Abstract
Hematopoietic cells arise from spatiotemporally restricted domains in the developing embryo. Although studies of non-mammalian animal and in vitro embryonic stem cell models suggest a close relationship among cardiac, endocardial, and hematopoietic lineages, it remains unknown whether the mammalian heart tube serves as a hemogenic organ akin to the dorsal aorta. Here we examine the hemogenic activity of the developing endocardium. Mouse heart explants generate myeloid and erythroid colonies in the absence of circulation. Hemogenic activity arises from a subset of endocardial cells in the outflow cushion and atria earlier than in the aorta-gonad-mesonephros region, and is transient and definitive in nature. Interestingly, key cardiac transcription factors, Nkx2-5 and Isl1, are expressed in and required for the hemogenic population of the endocardium. Together, these data suggest that a subset of endocardial/endothelial cells expressing cardiac markers serve as a de novo source for transient definitive hematopoietic progenitors.
The circulatory system is the first functional organ system that develops during mammalian life. The heart is highly modified muscular vessel and, like the aorta and other arteries, its muscular layer expresses the smooth muscle gene program at early stages1. The dorsal aorta is, however, not merely a conduit, but also a source for the third component of the circulatory system, the blood cells. During mammalian embryogenesis, hematopoiesis occurs in several major anatomical sites including the yolk sac, placenta, and the aorta-gonad-mesonephros (AGM) region that contains the dorsal aorta2–5. A common feature of these known de novo hemogenic sites is that the induction and generation of definitive hematopoietic cells is closely associated with the development of major arteries6–11. Hemodynamic stress and local nitric oxide (NO) also play a critical role in hematopoietic induction from the endothelium10,11. The endocardium shares all these properties with arterial endothelium including the arterial marker expression and exposure to the hemodynamic stresses and NO. However, despite all the structural, molecular, and hemodynamic similarities between the heart tube and the dorsal aorta, little is known about the hemogenic potential of the endocardium.
We have previously demonstrated that cardiac and endocardial/endothelial cells can arise from a single common progenitor cell expressing Flk1, Isl1 and Nkx2-5 during early mammalian cardiogenesis12. Notably, these early cardiac progenitors express multiple hematopoietic transcription factors, consistent with previous reports13, and endocardial cells express Flk1, Isl1 and Nkx2-5. However, the biological significance of hematopoietic genes in the developing mammalian heart is unknown, and it is unclear whether this represents a transient program that is subsequently repressed14, or, as in the aorta, a hematopoietic program is activated in the heart. As a close relationship among cardiac, endocardial and hematopoietic lineages has been suggested in fly, zebrafish, and embryonic stem cell in vitro differentiation models15–20, critical questions are when, where and how this hematopoietic gene program is in operation during in vivo mammalian cardiogenesis. Here, we report the hemogenic activity of the endocardium in developing mammalian heart and its Nkx2-5/Isl1-dependent mechanism.
Results
The early heart tube is a de novo site for hematopoiesis
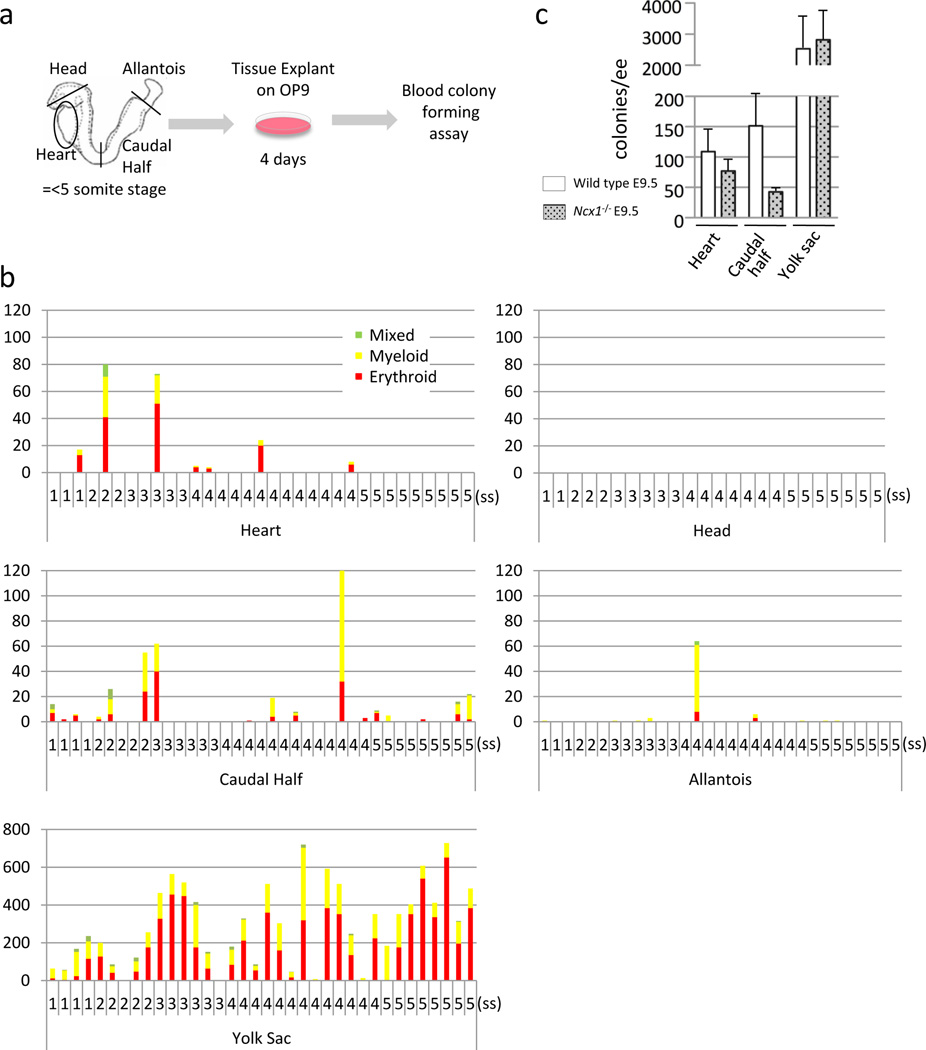
Defining the origin of blood cells in vivo is complicated by circulation. Once an effective heartbeat is initiated at around 8 somite stage (~E8.5), any blood cell may circulate and adhere to any vascular wall throughout the body. To examine whether the heart tube generates functional hematopoietic cells in situ, we employed two methods. First, we examined the colony forming activity of the heart tubes explanted at pre-circulation stages (1–5 somite stages) on an OP9 feeder layer, which supports myelo-erythroid differentiation21, followed by methylcellulose culture supplemented with hematopoietic growth factors (Fig. 1a). As a positive control, yolk sac yielded numerous colonies (Fig. 1b). The caudal half of the embryo (including dorsal aortae) and allantois (future placenta) also produced a much smaller but significant number of colonies, and the head explants produced no colonies at any stages. Interestingly, the heart tubes explanted from pre-circulation stages developed erythroid, myeloid and mixed lineage clonogenic progenitors (Fig. 1b). Second, to further confirm the in situ hemogenic activity of the heart tube, we utilized the Ncx1 knockout mouse model. Ncx1 is a sodium-calcium exchanger, of which expression is restricted to the myocardium. Ncx1 mutant embryos show normal morphogenesis and cardiac gene expression pattern until E9.5, but do not survive after E10.522. They have no heartbeat, and thus no systemic circulation, which makes them a suitable model for examining local hematopoietic emergence22. OP9 culture and subsequent colony assays revealed that the heart explants from Ncx1 mutants at E9.5 generated blood colonies in the absence of effective circulation (Fig. 1c). Together, these experiments suggest that the heart tube displays in situ hemogenic activity during embryogenesis.
