, 2.30) | 1.18 | (0.81, 1.70) | 0.502 | 1.18 | (0.84, 1.66) | 1.57 | (0.89, 2.78) | 0.400 |
| HR | XRCC2 | rs3218438 | 1.43 | (1.03, 1.98) | 0.94 | (0.72, 1.23) | 0.047 | 0.97 | (0.76, 1.24) | 1.62 | (1.04, 2.52) | 0.032 |
| HR | XRCC2 | rs3218408 | 1.00 | (0.78, 1.30) | 1.11 | (0.93, 1.34) | 0.495 | 1.18 | (0.99, 1.40) | 0.83 | (0.61, 1.13) | 0.049 |
| HR | XRCC2 | rs3218373 | 1.08 | (0.76, 1.54) | 1.02 | (0.77, 1.35) | 0.785 | 1.08 | (0.84, 1.39) | 0.89 | (0.57, 1.42) | 0.466 |
| HR | XRCC2 | rs2040639 | 0.93 | (0.75, 1.15) | 0.97 | (0.82, 1.13) | 0.854 | 0.93 | (0.80, 1.08) | 1.09 | (0.84, 1.41) | 0.340 |
| HR | XRCC3 | rs861539 | 0.90 | (0.72, 1.11) | 0.88 | (0.75, 1.04) | 0.936 | 0.89 | (0.77, 1.04) | 0.87 | (0.67, 1.14) | 0.852 |
| HR | XRCC3 | rs3212102 | 0.93 | (0.47, 1.86) | 0.85 | (0.50, 1.46) | 0.872 | 0.73 | (0.44, 1.23) | 1.20 | (0.56, 2.54) | 0.278 |
| HR | XRCC3 | rs3212090 | 1.08 | (0.86, 1.34) | 1.17 | (0.98, 1.39) | 0.646 | 1.20 | (1.03, 1.40) | 0.95 | (0.72, 1.26) | 0.133 |
| HR | XRCC3 | rs3212079 | 1.18 | (0.81, 1.74) | 0.80 | (0.58, 1.10) | 0.107 | 0.91 | (0.68, 1.22) | 0.94 | (0.59, 1.49) | 0.881 |
| HR | XRCC3 | rs861530 | 1.05 | (0.83, 1.33) | 0.98 | (0.82, 1.18) | 0.717 | 0.93 | (0.79, 1.10) | 1.25 | (0.93, 1.67) | 0.070 |
| HR | XRCC3 | rs1799794 | 0.99 | (0.75, 1.29) | 1.13 | (0.92, 1.39) | 0.372 | 1.02 | (0.84, 1.22) | 1.27 | (0.91, 1.77) | 0.226 |
| HR | XRCC3 | rs861528 | 0.88 | (0.69, 1.12) | 0.90 | (0.75, 1.08) | 0.900 | 0.91 | (0.77, 1.07) | 0.88 | (0.66, 1.17) | 0.878 |
aAmong all cases and controls; AA, homozygous major allele; Aa, heterozygous; aa, homozygous minor allele; numbers do not sum to total due to missing.
MMR, mismatch repair; BER, base excision repair; HR, homologous recombination.
p-value for interaction.
