Abstract
Transposons are DNA sequences capable of moving in genomes. Early evidence showed their accumulation in many species and suggested their continued activity in at least isolated organisms. In the past decade, with the development of various genomic technologies, it has become abundantly clear that ongoing activity is the rule rather than the exception. Active transposons of various classes are observed throughout plants and animals, including humans. They continue to create new insertions, have an enormous variety of structural and functional impact on genes and genomes, and play important roles in genome evolution. Transposon activities have been identified and measured by employing various strategies. Here, we summarize evidence of current transposon activity in various plant and animal genomes.
Keywords: somatic mutation, transposon, retrotransposon, polymorphism, genome dynamics, structural variation
INTRODUCTION
With rare exceptions, transposons are found in virtually all genomes. Although once regarded as junk DNA owing to their repetitive nature, decades of research have demonstrated that they represent an important evolutionary force in shaping the genomes (22, 24, 64). Nevertheless, it has been unclear to what extent transposons present today are immobile remnants from the past or active elements that continue to generate genomic diversity. Recently, with the increasing availability of whole-genome sequences from diverse organisms and the development of genomic techniques targeted for transposon studies, the existence of active transposons has proven to be widespread. This article focuses on evidence that transposons are active today and highlights the impact of recent insertions. Our goal here is to expose readers to the diversity of transposons present in the literature. When possible, we have chosen examples from commonly used model organisms in molecular biology and genetics. These include Arabidopsis thaliana, Oryza sativa (rice), Zea mays (corn), Caenorhabditis elegans, Drosophila melanogaster (fly), Mus musculus (mouse), Rattus norvegicus (rat), and Homo sapiens (human). Transposons are mobile genetic sequences with diverse structures and transposition mechanisms. A scheme for their classification is summarized in detail in References 34 and 131. In brief, they can be divided into RNA-based retrotransposons (Class 1) and DNA transposons (Class 2).
Retrotransposons use a copy-and-paste mechanism to propagate through an RNA intermediate. They are transcribed from the genome and then reverse transcribed into a new location via a transposon-encoded reverse transcriptase. Retrotransposons are further subdivided into orders. Some retrotransposons are highly related to retroviruses. They encode gag and pol proteins related to their viral equivalents and form viral particles during a distinct stage of the transposition cycle. In DNA, these elements are flanked by long terminal repeats (LTR) and are called LTR retrotransposons.
Other retrotransposons in metazoan genomes that lack LTRs (non-LTR retrotransposon) are separated into two groups, long interspersed elements (LINEs) and short interspersed elements (SINEs). LINEs are autonomous transposons of several kilobases in length. They generally encode a reverse transcriptase and endonuclease within the same open reading frame and are thought to be transcribed by RNA polymerase II. SINEs are nonautonomous retrotransposons that rely on LINE-encoded proteins for transposition; they do not encode proteins and thus depend on other elements for reverse-transcriptase activities. They comprise 50–200 bp sequences originating from small noncoding RNAs mostly derived from cellular RNAs involved in translation and related processes (transfer RNAs, ribosomal 7SL RNA, or 5S RNAs as well as others derived at least in part from retroviral sequences), and SINEs are also transcribed from polymerase III promoters.
By contrast, DNA transposons utilize a cut-and-paste mechanism to self-propagate. Most have terminal inverted repeats (TIR) and encode a transposase. There are also nonautonomous small DNA transposons, such as miniature inverted-repeat transposable elements (MITEs), that lack coding potential; these rely on autonomous DNA transposons for transposition. Most families of transposons leave target site duplications of a characteristic length upon integration into the genome. Helitrons, however, provide an exception. Discovered in 2001, this usual DNA transposon uses a rolling circle mechanism for transposition and frequently captures nearby genes or portions of them in the process (61).
HOW DO WE KNOW AN ELEMENT IS ACTIVE?
Detecting Activity
Recent transposon activity (Table 1) is recognized in genomes in two ways. The strongest evidence comes from identifying de novo insertions present in offspring and not parent(s). In earlier studies, these uncommon events were recognized only when they resulted in phenotypes. This is how transposable elements were first discovered in maize (84). Active excision of dissociation (Ds) sequence by activator (Ac) transposase from the region encoding an enzyme involved in pigment biosynthesis results in kernels of distinct coloration and patterns (128). Ds and Ac were later identified as nonautonomous and autonomous DNA transposons, respectively. The characterization of alleles resulting in spontaneous phenotypes in murine lab strains frequently demonstrates insertions of LTR retrotransposons (81). Examples of L1 LINE and Alu SINE retrotransposon insertions causing disease in humans are also known (19, 62). An Alu SINE retrotransposon insertion interrupting the exon of human factor VIII gene caused hemophilia. Although there are numerous examples of phenotype-causing insertions, these chance discoveries do not identify phenotypically silent integrations and thus do not readily lend themselves to calculating transposition frequency.
Table 1.
Transposon content and activities in various genomes
| Organism | Genome composition | Transposon activity | References | |
|---|---|---|---|---|
| Plants | ||||
| Physcomitrella patens | Moss; a model for evolutionary biology, manipulable genome | 480-Mb genome; more than half of the genome are transposon elements | High LTR activity; Helitron less active | (108) |
| Selaginella moellendorffii | Lycophyte and oldest extant vascular plant; useful model in evolutionary biology | 106-Mb genome; approximately one-third derived from transposon activity | LTRs predominate | (5) |
| Arabidopsis thaliana | Dicotyledonous flowering plant; its small size and short generation time led to its extensive use in genetics and developmental biology | 125-Mb genome; transposons account for approximately 10% | LTRs highly active; DNA transposons (mutator-like elements, MULEs and Basho) also active; LINEs and SINEs relatively quiescent | (50, 89, 97, 104, 139, 143) |
| Zea mays | Maize; organism in which transposons were discovered | 2.3-Gb genome; nearly 85% composed of transposable elements | LTRs are highly active; TIRs and Helitrons less active | (14, 46, 71–73, 79, 84, 88, 109–111, 122, 124, 128, 129, 140) |
| Oryza sativa | Rice; major calorie source for human | 420-Mb gnome; ~25% of the genome is of transposon origin | High LTR activity; TIRs less active; low LINEs and SINEs activity | (56, 58, 99, 137, 138) |
| Animals (invertebrates) | ||||
| Hydra magnipapillata | Freshwater cnidarian; a model for tissue patterning, transplantation, and regeneration | 1–1.5-Gb genome; transposons compose approximately 60% of the genome | CR1 non-LTR retrotransposons most abundant; followed by mariner and hAT DNA transposons | (18) |
| Caenorhabditis elegans | Nematode; a model for genetics, neurobiology, and development; manipulated by RNAi | 97-Mb genome; 12% of the genome is transposon derived | TIRs are highly active, particularly Tc1 and Tc3 transposons; LTRs less active | (9, 37, 49, 96) |
| Drosophila melanogaster | Fruit fly; extensively used model for development and genetics; meiotic recombination is female specific and tractable through use of balancer chromosomes | 180-Mb genome; 4%–9% of the genome transposon derived | TIRs are highly active; LTRs, LINEs, SINEs, and Helitrons less active | (6, 8, 10, 23, 27, 30, 35, 42, 43, 54, 66, 77, 80, 94, 98, 101, 114) |
| Animals (vertebrates) | ||||
| Takifugu rubripes | Pufferfish; relatively compact genome valuable in comparative genomics | 365-Mb genome; interspersed repeats comprise only 2.7% | Many types of transposable elements appear recently active, show low nucleotide divergence; LINE-like Mauis are the single-most common; SINE-FR and DNA transposons (Tc1_FR, Tc2_FR, and Chaplin) are also common | (3) |
| Anolis carolinensis | Lizard/green anole; the first nonavian reptilian genome sequenced | 2.2-Gb genome; ~20% transposons | Multiple families of non-LTRs are recently active. DNA transposons (hAT, Tc1, Helitron, Chapaev) show low divergence from consensus. LTR activity also present. | (92, 93, 100) |
| Ornithorhynchus anatinus | Platypus; a monotreme useful for studies of mammalian and reptilian evolution | 2.3-Gb genome; 50% interspersed repeats | LINE2 and companion SINEs (MIR and Mon-1) are highly active; no L1 activity | (126) |
| Myotis lucifugus | Brown bat | 2.3-Mb genome; ~25% transposons | TIRs are highly active; Helitrons second-most active; LINEs and SINEs are inactive | (95, 102, 105, 106) |
| Rattus norvegicus | Rat; first mammalian species domesticated for scientific research | 2.75-Gb genome; 40% interspersed repeats | LINEs and SINEs, followed by LTRs are highly active; 12% of the genome reflects rat-specific L1 insertions, not shared by mouse | (1, 40, 67, 125) |
| Mus musculus | Mouse; an especially genetically tractable mammalian model | 2.5-Gb genome; 37% interspersed repeats | LINEs, SINEs, and LTRs are highly active, with numerous species and strain-specific insertions characterized; ancestral repeats have a high rate of sequence divergence in mouse versus human | (1, 2, 44, 127, 142) |
| Homo sapiens | Human | 2.9-Gb genome; 46% interspersed repeats | LINEs and SINEs are highly active with L1, AluY, and SVA sequences polymorphic in populations; LTRs recently active; DNA transposons inactive | (7, 12, 13, 15, 19, 25, 31, 32, 47, 53, 55, 57, 63, 69, 112, 118, 134, 136) |
Abbreviations: LINE, long interspersed element; LTR, long terminal repeat; SINE, short interspersed element; TIR, terminal inverted repeat.
