Abstract
Dicer is a ribonuclease whose major role is to generate mature microRNAs although additional functions have been proposed. Deletion of Dicer leads to embryonic lethality in mice. To study the role of Dicer in adults, we generated mice in which administration of tamoxifen induces deletion of Dicer. Surprisingly, disruption of Dicer in adult mice induced lipid accumulation in the small intestine. To dissect the underlying mechanisms, we carried out miRNA, mRNA and proteomic profiling of small intestine. The proteomic analysis was done using mice metabolically labeled with heavy lysine (SILAC mice) for an in vivo readout. We identified 646 proteins of which 80 were upregulated >2-fold and 75 were downregulated. Consistent with the accumulation of lipids, Dicer disruption caused a marked decrease of microsomal triglyceride transfer protein, long-chain fatty acyl-CoA ligase 5, fatty acid binding protein, and very-long-chain fatty acyl-CoA dehydrogenase, among others. We validated these results using multiple reaction monitoring (MRM) experiments by targeting proteotypic peptides. Our data reveal a previously unappreciated role of Dicer in lipid metabolism. These studies demonstrate a systems biology approach by integrating mouse models, metabolic labeling, gene expression profiling and quantitative proteomics can be a powerful tool for understanding complex biological systems.
Keywords: Dicer, lipid, triglyceride, small intestine, SILAC, metabolic labeling, proteomics, microRNA, ribosome, multiple reaction monitoring
INTRODUCTION
Small non-coding RNA-mediated control of gene-expression has been demonstrated in a number of biological pathways. To date, three major classes of small non-coding RNAs—microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs—have been widely studied. The biogenesis of miRNAs and siRNAs involves precursor molecule processing by the ribonuclease III enzyme Dicer.1, 2 Dicer, a ubiquitously expressed and evolutionarily conserved protein, consists of a number of domains including DRBM, RNase III, PAZ, helicase and a DECH-box motif. To generate mature miRNAs, the PAZ domain of Dicer binds the 2-nucleotide 3’ overhang of precursor miRNAs while the two RNase III domains excise the loop portion of precursor miRNAs.3 Dicer associates with TRBP, another RNA binding protein, to load mature miRNAs onto the RNA-induced silencing complex, which mediates base-pairing between miRNAs and their cognate mRNA targets.
A variety of biological systems have been used to investigate the many functions of Dicer. Overexpression of Dicer has been shown to promote cell growth in interferon-defective Li-Fraumeni syndrome fibroblasts.4 Conversely, global deletion of Dicer in mice leads to lethality at early stages of embryonic development.5 Bernstein et al. deleted exon 22 of Dicer1 in mice to demonstrate that homozygous Dicer1−/− mouse embryos died as early as embryonic day 7.5. Wienholds et al. corroborated these findings in zebrafish by introducing nonsense mutations in the RNase III domain of Dicer.6 Growth of homozygous dicer1−/− fish embryos arrests on day 10 post-fertilization and fish die three weeks post-fertilization. To bypass this embryonic lethality caused by global deletion of Dicer, tissue-specific promoters for Cre recombinase were used to disrupt Dicer expression in different organs.7–9 Conditional ablation of Dicer increases apoptosis of myoblasts with a subsequent decrease in muscle mass.10 Liver-specific Dicer deficiency results in lipid accumulation and glycogen depletion in hepatocytes as well as autochthonous development of liver cancers.11 Dicer deletion has also been found to affect the complete differentiation of T and B lymphocytes.12, 13 Additionally, mouse embryonic stem cells that lack Dicer fail to properly silence their centromeric chromosomal region.14 A recent study in Caenorhabditis elegans also described a previously unexplored role of Dicer in DNA fragmentation during apoptosis.15 Mechanisms underlying most of the Dicer knockout phenotypes remain to be fully elucidated.
In instances where biological effects or pathways may be difficult to dissect, novel and integrated approaches are often the best way to gain more insight. For example, mass spectrometry-based quantification strategies, including label-free or in vitro labeling methods and in vivo metabolic labeling methods,16 represent powerful tools to address major biological questions. Indeed, many studies have adopted a label-free quantification approach owing to its flexibility in terms of multiplexing and cost-effectiveness.17, 18 Spectral counting and peak area ratios are among the commonly used features to quantify peptides in the label-free strategy.19, 20 However, label-free quantification is dependent on highly reproducible sample handling and LC conditions between samples—which is generally not practical for analyzing a large number of samples—and small changes between samples cannot be reliably quantified.21 Another commonly used in vitro labeling quantification method is isobaric tagging for relative and absolute quantification (iTRAQ).22 Although iTRAQ offers many advantages over the label-free approach, efficiency of labeling, co-elution of physicochemically similar peptides, and maximum multiplexing of 8 samples still limit its application. Further, iTRAQ quantification is generally not compatible with ion-trap mass analyzers. Thus, in vivo labeling offers the advantage of measuring changes by minimizing sample processing errors. Since 15N labeling generates complex spectra, Kruger et al. circumvented this problem by using 13C6-lysine metabolic labeling (SILAC) of mice to reveal the importance of kindlin-3 in structural integrity of red blood cells.23
To investigate the effects of Dicer deletion, we generated conditional knockout mice with a tamoxifen-regulated Cre-ERT2 system. Our studies reveal that global disruption of the Dicer gene in adult mice induces multiple changes that include abnormalities of bone marrow and small intestine. Here, we systematically evaluated global proteomic changes in the small intestine in the absence of functional Dicer. For in vivo quantification, we employed 13C6-lysine SILAC labeling of mice and found that Dicer deletion causes dysregulation of lipid metabolism, among other abnormalities. Further validation using multiple reactions monitoring (MRM) corroborated our findings in SILAC mice. Overall, our study demonstrates the strength of quantitative proteomic approaches such as SILAC in animal systems to study gene functions. Our findings indicate a previously unappreciated role for Dicer in lipid metabolism that can now be studied in greater detail.