Discussion
As expected, we observed an association between the MSH5 rs3131379/BAT3 rs3117582 known susceptibility locus and lung cancer risk. These genes lie in the highly complex human leukocyte antigen (HLA) region on 6p21.33. The HLA locus on 6p21.31 has also been reported to be associated with lung cancer risk among Japanese individuals [6]. Interestingly, one of the major findings from The Cancer Genome Atlas comprehensive genomic analysis of squamous cell cancers is the description of somatic loss-of-function mutations in the HLA-A class I major histocompatibility gene, which is also located in the 6p21.3 region [41]. Our other observations include an increased risk of lung cancer associated with a SNP in UNG, particularly among individuals who were already at increased risk because they carried at least one of the MSH5 rs3131379 A alleles; and an increased risk of lung cancer associated with certain SNPs in MSH2 and PCNA. Like SMUG1, TDG, and MBD4, UNG is a BER uracil-DNA glycosylase which repairs the mis-incorporation of the RNA constituent uracil. UNG binds to PCNA at replication foci, and is the major enzyme that removes uracil from U:A pairs. It may also be involved in short patch BER of uracil and pre-replication repair of U:G pairs [42]. MSH2 is typically involved in post-replicative MMR, forming heterodimers with MSH6 to repair base mismatches and small insertion deletion loops, and with MSH3 to repair larger insertion deletion loops [43]. MSH2 also binds to PCNA [42,44]. Lynch Syndrome, which is associated with a dramatically increased risk of colon, endometrial and ovarian cancers as well as several other cancer types, is characterized by mutations in MLH1, MSH2, and MSH6. Mutations in MSH2 confer particularly high risks, though this does not appear to be true for lung cancer [45]. PCNA performs a central role not only in DNA repair, but also in DNA replication and recombination. It forms a trimer that encircles DNA at replication forks, where it recruits other proteins [46].
While UNG and MSH2 perform distinct functions with respect to DNA repair, they have a similar and overlapping role in adaptive immunity [42]. The main function of the adaptive immune system, to recognize and remember specific pathogens, is performed through the differentiation of immunoglobulin (Ig) genes. In humans, this is achieved through two processes, somatic hypermutation (SHM) which yields antibody diversification, and class switch recombination (CSR), which produces the five Ig isotypes IgM, IgD, IgE, IgE, and IgA [42]. Both somatic hypermutation and class switch recombination are initiated by activation-induced cytidine deaminase (AID) which allows the introduction of uracil, forming key intermediate U:G pairings in Ig DNA. Recognition of the U:G pairs in specific regions of Ig by both UNG and MSH2 coupled with MSH6 allows for accumulation of mutations and diversification. Both UNG and MSH2 bind to PCNA, and additional DNA repair genes including APE1, POLN, POLB, and others are involved in the later steps, particularly for class switch recombination [47]. Mouse models deficient in either UNG or MSH2 result in mice able to produce antibodies at a level 2 to 3-fold lower than in wild type mice [48], and models deficient in UNG result in mice that develop B-cell lymphomas late in life [42]. However, deficiency in both UNG and MSH2 results in mice incapable of antibody gene diversification [48]. In humans, mutations in UNG alone result in the autosomal recessive hyper-IgM syndrome, a class switch recombination disorder characterized by IgG, IgA, and IgE deficiencies [49].
In the lung, innate and adaptive immunity launch inflammatory responses to a variety of insults such as particulate matter in cigarette smoke and other pollutants, microbial infections, and cell damage/injury. Chronic inflammation, and the interaction between innate and adaptive immune response, play central roles in cancer development [50]. Chronic pulmonary inflammation has been hypothesized to be an underlying mechanism for the increased risk of lung cancer associated with tobacco smoking, chronic obstructive pulmonary disease [51], silicosis, asbestosis [52], and lung infections (i.e., tuberculosis, pneumonia [53]), and the increased incidence of lung cancer among individuals with human immunodeficiency virus [54].