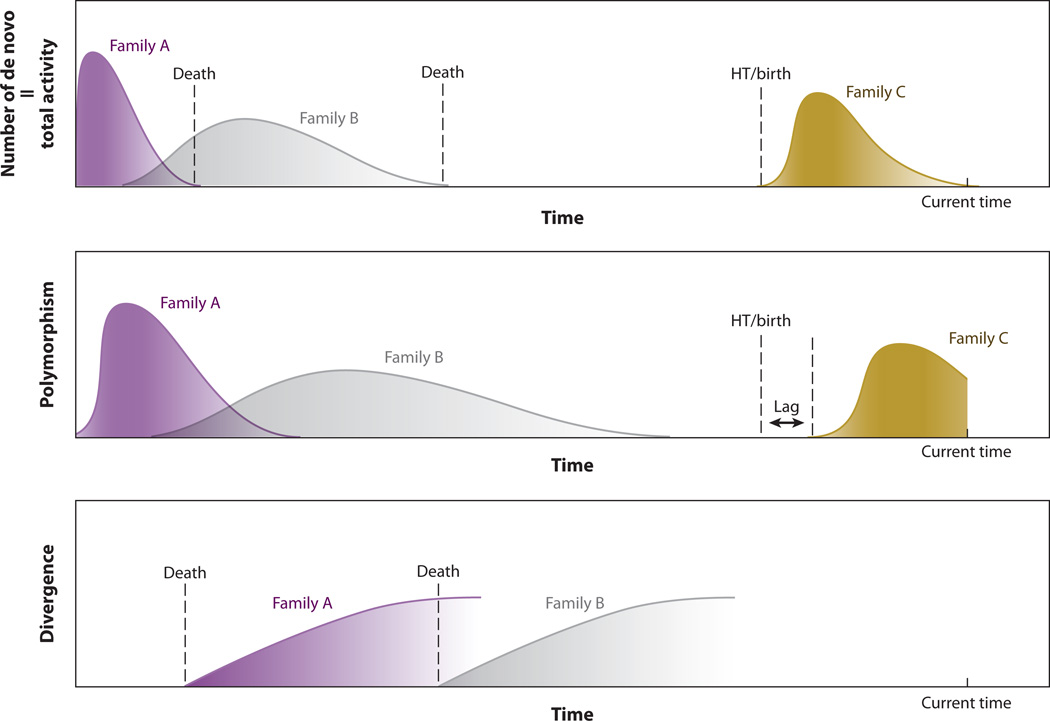
Another approach in detecting transposon activity is to identify polymorphic insertions between two individuals [transposon insertion polymorphisms (TIPs)] presumed to reflect insertions occurring after divergence from a common ancestor. For example, transposonintroduced restriction fragment length polymorphisms can differentiate natural isolates of Caenorhabditis elegans (49). The presence of polymorphic insertions between closely related species or individuals suggests that the element has been recently active and is most likely currently active (Figure 1).
Figure 1.
Transposon dynamics in genomes. Transposon activity in the genome varies throughout evolution of the host organism (see total activity track). Multiple families of the transposon can be active at the same time (overlapping peaks). Each family has a different activity tempo as shown by the different peak shapes. The birth of a transposon family can occur through horizontal transfer (HT), often seen with long terminal repeat (LTR) retrotransposons or DNA transposons, or with mutation of a preexisting family, e.g., by acquisition of a new promoter, which can occur in non-LTR retrotransposons. For simplicity, only HT is shown in this figure as an example. Transposon activity can be measured by various methods. Detecting the number of de novo insertions (which are found in offspring but not parents) in a population is the most direct way to measure the total activity. However, this method requires high-throughput screening of large sample sizes and is thus difficult to achieve. Polymorphic insertions between individuals reflect insertions that occur after diverging from a common ancestor. The number of polymorphic insertions in the population provides an indirect measurement of the activity. There is always a lag (middle panel), i.e., polymorphism, before new insertions can achieve an appreciable allele frequency and can be maintained/detected in the population. Polymorphism also remains in the population for some time after the death of the transposon family. Finally, transposon activity can also be inferred from sequence divergence among the elements within a family. At insertion, a transposon is assumed to be identical to a parent (template) copy, whereas older insertions will have passively accumulated more mutations through cycles of host replication. Therefore, if insertions with identical sequences are observed within a family, it is likely that the family remains active (bottom panel). Death of a transposon family occurs when the last remaining active copy of an element is lost through a mutational event(s) (top panel). Polymorphism of element copies will persist for some time following death of an element family, until either deleterious elements are eliminated by purifying selection and/or a population bottleneck and/or genetic drift eliminates variation. Transposon families are recognizable only for a certain period of time (the look-back time) after which sequence divergence becomes so extreme that the family is not recognizable by current sequence-analysis algorithms; this is indicated by the fading of the right boundary of the divergence curve (bottom panel).
Estimating Transposition Frequency
After establishing that an element is active, its activity can be quantified by measuring its transposition frequency. Numbers of polymorphic elements in combination with time from the common ancestor can be used to derive its transposition rate (86). Although this approach requires untested assumptions, it remains in use. This approach is problematic because both positive and negative changes in transposon number can occur by a multitude of mechanisms, for example, duplicative homologous recombination, which is entirely distinct from transposition at the molecular level. In addition, transposon copy number is not necessarily monotonically increasing, but may be in equilibrium with a number of processes that decrease transposon copy number, such as deletion or interelement recombination (123). Finally, it is important to recognize that transposition frequency is likely to vary over evolutionary time. Thus, the rate estimates made reflect average rates since divergence, and they may not reflect recent changes in activity.
Transposition assays using cell lines have become a gold standard for proving and quantifying current retrotransposon activity (11, 87). In these, a reporter cassette is added to a recombinant transposable element construct; this cassette is in antisense orientation with respect to the transposon and is interrupted by a sense-oriented intron (11, 87). The intron is spliced out of the transposon transcript, and the presence of daughter integrants is detected by expression of the now uninterrupted reporter from genomic DNA. Although invaluable for mechanistic studies, such assays do not necessarily reflect conditions affecting transposition in plants and animals, as the donor element copies are often plasmid borne and moreover are usually driven by heterologous promoters.
Inferring the Age of Transposable Elements
To refine when each transposition event occurred, one can use internal sequence consistency as a proxy for the age of a transposon insertion (12, 60, 74). The age of the youngest element in each family allows one to infer whether a family is still active, and, if it is not, to detect when the family lost its activity (Figure 1). At insertion, a transposon is assumed to be identical to a parent (template) copy, whereas older insertions will have accumulated a succession of mutations. Assuming a neutral model in terms of host selection for such intratransposon changes, one can calculate the age of an insertion by quantifying its divergence from a consensus sequence. For LTR retrotransposons, sequence divergence within individual element copies between the two LTRs has been proposed as an internal indicator of age, as these are usually identical to one another upon insertion (65, 109). In some organisms, one can also qualitatively corroborate relative ages of transposons by studying nested insertions, which are transposons inserted into preexisting transposons (20, 41).
Methods for Transposon Discovery
As molecular methods have advanced, tailored approaches for comparing highly repetitive genomic sequences have been developed. Initially, using a probe against elements of interest, researchers looked for differences in banding patterns across samples obtained by Southern blot (49). Transposon display similarly distinguishes between insertions on the basis of restriction enzyme cut-sites in the surrounding sequence. In this technique, digested genomic DNA is ligated with adaptors and used to template hemispecific polymerase chain reactions (PCRs) with a transposon terminal sequence primer and an adaptor sequence primer. When followed by electrophoresis, this method allows new insertions to appear as extra bands (120). Although both methods work well for analyzing activity of relatively low copy transposons or subsets of high copy-number repeats, they yield relatively low throughput when recovering specific insertion sites.
Modern genomic technologies have enabled comprehensive genomic screening, allowing easier identification of both de novo and polymorphic insertions. Features intrinsic to a transposon or its host organism lend each to specific methods of study, although some methods prove broadly applicable. For example, we recently applied a microarray-based method, originally established by us for mapping Ty1 retrotransposons in yeast, to L1(Ta) in humans (55, 130). In this method, genomic DNA is digested with multiple restriction enzymes, vectorette PCR is used to selectively amplify sequences adjacent to transposons of interest, and hybridization to genomic tiling microarrays is used to infer transposon locations. Others have developed similar L1(Ta)-based strategies that follow selective amplification with next generation sequencing (31, 57).
Because polymorphic transposons behave as indel structural variants, fosmid sequencing has also been used to study these elements. In this approach, genomic fragments of controlled length are inserted into fosmids, and the ends of the inserts are sequenced. Any deviation in length from the controlled size when the two sequenced ends are mapped back to the reference genome suggests an indel. Selecting for indels of the correct length enriches recovery of polymorphic insertions (7). Finally, transposon capture, a method that selects for terminal transposon sequences on custom microarrays prior to deep sequencing has revealed heterogeneity among human samples (4).
Meanwhile, recognition of sequences homologous to active elements or comparative studies across species have contributed to the rapid acceleration of in silico discovery of transposons both in sequencing traces and genome assemblies (69, 133, 136). In fact, analysis of existing sequences in repositories was one of the main approaches used to study human transposons before genome-wide transposon mapping techniques became available. Although draft genome assemblies are not complete, particularly regarding their incorporation of polymorphic repetitive elements (26), expanded efforts, such as the 1,000 Genomes Project in humans to capture intraspecies sequence differences (28), have produced prolific data on transposon variations (32, 115).
DISTRIBUTION OF RECENT TRANSPOSON ACTIVITY
Long Terminal Repeat Retrotransposons
Despite the presence of LTR retrotransposons in all eukaryotic genomes, their recent activity seems highest in plants. This is reflected in part by copy numbers seen in cross-species comparisons (Figure 2). In plants, although genome size may vary greatly as a consequence of polyploidy, different taxonomic groups often preserve a similar gene number, suggesting that other factors also regulate genome expansion. Retrotransposon numbers are highly correlated with plant genome size (Figure 2) (Arabidopsis thaliana versus maize) and compose the largest single component of most plant genomes (70, 111). Interestingly, many LTR retrotransposon insertions in plants have occurred within the past few million years. For example, in A. thaliana, more than 90% of copia superfamily LTR retrotransposon insertions are younger than the split between A. thaliana and its closest relative, Arabidopsis lyrata, 5.1–5.4 Mya (97). All retrotransposons in maize with intact LTRs were inserted within the past 4 Mya (110). Comparison of duplicated loci in plants confirms that recent bursts of retrotransposon activity have contributed significantly to genome size.