MATERIALS AND METHODS
Induction of Dicer knockout in adult animals and tissue necropsy for phenotyping
C57BL/6 transgenic mice were maintained in a pathogen-free environment. ROSA26-CreERT2 mice were crossed to Dicer1lox/lox mice to generate tamoxifen inducible deletion of exons 21 and 22. Briefly, 8-week-old female mice were administered tamoxifen (1 mg/day gavage) or vehicle control for five days. Dicer1lox/lox mice without ROSA26-CreERT2 were also administered tamoxifen and used as controls. Tissues were harvested after a 3-hour fasting period on day 8. Mice were perfused with 10% formalin prior to necropsy. For Oil Red O staining, the jejunum was harvested without formalin fixation and immediately immersed in O.C.T. Compound (Tissue-Tek) on dry ice. Tissue sections were rinsed with 70% ethanol, placed in Oil Red O stain for 10 minutes, washed twice in 70% ethanol and rinsed in water. Harris Hematoxylin was used for counterstaining. All animal studies were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine.
Quantitative RT-PCR (qRT-PCR)
qRT-PCR was used to confirm deletion of Dicer1 exons 21 and 22 and to validate microarray results. Total RNA was purified using RNeasy Mini Kit (Qiagen, Valencia, CA) after the integrity of RNA was checked (2100 Bioanalyzer, Agilent Technology, Santa Clara, CA). Total RNA was reverse-transcribed using SuperScript III Reverse Transcriptase with oligo(dT)20 primers (Invitrogen, Carlsbad, CA) for confirming Dicer1 deletion while RNA to cDNA EcoDry Premix Oligo dT primers (Clontech, Mountain View, CA) was used for validating the gene expression microarray results. Biological triplicate samples were analyzed using iQ™SYBR Green Supermix Assay (Bio-Rad, Hercules, CA). All primer sequences are listed in Supplementary Table 1. To validate miRNA microarray results, we analyzed biological triplicate samples employing TaqMan MicroRNA Assay (Life Technologies, Grand Island, NY). The relative abundance of transcripts was calculated using Cq-Log10 standard curve method with β-actin or snoRNA135 as controls.
Gene expression analysis
Total RNA was labeled and hybridized with Affymetrix GeneChip Mouse Exon 1.0 ST array for gene expression analysis as the manufacturer's instructions. Total RNA was labeled and hybridized with Affymetrix GeneChip miRNA array for miRNA analysis. All experiments were performed as biological triplicates. Analysis was carried out in GeneSpring GX 10 (Agilent, Santa Clara, CA). Benjamini-Hochberg method built in GeneSpring GX 10 was used for the multiple testing correction.
Metabolic labeling of mice
Stable isotope-labeled mouse food was obtained from Cambridge Isotope Laboratories (Mouse Feed Labeling Kit, catalog number: MLK-LYS-C). A twenty-eight-day-old female mouse (F0) was fed the diet containing 13C6-lysine and mated 14 days later. Heavy diet was continued until F1 pups were weaned. One F1 female mouse was maintained on heavy diet and propagated to F2. A littermate female mouse (F0) was fed light diet to generate unlabeled control mice. The labeling efficiency was monitored by mass spectrometric analysis of blood, liver and lung specimens collected at 10 weeks of age from F0 and 4 weeks of age from the F1 generation.
Sample preparation and in-solution digestion
Small intestine was harvested and homogenized in 2% SDS buffer using POLYTRON PT 1200 E homogenizer (KINEMATICA, Bohemia, NY) and sonicated. Proteins from control (heavy) and Dicer-deleted (light) tissues were mixed in equal proportions. In-solution digestion was modified from the previously described.24 Briefly, disulfide bonds were reduced with 5mM dithiothreitol (60 °C, 45 min) and alkylated with 20 mM iodoacetamide. Amicon Ultra 30 kDa centrifugal filters (Milipore, Billerica, MA) were used for changing the lysate buffer to 8 M urea solution. Proteins were digested using Lys-C (1:100 (w/w), Wako Chemical USA, Richmond, VA) at 37 °C for 4 hours. Following a 4-fold dilution, the samples were incubated with trypsin (1:20 (w/w), modified sequencing grade, Promega, Madison, WI) at 37 °C for 16 hours.
Strong cation exchange (SCX) chromatography and basic reverse phase liquid chromatography fractionation
SCX fractionation was modified from the previously described.24 Briefly, in-solution digests were dried and reconstituted in SCX solvent A (10 mM potassium phosphate buffer in 20% acetonitrile, pH 2.85). Peptides were loaded onto a POLYSULFOETHYL A column (100 × 4.6 mm, 300Å, 5µm, PolyLC, Columbia, MD) and resolved with an increasing gradient of SCX solvent B (solvent A containing 350 mM KCl, pH 2.85) over 70 minutes. The collected fractions were dried and desalted on a C18 column. For preparation of samples used in MRM analysis, basic RPLC fractionation was carried out on the XBridge C18 column (250 × 4.6 mm, 300Å, 5µm, Waters, Milford, MA) with mobile phase solvent A of 10 mM triethylammonium bicarbonate (TEABC, pH 9.5) and 10 mM TEABC with 90% acetonitrile (pH 9.5) as the mobile phase B increased from 5%, to 100% in 30 minutes, and persisted for 10 minutes.
LC-MS/MS analysis and data analysis
LC-MS/MS analysis of day-8 samples was carried out on a quadrupole time-of-flight mass spectrometer (Agilent's 6538 Ultra High Definition Accurate-Mass Q-TOF) equipped with an HPLC-chip cube: separation-150 mm × 75 µm, enrichment-9 mm 160 nl, Zorbax 300SB-C18 5 µm. The Q-TOF was operated at capillary voltage of 1800 V, fragmenter voltage of 150 V, drying gas temperature of 300°C, drying gas flow rate of 6 l/min, medium isolation width of 4 m/z and a collision energy slope of 3 V plus offset of 2 V. In each duty cycle, 6 precursors were chosen, and spectra were acquired in the range of m/z 350–1700. Precursors with a single charge or unknown charge state were excluded. Database search was carried out using Spectrum Mill MS Proteomics Workbench (Agilent Technologies, Santa Clara, CA) against the mouse RefSeq database. The search parameters were as follows: trypsin and Lys-C as proteolytic enzymes, up to one missed cleavage, precursor peptide mass range 500 ~ 8000 Da, peptide mass error tolerance of 20 ppm, fragment mass error tolerance of 100 ppm, carbamidomethylation of cysteine and SILAC 2 (Lys 0-6Da) as a fixed modification, oxidation of methionine, acetylation of protein N-termini and deamidation of glutamine and asparagine as variable modifications. The forward and reverse scores provided in .spo files generated by Spectrum Mill were used to calculate the interpolated false discovery fate (FDR). FDR of 1% was used to select proteins for further analysis.