Very few prior studies have interrogated variants in UNG, MSH2, and PCNA and lung cancer risk, and only in the context of candidate gene studies of DNA repair. Two comprehensive studies described in more detail below [8,15] that examined DNA repair pathway genotype data from samples assayed using the Illumina HumanHap300 BeadChip, did not observe differences in the genotype distribution between cases and controls for SNPs in MSH2, UNG or PCNA, but they did not directly measure the SNPs associated with risk in our study. A relatively small study of French Caucasian smokers (151 lung cancer cases and 172 hospital controls) [11] did not observe associations with MSH2 rs2303428 (G allele frequency 0.10 in cases, 0.12 in controls) or PCNA rs25406 (A allele frequency 0.40 in cases, 0.47 in controls (p=0.09)); data from this study is also included in the meta-analysis by Kazma et al. [15] described below. Two other studies examined associations with MSH2 rs2303428. A Korean study with 432 lung cancer patients matched to 432 healthy controls on age and gender observed that carriage of at least one C allele was associated with an increased risk of adenocarcinoma, compared to the TT genotype (adjusted OR, 1.52; 95% CI, 1.02-2.27; P = 0.04) [13], and while the confidence limit does not exclude 1, an elevated OR (1.29 (0.83—1.99)) was observed in a Taiwanese study of 156 NSCLC patients and 235 controls matched for age, gender and smoking [14]. A candidate SNP study of Caucasian smokers that included 343 NSCLC cases and 413 population-based controls matched on age, gender and smoking did not observe an association with PCNA rs25406 (MAF 0.38 in cases, 0.38 in controls; AG, and AA, versus GG, OR 0.73 (0.52–1.0) and 1.15 (0.74–1.79) [22]. These studies do not provide rigorous support for or against associations with the SNPs of interest, since a much larger sample size is needed in order to obtain stable risk estimates of the magnitude expected.
The largest and most comprehensive interrogations of DNA repair pathways and lung cancer risk were performed by Kazma et al. [15] and Yu et al. [8] Kazma et al. [15] included 1,655 SNPs in 211 DNA repair genes in 6,911 individuals pooled from four studies. Yu et al. [8] interrogated 1806 SNPs in 125 DNA repair genes in 1154 lung cancer cases and 1137 controls matched by smoking status. With the exception of MSH5 rs3131379, the SNPs that were associated with risk in our study are not present on the HumanHap300 BeadChip, but SNPs in LD with them (in HapMap-CEU) were not associated with risk in either study (UNG, rs2430682, in LD with rs246079 (r2=0.89); MSH2, rs2042649, in LD with rs2303428 (r2=1.0)). While they examined associations with SNPs in PCNA, none of the SNPs were in LD >0.52 with the SNP (rs25406) that we observed to be associated with risk in our study.
The variants (after MSH5 rs3131379) that were most strongly associated with lung cancer risk in Kazma et al. were in the genes UBE2N, SMC1L2, and POLB, with suggestive associations for variants in RAD52 and POLN [15]. Yu et al. observed associations with SNPs in XRCC4, but they were not replicated in a meta-analysis of these SNPs in four GWAS studies totaling ~12,000 cases and ~48,000 controls [8]. Other studies of DNA repair genes have reported associations with additional candidate SNPs. A hospital-based study of smokers including 722 cases and 929 controls interrogated 29 SNPs in the BER genes MPG, OGG1, PARP1, and XRCC1, one SNP in PARP1 and two SNPs in XRCC1 (rs1799782 and rs3213255) were associated with lung cancer risk [31]. Meta-analyses of selected SNPs have observed associations with OGG1 Ser326Cys rs1052133 [20,55,56] and XRCC3 T241M rs861539 [20]. Of the genes reported to be associated with lung cancer risk in prior studies, we only examined variants in POLB, XRCC1, OGG1 and XRCC3, and they were not related to risk in our study.
An important difference between our study and the meta-analysis by Kazma et al. is the prevalence of smoking, because Kazma et al. specifically limited their analysis to studies that included both smokers and non-smokers in order to evaluate interactions between SNPs and smoking. In our study, among the controls, none were never smokers, 27.6% were former, and 72.4% were current smokers, whereas in Kazma et al., 38.8% of the controls were never smokers, 25.6% were former, and 34.5% were current smokers. The distribution was identical in the cases and controls for our study because they were matched on smoking exposure, but the distribution in cases from Kazma et al. --9.7% never, 20.5% former, and 68.9% current--differed considerably from the distribution in their controls. It is possible that there are underlying differences in the distribution of genotypes in the controls due to differences in smoking exposure. A well-documented example of this is genetic variation in the nicotinic acetylcholine receptor gene cluster on chromosome 15q25.1. For rs1051730 (which is in complete LD with rs16969968 (r2=1.0, HapMap-CEU)), the frequency of the T allele increases with increasing numbers of cigarettes smoked, with a large difference in frequency between individuals smoking 1-10 cigarettes per day (T allele frequency 0.305) and 31 or more cigarettes per day (T allele frequency 0.391) [57]. Matching controls to cases based on cigarette smoking (as we did) is arguably an advantage when attempting to identify genetic factors that might differentiate between the ~20 % of smokers who develop lung cancer from the ~80 % who do not.