Figure 2.
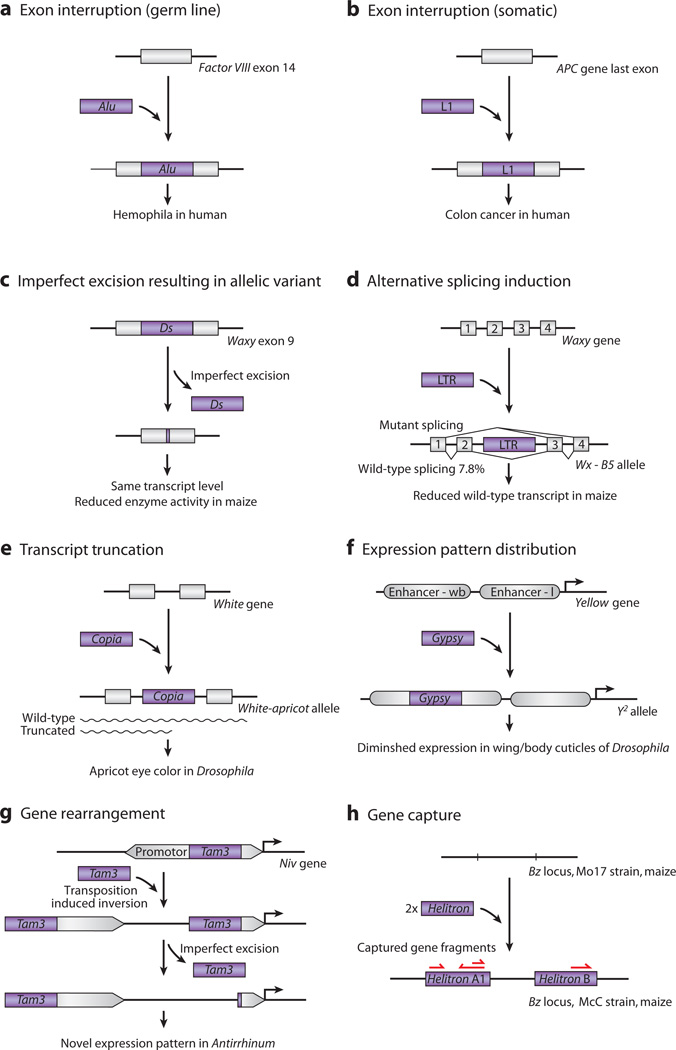
Functional impact of de novo insertions. (a) Exon interruption (germ line). A heritable human Alu SINE retrotransposon insertion interrupting exon 14 of human factor VIII gene causes hemophilia (117). (b) Exon interruption (somatic). A somatic insertion of a human L1 retrotransposon leads to loss of function of the APC gene, thereby promoting colon cancer development (85). (c) Long terminal repeat (LTR) recombination resulting in allelic variant. Recombination between the two LTRs of a Gret retrotransposon at the VvmybA1 locus rescued its gene expression. This recombined allele, VvmybA1b, reconstituted the expression of skin pigment in red grapes (68). (d) Alternative splicing induction. Insertion of an LTR retrotransposon in the waxy locus leads to exon skipping and kernel color changes in maize due to reduced transcript level (122). (e) Transcript truncation. A copia LTR retrotransposon insertion in Drosophila causes a hypomorphic white allele (white-apricot, w[a]). The intronic insertion causes truncated and nonfunction transcripts, with some read-through transcripts remaining functional; the result is the apricot phenotype (78). (f) Gene silencing. In the Melon genome, a hAT family DNA transposon inserted into the second intron of CmWIP1 gene leads to the spread of DNA methylation to the promoter region and subsequent gene silencing. Organisms with this insertion develop female flowers because of repressed CmWIP1 gene expression (82). (g) Gene rearrangement. A Tam3 DNA transposon–related inversion at the niv locus in Antirrhinum. The inversion results from DNA breaks on opposite ends of replicated copies of Tam3 rather than on opposite ends of a single copy. Their recombination with the upstream sequence leads to an inversion with altered niv promoter sequences and reduced expression. Excision of the proximal Tam3 causes another allele with increased and novel patterning of anthocyanin pigment (21). (h) Gene capture. Helitron DNA transposons inserted at the bronze (bz) locus in maize have also captured several neighboring genes, leading to their duplication and significant noncolinearity at this locus in different strains. Functional consequences have not been shown (124).
Purifying selection and nonrandom distribution of LTR retrotransposon are observed in A. thaliana, prompting Vitte & Panaud (123) to propose that its compact genome selects against insertions into euchromatin (97). There are seemingly far fewer constraints in genomes whose organization is relatively dispersed. Emerging models suggest varying balances are struck between transposon activity and intra/interelement recombination-mediated deletions to determine genome size in plants (e.g., A. thaliana versus maize) (123).
Intraspecies genome comparison has also provided insight into plant transposon activities. The 100-kb bronze (bz) locus from eight inbred maize lines showed extensive lack of colinearity (124). Intergenic regions were not alignable because of their dramatically different TIPs. These include exonic LTR retrotransposons, which suggests functional impact. The first genome-wide identification of TIPs in plant was performed in rice (Oryza sativa) (56). Sequence comparison of two rice subspecies, indica and japonica, revealed more than 50% of indels larger than 100 bp are due to TIPs, the majority being LTR retrotransposons. Plant retrotransposons predominantly accumulate in centromeres and intergenic regions; however, more than 10% of rice TIPs are within genes, and a subset of these are exonic. Many TIPs have been shown to affect host gene structure and expression. Spontaneous mutations due to retrotransposon insertions are frequently found in plants (90, 122). Collectively, these data indicate plant LTR retrotransposons are an ongoing source of genetic variation and important drivers of intraspecies diversity.
LTR retrotransposons account for only 0.4% of the C. elegans genome (37). However, they remain active in the organism: Full-length elements have been found, and retrotransposon activity has been documented. In a study of molecular mechanisms underlying C. elegans strain differences in copulatory plug formation, Palopoli et al. (96) discovered a retrotransposon insertion interrupting an exon of the plg-1 gene.
In Drosophila, transposon insertions are estimated to account for half of all spontaneous phenotype-inducing mutations (35). This has been verified using mutation accumulation (MA) experiments whereby inbred lines are allowed to accumulate spontaneous insertions over almost 100 generations (54). By comparing the latest generations to cryopreserved controls, insertion-dependent impacts on fitness can be addressed. Coupled with nesting analyses indicating nearly all LTR retrotransposons in the fly are fewer than 100,000 years old (8), genome-wide insertions rates are estimated for each element copy at one in 103 to 106 generations (27, 94).
Although horizontal transmission between individuals is the exception, not the rule, for retrotransposons (23), gypsy elements are infectious in Drosophila (66, 114). Gypsy is a superfamily of LTR retrotransposons approximately 7.5kb in length that is widely distributed among animals, fungi, protista, and plants. Its activity can be transferred among Drosophila strains by microinjection of egg plasma into embryos or by exposing larvae to isolated viral particles. Thus, interspecies transfer is highly likely (6, 80). The majority of LTR retrotransposons in vertebrates belong to the endogenous retroviral (ERV) superfamily. These make up approximately 10% of the genome in mouse, rat, and human; however, activity levels differ dramatically between rodents and humans (40, 142). LTR retrotransposons result in 15% of all spontaneous germ-line mutations in murine lab strains (127). Most mutagenic insertions in mouse are from the intracisternal A particle (IAP) and MusD/ETn elements. IAP is a family of LTR retrotransposon approximately 6.5kb in length, whereas MusD/ETn are two families of retrotransposons with almost identical LTRs. The protein coding region of MusD supports the transposition of nonautonomous ETn. Sequence analysis of three inbred strains revealed extensive polymorphisms in these elements; 60% of IAP and 25% of MusD/ETn copies identified are absent from at least one of the three strains tested (142). Approximately one-fifth (700 copies) of these element copies reside in introns; some disrupt splicing and lower expression levels of host genes.
In contrast, reports of polymorphic LTR retrotransposons in the human genome remain scarce (118), and active source elements are unknown. Of the human ERVs (HERVs), the youngest element in humans is HERV-K113, which is estimated to have inserted approximately 200,000 years ago (118). There are no reports of disease-causing mutations of HERVs. Nevertheless, successful reconstruction of a functional retrovirus from the HERV-K consensus sequence is possible, suggesting the human genome contains sequences sufficient to generate an active element (25).
Non–Long Terminal Repeat Retrotransposons
In comparison with LTR retrotransposons, non-LTR retrotransposon activities have a patchier species distribution. Studies have suggested activity in most plants and animals. Nevertheless, they are clearly absent from a few organisms. Birth and death rates of transposable element families are quite high and the selective pressure to “transpose or die” is significant. This represents a constant battle between the hosts and the different transposon families. The outcome is usually similar among species from closely related taxonomic clades, with the exception of bat. Bats have drawn special attention due to their completely opposite transposon activity pattern compared to neighboring species in the phylogenetic tree. This is illustrated here and in subsequent sections. Factors that determine the fate of each transposon family in different hosts are exciting areas of future research. Non-LTR retrotransposon activity is minimal in plants. Comparison of subspecies pairs either in rice or Arabidopsis showed that recent transposition frequencies are two to three orders of magnitudes lower than for other transposons, including LTR retrotransposons (56, 143). In addition, sequence-divergence estimates indicate the median age of non-LTR retrotransposons in Drosophila melanogaster is 782,400 years (8). This dates more than 80% of present-day insertions subsequent to divergence from Drosophila simulans. In flies, MA experiments can be used to assess not only transposon activity over generations, but also the fitness impact of these new insertions. Consistent with estimations from sequence-divergence studies, in vivo transposition rates of the youngest non-LTR superfamilies, Doc and I, as measured in MA experiments are between 4.2×10−5 and 8.0×10−4 insertions per element per generation (94).