MRM assays
Small intestine tissue from four Dicer knockout mice and four controls were lysed in 8 M urea solution. Lysates were digested with Lys-C/trypsin and fractionated by basic RPLC. Fractions were analyzed separately on Agilent 6538 Ultra High Definition Accurate-Mass Q-TOF LC/MS. Proteins identified from the SILAC mice were selected as the candidates for MRM-based validation. Protein sequences of seven differentially regulated proteins in Dicer knockout mice were used as the input in Skyline (version 0.7.0.2556) to select the proteotypic peptides. Tryptic peptides with complete digestion and absence of cysteine and methionine residues were given preference for selection. For peptides containing histidine residues, +2 charged fragment ions were also included in the transition list. The majority of the selected precursors were with charge +2. At least two peptides with a minimum of 5 transitions were selected for each of the seven proteins.
Quantification validation using MRM
For MS-based validation of differentially expressed proteins in the small intestine of Dicer knockout mice, selected peptides from seven proteins were monitored on Agilent's 6430 triple quadrupole mass spectrometer. MassHunter Optimizer software (version B.04.00, Agilent, Santa Clara, CA) was used for optimization of dwell time and collision energy to specify transitions of selected peptides. Titration curve of intensity was plotted for increasing amount of the standard heavy peptides. Sample was loaded and resolved onto ZORBAX 300SB-C18 chip using 1200 series HPLC. The triple quadrupole mass spectrometer was also interfaced with the HPLC-chip cube. A 35 min gradient with increasing solvent B (3%–100%) was applied to resolve peptides. Data were acquired using MassHunter, and the relative abundance was calculated by comparing the total intensity of 5 transitions.
RESULTS & DISCUSSION
Dicer is a ubiquitously expressed protein involved in the biogenesis of miRNAs. However, Dicer also plays an important role in transcriptional silencing and in the formation of heterochromatin.25, 26 Here, we employed metabolic labeling in the context of tamoxifen-inducible disruption of the Dicer gene to systematically investigate the function of Dicer in adult mammalian tissues.
Generation of inducible Dicer knockout mice to bypass embryonic lethality
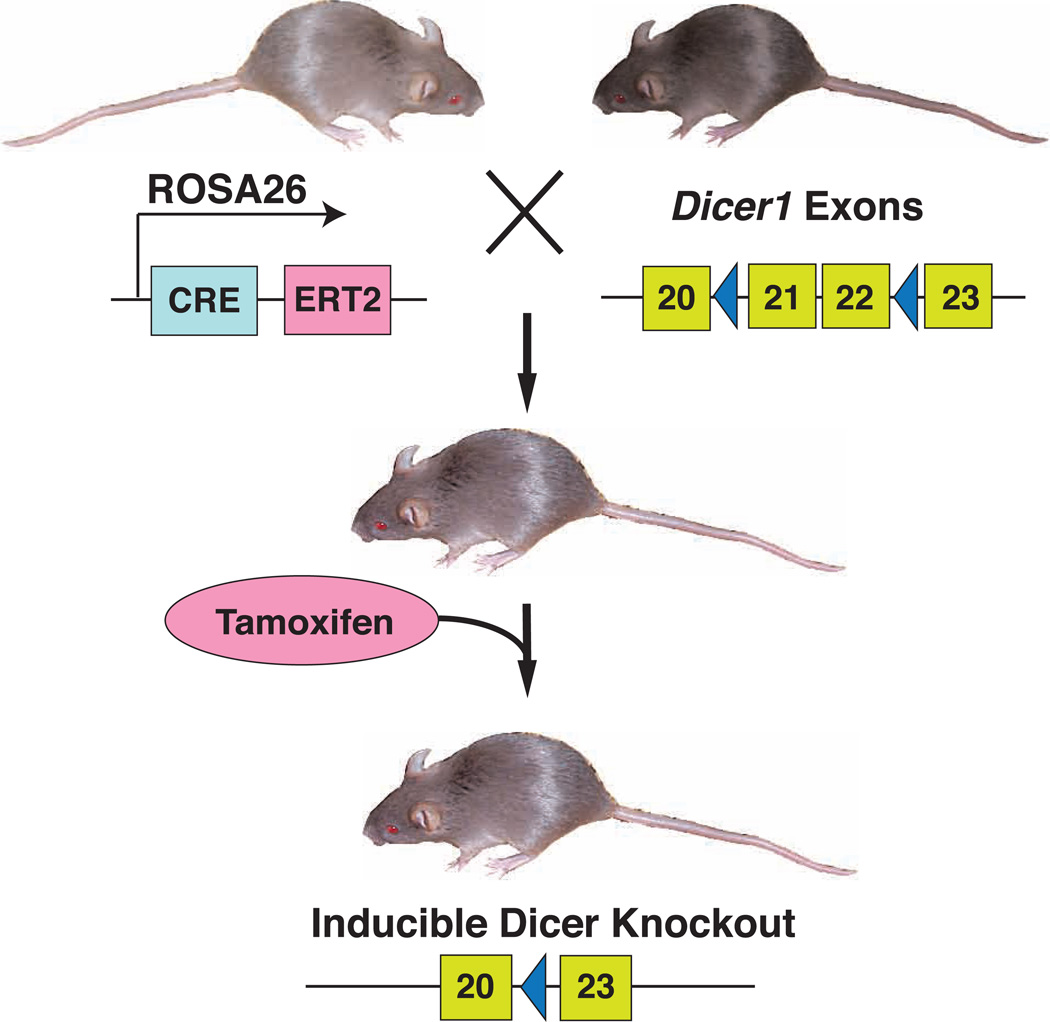
To overcome embryonic lethality resulting from global disruption of the Dicer gene during development, we used the Cre-loxP system to disrupt the expression of floxed Dicer alleles in adult mice.27 For this purpose, tamoxifen responsive ROSA26-driven Cre recombinase (Cre-ERT2) was used to specifically delete floxed exons 21 and 22 of the Dicer gene (Figure 1). In the absence of tamoxifen, double-transgenic adult mice (CRE-ERT2:Dicer1lox/lox) were phenotypically normal and fertile. Tamoxifen administration facilitates the nuclear import of Cre recombinase, thereby triggering excision of floxed Dicer1 exons 21 and 22. qRT-PCR analysis confirmed the disruption of Dicer based on monitoring of the junctional region spanning exons 21 and 22. Excision of the Dicer floxed allele was ~91%. Thus, the inducible knockout of Dicer in adult animals enabled us to address its homeostatic roles in fully differentiated tissues.
