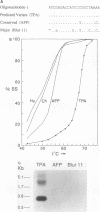
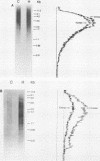
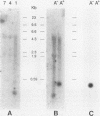
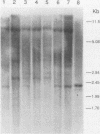
Abstract
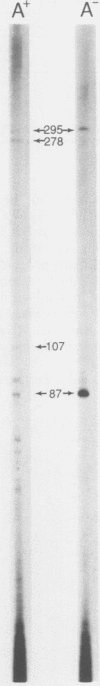
DNA base sequence comparisons indicate that a subfamily of recently transposed human Alu repeats are distinguished from most Alu repeats by diagnostic sequence differences. Using an oligonucleotide hybridization probe that incorporates these sequence features, we found that there was an expansion of this Alu subfamily following the divergence of humans and African apes. This oligonucleotide was used to select human genomic clones containing representatives of this subfamily. One representative member of this subfamily was evidently absent from the corresponding chimpanzee locus and was associated with a restriction fragment length polymorphism in the human genome. This apparently polymorphic member had all the diagnostic sequence features that initially predicted the existence of a newly expanding Alu subfamily. A transpositionally active sequence variant should also be transcriptionally active in at least some cell types or tissues. Northern (RNA) blot hybridization, primer extension, and RNA sequence analysis demonstrated the existence of different-length polyadenylated and nonpolyadenylated transcripts corresponding to this subfamily. Evidence for 3' processing and subcellular localization of these transcripts is discussed. Most of the nearly one million human Alu repeats are pseudogenes with respect to coding for either an RNA product or new family members; a select and identifiable subset of Alu repeats serve as transcriptionally and transpositionally competent source genes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adeniyi-Jones S., Zasloff M. Transcription, processing and nuclear transport of a B1 Alu RNA species complementary to an intron of the murine alpha-fetoprotein gene. Nature. 1985 Sep 5;317(6032):81–84. doi: 10.1038/317081a0. [DOI] [PubMed] [Google Scholar]
- Andrews P. G., Kole R. Alu RNA transcribed in vitro binds the 68-kDa subunit of the signal recognition particle. J Biol Chem. 1987 Feb 25;262(6):2908–2912. [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Baron W. F., Stout D. B., Davidson E. H. Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4770–4774. doi: 10.1073/pnas.85.13.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen S. J., Rajput B., Reich E. The human tissue plasminogen activator gene. J Biol Chem. 1986 May 25;261(15):6972–6985. [PubMed] [Google Scholar]
- Deininger P. L., Jolly D. J., Rubin C. M., Friedmann T., Schmid C. W. Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol. 1981 Sep 5;151(1):17–33. doi: 10.1016/0022-2836(81)90219-9. [DOI] [PubMed] [Google Scholar]
- Deininger P. L., Slagel V. K. Recently amplified Alu family members share a common parental Alu sequence. Mol Cell Biol. 1988 Oct;8(10):4566–4569. doi: 10.1128/mcb.8.10.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou-Pachnis A., Tsichlis P. N. Insertion of an Alu SINE in the human homologue of the Mlvi-2 locus. Nucleic Acids Res. 1985 Dec 9;13(23):8379–8387. doi: 10.1093/nar/13.23.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs P. E., Zielinski R., Boyd C., Dugaiczyk A. Structure, polymorphism, and novel repeated DNA elements revealed by a complete sequence of the human alpha-fetoprotein gene. Biochemistry. 1987 Mar 10;26(5):1332–1343. doi: 10.1021/bi00379a020. [DOI] [PubMed] [Google Scholar]
- Hwu H. R., Roberts J. W., Davidson E. H., Britten R. J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinek W., Leinwand L. Low molecular weight RNAs hydrogen-bonded to nuclear and cytoplasmic poly(A)-terminated RNA from cultured Chinese hamster ovary cells. Cell. 1978 Sep;15(1):205–214. doi: 10.1016/0092-8674(78)90095-8. [DOI] [PubMed] [Google Scholar]