LINE-1 (L1) is the most common superfamily of autonomous retrotransposons in mammals. They are highly active, except in megabats and sigmodontine rodents (16, 45) in which L1s appear to have gone extinct. Degenerate PCR isolated only inactive L1 sequences from 18 South American Sigmodontinae species and 11 megabat species. All elements lacked intact open reading frames or accumulated mutations at conserved amino acid positions. In contrast to the singular successive waves of L1 activity seen in many mammals, phylogenetic analysis of L1 revealed two distinct lineages were active in megabats before a simultaneous extinction. L1 of both taxonomies showed similar average sequence divergence (0.089 and 0.088 per site) from their ancestor with very low variance (0.00040, 0.00050). Determining what kind of change in host defense may have prompted this and the consequences of L1 extinction for megabats and sigmodontine rodents are interesting areas for further study.
Mice and rats, by contrast, show extensive L1 activity despite their relationship to sigmodontine rodents (2). By comparing whole-genome shotgun traces from four different inbred mouse strains against the reference mouse genome, Akagi et al. (2) discovered that 85% of intermediate-sized structural variants of 100 bp to 10 kb resulted from recent retrotransposition events, mostly of the L1 superfamily. The authors estimated that up to 5% of the 666,328 reference L1s are polymorphic, that is, absent from at least one of the four inbred stains. Genomic distribution in mouse is not random; antisense L1s are especially underrepresented within genes, suggesting that they may impose deleterious effects on host gene function and be selected against. Congruent with this, examination of a subset of full-length, intronic antisense polymorphic L1 revealed that 19% produced fusion transcripts. In vitro retrotransposition assays and sequence analyses together suggest that the mouse genome contains approximately 3,000 active L1 elements (44). Although similar studies remain to be done in rats, on the basis of the reference genome alone, L1 appears to have higher activity in this lineage. In contrast to mouse-specific insertions, which account for 10% of that genome, 150,000 copies of elements from six rat-specific L1 subfamilies compose approximately 12% of the rat genome (40, 67).
Given the presence and activity of LINEs, it is not surprising to find L1-dependent SINE activity in mouse and rat. More than 300,000 SINE elements have inserted into both genomes since they diverged (40). SINE polymorphisms are also observed among different inbred mouse strains (2). The same four families of SINE elements exist in the two species; however, the relative activity of each is distinct. This could reflect stochastic effects of where “source” or parental elements landed in each genome or could result from differences in host defense mechanisms. There is biased insertion of SINEs into gene-rich regions, and analysis of orthologous sites revealed accumulation of species-specific SINEs is highly correlated between mouse and rat (40). Thus, SINEs have either displayed a common site-specific insertion preference (distinct from L1 because L1 is depleted from gene regions) or have responded to similar selection forces. The latter seems more likely given that younger SINEs are much more similar to L1s in insertion distribution than older ones.
Four recent genome-wide studies have reinforced that L1s are highly polymorphic in humans (7, 31, 55, 57) (Figure 2). These are pre-Ta and Ta L1 subfamilies, shown in several clinical case reports to cause phenotypes by insertion mutations (15, 19). More than 100 active L1s were estimated to be present in a diploid human genome (13). Using TIP-chip, we demonstrated that there are many insertions missing from existing whole-genome sequence assemblies and that even those assemblies constructed using long Sanger sequencing reads incompletely capture these high copy-number repeats. This is consistent with the observation that polymorphic insertions are underrepresented in the primary reference human genome build and tailored techniques are required to fully capture the polymorphic insertions for ascertainment. In agreement with Ewing et al. (31), who performed deep sequencing of libraries enriched for 3′ L1–containing fragments, we estimated more than one-third of human-specific L1s differ between two unrelated individuals. Ewing et al. (31) extrapolated to predict that 3,000–10,000 novel L1s of allele frequency >5% will be identified in human populations. Using a different approach that uniquely lent itself to recovery of full-length L1s, Beck et al. (7) isolated 68 6-kb L1s from the fosmid libraries of six individuals and found the majority were active in transposition assays. In contrast to mouse genes, antisense enrichment of L1s is observed in human genes (55). Mouse and human L1 elements are divergent in their sequences, especially in the 5′ untranslated and open reading frame 1 regions; therefore, different selection pressures or insertional biases are expected. Our group and the Kazazian group independently estimated nonparental L1 retrotransposition events in the germ line at between ~1 in 95 and 1 in 270 births (31, 55).
Alu is a family of nonautonomous non-LTR retrotransposons that is approximately 300 bp in length and has undergone massive expansion in the primate lineage. AluY is the youngest Alu family in humans, followed by AluS. Although evidences from disease reports and early polymorphism studies have indicated that both AluS and AluY are active in humans, AluY contains most of the activity. All subfamilies of AluY are active in humans (53, 134). Analysis of eight genomes generated by paired-end deep sequencing have revealed that 89% of the new Alu insertions, as compared with the reference genome, belong to the AluY family, and almost half of them are only present in one individual, suggesting a high degree of polymorphism (53). It is estimated that approximately 1 de novo Alu insertion arises in every 21 births (136).
Finally, SVA is another active retrotransposon in human and is represented by disease-causing insertions. At ~1200 bp, SVA’s length falls between that of LINEs and SINEs, but as a nonautonomous transposon, its transposition cycle is more like a SINE. It is a composite transposable element found in primates and named for its component SINE-R, variable number tandem repeat (VNTR), and Alu portions. SVA is active in cell-culture assays. Moreover, 40% of the 2,700 reference genome copies are estimated to be polymorphic (47, 48, 103).
Cut and Paste DNA Transposons
DNA transposons are active in plants and lower-order animals. Activity is absent in mammals, except in bats. By contrast, the ratio of DNA transposon- and retrotransposon-induced TIPs is very different between plant species. Comparison of contigs of two A. thaliana (a dicotyledonous plant) lines for TIPs revealed 40% retrotransposons and 59.6% DNA transposons, including 19.9% MULE superfamily members and 26.5% Helitrons, which are discussed below (143). In contrast, retrotransposon activity in rice and other monocotyledonous plants is higher. Between rice strains, TIPs account for 14% of sequence differences; 80% are retrotransposons, whereas only 15%–20% are DNA transposons (56). The two most active DNA transposon superfamilies in rice are CACTA (5%–10%) and MULE (6%–7%). Members of almost every superfamily of DNA transposon have been identified in analysis of TIPs in the bz locus between strains of maize, suggesting that they are active (124).
A high-copy DNA transposon with only TIRs, but no coding potential, was first described in plants (14). These are usually several hundred base pairs in length and are therefore named MITEs. MITEs were later found in other eukaryotic genomes, including insects, fish, and humans (23). Through homology searches, some MITEs are recognizable as deletion derivatives of autonomous DNA transposons. However, the origins of many are unknown. Using transposition assays to test for cross mobilization, Yang et al. (138) identified the related autonomous element for Stowaway, a family of MITE in rice. With mutagenesis, they demonstrated that the autonomous Marinerlike element Osmars harbors a repression motif missing from the Stowaway-like MITEs that it mobilizes. This explains the relative success of MITEs and supports amodel of their evolution from their ancestral DNA transposons.
mPing was the first active MITE to be identified in any organism (58). Its activity is best characterized in rice. Although mPing is apparently stable in some strains, having only ~50 copies in the genome, dozens of new insertions are found in each generation of the high-copy strain, accumulating to >1,000 copies (139). As suggested by de novo events, mPing can insert into both exons and introns. However, these insertions are selected against. Of all new insertions, 76% are within 5 kb of a gene, whereas only 22% of old insertions are found in those regions (89). The observed selection implies that these new insertions have a significant functional negative impact on host fitness.
Most transpositions observed in C. elegans reflect DNA transposon activity. These were primarily discovered by recognizing polymorphisms among different stains or identifying de novo insertions in spontaneous mutants (reviewed in 9). The P element is perhaps the best-characterized active DNA transposon in D. melanogaster. It is known to create phenotypes, and therefore is frequently used in mutagenesis screens (10). The element is a recent invader of wild D. melanogaster populations through horizontal transfer (30, 80). In a systematic survey of the D. melanogaster genome, 40% of the potentially active DNA transposon families had lower than expected divergence among different Drosophila species, consistent with horizontal transfer. Surveys of transposable elements in natural populations using PCR or in situ hybridization of giant salivary-gland polytene chromosomes suggest that more than two-thirds of the DNA transposon sites are polymorphic (77).
A high level of contemporary DNA transposon activity has also been observed in the bat Myotis lucifugus, despite the apparent extinction of these elements in virtually all other mammals studied (105, 106). Sequence divergence indicates nine different families were active in bats over the past 40 million years (95, 105, 106). A large fraction of copies of the youngest family are 100% identical, indicating ongoing transposition. What is special about bat that allows DNA transposons to remain active will be an interesting topic for investigation. It is possible that teleologic benefits promote DNA transposon activity in bats or that evolutionary pressures existing to extinguish these mobile DNAs in other species are less relevant in bats.