Figure 1.
Generation of a conditional Dicer knockout mouse model. Mice with ROSA26:Cre-ERT2 were crossed with Dicer1lox/lox mice to generate ROSA26-Cre-ERT2: Dicer1lox/lox mice. Upon administration of tamoxifen, Cre induced DNA recombination between two loxP sites (blue triangles) flanking Dicer exons 21 and 22, excising that portion of the allele.
Dicer disruption alters the small intestine
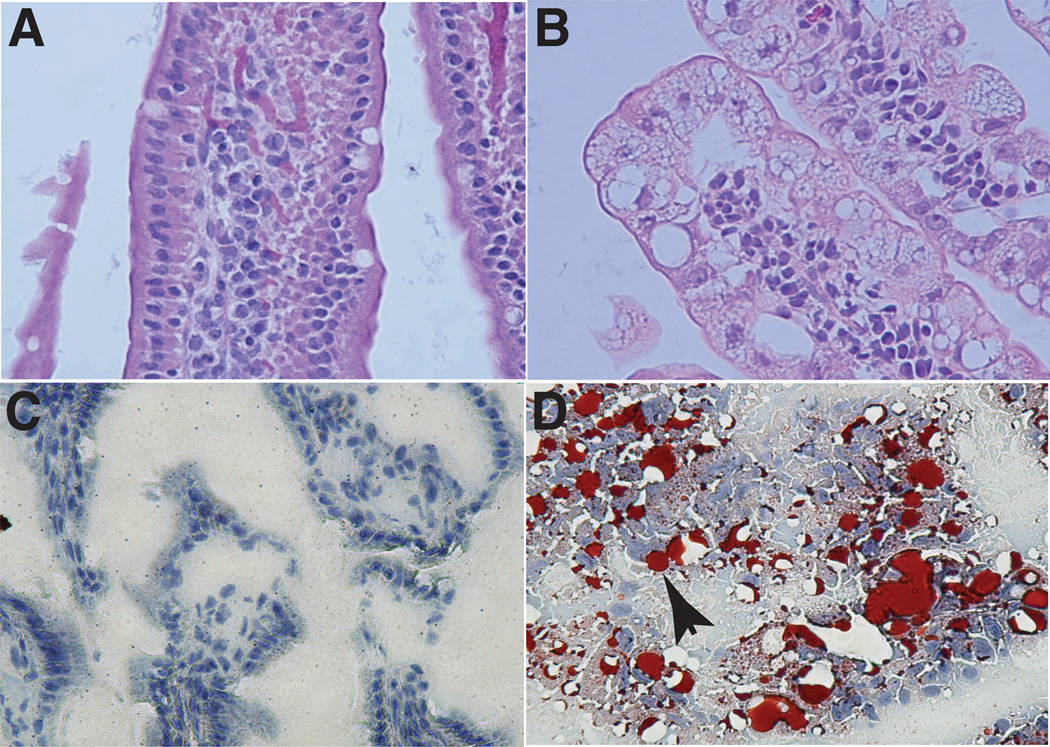
Upon homozygous deletion of Dicer1 in Cre-ERT2:Dicer1lox/lox mice, we expected a poor survival rate and severe phenotypic changes. We monitored the mice daily and no gross physical abnormalities were observed in the first 6 days. Beginning day 7, half of the tamoxifen-treated mice developed diarrhea and 80% (4/5) of these mice died at day 10 post Dicer deletion. Histopathological examination of day-8 Dicer-deleted mice revealed no overt abnormalities in the lung, heart, liver, kidney or muscle. In contrast, the small intestinal epithelium showed villous distortion, abnormal cytoplasmic vacuolation, and enlarged nucleoplasm with prominent nucleoli in enterocytes (Figure 2, A, B). Bone marrow failure was also observed with a reduction in myeloid cells (Figure S1). Further, Dicer knockout mice showed evidence of splenic and thymic cortical atrophy. Given the striking pathology of the small intestine, we decided to examine in greater detail the role Dicer plays in maintaining intestinal tissue homeostasis.
Figure 2.
Pathological changes in the small intestine observed in Dicer knockout mice. A: Normal villi in control mice; B: Abnormal vacuolation in the enterocytes, increased nucleoplasm and enlarged nucleoli in Dicer knockout mice; C: Oil Red O staining in control mice; D: Oil Red O staining in Dicer knockout mice showing neutral lipid accumulation (arrowhead) in vacuoles of varying sizes in enterocytes.
Dicer knockout in adult animals causes abnormal lipid accumulation in small intestine
Upon histological examination, extensive and abnormal vacuolation of enterocytes was evident in the small intestine of Dicer knockout but not control mice. Although vacuolation of enterocytes could result from a variety of reasons, we hypothesized that it likely results from aberrant lipid accumulation. To confirm this, we used Oil Red O staining to detect the presence of neutral lipids. As seen in panels C and D in Figure 2, the vacuolated areas were prominently stained by Oil Red O, indicating abnormal accumulation of lipid droplets in the enterocytes. A similar phenotype has been documented in humans with abetalipoproteinemia, where lipid droplets accumulate abnormally in the small intestinal epithelium. In contrast, the small intestine of control mice harbored little, if any, neutral lipid droplets. These droplets could contain any of the neutral lipids such as triglycerides, free fatty acids and cholesterol esters. Despite the aberrant lipid droplet accumulation observed in the enterocytes, the specific defect in lipid metabolism that was altered in the Dicer knockout mice was not apparent (i.e., whether the defect was in lipid absorption, catabolism, packaging and/or transport). To gain mechanistic insights into intestinal pathology of the Dicer knockout mouse model, we combined inducible Dicer deletion and SILAC mice to measure global proteomic changes resulting from disruption of the Dicer gene. This in vivo labeling strategy also enabled us to study the microenvironment of the small intestine, which cannot be recapitulated by in vitro studies.