In conclusion, we observed associations with SNPs in UNG, MSH2, and PCNA, all of which are involved both in DNA repair pathways and also in adaptive immunity, and the associations with the UNG variants were stronger among individuals carrying the documented MSH5/BAT3 lung cancer susceptibility allele, which was also associated with risk in our study. We were unable to confirm associations reported in prior studies with POLB, XRCC1, OGG1 and XRCC3 SNPs, and we did not evaluate variation in UBE2N, SMC1L2, RAD52, or POLN. Our study was not large enough to be able to reliably identify the presence of true weak associations, and is limited by having genotype data on only two of the five documented lung cancer susceptibility loci described to date in Caucasian populations [5]. However, our study differs from most other prior studies because it is prospective in nature and includes only heavy smokers, with cases and controls matched on smoking history. Because lung cancer is so rapidly fatal, case control study response proportions can be very low, and our study is likely to have a more representative case group, specific to smoking-associated lung cancer, than case-control studies. Furthermore, no prior studies have reported pathway-based SNP results stratified by know lung cancer susceptibility loci. The patterns of associations observed should be viewed as hypothesis-generating, requiring follow up in other studies of smoking-related lung cancer.
Acknowledgements
We thank the CARET participants, along with the CARET investigators and staff at all participating trial institutions, for their effort and contribution to this research.
Abbreviations
- APEX1
apurinic/apyrimidinic-endonuclease-1
- CARET
β-Carotene and Retinol Efficacy Trial
- CI
confidence interval
- LIG3
ligase III
- MBD4
methyl binding domain 4
- MLH1
MutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli)
- MPG
N-methylpurine-DNA glycosylase
- MSH2
mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli)
- MSH4
mutS homolog 4, colon cancer, nonpolyposis type 1 (E. coli)
- MSH5
mutS homolog 5, colon cancer, nonpolyposis type 1 (E. coli)
- MSH6
mutS homolog 6, colon cancer, nonpolyposis type 1 (E. coli)
- MUTYH
mutY homolog (E. coli)
- NEIL1
nei endonuclease VIII-like 1 (E. coli)
- NEIL2
nei endonuclease VIII-like 2 (E. coli)
- NTHL1
nth endonuclease III-like 1 (E. coli)
- OGG1
8-oxo-guanine glycosylase-1
- OR
odds ratio
- PCNA
proliferating cell nuclear antigen
- PNKP
polynucleotide kinase 3’-phosphatase
- POLB
polymerase (DNA directed), beta
- POLI
polymerase (DNA directed) iota
- PPP1R13L
protein phosphatase 1, regulatory subunit 13 like
- RAD18
RAD18 homolog (S. cerevisiae)
- SD
standard deviation
- SMUG1
single strand selective monofunctional uracil-DNA glycosylase
- SNP
single nucleotide polymorphism
- TDG
thymine/uracil mismatch DNA glycosylase
- UNG
uracil-DNA glycosylase
- XRCC1
X-ray repair complementing defective repair in Chinese-hamster cells 1
- XRCC2
X-ray repair complementing defective repair in Chinese-hamster cells 2
- XRCC3
X-ray repair complementing defective repair in Chinese-hamster cells 3
Financial support