Helitrons
Helitrons were discovered in C. elegans, rice, and Arabidopsis through in silico genome analysis (61); they are studied extensively in maize. The first examples of de novo Helitron insertions were isolated from two maize mutants (46, 73). Helitrons contribute substantially to a lack of colinearity in locus alignments across maize inbred lines (72). Gene annotation differences at the bz locus between two inbred maize lines reflect fragments captured/transduced by two Helitron transposons (124) (Figure 3h). In a high-throughput study, nonshared genes between two inbred maize lines were isolated by hybridizing exon probes to contigs. Of the nine polymorphic, gene-containing insertions examined, eight resulted from helitron transposition (88). This suggests Helitrons could be a principal driver of changing gene content in plants and important for the evolution of new proteins through exon shuffling; however, evidence for a high frequency of gene capture is currently limited, and functional genes derived from this process have not been found. Analysis of the A. thaliana genome revealed that although 39% of Helitrons within the Basho family, a nonautonomous Helitron, contain gene fragments, these are derived from just five independent gene capture events (50). Finally, brown bats are notable among mammals for their recent Helitron activity. Helitrons have amplified during the past 30–36 million years to 3% of the M. lucifugus genome (102). Although indirect lines of evidence suggest their activity, isolation of an active, autonomous Helitron remains to be demonstrated.
Figure 3.
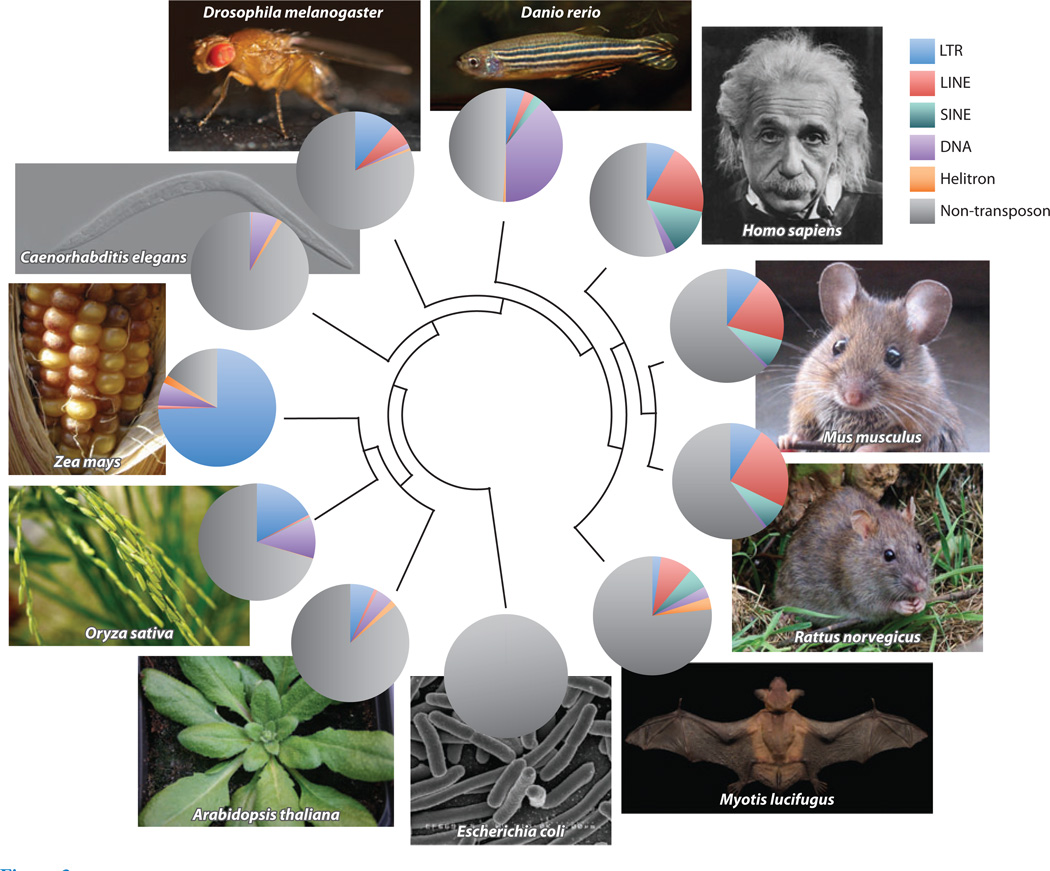
Transposon compositions in different species. In clockwise order, species shown are Homo sapiens, Mus musculus, Rattus norvegicus, Myotis lucifugus, Escherichia coli, Arabidopsis thaliana, Oryza sativa, Zea mays, Caenorhabditis elegans, Drosophila melanogaster, and Danio rerio. The phylogenetic tree in the center describes the evolutionary relationships among them. The pie charts illustrate the fraction of the genome accounted for by different transposon classes (113). Although the bat Myotis lucifugus is a mammal, its transposon composition is distinct, even among bats. Data on non-Myotis bats provided by D.A. Ray & H. Pagán, personal communication (105, 106).
SOMATIC TRANSPOSITION EVENTS
The first evidence of somatic transposon activity, in which insertions are present in only a subset of cells within an individual organism, came from maize with variable kernel colors (128). In plants that use vegetative reproduction, somatic transposon activities are especially important, as these insertions can be inherited and subsequently expanded in the population. Recently, a somatic DNA transposon insertion in the grapevine cultivar Carignan has been shown to cause a delayed flowering and altered fruit-clustering phenotype (33). Sequence analysis demonstrated this somatic insertion was the sole polymorphism in the 4-kb proximal interval of VvTFL1A gene that segregated with the phenotype. Inserted in the promoter region, the transposable element behaves as a cis-acting enhancer and upregulates VvTFL1A gene expression (33). Whether somatic transposon activity is a universal phenomenon in plants and how these activities may be selected for as a mechanism for generating genetic diversity at a population level will be interesting questions to address in the future.
In C. elegans, certain strains have defense mechanisms that prevent transposon activity in the germ line while allowing somatic insertions to occur (29). By contrast, hybrid dysgenesis provides early evidence suggesting that activities of Drosophila P-element and I factor, an L1-like non-LTR retrotransposon, are restricted to the germ line (17, 75). Recently, however, the Eickbush laboratory has demonstrated somatic activity of R2, an autonomous non-LTR retrotransposon, in Drosophila simulans (141). In one study, they characterized de novo R2 insertions in the offspring of a single female fly and identified the same insertion in multiple flies, consistent with germ-line mosaicism created by a somatic transposition event (141). In a subsequent study, using a PCR assay to amplify transposon junctions across multiple tissues of individual flies or larvae, they revealed insertions that were restricted to certain tissues (141). These insertions were often identifiable in multiple tissue types, thus suggesting that the somatic integration occurred early in fly development.
Examples of somatic transposon activities in mammals are also accumulating. Initial clues came from two phenotype-causing insertions. An early report documented an L1 insertion disrupting an exon of the APC gene (Figure 3b) as present only in a colon cancer tumor sample and not constitutional in the patient (85). An X-linked choroideraemia male patient was also found to have inherited a disease-causing L1 insertion that was later found to be mosaic in somatic cells of his mother (121). Using transgenic mouse models with either mouse or human L1 carrying the endogenous promoter, Kano et al. (59) showed that L1 RNA from these transgenic animals can be carried over to embryos that do not inherit the transgene but show tissue mosaicism for new insertions. Interestingly, although human embryonic stem cells support retrotransposition from an engineered L1 construct, reactivation of retrotransposon transcription was also demonstrated in induced pluripotent cells (38, 132).
Recently, Iskow et al. (57) isolated a number of tumor-specific L1 insertions in lung cancers, suggesting somatic instability may play roles in oncogenesis. By contrast, the Gauge laboratory has demonstrated both in vivo and in vitro that de novo insertions from an engineered human L1 element occur in the mouse brain (112), suggesting that somatic L1 insertions may also play a role in generating neuronal diversity. Faulkner’s group has provided evidence for the presence of L1 somatic activities in distinct regions of individual human brains by selectively using transposon capture prior to deep sequencing (4). They further demonstrated new integrations of both Alu and SVA in brain cells. These studies suggest that somatic transposon activity in animals may be more common than previously anticipated, although it is important to emphasize that the number of individual retrotransposition events observed in this study and the sequencing reads that support them are both quite small. The seemingly widespread nature of somatic transposon activity in animals is intriguing, as most of these somatic insertions have no potential to enter the germ line, making one wonder if these activities represent a functional impact on brain behavior or whether it is a low-level background of somatic hopping that is of little or no consequence.
SPECIES DISTRIBUTION OF TRANSPOSABLE ELEMENTS
Transposon activities are extremely variable across different taxa. Variables affecting transposon copy number in genomes have long been areas of interest. Genome-wide transposon profiles of closely related species with dramatically different transposon distribution and activities have provided an excellent opportunity to further our understanding in these dynamics. In Helianthus (sunflower) species, H. anomalus, H. deserticola, and H. paradoxus are products of an ancient hybridization event between H. annuus and H. petiolaris. In contrast to their parental origins, these hybrid taxa occupy abiotically extreme habitats. Interestingly, their Ty3/gypsy-like superfamily of LTR retrotransposon has amplified from 5.6- to 23.6-fold in copy number during hybridization, resulting in a genome size that is at least 50% larger than their parental species (119). These observations agree with McClintock’s (84) initial hypothesis in which transposon activations are a kind of “response to genomic shock” that helps generate genetic diversity.
Another elegant observation came from studies of two closely related Arabidopsis species, A. thaliana and A. lyrata. The former is relatively devoid of transposable elements among most flowering plants and is primarily a self-pollinator, whereas the latter has three times more transposon insertions in its genomes and is largely an outcrosser. This suggests that the dominating mechanism in controlling transposon copy number should differ between the two species. Genome-wide correlation studies of A. thaliana have revealed that transposon copies targeted by small interfering RNA (siRNA) for methylation are associated with reduced neighboring gene expression (51). In contrast to unmethylated transposon copies near genes, these silenced insertions are under purifying selection. Although epigenetic silencing of transposon insertions reduces their activities, these elements are further selected against because of their deleterious effects on nearby gene expression. By comparing transposable element targeting siRNA in A. thaliana and A. lyrata, researchers demonstrated that a higher fraction of transposable elements in A. thaliana are targeted by uniquely mapped siRNAs, a proxy for more efficient silencing of transposon expression (52). By contrast, gene expressions in A. lyrata are less affected by neighboring methylated transposon insertions (52); therefore, these insertions should be under less selection pressure. These studies suggest that the efficiency of siRNA silencing and robustness of gene expression in genomes may shape species-specific transposon activity patterns.