Effect of Dicer deletion on miRNA profiles
Because Dicer is responsible for generating mature miRNAs, we predicted that Dicer knockout can deplete the whole repertoire of miRNAs. Thus, we carried out microRNA microarray profiling (as biological triplicates) and identified 150 miRNAs with the p-value cutoff of 0.05, expressed in the small intestine of Dicer knockout and control mice (GEO Accession #: GSE34909). Interestingly, only 24 miRNAs were downregulated ≥2-fold on day 8 after Dicer deletion. The most downregulated miRNAs were mmu-miR-215 (75.5-fold) and mmu-miR-194 (18.2-fold). Quite unexpectedly, we also observed 18 miRNAs to be upregulated ≥2-fold, including mmu-miR-195 (3.8-fold of control) and mmu-miR-199b (4.0-fold of control) (Table 1). qRT-PCR experiments carried out to validate the changes in a subset of miRNA microarray results revealed the same trend as microarray results (Figure S2).
Table 1.
A list of miRNAs exhibiting a significant change of expression level in mouse small intestine upon deletion of Dicer.
| miRNA | miRNA Abundance* (Knockout/ Control) |
|---|---|
| mmu-miR-215 | 0.01 |
| mmu-miR-194 | 0.05 |
| mmu-miR-31 | 0.08 |
| mmu-miR-429 | 0.11 |
| mmu-miR-192 | 0.13 |
| mmu-miR-203 | 0.13 |
| mmu-miR-200c | 0.17 |
| mmu-miR-182 | 0.19 |
| mmu-miR-200a | 0.26 |
| mmu-miR-93 | 0.27 |
| mmu-miR-425 | 0.28 |
| mmu-miR-200b | 0.30 |
| mmu-miR-18a | 0.30 |
| mmu-miR-106a | 0.31 |
| mmu-miR-17 | 0.34 |
| mmu-miR-20b | 0.35 |
| mmu-miR-20a | 0.36 |
| mmu-miR-185 | 0.40 |
| mmu-miR-423-3p | 0.43 |
| mmu-miR-107 | 0.44 |
| mmu-miR-200b* | 0.44 |
| mmu-miR-106b | 0.45 |
| mmu-miR-103 | 0.46 |
| mmu-miR-151-3p | 0.46 |
| mmu-miR-214 | 2.02 |
| mmu-miR-455 | 2.02 |
| mmu-let-7e | 2.04 |
| mmu-miR-27a | 2.15 |
| mmu-miR-126-3p | 2.17 |
| mmu-miR-99b | 2.17 |
| mmu-miR-705 | 2.18 |
| mmu-miR-21 | 2.37 |
| mmu-miR-152 | 2.39 |
| mmu-miR-125a-5p | 2.81 |
| mmu-miR-146b | 2.95 |
| mmu-miR-132 | 3.18 |
| mmu-miR-125b-5p | 3.58 |
| mmu-miR-199a-3p | 3.69 |
| mmu-miR-195 | 3.80 |
| mmu-miR-199b | 3.96 |
| mmu-miR-1224 | 5.03 |
| mmu-miR-762 | 8.11 |
Changes are rounded off to two significant figures.
Generation of SILAC mice for quantitative proteomics
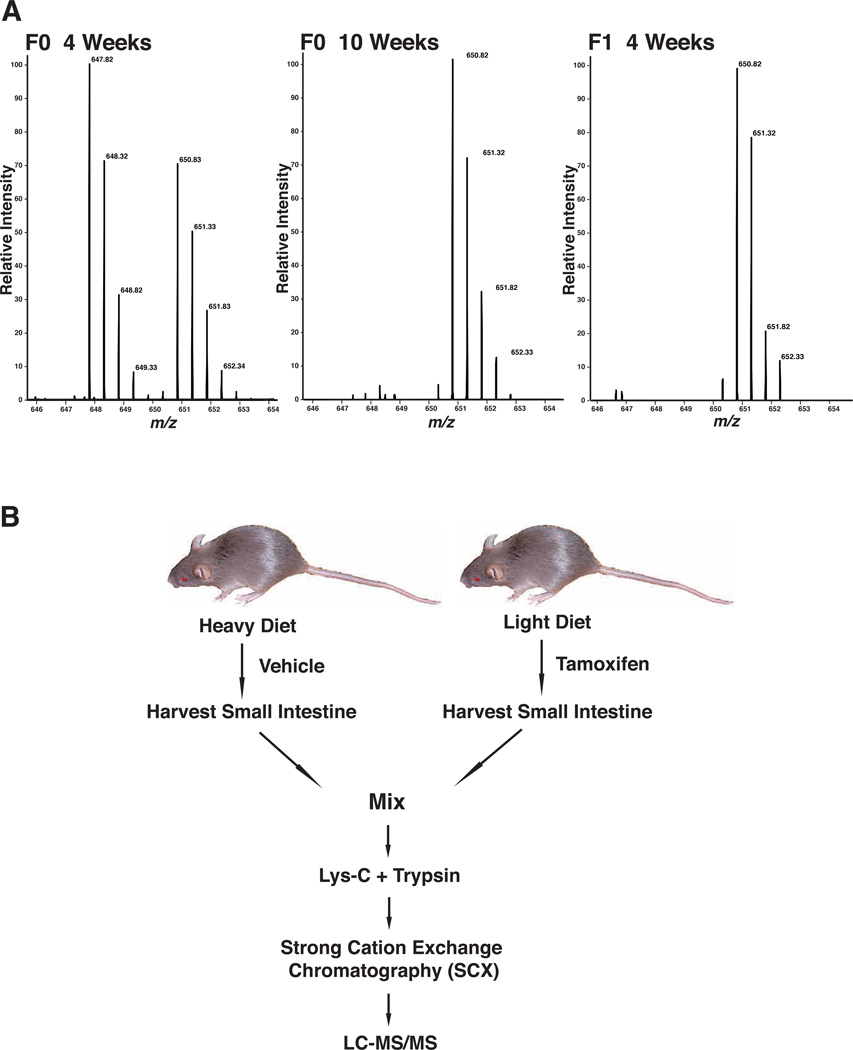
Previous studies to examine differential regulation of protein abundance by miRNAs have shown that the magnitude of the effect observed is often subtle. Although both in vitro labeling as well as label-free approaches can provide quantitative data, they are not generally as reliable as in vivo labeling approaches such as SILAC if the changes are not dramatic. Therefore, we decided to use SILAC mice for our analysis. In this approach, proteins are labeled in vivo by feeding mice a 13C6-lysine diet. We observed incorporation of 13C6-lysine within two generations of mice fed continuously with the 13C6-lysine diet as assessed by mass spectrometric analysis of proteins extracted from their blood, liver, and lung. After 10 weeks on this diet, the labeling efficiency of proteins in F0 mice (as indicated by percent incorporation in proteins) ranged from ~88% to 99%. After a 4 week 13C6-lysine diet through the F1 generation, the labeling efficiency of proteins was >97%, indicating near complete incorporation of exogenously fed 13C6-lysine (Figure 3A and Supplementary Table 2).