Figure 1. The heart tube is a de novo source for hematopoietic cells.
- Schematic representation of the colony forming assay from ex vivo organ explant at pre-circulation stages. The heart tube, head, allantois, caudal half (including future AGM region) and yolk sac were dissected at somite stages 1–5, before the formation of effective circulatory loop. Tissues were washed in 3 changes of PBS and cultured on an OP9 feeder layer for 4 days, followed by methylcellulose culture in the presence of hematopoietic growth factors.
- Hematopoietic colonies retrieved from various tissues at various somite stages. Each column represents colonies from one tissue. The heart tubes displayed hematopoietic activity whereas the head explants did not. Note the difference in the scale in the yolk sac.
- Colonies from Ncx1 mutant embryos that lack heartbeat, showing the hematopoietic activity in the heart tube in the absence of effective heartbeat. Mean±SEM.
CD41 is expressed in a subset of the endocardial cells
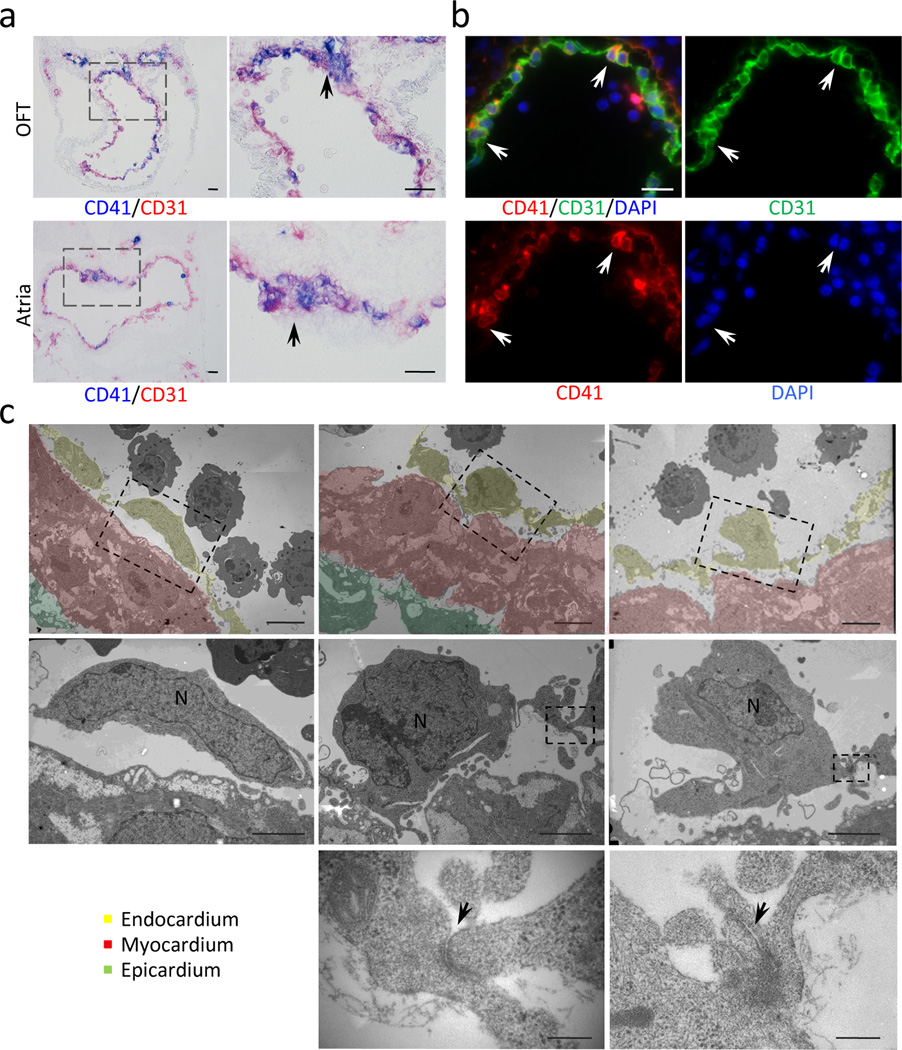
We hypothesized that the de novo hematopoietic activity of the heart tube arises from endocardium because it shares many of the properties with the endothelium in the dorsal aorta. To examine the hemogenic potential of the endocardium, we first analyzed the local expression of CD41 (integrin alpha2b, GpIIB), an early surface marker for nascent hematopoietic progenitor cells. Immunostaining revealed that CD41 is expressed in a subset of CD31+ endocardial cells in the outflow tract and atria (Fig. 2a, b, arrows). CD31+/CD41+ endocardial cells were rounded up and scattered circumferentially in atria and less frequently in ventricles. Clusters of CD31+/CD41+ cells were found in the endocardial cells in the outflow cushion, atrioventricular (AV) cushion, aortic sac, aortic arches, and the dorsal atrial wall near the dorsal mesocardium where the common atrial chamber and future pulmonary vein are in the process of establishing connection (Supplementary Fig. S1). CD41+ cells were identified even at E8.25 in the endocardium, but not until around E10.5 in the AGM (Supplementary Fig. S2), indicating that the CD41+ putative hematopoietic progenitor cells emerge earlier in endocardium than in the endothelium of the AGM. Although CD41 also labels megakaryocytes/platelets, the majority of CD41+ cells in the endocardium were non-megakaryocyte/platelets at these stages, as they were negative for GP1bβ (Supplementary Fig. S3). Co-staining using an antibody against phosphorylated histone H3 (pH3) suggests that CD31+/CD41+ cells proliferate in situ on the endocardial wall (Supplementary Fig. S4). Furthermore, electron microscopic analysis of the embryonic hearts at E9.5–10.5 revealed that some of the endocardial cells are rounded up (Fig. 2c, middle panels) and protrude into the cardiac lumen (Fig. 2c, right panels). These cells have relatively large nuclei, and are connected to adjacent endocardial cells by adherens junctions (Fig. 2c, bottom panels), suggesting that they are not circulating blood cells attaching to the endocardial wall, but are endocardial cells emerging into circulation. Together, these data indicate that these putative hematopoietic progenitors are generated in situ in the endocardium.
Figure 2. A subset of endocardial cells express CD41 and undergo budding into the heart lumen.
- Immunohistochemistry for CD41 (blue) and CD31 (red) in outflow tract (OFT) and atria at E9.5. CD41+ endocardial cells are clustered in the outflow cushion and the dorsal wall of the atria, and also scattered in the entire circumference (arrows). Scale bar = 100µm (lower magnification), 50µm (higher magnification).