Active transposable element types in mammals [with the exclusion of vesper bats, a taxonomic family (Order, Chiroptera; Family, Vespertilionidae)] of three hundred bat species, also known as evening or common bats) are peculiar relative to other eukaryotic clades. All nonvesper bat mammalian genomes, including the primitive monotreme platypus genome, are typified by a single dominant lineage of LINE (and corresponding nonautonomous SINEs) (36, 126). In contrast, in invertebrates, fish, and amphibians, active retrotransposons as well as DNA transposons are found, and among the retrotransposons there are often multiple active transposable element families in parallel (36).
What could explain this difference? Thus far, no unifying explanation has been forwarded, so we have considered covariates. One biological feature that may shape susceptibility to transposable elements is internal fertilization (A. Smit, personal communication). Internal fertilization may minimize exposure of mammalian gametes (and embryos) to horizontal transfer of DNA transposons and/or viral precursors of retrotransposons. In the absence of a continual introduction of new elements at this stage, it is possible that these transposable elements cannot effectively establish an endogenous presence in the population. Although an attractive hypothesis, the life cycles of some animals suggest fertilization may not be the principal determinant. Some reptiles and vesper bats are internally fertilized and also show a more fish/amphibian-like pattern of transposable element activity.
We speculate that body temperature may also be an important factor, and reptiles as well as hibernating mammals that sustain significant body-temperature decrements are relatively susceptible to DNA transposon activity and permissive of more retrotransposon lineages. A few experimental systems have indicated temperature effects directly on transposition (76), although relationships between body temperature and genome-level features (size, CG content, etc.) are complex. Tests of these hypotheses and critical reevaluation will be required as other genomes are published.
FUNCTIONAL IMPACT OF RECENT TRANSPOSON INSERTIONS
Examples of the functional impact of recent transposon insertions are particularly prevalent in plants, not surprising given that transposons are a dominant component of most plant genomes. Recently, it has been shown that a number of phenotypes selected for during domestication were caused by transposon insertions. Examples include an insertion disrupting the homozygous pigment-production pathway in white grapes (68) (Figure 3c) and a gene-duplication event mediated by a retrotransposon that resulted in an elongated fruit shape in tomato (135). Modern domesticated maize has substantially fewer branches compared with teosinte, three closely related subspecies of maize (Zea mays) of whom they share common ancestor with. Previous association studies have placed the causal variant of this phenotype at ~65kb upstream of the tb1 gene, a repressor of branch formation (116). Two polymorphic transposon insertions were later shown to lie within this region. The allele with both insertions is present in >95% of domesticated maize populations but only in <5% of the teosinte. By testing different DNA fragments from this allele for their ability to drive expression in luciferase assays, Studer et al. (116) isolated one transposon insertion as the functional element involved in upregulation of tb1 gene expression. As illustrated by this example, phenotype-causing insertions are often in complete linkage disequilibrium with other polymorphisms. Thus, association signals between variants and phenotypes should always be interpreted with caution. Further functional studies are required to isolate the causative variant.
González et al. (42) performed interesting studies on the impact of recent transposon insertions on Drosophila adaptation. D. melanogaster was challenged by environmental change when it expanded from sub-Saharan Africa to a worldwide distribution 10,000–16,000 years ago. Identifying young insertions and assaying allelic frequencies in different regions of the world, the authors identified 13 potentially adaptive insertions, including both DNA transposons and retrotransposons. A partial selective sweep localized to insertion flanking sequences suggested the insertions were the critical factor. Follow-up studies revealed a majority of these insertions resulted in regulatory changes in nearby genes (43). The authors then extrapolated these findings to estimate that one transposable element–induced adaptation occurs every 200–1,250 years. These studies argue that modern transposon activities are functionally relevant in generating intraspecies phenotypic diversity.
As indicated above (see section “Species Distribution of Transposable Elements”), transposon insertion silencing by DNA methylation can affect nearby gene expression. Interestingly, this side effect of silencing the transposable element on genes has been co-opted in melon for sex determination (82). In melon, expression of CmWIPI results in unisexual male flower development, whereas repression of CmWIPI leads to female flower development. An allele of CmWIPI with a DNA transposon inserted within the second intron is not expressed because methylation spreads to its promoter region (Figure 3f). Recombinant plants that lost the insertion reexpress the gene product, implicating the transposon insertion as the causative variant in initiating epigenetic silencing. With genome-wide transposon profiling technologies available, it is now possible to ask if transposon-induced heterochromatin spreading is also present in mammals. Rebollo et al. (107) compared two mouse embryonic stem cell lines with transposon dimorphisms at a number of loci for disparate heterochromatin histone markers in their flanking genomic regions. They demonstrated the presence of a heterochromatin spreading effect that varied across different transposon families, with IAP elements being the strongest, followed by ETn/MusD transposons. LINE elements, by contrast, did not induce heterochromatin formation. In rare instances, these IAP insertions are close to coding regions and result in gene transcriptional silencing (107). These studies begin to reveal diverse functional consequences of recent transposon insertions, which in aggregate extend far beyond the disruption of open reading frames. As host fitness is affected, these elements could face selective pressures and their presence or absence could become integral to the species.
CONCLUSION
Genomes are continuously evolving, and transposons have been central sources of genome variation throughout taxa, creating inter/intraspecies diversity and enhanced potential for adaptation to changing environments. Mobile DNAs are still active today in virtually every organism examined. They continue to play major roles in generating genome diversity and imposing significant intragenomic selection pressures through their effects on gene expression. Insertion polymorphisms within species and interspecies differences will help illuminate studies of population genetics, transposition mechanisms, and the functional impact of insertions.
Two critical properties of transposon insertions make them ideal markers in population genetics studies. First, they are homoplasy free, meaning that an insertion event is uniquely identified by its location and associated sequence (e.g., length of the insertion and target-site duplication). Regardless of the individual, the allele can be safely assumed to derive from the same insertion event. Second, the ancestral allele is unambiguous (with the exception of DNA transposons that may be due to precise excision). These attributes allow exploitation of polymorphic insertions in studying population structure and understanding speciation.
As each transposon family shows variable activity in different organisms, complex relationships will characterize each unique organism and combination of transposable elements. As host interactions are explored, outlier taxa, such as bats, with transposon activity patterns distinct from other species in the same class may be instructive. Using synthetic or natural genomes (see sidebar, Organisms Without Transposable Elements), studies of organisms lacking transposons will also provide important insights into transposition mechanisms and host responses.
Finally, phenotypic traits with complex genetic components may be more likely to be caused by “soft” forms of genetic variations that alter gene expression (83), making TIPs attractive candidates. Transposon insertions can have a functional impact even when inserted outside of coding regions (Figure 3c–e,g). They often create hypomorphic alleles instead of completely abolishing gene function. The roles of these polymorphic insertions in gene regulation and complex traits will be of great interest in disease and quantitative-trait genetics.
ORGANISMS WITHOUT TRANSPOSABLE ELEMENTS.
Most eukaryotes studied to date have genomes with numerous examples of mobile DNA insertion. We know of one exception apparently lacking transposons: Ashbya gossypii, a filamentous fungus with a streamlined genome having only one copy of a Ty3-like open reading frame (39; F. Dietrich, personal communication), and they are rare in other pregenome duplication yeasts/fungi such as Kluveromuyces lactis (91). Could this be a lineage generally refractory to DNA invaders?
SUMMARY POINTS.
With rare exceptions, transposons are present in every genome studied so far (see sidebar).
Transposons are still active today. However, their activity level varies across different species.
Somatic transposon activities can be observed in various organisms, including both plants and animals.
Transposon insertions can affect gene expression and structure; therefore, they play important roles in evolution and often represent targets of selection, most commonly negative but occasionally positive.
FUTURE ISSUES.
What is the transposon distribution/activity in genomes that have not yet been sequenced?
Why are transposon distributions different across various genomes? What factors regulate transposon activities?
How common are somatic transposition events? In what cell types and developmental stages do they occur?
What is the full range of functional impacts of transposon insertions? What are their roles in generating phenotypic diversity?
Glossary
- Autonomous transposons
elements that encode one or more proteins required for movement
- Rolling circle
a replication process used by many eukaryotic viruses to multiply their circular genome; the replication intermediates are circular with a tail of newly made DNA
- Transposon insertion polymorphism (TIP)
the presence and absence of a transposon insertion at a certain location in the genome
- Neutral model (of selection)
changes in allele pools that are the result of random events instead of any advantageous or deleterious effect on the species
- Indel
insertion and deletion. This is a class of sequence variation that results in a net gain or loss of nucleotides at the position
- Purifying selection
also called negative selection. This refers to an evolutionary process that selectively removes deleterious alleles
- Euchromatin
a loosely packed form of chromatin that is gene rich and usually (but not always) associated with active transcription
- Source element
the parental transposon insertion that gave rise to the copy in the new location
- Megabat
a taxonomic family of bats (Order, Chiroptera; Family, Pteropodidae); also called fruit bats, old world fruit bats, or flying foxes
- Sigmodontine
a taxonomic subfamily that includes both rats and mice
- Degenerate PCR
PCR using a primer mixture instead of two specific primers. Useful when the exact sequence to be amplified is unknown
- Orthologous
homologous (similar) sequences in two different species; shared in an ancestor and separated by the speciation event
- Nonautonomous transposons
transposons dependent on an autonomous element to effect transposition; examples include SINEs and MITEs
- Vegetative reproduction
asexual reproduction occurring in some plants; new organisms grow from pieces of an existing plant without intermediate seeds or spores
- Germ-line mosaicism
having two or more genetically different pools of sperm or oocytes in an individual
- Methylation
a type of chemical modification that occurs on DNA, usually on the cytosine nucleotide. DNA methylation is associated with silencing of the region
- Monotreme
a taxonomic order of mammals that lay eggs for reproduction
- Adaptive insertions
insertions that increase the fitness and survival of a species
- Partial selective sweep
sequence signature resulting from a beneficial mutation that is nearly but not completely fixed in a population
- Heterochromatin
a tightly packed form of chromatin that results in gene silencing. This is in contrast to euchromatin
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Cheng Ran Lisa Huang, Email: chuang36@jhmi.edu.