This work is supported by R01 CA111703 and U01 CA63673 from the National Cancer Institute of the U.S. National Institutes of Health (NIH).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Alberg AJ, Ford JG, Samet JM American College of Chest Physicians. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition) Chest. 2007;132:29S–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 3.Hang B. Formation and repair of tobacco carcinogen-derived bulky DNA adducts. J Nucleic Acids. 2010;2010:709521. doi: 10.4061/2010/709521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J, Chatterjee N, Rotunno M, Wang Y, Pesatori AC, Consonni D, Li P, Wheeler W, Broderick P, Henrion M, Eisen T, Wang Z, Chen W, Dong Q, Albanes D, Thun M, Spitz MR, Bertazzi PA, Caporaso NE, Chanock SJ, Amos CI, Houlston RS, Landi MT. Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2:131–139. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeboller H, Risch A, McKay JD, Wang Y, Dai J, Gaborieau V, McLaughlin J, Brenner D, Narod SA, Caporaso NE, Albanes D, Thun M, Eisen T, Wichmann HE, Rosenberger A, Han Y, Chen W, Zhu D, Spitz M, Wu X, Pande M, Zhao Y, Zaridze D, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Krokan HE, Gabrielsen ME, Skorpen F, Vatten L, Njolstad I, Chen C, Goodman G, Lathrop M, Benhamou S, Vooder T, Valk K, Nelis M, Metspalu A, Raji O, Chen Y, Gosney J, Liloglou T, Muley T, Dienemann H, Thorleifsson G, Shen H, Stefansson K, Brennan P, Amos CI, Houlston R, Landi MT Transdisciplinary Research in Cancer of the Lung (TRICL) Research Team. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohno T, Kunitoh H, Shimada Y, Shiraishi K, Ishii Y, Goto K, Ohe Y, Nishiwaki Y, Kuchiba A, Yamamoto S, Hirose H, Oka A, Yanagitani N, Saito R, Inoko H, Yokota J. Individuals susceptible to lung adenocarcinoma defined by combined HLA-DQA1 and TERT genotypes. Carcinogenesis. 2010;31:834–841. doi: 10.1093/carcin/bgq003. [DOI] [PubMed] [Google Scholar]
- 7.Park SH, Lee GY, Jeon HS, Lee SJ, Kim KM, Jang SS, Kim CH, Lee WK, Kam S, Park RW, Kim IS, Jung TH, Park JY. -93G-->A polymorphism of hMLH1 and risk of primary lung cancer. Int J Cancer. 2004;112:678–682. doi: 10.1002/ijc.20359. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Zhao H, Wang LE, Han Y, Chen WV, Amos CI, Rafnar T, Sulem P, Stefansson K, Landi MT, Caporaso N, Albanes D, Thun M, McKay JD, Brennan P, Wang Y, Houlston RS, Spitz MR, Wei Q. An analysis of single nucleotide polymorphisms of 125 DNA repair genes in the Texas genome-wide association study of lung cancer with a replication for the XRCC4 SNPs. DNA Repair (Amst) 2011;10:398–407. doi: 10.1016/j.dnarep.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih CM, Chen CY, Lee IH, Kao WT, Wang YC. A polymorphism in the hMLH1 gene (-93G-->A) associated with lung cancer susceptibility and prognosis. Int J Mol Med. 2010;25:165–170. [PubMed] [Google Scholar]
- 10.An Y, Jin G, Wang H, Wang Y, Liu H, Li R, Wang H, Qian J, Sun W, Wang Y, Ma H, Miao R, Hu Z, Jin L, Wei Q, Shen H, Huang W, Lu D. Polymorphisms in hMLH1 and risk of early-onset lung cancer in a southeast Chinese population. Lung Cancer. 2008;59:164–170. doi: 10.1016/j.lungcan.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Michiels S, Danoy P, Dessen P, Bera A, Boulet T, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28:1731–1739. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- 12.Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Gioia-Patricola L, Hall J, Boffetta P, Hung RJ, Brennan P. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006;66:11062–11069. doi: 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 13.Jung CY, Choi JE, Park JM, Chae MH, Kang HG, Kim KM, Lee SJ, Lee WK, Kam S, Cha SI, Kim CH, Han SB, Jung TH, Jeon SH, Park JY. Polymorphisms in the hMSH2 gene and the risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:762–768. doi: 10.1158/1055-9965.EPI-05-0834. [DOI] [PubMed] [Google Scholar]