- Jurka J., Smith T. A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4775–4778. doi: 10.1073/pnas.85.13.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop B. F., Miyamoto M. M., Embury J. E., Goodman M., Czelusniak J., Slightom J. L. Nucleotide sequence and evolution of the orangutan epsilon globin gene region and surrounding Alu repeats. J Mol Evol. 1986;24(1-2):94–102. doi: 10.1007/BF02099956. [DOI] [PubMed] [Google Scholar]
- Labuda D., Striker G. Sequence conservation in Alu evolution. Nucleic Acids Res. 1989 Apr 11;17(7):2477–2491. doi: 10.1093/nar/17.7.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R., Zasloff M., Plotz P., Adeniyi-Jones S. Pathway of B1-Alu expression in microinjected oocytes: Xenopus laevis proteins associated with nuclear precursor and processed cytoplasmic RNAs. Mol Cell Biol. 1988 Oct;8(10):4433–4440. doi: 10.1128/mcb.8.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson K. E., Matera A. G., Deka N., Schmid C. W. Transcription of a human transposon-like sequence is usually directed by other promoters. Nucleic Acids Res. 1987 Jul 10;15(13):5199–5215. doi: 10.1093/nar/15.13.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson K. E., Schmid C. W. Transcriptional inactivity of Alu repeats in HeLa cells. Nucleic Acids Res. 1986 Aug 11;14(15):6145–6158. doi: 10.1093/nar/14.15.6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y. The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol. 1988;27(3):194–202. doi: 10.1007/BF02100074. [DOI] [PubMed] [Google Scholar]
- Rinehart F. P., Ritch T. G., Deininger P. L., Schmid C. W. Renaturation rate studies of a single family of interspersed repeated sequences in human deoxyribonucleic acid. Biochemistry. 1981 May 26;20(11):3003–3010. doi: 10.1021/bi00514a003. [DOI] [PubMed] [Google Scholar]
- Sawada I., Schmid C. W. Primate evolution of the alpha-globin gene cluster and its Alu-like repeats. J Mol Biol. 1986 Dec 20;192(4):693–709. doi: 10.1016/0022-2836(86)90022-7. [DOI] [PubMed] [Google Scholar]
- Sawada I., Willard C., Shen C. K., Chapman B., Wilson A. C., Schmid C. W. Evolution of Alu family repeats since the divergence of human and chimpanzee. J Mol Evol. 1985;22(4):316–322. doi: 10.1007/BF02115687. [DOI] [PubMed] [Google Scholar]
- Schoeniger L. O., Jelinek W. R. 4.5S RNA is encoded by hundreds of tandemly linked genes, has a short half-life, and is hydrogen bonded in vivo to poly(A)-terminated RNAs in the cytoplasm of cultured mouse cells. Mol Cell Biol. 1986 May;6(5):1508–1519. doi: 10.1128/mcb.6.5.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986 Mar 6;320(6057):81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- Skowronski J., Fanning T. G., Singer M. F. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988 Apr;8(4):1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppa-Lyonnet D., Carter P. E., Meo T., Tosi M. Clusters of intragenic Alu repeats predispose the human C1 inhibitor locus to deleterious rearrangements. Proc Natl Acad Sci U S A. 1990 Feb;87(4):1551–1555. doi: 10.1073/pnas.87.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuchet G., Chebloune Y., Savatier P., Lachuer J., Faure C., Verdier G., Nigon V. M. Recent insertion of an Alu sequence in the beta-globin gene cluster of the gorilla. J Mol Evol. 1987;25(4):288–291. doi: 10.1007/BF02603112. [DOI] [PubMed] [Google Scholar]
- Ullu E., Murphy S., Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell. 1982 May;29(1):195–202. doi: 10.1016/0092-8674(82)90103-9. [DOI] [PubMed] [Google Scholar]
- Ullu E., Weiner A. M. Upstream sequences modulate the internal promoter of the human 7SL RNA gene. 1985 Nov 28-Dec 4Nature. 318(6044):371–374. doi: 10.1038/318371a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Willard C., Nguyen H. T., Schmid C. W. Existence of at least three distinct Alu subfamilies. J Mol Evol. 1987;26(3):180–186. doi: 10.1007/BF02099850. [DOI] [PubMed] [Google Scholar]