- Immunofluorescent staining for CD41 (red), CD31 (green) and DAPI (blue) in left atrium at E9.5. Arrows indicate scattered CD31+ endocardial cells co-expressing CD41. Scale bar = 20µm.
- Electron microscopic analysis of E10.0–10.5 atrium. Bottom panels show higher magnification of the broken-lined area in upper panels. Left panels indicate regular flat endocardial cells. Center panels are representative images of endocardial cells rounded up. Endothelial cells occasionally protrude into the cardiac lumen (right panels). The bottom panels are higher magnification of adherens junctions in the dotted area in the middle panels (arrows). Scale bar = 2µm (upper panels), 1µm (middle panels) and 100nm (bottom panels). Yellow = endocardial layer. Red = myocardial layer.
The endocardial cells display hemogenic activity
To examine hemogenic activity of the endocardium, we performed a blood colony forming assay using FACS purified endocardial/endothelial cells from embryonic tissues as previously described23,24. As a subset of circulating hematopoietic progenitor cells express CD31, the local endocardial cells at E9.5 and 10.5 were sorted as a CD31+/CD41−/CD45− population (Supplementary Fig. S5a). The CD31+/CD41−/CD45− cell population in the heart at these stages represents endocardial cells, as coronary vessels are not yet formed. Microarray analysis from CD31+/CD41−/CD45− cells in the heart, caudal half and yolk sac identified 23,922 gene probes expressed in the endocardium, 19,387 of which are expressed in common and 1829 of which are specific to the endocardium (Supplementary Fig. S5b). The CD31+/CD41−/CD45− population of the endocardium shared expression of hematopoietic genes with other endothelial populations including Cbfa2t3, Kit, Klf1, Mll1, Pbx1 and Scl/Tal1 (Supplementary Fig. S5c, Supplementary Table S1). Within 20,966 gene probes commonly expressed in the heart tube and caudal half, 4631 probes were expressed within 1.5-fold from each other. These similarly expressed probes included 481 transcription factors (Supplementary Table S2). Gene ontology (GO) analyses of these probe sets showed that hematopoiesis-related transcription factors are significantly enriched in this group (Supplementary Fig. S5d). PCR suggests that Runx1, cMyb, Klf1 and Scl are expressed in the endocardium (Supplementary Fig. S5e). As non-hemogenic tissue is not included in the analysis, it is possible that all the endothelial cells at this stage express some level of hematopoietic transcription factors. However, OP9/colony assays revealed that myeloid colonies and rare erythroid colonies develop from FACS-purified endocardium (Supplementary Fig. S6a, b, d). Direct colony assay without OP9 pre-culture yielded no colony from the heart or AGM (Supplementary Fig. S6c, indicating that CD31+/CD41−/CD45− cells are not yet committed to the hematopoietic progenitor fate and that there is no contamination of circulating hematopoietic progenitors with this sorting method. Together, these data suggest that the endocardium is capable of generating functional hematopoietic cells ex vivo, and support the idea that endocardium is a novel site of de novo hematopoiesis in the developing embryo.
CD41+ endocardial cells arise from Nkx2-5+ cells
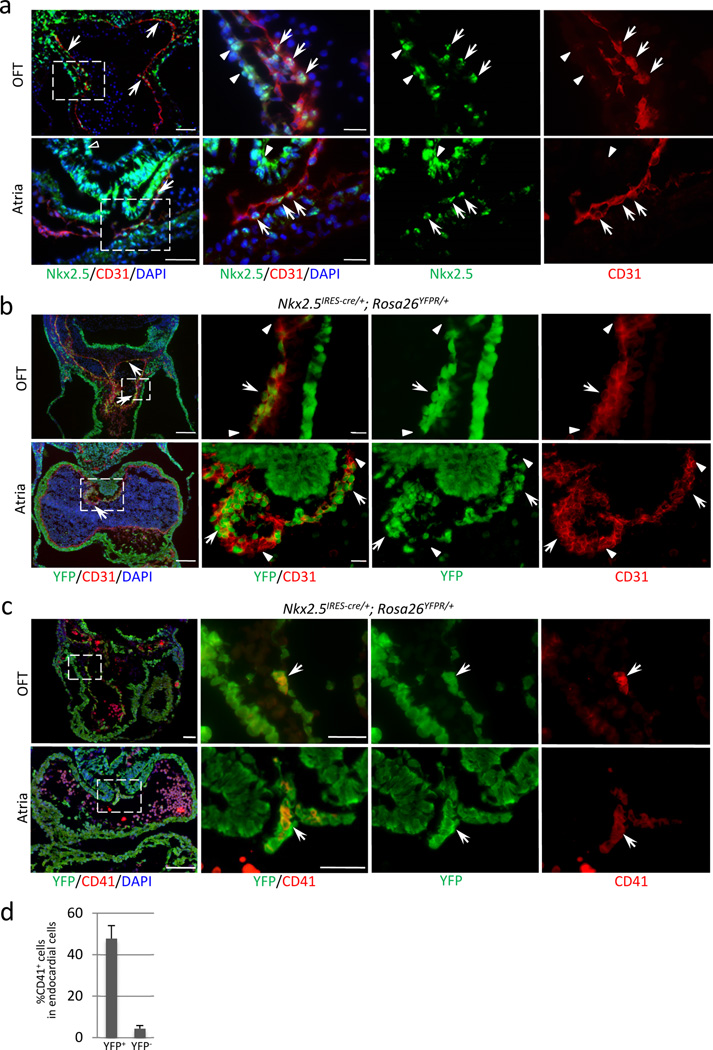
One of the few genes known to be differentially expressed in the endocardium is a homeobox transcription factor, Nkx2-525,26. Previous reports suggest that Nkx2-5+ cells contribute to the endocardial cells and that Nkx2-5 mRNA is expressed in the endocardium25,26. While the function of Nkx2-5 in the myocardium is extensively studied, little is known about its function in the endocardium/endothelium26. Immunostaining revealed that Nkx2-5 is expressed in a subset of endocardial cells in the outflow and AV cushion, the dorsal wall of the atria, and the aortic sac, although the expression level was significantly weaker than that in the myocardium (Fig. 3a). Consistently, lineage tracing using Nkx2-5IRES-Cre/+; R26YFPR/+ showed clusters of YFP+ cells in the outflow cushion endocardium (Fig. 3b, upper panels) and the atrial endocardium near the dorsal mesocardium (Fig. 3b, lower panels). Isolated YFP+ cells were also found scattered in other parts of the atrial and outflow endocardium. Because this specific distribution pattern of Nkx2-5-positive/derived cells corresponded to that of CD31+/CD41+ cells in the endocardium (Fig.2a, b, and Supplementary Fig. S1), we examined whether CD41 is expressed in YFP+ cells in the endocardium. Double staining for YFP/CD41 in Nkx2-5IRES-Cre/+; R26YFPR/+ hearts revealed that the majority of endocardial CD41+ cells arise from Nkx2-5-derived endocardium (Fig. 3c, d). These data suggest that Nkx2-5-positive endocardium represents the hemogenic subpopulation in the endocardium. Nkx2-5-positive/derived cells are also found in the yolk sac endothelium with variation in frequency, but not in the AGM endothelium (Supplementary Fig. S7).