- 14.Hsu HS, Lee IH, Hsu WH, Kao WT, Wang YC. Polymorphism in the hMSH2 gene (gISV12-6T > C) is a prognostic factor in non-small cell lung cancer. Lung Cancer. 2007;58:123–130. doi: 10.1016/j.lungcan.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 15.Kazma R, Babron MC, Gaborieau V, Génin E, Brennan P, Hung RJ, McLaughlin JR, Krokan HE, Elvestad MB, Skorpen F, Anderssen E, Vooder T, Välk K, Metspalu A, Field JK, Lathrop M, Sarasin A, Benhamou S ILCCO consortium. Lung Cancer and DNA Repair Genes: Multilevel Association Analysis from the International Lung Cancer Consortium. Carcinogenesis. 2012 May;33:1059–64. doi: 10.1093/carcin/bgs116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misra RR, Ratnasinghe D, Tangrea JA, Virtamo J, Andersen MR, Barrett M, Taylor PR, Albanes D. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003;191:171–178. doi: 10.1016/s0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- 17.Hung RJ, Brennan P, Canzian F, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Chabrier A, Borel S, Hall J, Boffetta P. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 18.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Popanda O, Schattenberg T, Phong CT, Butkiewicz D, Risch A, Edler L, Kayser K, Dienemann H, Schulz V, Drings P, Bartsch H, Schmezer P. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis. 2004;25:2433–2441. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- 20.Hung RJ, Christiani DC, Risch A, Popanda O, Haugen A, Zienolddiny S, Benhamou S, Bouchardy C, Lan Q, Spitz MR, Wichmann HE, LeMarchand L, Vineis P, Matullo G, Kiyohara C, Zhang ZF, Pezeshki B, Harris C, Mechanic L, Seow A, Ng DP, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, Fabianova E, Mates D, Foretova L, Janout V, Bencko V, Caporaso N, Chen C, Duell EJ, Goodman G, Field JK, Houlston RS, Hong YC, Landi MT, Lazarus P, Muscat J, McLaughlin J, Schwartz AG, Shen H, Stucker I, Tajima K, Matsuo K, Thun M, Yang P, Wiencke J, Andrew AS, Monnier S, Boffetta P, Brennan P. International Lung Cancer Consortium: pooled analysis of sequence variants in DNA repair and cell cycle pathways. Cancer Epidemiol Biomarkers Prev. 2008;17:3081–3089. doi: 10.1158/1055-9965.EPI-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73:138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 23.Zhong D, Li G, Long J, Wu J, Hu Y. The hOGG1Ser326Cys polymorphism and increased lung cancer susceptibility in Caucasians: an updated meta-analysis. Sci Rep. 2012;2:548. doi: 10.1038/srep00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu J, Zhang S, Chen D, Wang H, Wu W, Wang X, Lei Y, Wang J, Qian J, Fan W, Hu Z, Jin L, Shen H, Huang W, Wei Q, Lu D. Functional characterization of a promoter polymorphism in APE1/Ref-1 that contributes to reduced lung cancer susceptibility. FASEB J. 2009;23:3459–3469. doi: 10.1096/fj.09-136549. [DOI] [PubMed] [Google Scholar]
- 25.Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen KY, Chen YM, Huang MS, Hu CY, Chen CJ, Hsiung CA. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:223–229. doi: 10.1158/1055-9965.EPI-08-0749. [DOI] [PubMed] [Google Scholar]