Figure 3.
Strategy for proteomic analysis of Dicer knockouts using SILAC mice. A: Serial examinations of a representative peptide from hemoglobin (DFTPAAQAAFQK) from blood of SILAC mice at different time points analyzed by mass spectrometry; B: Strategy for proteomic analysis of mice with inducible deletion of Dicer. Mice fed a light diet were administered tamoxifen to induce deletion of Dicer while mice on heavy diet were administered the vehicle alone. The small intestine was harvested on day 8 after initiation of tamoxifen. Harvested samples were subjected to in-solution digestion by trypsin/Lys-C and the peptides fractionated by strong cation exchange chromatography prior to LC-MS/MS analysis.
Proteomic changes induced by Dicer gene ablation in small intestine
SILAC mice were treated with vehicle control while unlabeled mice were administered tamoxifen to induce deletion of Dicer. Small intestine tissue from the two groups of mice was harvested on day 8 and equal amounts of tissue lysates were mixed from SILAC and light mice. The pooled lysates were digested with Lys-C followed by trypsin. The digests were subsequently separated by SCX to generate 24 fractions. These fractions were analyzed by LC-MS/MS (Figure 3B).
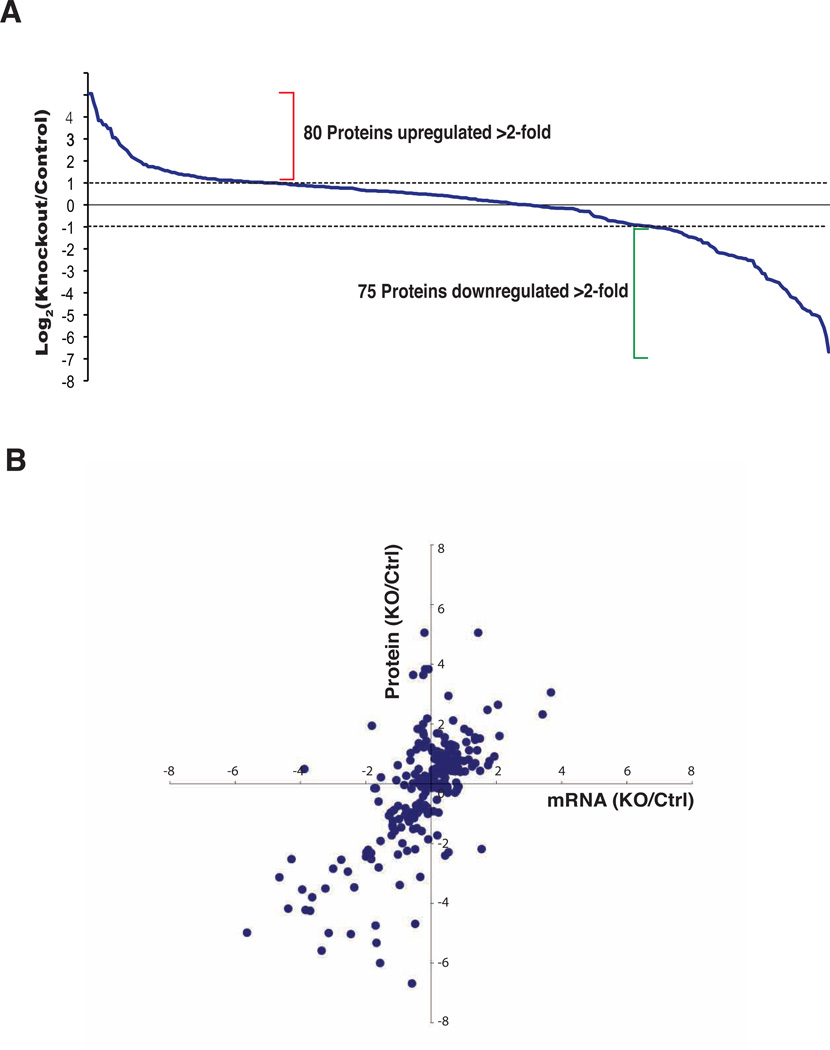
The data were searched against a mouse RefSeq database and a reversed protein sequence database using Spectrum Mill. Peptides scoring better than the 1% FDR cut-off were considered for further analysis. In all, we identified 3,087 peptides corresponding to 646 protein groups (Supplementary Table 3). Because only lysine containing peptides could be quantified, we obtained fold changes for ~54% of the identified peptides (derived from 313 proteins) (Figure 4A).
Figure 4.