Figure 3. Expression of CD41 in Nkx2-5-positive subset of endocardial cells.
- Immunofluorescent staining for Nkx2-5 (green) and CD31 (red) at E9.5. Clusters of Nkx2-5+ cells are found in the endocardium of outflow cushion, atrioventricular cushion, and the dorsal wall of the atria (white arrows). Note that myocardium shows stronger level of Nkx2-5 expression (white arrowheads). Endodermal cells are also positive for Nkx2-5 (left lower panel, open arrowhead). Scale bar = 50µm (lower magnification), 20µm (higher magnification).
- Heart section of Nkx2-5IRES-Cre/+; R26YFPR/+ embryo at E10.5 stained for YFP (green) and CD31 (red). Nkx2-5-derived endocardial cells (YFP/CD31 double positive) were clustered in the outflow cushion and the dorsal wall of the atrium, and also found scattered along the perimeter (arrows). This specific distribution pattern corresponds to that of CD41+ cells (Fig. 2a, b). Arrowheads indicate non-Nkx2-5-derived endocardial cells. Scale bar = 100µm (lower magnification), 20µm (higher magnification).
- Heart section of Nkx2-5IRES-Cre/+; R26YFPR/+ embryo at E10.5 stained for YFP (green) and CD41 (red). Nkx2-5+ cells give rise to CD41+ cells in the endocardium (YFP/CD41 double positive, arrows). Scale bar = 100µm (lower magnification), 20µm (higher magnification).
- Percentage of CD41+ cells in Nkx2-5-derived fraction of endocardium at E9.5. 48% of Nkx2-5-lineage labeled endocardial cells express CD41 whereas 4.4% are CD41-positive in non-Nkx2-5-derived endocardial cells. The graph represents the average of three independent experiments. Mean±SEM.
Nkx2-5+ cells contribute to definitive hematopoiesis
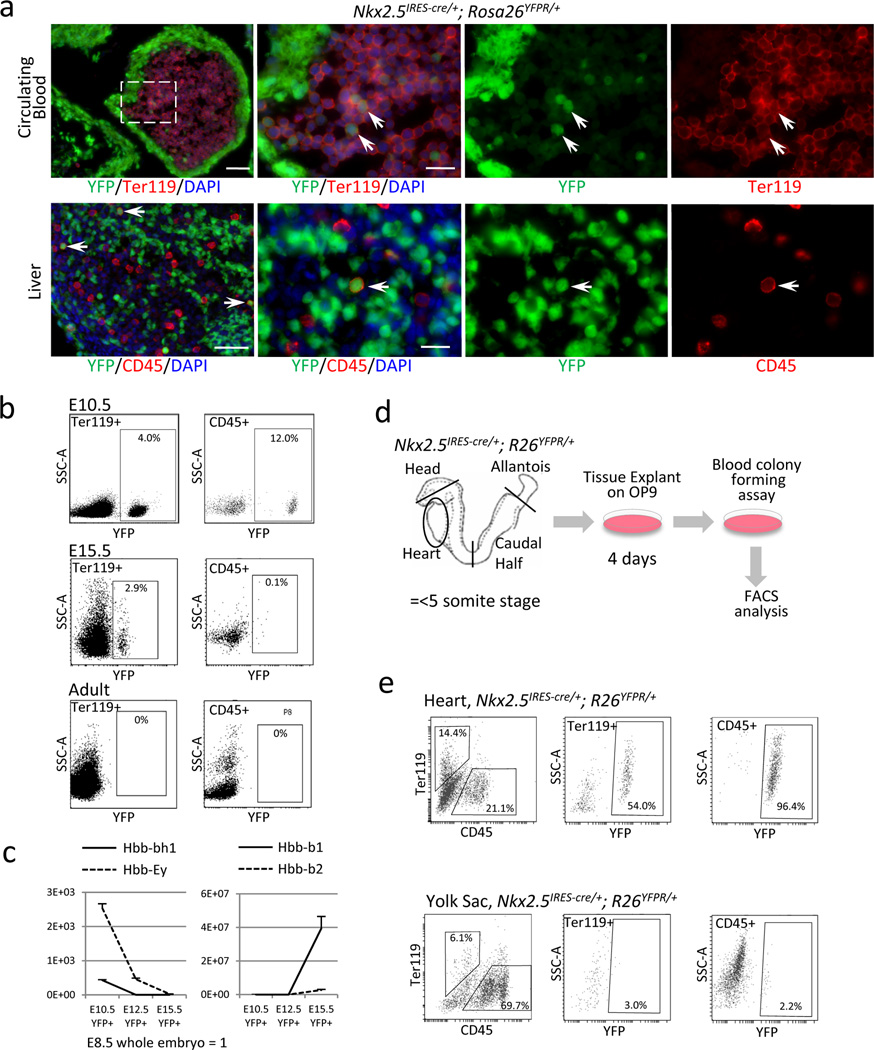
To characterize the developmental nature of Nkx2-5-derived hematopoietic population, we first examined the contribution of Nkx2-5+ cells to the peripheral circulation. Examination of the peripheral blood in Nkx2-5IRES-Cre/+; R26YFPR/+ embryos identified YFP-labeled cells in the circulating blood (Fig. 4a, upper panels, arrows). YFP-labeled CD45+ cells also repopulated the liver (Fig. 4a, lower panels, arrows). FACS analysis of the peripheral blood in Nkx2-5IRES-Cre/+; R26YFPR/+ embryos revealed that Nkx2-5+ cells contributed to 4.0% of peripheral Ter119+ (erythroid) cells and 12.0% of peripheral CD45+ (non-erythroid) cells at E10.5. At E15.5, the percentage of Nkx2-5-derived Ter119+ and CD45+ became 2.9% and 0.1%, respectively (Fig. 4b). However, Nkx2-5-derived blood cells were not identified in the peripheral circulation in adult Nkx2-5IRES-Cre/+; R26YFPR/+ mice. Expression analysis by qPCR for β-globin genes indicates that Nkx2-5-derived erythroids were initially positive for Hbb-bh1 and Ey at E10.5, and underwent maturational globin switching to Hbb-b1 and Hbb-b2 by E15.5 (Fig. 4c). FACS analyses of E9.5 Nkx2-5IRES-Cre/+; R26YFPR/+ embryos suggested that about 16% and 7.5% of CD41+/c-kit+ cells in the heart and the yolk sac, respectively, were derived from Nkx2-5+ cells (Supplementary Fig. S8). To examine whether Nkx2-5+ cells contribute to the definitive erythroids, circulating Ter119+ cells were separated using forward and side scatter27. Hoechst staining from primitive (P2) and definitive (P1) erythroids showed distinct nucleated and enucleated cells (Fig. 4d) as previously demonstrated27. As shown in right panels of Fig. 4d, Nkx2-5+ cells contributed to both primitive nucleated and definitive enucleated cells. qPCR from YFP+ nucleated primitive and YFP+ enucleated definitive erythroids showed typical Hbb-Ey and Hbb-b1/b2 dominant pattern of β-globin expression, respectively (Fig. 4d, bottom panel). Furthermore, analyses of Runx1 mutants (Runx1lz/rd) revealed that Runx1 is required for the formation of the hematopoietic colonies, although not for the specification of CD41+ cells in the endocardium (Supplementary Fig. S9). Together, these data suggest that Nkx2-5+ hemogenic endocardial/endothelial cells contribute to transient definitive erythroid/myeloid progenitors28–33. This hematopoietic population could also be isolated from Nkx2-5-GFP transgenic mouse and ES cell lines. As shown in Supplementary Fig. S10, GFP+/CD41+ cells at day 4.5 and 5.5 EBs were successfully developed into erythroid, myeloid and mixed colonies, suggesting that pluripotent cells can serve as alternative sources for Nkx2-5+ hematopoietic cells.