Kathleen H. Burns, Email: kburns9@jhmi.edu.
Jef D. Boeke, Email: jboeke@jhmi.edu.
LITERATURE CITED
- 1.Adey NB, Schichman SA, Graham DK, Peterson SN, Edgell MH, Hutchison CA. Rodent L1 evolution has been driven by a single dominant lineage that has repeatedly acquired new transcriptional regulatory sequences. Mol. Biol. Evol. 1994;11:778–789. doi: 10.1093/oxfordjournals.molbev.a040158. [DOI] [PubMed] [Google Scholar]
- 2.Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J, et al. Whole-genome shotgun assembly and analysis of the genome of fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 4.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, et al. The selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartolomé C, Bello X, Maside X. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10:R22. doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergman CM, Bensasson D. Recent LTR retrotransposon insertion contrasts with waves of non-LTR insertion since speciation in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:11340–11345. doi: 10.1073/pnas.0702552104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessereau JL. Transposons in C. elegans. WormBook Online Review C.elegans Biol. 2006 doi: 10.1895/wormbook.1.70.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bingham PM, Kidwell MG, Rubin GM. The molecular basis of PM hybrid dysgenesis: The role of the P element, a P-strain-specific transposon family. Cell. 1982;29:995–1004. doi: 10.1016/0092-8674(82)90463-9. [DOI] [PubMed] [Google Scholar]
- 11.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Tyelements transpose through anRNAintermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 12.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 13.Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bureau TE, Wessler SR. Tourist: A large family of small inverted repeat elements frequently associated with maize genes. Plant Cell Online. 1992;4:1283–1294. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Callinan PA, Batzer MA. Retrotransposable elements and human disease. Genome Dis. 2006;1:104–115. doi: 10.1159/000092503. [DOI] [PubMed] [Google Scholar]
- 16.Cantrell MA, Scott LA, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics. 2008;178:393–404. doi: 10.1534/genetics.107.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaboissier M, Busseau I, Prosser J, Finnegan D, Bucheton A. Identification of a potential RNA intermediate for transposition of the LINE-like element I factor in Drosophila melanogaster. EMBO J. 1990;9:3557. doi: 10.1002/j.1460-2075.1990.tb07566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, et al. The dynamic genome of hydra. Nature. 2010;464:592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JM, Stenson PD, Cooper DN, Férec C. A systematic analysis of LINE-1 endonucleasedependent retrotranspositional events causing human genetic disease. Hum. Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 20.Churakov G, Grundmann N, Kuritzin A, Brosius J, Makałowski W, Schmitz J. A novel web-based TinT application and the chronology of the primate Alu retroposon activity. BMC Evol. Biol. 2010;10:376. doi: 10.1186/1471-2148-10-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coen ES, Carpenter R, Martin C. Transposable elements generate novel spatial patterns of gene expression in antirrhinum majus. Cell. 1986;47:285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- 22.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig N, Craigie R, Gellert M, Lambowitz A. Mobile DNA II. Washington, D.C.: American Society of Microbiology Press; 2002. [Google Scholar]
- 24.Deininger PL, Moran JV, Batzer MA, Kazazian HH. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, et al. Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolgin E. Human genomics: the genome finishers. Nature. 2009;462:843–845. doi: 10.1038/462843a. [DOI] [PubMed] [Google Scholar]
- 27.Domínguez A, Albornoz J. Rates ofmovement of transposable elements in Drosophila melanogaster. MGG. 1996;251:130–138. doi: 10.1007/BF02172910. [DOI] [PubMed] [Google Scholar]
- 28.Durbin RM, Altshuler DL, Abecasis GR, Bentley DR, Chakravarti A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmons SW, Yesner L. High-frequency excision of transposable element tc 1 in the nematode Caenorhabditis elegans is limited to somatic cells. Cell. 1984;36:599. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- 30.Engels WR. The origin of P elements in Drosophila melanogaster. Bioessays. 1992;14:681–686. doi: 10.1002/bies.950141007. [DOI] [PubMed] [Google Scholar]
- 31.Ewing AD, Kazazian HH. High-throughput sequencing reveals extensive variation in humanspecific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ewing AD, Kazazian HH. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2010;6:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez L, Torregrosa L, Segura V, Bouquet A, Martinez-Zapater JM. Transposon-induced gene activation as a mechanism generating cluster shape somatic variation in grapevine. Plant J. 2010;61:545–557. doi: 10.1111/j.1365-313X.2009.04090.x. [DOI] [PubMed] [Google Scholar]
- 34.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finnegan DJ. Transposable elements. Curr. Opin. Genet. Dev. 1992;2:861–867. doi: 10.1016/s0959-437x(05)80108-x. [DOI] [PubMed] [Google Scholar]
- 36.Furano AV, Duvernell DD, Boissinot S. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Ganko EW, Bhattacharjee V, Schliekelman P, McDonald JF. Evidence for the contribution of LTR retrotransposons to C. elegans gene evolution. Mol. Biol. Evol. 2003;20:1925–1931. doi: 10.1093/molbev/msg200. [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Perez JL, Marchetto MCN, Muotri AR, Coufal NG, Gage FH, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 39.Gattiker A, Rischatsch R, Demougin P, Voegeli S, Dietrich FS, et al. Ashbya genome database 3.0: a cross-species genome and transcriptome browser for yeast biologists. BMC Genomics. 2007;8:9. doi: 10.1186/1471-2164-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, et al. Genome sequence of the brown norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 41.Giordano J, Ge Y, Gelfand Y, Abrusán G, Benson G, Warburton PE. Evolutionary history of mammalian transposons determined by genome-wide defragmentation. PLoS Computat. Biol. 2007;3:e137. doi: 10.1371/journal.pcbi.0030137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 2008;6:e251. doi: 10.1371/journal.pbio.0060251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gonzalez J, Macpherson JM, Petrov DA. A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol. Biol. Evol. 2009;26:1949–1961. doi: 10.1093/molbev/msp107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodier JL, Ostertag EM, Du K, Kazazian HH. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grahn R, Rinehart T, Cantrell M, Wichman H. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 2005;110:407–415. doi: 10.1159/000084973. [DOI] [PubMed] [Google Scholar]
- 46.Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. A novel class of helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol. Biol. 2005;57:115–127. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- 47.Hancks DC, Kazazian HH., Jr SVA retrotransposons: evolution and genetic instability. Semin. Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition ofmarked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollister JD, Gaut BS. Population and evolutionary dynamics of helitron transposable elements in Arabidopsis thaliana. Mol. Biol. Evol. 2007;24:2515–2524. doi: 10.1093/molbev/msm197. [DOI] [PubMed] [Google Scholar]
- 51.Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19:1419–1428. doi: 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA. 2011;108:2322–2327. doi: 10.1073/pnas.1018222108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hormozdiari F, Alkan C, Ventura M, Hajirasouliha I, Malig M, et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2010;21:840–849. doi: 10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Houle D, Nuzhdin SV. Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 2004;83:7–18. doi: 10.1017/s0016672303006505. [DOI] [PubMed] [Google Scholar]
- 55.Huang CRL, Schneider AM, Lu Y, Niranjan T, Shen P, et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang X, Lu G, Zhao Q, Liu X, Han B. Genome-wide analysis of transposon insertion polymorphisms reveals intraspecific variation in cultivated rice. Plant Physiol. 2008;148:25–40. doi: 10.1104/pp.108.121491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, et al. An active DNA transposon family in rice. J. Comp. Physiol. A. 1986;159:801–811. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 59.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kapitonov V, Jurkal J. The age of Alu subfamilies. J. Mol. Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- 61.Kapitonov VV, Jurka J. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 2007;23:521–529. doi: 10.1016/j.tig.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 62.Kazazian HH. Mobile elements and disease. Curr. Opin. Genet. Dev. 1998;8:343–350. doi: 10.1016/s0959-437x(98)80092-0. [DOI] [PubMed] [Google Scholar]
- 63.Kazazian HH, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 64.Kidwell MG, Lisch DR. Transposable elements and host genome evolution. Trends Ecol. Evol. 2000;15:95–99. doi: 10.1016/s0169-5347(99)01817-0. [DOI] [PubMed] [Google Scholar]
- 65.Kijima TE, Innan H. On the estimation of the insertion time of LTR retrotransposable elements. Mol. Biol. Evol. 2010;27:896–904. doi: 10.1093/molbev/msp295. [DOI] [PubMed] [Google Scholar]
- 66.Kim A, Terzian C, Santamaria P, Pélisson A, Purd’Homme N, Bucheton A. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirilyuk A, Tolstonog GV, Damert A, Held U, Hahn S, et al. Functional endogenous LINE-1 retrotransposons are expressed and mobilized in rat chloroleukemia cells. Nucleic Acids Res. 2008;36:648–665. doi: 10.1093/nar/gkm1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 69.Konkel MK, Wang J, Liang P, Batzer MA. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene. 2007;390:28–38. doi: 10.1016/j.gene.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 70.Kumar A, Bennetzen JL. Plant retrotransposons. Annu. Rev. Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- 71.Lai J, Li Y, Messing J, Dooner HK. Gene movement by helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA. 2005;102:9068. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lal SK, Georgelis N, Hannah CL. Helitrons: their impact on maize genome evolution and diversity. Handbook Maize. 2009;III:329–339. [Google Scholar]