Proteomic and transcriptomic changes caused by knockout of Dicer. A: Proteomic changes between Dicer knockout mice and controls. Of the 313 quantifiable proteins in day-8 samples, 80 proteins were upregulated >2-fold and 75 proteins were downregulated to <0.5 in Dicer knockout mice as compared to control mice; Dashed lines indicate a two-fold change in abundance of either direction; B: Comparison of relative changes in mRNA and protein abundance. Fold-changes (Dicer knockout/control) in mRNA and protein abundance are expressed as log2 values.
As Dicer generates mature miRNAs which suppress translation of mRNAs, we expected Dicer knockout to exhibit a widespread increase of protein abundance. To our surprise, we did not observed any major change in abundance in half of the proteins. We found 80 proteins to be upregulated (≥2 fold) and 75 proteins to be downregulated (≥2 fold) in the small intestine (Figure 4A). Representative lists of upregulated and downregulated proteins are provided in Table 2. Strikingly, 34 out of 80 upregulated proteins were ribosomal protein subunits. Additionally, proteins relevant to ribosomal biogenesis such as ribosomal RNA 2’-O-methyltransferase, nucleolin and nucleophosmin were also upregulated. Despite synchronized upregulation of proteins involved in ribosomal biogenesis and protein constituents of the translation machinery (Table 2), processes expected to increase overall protein synthesis, we still observed downregulation of 75 proteins in our study, suggesting production of these 75 proteins was significantly compromised. As expected from the abnormal lipid phenotype of Dicer knockout mice, enzymes that are involved in lipid metabolism appeared to be one of the prominent classes of proteins that were among these 75 proteins.
Table 2.
A representative list of proteins with altered protein abundance in mice with deletion of Dicer.
| Gene Symbol | GI Accession# |
Protein | Number of Peptides |
Protein Abundance (Knockout/ Control) |
|---|---|---|---|---|
| Fbn1 | 118197277 | Fibrillin-1 | 5 | 33.33 |
| Rpl12 | 160333553 | 60S ribosomal protein L12 | 4 | 11.11 |
| Rpl8 | 6755358 | 60S ribosomal protein L8 | 4 | 7.69 |
| Anxa1 | 124517663 | Annexin A1 | 7 | 5.00 |
| Ahnak | 61743961 | AHNAK nucleoprotein isoform 1 | 10 | 3.57 |
| Eef1g | 110625979 | Elongation factor 1-gamma | 9 | 3.33 |
| Rps11 | 21426889 | 40S ribosomal protein S11 | 4 | 2.38 |
| LOC100043000 | 149270989 | 60S ribosomal protein L3 | 4 | 2.33 |
| Flna | 125347376 | Filamin-A | 48 | 2.27 |
| Rplp2 | 83745120 | 60S acidic ribosomal protein P2 | 4 | 2.27 |
| Eprs | 82617575 | Bifunctional aminoacyl-trna synthetase | 4 | 2.27 |
| Rplp0 | 6671569 | 60S acidic ribosomal protein P0 | 7 | 2.22 |
| LOC100045859 | 149255974 | 40S ribosomal protein S25 | 4 | 2.22 |
| Ncl | 84875537 | Nucleolin | 10 | 2.17 |
| Rpl13a | 31981945 | 60S ribosomal protein l13a | 4 | 2.17 |
| Eef2 | 33859482 | Elongation factor 2 | 13 | 2.04 |
| LOC100043269 | 149265702 | 40S ribosomal protein S19 | 5 | 2.04 |
| Rpl14 | 13385472 | 60S ribosomal protein L14 | 4 | 2.04 |
| Rps20 | 13385652 | 40S ribosomal protein S20 | 4 | 2.04 |
| LOC100046628 | 149251776 | Nucleophosmin | 4 | 2.04 |
| Atp5c1 | 163838641 | ATP synthase subunit gamma | 3 | 0.49 |
| Acat1 | 21450129 | Acetyl-CoA acetyltransferase | 1 | 0.38 |
| Aldh2 | 6753036 | Aldehyde dehydrogenase | 9 | 0.33 |
| Akr1c12 | 85719330 | Aldo-keto reductase family 1, member C12 | 4 | 0.23 |
| Acsl5 | 58218988 | Long-chain-fatty-acid--CoA ligase 5 | 7 | 0.22 |
| Casp6 | 157951736 | Caspase-6 precursor | 2 | 0.20 |
| Aldoa | 6671539 | Fructose-bisphosphate aldolase A | 5 | 0.19 |
| Galm | 28892785 | Galactose mutarotase | 3 | 0.17 |
| Cat | 157951741 | Catalase | 3 | 0.10 |
| Mttp | 254540023 | Microsomal triglyceride transfer protein large subunit | 14 | 0.09 |
| Slc5a1 | 261824023 | Sodium/glucose cotransporter 1 | 2 | 0.07 |
| Adh6a | 124286789 | Alcohol dehydrogenase 6A (class V) | 6 | 0.06 |
| Fbp1 | 9506589 | Fructose-1,6-bisphosphatase 1 | 2 | 0.06 |
| Fabp1 | 8393343 | Fatty acid-binding protein-1 | 4 | 0.03 |
| Adh1 | 6724311 | Alcohol dehydrogenase 1 | 4 | 0.03 |
| Dak | 21703976 | Dihydroxyacetone kinase/FAD-AMP lyase | 12 | 0.03 |
| Sis | 124487275 | Sucrase-isomaltase | 22 | 0.03 |
| Aldob | 21450291 | Fructose-bisphosphate aldolase B | 15 | 0.02 |
| Akp3 | 110347479 | Intestinal-type alkaline phosphatase precursor | 5 | 0.02 |
Effect of Dicer deletion on mRNA profiles
Considering deletion of Dicer causes a measurable change in the profile of miRNAs (see above) that interact with mRNAs and regulate protein abundance, we also performed transcriptomic analysis using DNA microarrays. Expression levels of 16,526 genes were quantifiable between Dicer deleted and control mice. After applying a p-value cutoff of 0.05 and correcting for multiple hypothesis testing, a total of 1,689 genes were found to exhibit >2-fold change in mRNA expression (GEO Accession #: GSE34905). We validated the changes at the mRNA level on a subset of transcripts using qRT-PCR, which confirmed the same trend as was observed in the microarray experiments (Figure S3). Overlaying the data from transcriptomic and proteomic analyses revealed that 219 genes have both mRNA and protein quantification values for direct comparison. Of these, ~76% exhibited a change in mRNA and protein levels in the same direction (Figure 4B, the correlation coefficient=0.68) while almost 15% genes showed a decrease in mRNA but an increase in protein abundance. Conversely, ~10% of the genes showed an increase in mRNA but a decrease in protein abundance. We did not observe any significant correlation between miRNA and their mRNA target levels and between miRNA and their protein target levels (the correlation coefficients are both <0.10). These data reveal a complex regulation of mRNA and protein levels by alterations in miRNA levels and presumable other functions of Dicer.