Figure 4. A subset of erythroids and myeloids in the peripheral circulation arise from Nkx2-5+ endocardial/endothelial cells.
- Peripheral blood and liver in Nkx2-5IRES-Cre/+; R26YFPR/+ E10.5 embryo stained for YFP (green) and Ter119 (red). Double positive cells (arrows) indicate circulating erythroids (upper panels) and CD45+ cells repopulating in the liver (lower panels) are positive for YFP. Note that some endodermal cells in the liver are also labeled by YFP reporter. Scale bar = 10µm.
- Peripheral blood cells from Nkx2-5IRES-Cre/+; R26YFPR/+ embryos at E10.5, E15.5 and adult stages were analyzed for YFP, CD45 and Ter119 by FACS. Nkx2-5+ cells contribute to 4.0% of Ter119+ cells and 12.0% of CD45+ cells at E10.5, and 2.9% and 0.1%, respectively, at E15.5. No YFP+ blood cells are identified in the adult peripheral circulation.
- Expression analyses of β-globins in Nkx2-5-derived peripheral blood at E10.5, 12.5 and 15.5 by qPCR from YFP+ circulating cells of Nkx2-5IRES-Cre/+; R26YFPR/+ embryos. Hbb-bh1 (solid line, left) was first downregulated by E12.5 followed by downregulation of Ey (dashed line, left) by E15.5. Hbb-b1 and b2 were strongly upregulated by E15.5.
- Primitive and definitive erythroids were sorted from E15.5 control and Nkx2-5IRES-Cre/+; R26YFPR/+ embryos based on forward and side scatters (top left panels). FACS analyses for Hoechst and YFP revealed that Nkx2-5+ cells contribute to both nucleated primitive erythroids (solid blue line) and enucleated definitive erythroids (solid red line, top right panels). qPCR analysis for β-globins revealed that Nkx2-5-derived primitive and definitive erythroids dominantly express Hbb-Ey and Hbb-b1/b2, respectively (bottom panel).
- Schematic representation of the colony forming assay from the explant of Nkx2-5-lineage labeled organs at pre-circulation stages. Tissues were washed in 3 changes of PBS and cultured on OP9 feeder for 4 days, followed by methylcellulose culture in the presence of hematopoietic growth factors.
- FACS analysis of the colonies retrieved from Nkx2-5-lineage labeled heart tubes and yolk sacs at pre-circulation stages. Nkx2-5-derived cells constitute 54.0% of Ter119+ cells and 96.4% of CD45+ cells in the colonies from the heart tubes (left), but only 3.0% and 2.2%, respectively, in those from the yolk sac (right).
As Nkx2-5 lineage tracing experiments by themselves cannot distinguish Nkx2-5+ cells from endocardium or yolk sac endothelium, we examined whether Nkx2-5+ endocardial cells contribute to Ter119+ and/or CD45+ cells by pre-circulation stage explant assay using Nkx2-5IRES-Cre/+; R26YFPR/+ embryos (Fig. 4e). FACS analysis of the colonies from Nkx2-5IRES-Cre/+; R26YFPR/+ embryos indicates that a majority of the colonies from the heart tube arise from Nkx2-5-positive cells. As shown in Fig. 4f, Nkx2-5-derived cells constitute 54.0% of Ter119+ cells and 96.4% of CD45+cells in the colonies from the heart tubes, whereas only 3.0% and 2.2%, respectively, are labeled in the colonies from the yolk sac. The difference in the percentage of YFP+ fraction in the heart tubes and the yolk sac further support that the heart-derived colonies are not the result of contamination from the yolk sac during the procedure. Taken together, these data suggest that Nkx2-5+ endocardial cells give rise to transient hematopoietic progenitors for both Ter119+ erythroid and CD45+ non-erythroid cells independently from the yolk sac.
Endocardial hematopoiesis is regulated by Nkx2-5 and Isl1
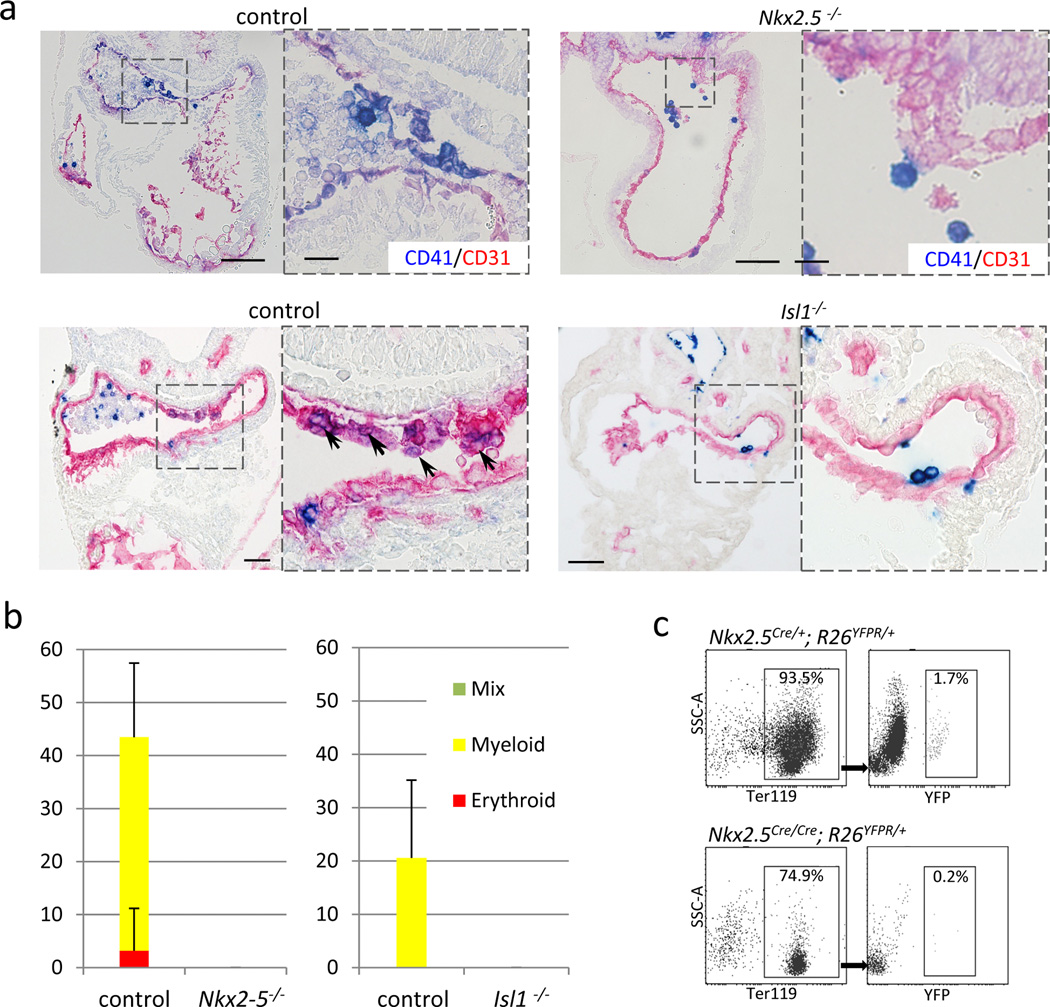
Specific expression of Nkx2-5 in the hemogenic endocardium leads to the question of whether or not Nkx2-5 expression is required for the hemogenic potential of the endocardium. Nkx2-5 null embryos die at around E10.5 with cardiac looping defects, and also develop impaired angiogenesis and hematopoiesis34,35. It was previously speculated that these hemato-vascular defects are secondary to the myocardial phenotype35. However, the identification of Nkx2-5-positive/derived endocardium and yolk sac endothelium raises the possibility that Nkx2-5 is directly involved in embryonic hematopoiesis. Indeed, ectopic expression of NKX2-5 is found in the leukemic cells in children with T- and B-cell acute lymphoblastic leukemia with t(5;14) translocation36,37. Given that the molecular pathways regulating on cogenesis often recapitulate aberrations of processes governing embryogenesis, these