- 73.Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. The maize genome contains a helitron insertion. Plant Cell Online. 2003;15:381–391. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 75.Laski FA, Rio DC, Rubin GM. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 76.Lawler JF, Jr, Haeusser DP, Dull A, Boeke JD, Keeney JB. Ty1 defect in proteolysis at high temperature. J. Virol. 2002;76:4233–4240. doi: 10.1128/JVI.76.9.4233-4240.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee YCG, Langley CH. Transposable elements in natural populations of Drosophila melanogaster. Philos. TransRSoc. B: Biol. Sci. 2010;365:1219–1228. doi: 10.1098/rstb.2009.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levis R, O’Hare K, Rubin GM. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell. 1984;38:471–481. doi: 10.1016/0092-8674(84)90502-6. [DOI] [PubMed] [Google Scholar]
- 79.Lisch D, Jiang N. Mutator and MULE transposons. Handbook Maize. 2009;III:277–306. [Google Scholar]
- 80.Loreto E, Carareto C, Capy P. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 81.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, VanDeLagemaat LN,Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 83.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 84.McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 85.Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643. [PubMed] [Google Scholar]
- 86.Mills RE, Bennett EA, Iskow RC, Luttig CT, Tsui C, et al. Recently mobilized transposons in the human and chimpanzee genomes. AmJHum. Genet. 2006;78:671–679. doi: 10.1086/501028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 88.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 89.Naito K, Cho E, Yang G, Campbell MA, Yano K, et al. Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl. Acad. Sci. USA. 2006;103:17620. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakatsuka T, Nishihara M, Mishiba K, Hirano H, Yamamura S. Two different transposable elements inserted in flavonoid 3′, 5′-hydroxylase gene contribute to pink flower coloration in Gentiana scabra. Mol. Genet. Genomics. 2006;275:231–241. doi: 10.1007/s00438-005-0083-7. [DOI] [PubMed] [Google Scholar]
- 91.Neuvéglise C, Feldmann H, Bon E, Gaillardin C. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 2002;12:930–943. doi: 10.1101/gr.219202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Novick PA, Basta H, Floumanhaft M, McClure MA, Boissinot S. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis showsmore similarity to fish than mammals. Mol. Biol. Evol. 2009;26:1811–1822. doi: 10.1093/molbev/msp090. [DOI] [PubMed] [Google Scholar]
- 93.Novick PA, Smith JD, Floumanhaft M, Ray DA, Boissinot S. The evolution and diversity of DNA transposons in the genome of the lizard Anolis carolinensis. Genome Biol. Evol. 2011;3:1–14. doi: 10.1093/gbe/evq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nuzhdin SV, Mackay TF. The genomic rate of transposable element movement in Drosophila melanogaster. Mol. Biol. Evol. 1995;12:180–181. doi: 10.1093/oxfordjournals.molbev.a040188. [DOI] [PubMed] [Google Scholar]
- 95.Pace JK, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. USA. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, et al. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pereira V. Insertion bias and purifying selection of retrotransposons in the Arabidopsis thaliana genome. Genome Biol. 2004;5:R79. doi: 10.1186/gb-2004-5-10-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Gonzalez CE, Eickbush TH. Rates of R1 and R2 retrotransposition and elimination from the rDNA locus of Drosophila melanogaster. Genetics. 2002;162:799. doi: 10.1093/genetics/162.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Picault N, Chaparro C, Piegu B, Stenger W, Formey D, et al. Identification of an active LTR retrotransposon in rice. Plant J. 2009;58:754–765. doi: 10.1111/j.1365-313X.2009.03813.x. [DOI] [PubMed] [Google Scholar]
- 100.Piskurek O, Nishihara H, Okada N. The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene. 2009;441:111–118. doi: 10.1016/j.gene.2008.11.030. [DOI] [PubMed] [Google Scholar]
- 101.Poulter R, Goodwin TJD, Butler MI. Vertebrate helentrons and other novel helitrons. Gene. 2003;313:201–212. doi: 10.1016/s0378-1119(03)00679-6. [DOI] [PubMed] [Google Scholar]
- 102.Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci. USA. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Raiz J, Damert A, Chira S, Held U, Klawitter S, et al. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40:1666–1683. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rangwala SH, Richards EJ. The structure, organization and radiation of sadhu non-long terminal repeat retroelements in Arabidopsis species. Mob. DNA. 2010;1:10. doi: 10.1186/1759-8753-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ray DA, Feschotte C, Pagan HJT, Smith JD, Pritham E, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ray DA, Pagan HJT, Thompson ML, Stevens RD. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol. Biol. Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- 107.Rebollo R, Karimi MM, Bilenky M, Gagnier L, Miceli-Royer K, et al. Retrotransposon-induced heterochromatin spreading in themouse revealed by insertional polymorphisms. PLoSGenet. 2011;7:e1002301. doi: 10.1371/journal.pgen.1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. The physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 109.SanMiguel P, Tikhonov A, Jin YK,Motchoulskaia N, Zakharov D, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 110.SanMiguel P, Vitte C. The LTR-retrotransposons of maize. Handbook Maize. 2009;III:307–327. [Google Scholar]
- 111.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 112.Singer T, McConnell MJ, Marchetto MCN, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smit A, Hubley R, Green P. RepeatMasker open-3.0. 1996–2011. < http://www.repeatmasker.org>. [Google Scholar]
- 114.Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG. An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 115.Stewart C, Kural D, Strömberg MP, Walker JA, Konkel MK, et al. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet. 2011;7:e1002236. doi: 10.1371/journal.pgen.1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011;43:1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sukarova E, Dimovski A, Tchacarova P, Petkov G, Efremov G. An Alu insert as the cause of a severe form of hemophilia A. Acta Haematol. 2000;106:126–129. doi: 10.1159/000046602. [DOI] [PubMed] [Google Scholar]
- 118.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 119.Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006;16:R872–R873. doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 120.Va D, De N, Broeck N, Maes T, Sauer M, Zethof J. Transposon display identifies individual transposable elements in high copy number lines. Plant J. 1998;13:121–129. doi: 10.1046/j.1365-313X.1998.00004.x. [DOI] [PubMed] [Google Scholar]
- 121.van den Hurk JAJM, Meij IC, del Carmen Seleme M, Kano H, Nikopoulos K, et al. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 122.Varagona MJ, Purugganan M, Wessler SR. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell Online. 1992;4:811. doi: 10.1105/tpc.4.7.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet. Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- 124.Wang Q, Dooner HK. Remarkable variation in maize genome structure inferred from haplotype diversity at the bz locus. Proc. Natl. Acad. Sci. USA. 2006;103:17644–17649. doi: 10.1073/pnas.0603080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang Y, Liška F, Gosele C, Šedová L, Křen V, et al. A novel active endogenous retrovirus family contributes to genome variability in rat inbred strains. Genome Res. 2010;20:19. doi: 10.1101/gr.100073.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Warren WC, Hillier LDW, Graves JAM, Birney E, Ponting CP, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 128.Wessler SR. Phenotypic diversity mediated by themaize transposable elements ac and spm. Science. 1988;242:399. doi: 10.1126/science.2845581. [DOI] [PubMed] [Google Scholar]
- 129.Wessler S, Baran G, Varagona M, Dellaporta S. Excision of ds produces waxy proteins with a range of enzymatic activities. EMBO J. 1986;5:2427. doi: 10.1002/j.1460-2075.1986.tb04517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wheelan SJ, Scheifele LZ, Martinez-Murillo F, Irizarry RA, Boeke JD. Transposon insertion site profiling chip (TIP-chip) Proc. Natl. Acad. Sci. USA. 2006;103:17632–17637. doi: 10.1073/pnas.0605450103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 132.Wissing S, Muñoz-Lopez M, Macia A, Yang Z, Montano M, et al. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Witherspoon DJ, Marchani EE, Watkins WS, Ostler CT, Wooding SP, et al. Human population genetic structure and diversity inferred from polymorphic L1 (LINE-1) and alu insertions. Hum. Hered. 2006;62:30–46. doi: 10.1159/000095851. [DOI] [PubMed] [Google Scholar]
- 134.Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB. Mobile element scanning (ME-scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 136.Xing J, Zhang Y, Han K, Salem AH, Sen SK, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu Z, Ramakrishna W. Retrotransposon insertion polymorphisms in six rice genes and their evolutionary history. Gene. 2008;412:50–58. doi: 10.1016/j.gene.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 138.Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. Tuned for transposition: molecular determinants underlying the hyperactivity of a stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- 139.Yang G, Zhang F, Hancock CN,Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2007;104:10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang J, Peterson T, Peterson PA. Transposons Ac/Ds, En/Spm and their relatives in maize. Handbook Maize. 2009;III:251–276. [Google Scholar]
- 141.Zhang X, Zhou J, Eickbush TH. Rapid R2 retrotransposition leads to the loss of previously inserted copies via large deletions of the rDNA locus. Mol. Biol. Evol. 2008;25:229–237. doi: 10.1093/molbev/msm250. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Maksakova IA, Gagnier L, Van De Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ziolkowski PA, Koczyk G, Galganski L, Sadowski J. Genome sequence comparison of col and ler lines reveals the dynamic nature of Arabidopsis chromosomes. Nucleic Acids Res. 2009;37:3189–3201. doi: 10.1093/nar/gkp183. [DOI] [PMC free article] [PubMed] [Google Scholar]