- 26.Ito H, Matsuo K, Hamajima N, Mitsudomi T, Sugiura T, Saito T, Yasue T, Lee KM, Kang D, Yoo KY, Sato S, Ueda R, Tajima K. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg-399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25:1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- 27.Ryk C, Kumar R, Thirumaran RK, Hou SM. Polymorphisms in the DNA repair genes XRCC1, APEX1, XRCC3 and NBS1, and the risk for lung cancer in never- and ever-smokers. Lung Cancer. 2006;54:285–292. doi: 10.1016/j.lungcan.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhou B, Shan H, Su Y, Xia K, Shao X, Mao W, Shao Q. The association of APE1 -656T > G and 1349 T > G polymorphisms and cancer risk: a meta-analysis based on 37 case-control studies. BMC Cancer. 2011;11:521. doi: 10.1186/1471-2407-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez-Cima MF, Gonzalez-Arriaga P, Garcia-Castro L, Pascual T, Marron MG, Puente XS, Tardon A. Polymorphisms in XPC, XPD, XRCC1, and XRCC3 DNA repair genes and lung cancer risk in a population of northern Spain. BMC Cancer. 2007;7:162. doi: 10.1186/1471-2407-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Guan W, Li MX, Zhong ZY, Qian CY, Yang XQ, Liao L, Li ZP, Wang D. Genetic polymorphism of DNA base-excision repair genes (APE1, OGG1 and XRCC1) and their correlation with risk of lung cancer in a Chinese population. Arch Med Res. 2011;42:226–234. doi: 10.1016/j.arcmed.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 31.Buch SC, Diergaarde B, Nukui T, Day RS, Siegfried JM, Romkes M, Weissfeld JL. Genetic variability in DNA repair and cell cycle control pathway genes and risk of smoking-related lung cancer. Mol Carcinog. 2011;51(Suppl 1):E11–20. doi: 10.1002/mc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ricceri F, Matullo G, Vineis P. Is there evidence of involvement of DNA repair polymorphisms in human cancer? Mutat Res. 2012;736:117–121. doi: 10.1016/j.mrfmmm.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Sakoda LC, Loomis MM, Doherty JA, Julianto L, Barnett MJ, Neuhouser ML, Thornquist MD, Weiss NS, Goodman GE, Chen C. Germ line variation in nucleotide excision repair genes and lung cancer risk in smokers. Int J Mol Epidemiol Genet. 2012;3:1–17. [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL Jr, Omenn GS, Valanis B, Williams JH Jr. The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J Natl Cancer Inst. 2004;96:1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 35.Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, Keogh JP, Meyskens FL, Valanis B, Williams JH, Barnhart S, Hammar S. Effects of a combination of beta carotene and vitamin A on lung cancer and cardiovascular disease. N Engl J Med. 1996;334:1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 36.Omenn GS, Goodman GE, Thornquist MD, Balmes J, Cullen MR, Glass A, Keogh JP, Meyskens FL Jr, Valanis B, Williams JH Jr, Barnhart S, Cherniack MG, Brodkin CA, Hammar S. Risk factors for lung cancer and for intervention effects in CARET, the Beta-Carotene and Retinol Efficacy Trial. J Natl Cancer Inst. 1996;88:1550–1559. doi: 10.1093/jnci/88.21.1550. [DOI] [PubMed] [Google Scholar]
- 37.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, Collisson EA, Cope L, Creighton CJ, Getz G, Herman JG, Johnson BE, Kucherlapati R, Ladanyi M, Maher CA, Robertson G, Sander C, Shen R, Sinha R, Sivachenko A, Thomas RK, Travis WD, Tsao MS, Weinstein JN, Wigle DA, Baylin SB, Govindan R, Meyerson M Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA--general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 44.Hagen L, Pena-Diaz J, Kavli B, Otterlei M, Slupphaug G, Krokan HE. Genomic uracil and human disease. Exp Cell Res. 2006;312:2666–2672. doi: 10.1016/j.yexcr.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Dowty JG, Win AK, Buchanan DD, Lindor NM, Macrae FA, Clendenning M, Antill YC, Thibodeau SN, Casey G, Gallinger S, Le Marchand L, Newcomb PA, Haile RW, Young GP, James PA, Giles GG, Gunawardena SR, Leggett BA, Gattas M, Boussioutas A, Ahnen DJ, Baron JA, Parry S, Goldblatt J, Young JP, Hopper JL, Jenkins MA. Cancer risks for MLH1 and MSH2 mutation carriers. Hum Mutat. 