clinical observations support the idea that Nkx2-5 plays a direct role during hematopoiesis. To examine the role of Nkx2-5 in endocardial hematopoiesis, we analyzed Nkx2-5 knockout embryos. At E9.5, Nkx2-5-null hearts showed a significantly reduced number of CD41+ cells in the endocardium (Fig 5a, top panels). Lineage tracing of Nkx2-5-positive cells in Nkx2-5-null background (Nkx2-5Cre/Cre; R26YFPR/+ embryos) revealed that Nkx2-5-expressing endocardial cells were found in the endocardial layer (Supplementary Fig. S11), suggesting that Nkx2-5 is required not for the migration of the hemogenic endocardial cells to the cushion but for the specification of hematopoietic progenitor cells from the endocardium. Colony forming assays with sorted endocardial cells showed that no blood colonies are generated from Nkx2-5 knockout endocardium at E9.5 (Fig. 5b, left). Furthermore, the number of Nkx2-5-derived Ter119+ cells was significantly reduced in the peripheral circulation in Nkx2-5-null background (Nkx2-5Cre/Cre; R26YFPR/+ embryos) at E9.5 (Fig. 5c). These data suggest that the expression of Nkx2-5 is required for the emergence of hematopoietic progenitors in the endocardium.
Figure 5. Figure 5. Endocardial hematopoiesis depends on Nkx2-5 and Isl1.
- Nkx2-5 mutant heart at E9.5 stained for CD41 (blue) and CD31 (red). CD41+ cells were hardly found in the endocardium of Nkx2-5 mutants. Note that Nkx2-5 mutant also develops known cardiac defects including a single ventricle with poorly developed trabeculae, open atrioventricular canal, and lack of epithelial-mesenchymal transformation in the endocardial cushion. Scale bar = 100µm.
- Nkx2-5-derived endocardial cells in Nkx2-5 hetero (left panels) and null (right panels) background. Nkx2-5-driving cells were identified in the cushion endocardium even in the absence of Nkx2-5.
- Blood colony formation assay from FACS-purified endocardial cells. No colonies were formed from Nkx2-5 mutant endocardial cells. Data represent average of three independent experiments.
- Comparison of the peripheral blood cells derived from Nkx2-5-expressing cells in Nkx2-5-null and its control backgrounds at E10.0. YFP-labeled red blood cells (Ter119+) were found less frequently (0.2%) in Nkx2-5Cre/Cre; R26YFPR/+ embryos compared with control littermates (1.7%), suggesting that expression of Nkx2-5 is required for the hemogenic activity of the Nkx2.5+ endocardial/endothelial cells in vivo.
Another key cardiac transcription factor, Isl1, is also expressed in the endocardium of outflow cushion and atria (Supplementary Fig. S12)38,39. Isl1 mutants, like Nkx2-5 mutants, showed much less CD41 expression and no blood colony formation from the endocardium than control littermates at E9.5 (Fig. 5a, bottom panels; Fig. 5b, right). These data suggest that Isl1 is also required for the emergence of CD41+ cells in the endocardium.
Discussion
In conclusion, we have demonstrated that Nkx2-5-positive subset of the endocardium and yolk sac endothelium gives rise to progenitors for transient definitive hematopoiesis that contribute to circulating erythroid/myeloid cells until late gestational stages via an Nkx2-5 and Isl1-dependent mechanism. Given that the endocardial cells may be heterogeneous in their origins40–43, an intriguing possibility is that the hemogenic endocardial cells are derived from multipotent Flk1/Isl1/Nkx2-5-positive cardiovascular progenitors12 through endocardial intermediates. Indeed, hemogenic endocardial cells express all these markers (Fig. 3; Supplementary Fig. S12)38,39, and early cardiac progenitors express multiple hematopoietic transcription factors12,13. Alternative possibility is that yolk sac-derived Runx1+ precursors contribute to hemogenic endocardial cells as in the dorsal aorta44. The identification of hemogenic endocardial cells in this study potentially adds a hematopoietic component into the cardiovascular lineage tree and places Nkx2-5 at the center of diverse cardio-vasculo-hematopoietic lineages. Finally, it leads to a better understanding of the origins and roles of transient definitive hematopoiesis during embryogenesis and leukemogenesis.
Methods
Mouse models
Genetically modified Nkx2-5-null, Nkx2-5-IRES-Cre, Nkx2-5-Cre, NCX1-null, Isl1-null, Runx1-rd, and Runx1-lz mouse lines are described previously22,25,45–49. Animals were maintained in accordance with the guidelines of the UCLA Animal Research Committee.
Preparation of frozen tissue sections
Mouse embryos were isolated in cold PBS and fixed in 4% PFA for 2~3 hrs, followed by equilibration in 30% sucrose in PBS solution overnight. The tissues were placed in 1:1 30% sucrose/OCT (Tissue-Tek, Electron Microscopy Sciences) solution for 1–2 hr, in 100% OCT compound for 1 hr at 4°C, and embedded in 100% OCT compound, carefully oriented in Cryomolds (Ted Pella). The blocks were immediately frozen on dry ice with isopropanol and stored at -80°C. The sections were cut 7–10 µm with a Leica CM3050 S cryostat. The following primary antibodies were used: rat CD41 (1:50, BD PharMingen), Phospho-Histone 3 (1:200, Millipore), Ter119 (1:50 eBioscience), and PECAM-1/CD31 (1:200, BD PharMingen) were used on fixed frozen sections. Tyramide amplification (Invitrogen) was applied to CD41, and Ter119 stainings. The following secondary antibodies were used: Biotinylated IgG antibodies (Vector Laboratories) for colorimetric staining, Alexa Fluor 488 (green), Alexa Fluor 594 (red)-conjugated secondary antibodies specific to the appropriate species were used (1:500; Invitrogen) for fluorescent staining. Sections were mounted with antifade mounting medium with DAPI (Invitrogen), and analyzed by using AxioImager D1 (Carl Zeiss Microimaging, Inc)
Electron microscopic analysis
Tissues were fixed in 1% glutaraldehyde, 4% paraformaldehyde in PBS and washed. After postfixation in 1% OsO4 in PB for 1 hour, the tissues were dehydrated in a graded series of ethanol, treated with propylene oxide and embedded in Eponate 12 (Ted Pella). Approximately 60–70 nm thick sections were cut on a Reichert-Jung Ultracut E ultramicrotome and picked up on formvar coated copper grids. The sections were stained with uranyl acetate and Reynolds lead citrate and examined on a JEOL 100CX electron microscope at 80kV.
Flow cytometry for sorting cells of mouse embryonic tissues
We harvested tissues from mouse embryos of several developmental stages (E9.5-10.5). Isolated tissues were washed three times and incubated at 37°C in a dissociation enzyme solution with occasinal pipetting to a single-cell suspension. The enzyme solution contained 1% Penicillin/Streptomycin (Invitrogen, 15140-122), 10%Fetal Bovine Serum (Hyclone), collagenase 2mg/ml (Worthington, CLS-2), dispase 0.25mg/ml (Gibco, 17105-041), DNAase I (Invitrogen in PBS. The cells were analyzed and sorted by a BD FACSAria with the following rat anti-monoclonal antibodies: CD41-PE-Cy7 (eBioscience), CD45APC (BD Pharmingen), Ter119 (eBioscience), ckit-APC-Cy7 (Biolegend), CD31-PE (BD Pharmingen). 7-AAD (7-amino-actinomycinD, BD Bioscinces) staining was used to exclude non-viable cells.
Hematopoietic colony-forming assays
Sorted cells, as described above, were cultured on OP9 stromal cells for 4 days in 48-well plates in 500µl of α-MEM (GIBCO/Invitrogen) containing 20% fetal bovine serum (Hyclone), 1% penicillin/streptomycin and supplemented with stem cell factor (SCF, 50 ng/ml), interleukin-3 (IL3, 5 ng/ml), interleukin-6 (IL6, 5 ng/ml), Thrombopoietin (TPO, 5 ng/ml), and Flt-3 Ligand (Flt-3L, 10 ng/ml). The cells were then dissociated mechanically from stroma by transfer pipette and filtered to remove the stromal cells (celltreck). The filtered cells were transferred into 1.5 ml methylcellulose with SCF, IL-6, IL-3, and EPO (MethoCult 3434, Stem Cell Technologies) supplemented with TPO (5 ng/ml) to determine multilineage potential. Colonies were scored 7-10 days later.
Embryonic stem cell culture
Nkx2-5-EmGFP ES cell line50 was maintained on irradiated feeder. After the differential plating of feeder for 45min on gelatin-coated dish, 100 non-adherent cells were collected in 10µl of IMDM-based hanging drop containing 15% FBS, MTG, hTransferrin, ascorbic acid (25µg/ml), hBMP (45ng/ml), mSCF (50ng/ml) and mVEGF (10ng/ml). EBs in hanging drops were plated on day 2 on ultra low attachment plate, and cultured in the same media until the FACS sorting.
Organ explant culture
Fetal organs were dissected out, washed three times and mechanically dissociated to small pieces. They were cocultured on mouse OP9 stromal cells in the same media used for the hematopoietic colony-forming assay. After 4 days of culture, they were transferred into methylcellulose in the same way as described in the hematopoietic colony-forming assay method.
Gene expression analysis by microarray
RNeasy Micro Kit (Qiagen) was used to isolate total RNA from sorted cells from biological replicates of each cell population. RNA amplified by Nugen kit was hybridized on Affymetrix MOE 430 2.0 mouse gene expression microarrays. We used the R package Limma 51 provided through the open source project Bioconductor (version 2.7) 52 for assessing differential expression. To obtain a final list of differentially expressed genes, thresholds for selecting significant genes were set at a relative fold difference > 2 fold and p-value <0.01 on the list of probe sets reported by Limma. To calculate absolute mRNA expression levels, we used the RMA (Robust Multiarray Averaging 53) method provided through R library Affy to obtain background adjusted, quantile normalized and probe level data summarized values for all probe sets. For genes with multiple probe sets, in order to avoid over counting, we chose the probe set with the lowest p-value. Official gene symbols for probe sets were obtained using the Bioconductor annotation database mouse4302.db. Differentially up and down-regulated gene sets were uploaded into the online DAVID interface to identify statistically significantly over-represented GO biological process categories. We used the MAS5.0 algorithm 54 through the Affy for calculating PMA detection calls for each array sample. The algorithm employs a signed rank test to consider the significance of the difference between the PM (Perfect Match) and MM (MisMatch) values for each probeset and returns a detection p-value that is used to flag a transcript as 'Present', 'Marginal' or 'Absent' (P/M/A).
Gene expression analysis by qRT-PCR
RNA was extracted from the sorted cells using with RNeasy Micro kit (Qiagen). RNA was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (BioRad). qRT-PCR was performed using Lightcycler 480 SYBR Green I (Roche Applied Science). Forward and reverse primer sequences are as follows:
Hbb-bh1; aggcagctatcacaagcatctg, aacttgtcaaagaatctctgagtccat
Hbb-Ey; caagctacatgtggatcctgagaa, tgccgaagtgactagccaaa
Hbb-b1; cgtttgcttctgattctgttg, ctagatgcccaaaggtcttc
Hbb-b2; aaaggtgaaccccgatgaag, tgtgcatagacaatagcaga
Gapdh; gaagggcatcttgggctacac, gcagcgaactttattgatggtatt
Supplementary Material
Acknowledgements
We thank UCLA EM Core for the assistance with electron microscopic analysis, and the BSCRC Flow Cytometry Core for FACS sorting. This work is funded by BSCRC, AHA (IRG4870007, GRNT9420039) and NIH (R21HL109938) to A.N, and NIH (R01HL097766) to HKAM. A.A. is supported by HHMI undergraduate fellowship. Y.N. is supported by fellowship grants from Japanese Circulation Society and Uehara Memorial Foundation. A.W.H. is supported by NIH graduate fellowship. Isl1 mutant, Runx1 mutants and Nkx2-5-GFP mouse ES cell line are kind gift from Drs. Sylvia Evans, Nancy A. Speck, and Edward C. Hsiao. We also thank Drs. Utpal Banerjee, Luisa Iruela-Arispe, Kenneth Dorshkind and Shin-ichi Nishikawa for fruitful discussion of the data. S.J.C was supported, in part, by the Indiana University Department of Pediatrics (Neonatal-Perinatal Medicine) and NIH (P30DK090948).