2013 Mar;34:490–7. doi: 10.1002/humu.22262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dieckman LM, Freudenthal BD, Washington MT. PCNA Structure and Function: Insights from Structures of PCNA Complexes and Post-translationally Modified PCNA. Subcell Biochem. 2012;62:281–299. doi: 10.1007/978-94-007-4572-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kracker S, Durandy A. Insights into the B cell specific process of immunoglobulin class switch recombination. Immunol Lett. 2011;138:97–103. doi: 10.1016/j.imlet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Rada C, Di Noia JM, Neuberger MS. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell. 2004;16:163–171. doi: 10.1016/j.molcel.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 50.de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6:24–37. doi: 10.1038/nrc1782. [DOI] [PubMed] [Google Scholar]
- 51.Brusselle GG, Joos GF, Bracke KR. New insights into the immunology of chronic obstructive pulmonary disease. Lancet. 2011;378:1015–1026. doi: 10.1016/S0140-6736(11)60988-4. [DOI] [PubMed] [Google Scholar]
- 52.Maeda M, Nishimura Y, Kumagai N, Hayashi H, Hatayama T, Katoh M, Miyahara N, Yamamoto S, Hirastuka J, Otsuki T. Dysregulation of the immune system caused by silica and asbestos. J Immunotoxicol. 2010;7:268–278. doi: 10.3109/1547691X.2010.512579. [DOI] [PubMed] [Google Scholar]
- 53.Brenner DR, Boffetta P, Duell EJ, Bickeboller H, Rosenberger A, McCormack V, Muscat JE, Yang P, Wichmann HE, Brueske-Hohlfeld I, Schwartz AG, Cote ML, Tjonneland A, Friis S, Le Marchand L, Zhang ZF, Morgenstern H, Szeszenia-Dabrowska N, Lissowska J, Zaridze D, Rudnai P, Fabianova E, Foretova L, Janout V, Bencko V, Schejbalova M, Brennan P, Mates IN, Lazarus P, Field JK, Raji O, McLaughlin JR, Liu G, Wiencke J, Neri M, Ugolini D, Andrew AS, Lan Q, Hu W, Orlow I, Park BJ, Hung RJ. Previous lung diseases and lung cancer risk: a pooled analysis from the International Lung Cancer Consortium. Am J Epidemiol. 2012;176:573–585. doi: 10.1093/aje/kws151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engels EA. Inflammation in the development of lung cancer: epidemiological evidence. Expert Rev Anticancer Ther. 2008;8:605–615. doi: 10.1586/14737140.8.4.605. [DOI] [PubMed] [Google Scholar]
- 55.Hung RJ, Hall J, Brennan P, Boffetta P. Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review. Am J Epidemiol. 2005;162:925–942. doi: 10.1093/aje/kwi318. [DOI] [PubMed] [Google Scholar]
- 56.Wei W, He XF, Qin JB, Su J, Li SX, Liu Y, Zhang Y, Wang W. Association between the OGG1 Ser326Cys and APEX1 Asp148Glu polymorphisms and lung cancer risk: a meta-analysis. Mol Biol Rep. 2012;39:11249–11262. doi: 10.1007/s11033-012-2035-8. [DOI] [PubMed] [Google Scholar]
- 57.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A, Stacey SN, Bergthorsson JT, Thorlacius S, Gudmundsson J, Jonsson T, Jakobsdottir M, Saemundsdottir J, Olafsdottir O, Gudmundsson LJ, Bjornsdottir G, Kristjansson K, Skuladottir H, Isaksson HJ, Gudbjartsson T, Jones GT, Mueller T, Gottsater A, Flex A, Aben KK, de Vegt F, Mulders PF, Isla D, Vidal MJ, Asin L, Saez B, Murillo L, Blondal T, Kolbeinsson H, Stefansson JG, Hansdottir I, Runarsdottir V, Pola R, Lindblad B, van Rij AM, Dieplinger B, Haltmayer M, Mayordomo JI, Kiemeney LA, Matthiasson SE, Oskarsson H, Tyrfingsson T, Gudbjartsson DF, Gulcher JR, Jonsson S, Thorsteinsdottir U, Kong A, Stefansson K. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]