Footnotes
Author Contributions
H.N. and A.N. designed the project, H.N., X.L., A.A., Y.N., B.v.H., JH.S. and A.W.H. performed experiments, H.N., Y.N., R.S., S.J.C., R.P.H., M.P., and A.N. analyzed and interpreted the data, R.J.S., S.J.C., R.P.H., and M.P. contributed materials, and H.N. and A.N. prepared the manuscript.
Author Information
The authors declare no competing financial interests.
Accession Codes
All original microarray data have been deposited in NCBI’s Gene Expression Omnibus under accession number GSE40260.
References
- 1.Ji RP, et al. Onset of cardiac function during early mouse embryogenesis coincides with entry of primitive erythroblasts into the embryo proper. Circ Res. 2003;92:133–135. doi: 10.1161/01.res.0000056532.18710.c0. [DOI] [PubMed] [Google Scholar]
- 2.Dieterlen-Lievre F. Emergence of haematopoietic stem cells during development. C R Biol. 2007;330:504–509. doi: 10.1016/j.crvi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshimoto M, Porayette P, Yoder MC. Overcoming obstacles in the search for the site of hematopoietic stem cell emergence. Cell Stem Cell. 2008;3:583–586. doi: 10.1016/j.stem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zovein AC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertrand JY, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisset JC, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- 9.Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- 10.Adamo L, et al. Biomechanical forces promote embryonic haematopoiesis. Nature. 2009;459:1131–1135. doi: 10.1038/nature08073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.North TE, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–748. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moretti A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Masino AM, et al. Transcriptional regulation of cardiac progenitor cell populations. Circ Res. 2004;95:389–397. doi: 10.1161/01.RES.0000138302.02691.be. [DOI] [PubMed] [Google Scholar]
- 14.Caprioli A, et al. Nkx2-5 represses gata1 gene expression and modulates the cellular fate of cardiac progenitors during embryogenesis. Circulation. 2011;123:1633–1641. doi: 10.1161/CIRCULATIONAHA.110.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Adhami MA, Kunz YW. Ontogenesis of haematopoietic sites in brachydanzo rerio (Hamilton-Buchanan) (Teleostei) Develop, Growth and Differ. 1977;19:171–179. doi: 10.1111/j.1440-169X.1977.00171.x. [DOI] [PubMed] [Google Scholar]
- 16.Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aorta-gonadal-mesonephros mesoderm. Nat Genet. 2004;36:1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- 17.Han Z, Olson EN. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development. 2005;132:3525–3536. doi: 10.1242/dev.01899. [DOI] [PubMed] [Google Scholar]
- 18.Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen VC, Stull R, Joo D, Cheng X, Keller G. Notch signaling respecifies the hemangioblast to a cardiac fate. Nat Biotechnol. 2008;26:1169–1178. doi: 10.1038/nbt.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterkin T, Gibson A, Patient R. Common genetic control of haemangioblast and cardiac development in zebrafish. Development. 2009;136:1465–1474. doi: 10.1242/dev.032748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 22.Koushik SV, et al. Targeted inactivation of the sodium-calcium exchanger (Ncx1) results in the lack of a heartbeat and abnormal myofibrillar organization. Faseb J. 2001;15:1209–1211. doi: 10.1096/fj.00-0696fje. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimoto M, et al. Embryonic day 9 yolk sac and intra-embryonic hemogenic endothelium independently generate a B-1 and marginal zone progenitor lacking B-2 potential. Proc Natl Acad Sci U S A. 2011;108:1468–1473. doi: 10.1073/pnas.1015841108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, et al. Mouse embryonic head as a site for hematopoietic stem cell development. Cell Stem Cell. 2012;11:663–675. doi: 10.1016/j.stem.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- 26.Ferdous A, et al. Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc Natl Acad Sci U S A. 2009;106:814–819. doi: 10.1073/pnas.0807583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kingsley PD, et al. "Maturational" globin switching in primary primitive erythroid cells. Blood. 2006;107:1665–1672. doi: 10.1182/blood-2005-08-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertrand JY, et al. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palis J, et al. Spatial and temporal emergence of high proliferative potential hematopoietic precursors during murine embryogenesis. Proc Natl Acad Sci U S A. 2001;98:4528–4533. doi: 10.1073/pnas.071002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 31.Wong PM, Chung SW, Reicheld SM, Chui DH. Hemoglobin switching during murine embryonic development: evidence for two populations of embryonic erythropoietic progenitor cells. Blood. 1986;67:716–721. [PubMed] [Google Scholar]
- 32.McGrath KE, et al. A transient definitive erythroid lineage with unique regulation of the beta-globin locus in the mammalian embryo. Blood. 2011;117:4600–4608. doi: 10.1182/blood-2010-12-325357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen MJ, et al. Erythroid/Myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- 36.Nagel S, Kaufmann M, Drexler HG, MacLeod RA. The cardiac homeobox gene NKX2-5 is deregulated by juxtaposition with BCL11B in pediatric T-ALL cell lines via a novel t(5;14)(q35.1;q32.2) Cancer Res. 2003;63:5329–5334. [PubMed] [Google Scholar]
- 37.Su X, et al. Transcriptional activation of the cardiac homeobox gene CSX1/NKX2-5 in a B-cell chronic lymphoproliferative disorder. Haematologica. 2008;93:1081–1085. doi: 10.3324/haematol.12595. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y, et al. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304:286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snarr BS, Kern CB, Wessels A. Origin and fate of cardiac mesenchyme. Dev Dyn. 2008;237:2804–2819. doi: 10.1002/dvdy.21725. [DOI] [PubMed] [Google Scholar]
- 40.Noden DM. Origins and patterning of avian outflow tract endocardium. Development. 1991;111:867–876. doi: 10.1242/dev.111.4.867. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg LM, Markwald RR. Molecular regulation of atrioventricular valvuloseptal morphogenesis. Circ Res. 1995;77:1–6. doi: 10.1161/01.res.77.1.1. [DOI] [PubMed] [Google Scholar]
- 42.Noden DM, Poelmann RE, Gittenberger-de Groot AC. Cell origins and tissue boundaries during outflow tract development. Trends Cardiovasc Med. 1995;5:69–75. doi: 10.1016/S1050-1738(99)80002-4. [DOI] [PubMed] [Google Scholar]
- 43.Baldwin HS. Early embryonic vascular development. Cardiovasc Res. 1996;31:E34–E45. Spec No. [PubMed] [Google Scholar]
- 44.Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- 45.Wang Q, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.North T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- 47.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 48.Pashmforoush M, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117:373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 49.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433:647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsiao EC, et al. Marking embryonic stem cells with a 2A self-cleaving peptide: a NKX2-5 emerald GFP BAC reporter. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth GK. In: Bioinformatics and Computational Biology Solutions using R and Bioconductor. Carey S, Gentleman R, Dudoit R, Irizarry R, Huber W, editors. Springer; 2005. [Google Scholar]
- 52.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 54.Liu WM, et al. Analysis of high density expression microarrays with signed-rank call algorithms. Bioinformatics. 2002;18:1593–1599. doi: 10.1093/bioinformatics/18.12.1593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.