Impaired lipid metabolism in small intestine of Dicer knockout mice
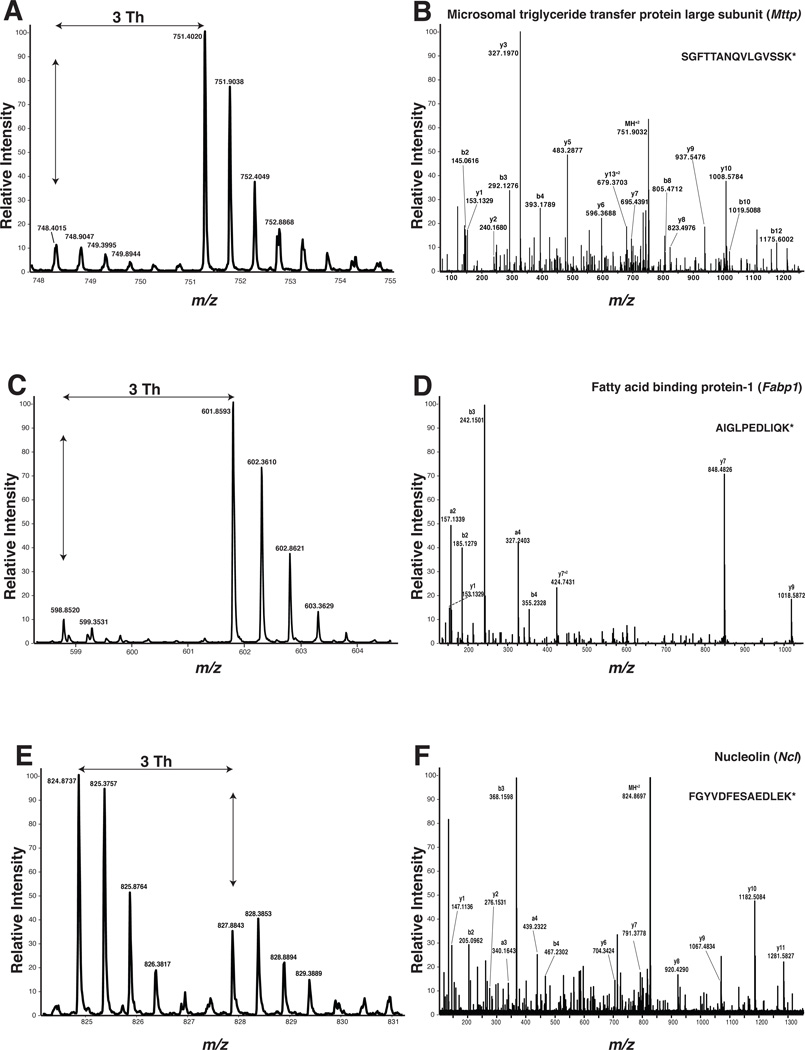
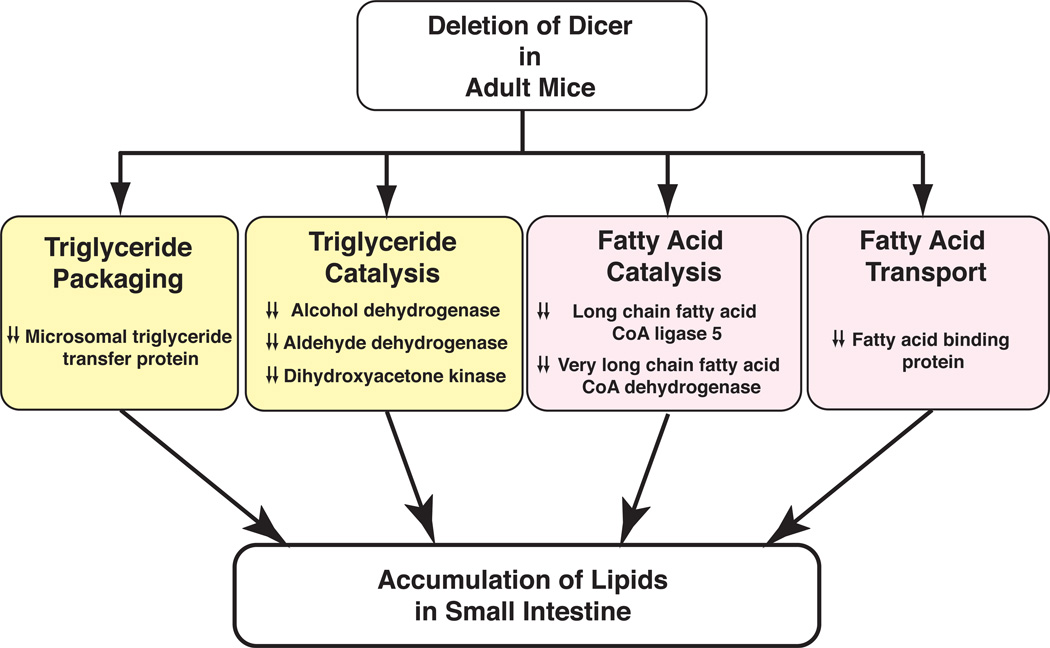
When we examined the repertoire of proteins whose abundance was altered in the small intestine of Dicer knockout mice, we used Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com/products/pathways_analysis.html) and found that proteins involved directly or indirectly in lipid metabolism constituted a major fraction (16 out of 75 ≥2-fold, 14 out of 47 ≥4-fold) of the downregulated proteins observed in the small intestine of Dicer knockout mice. The list included proteins involved in lipid transport and catabolism (Table 3 and Supplementary Table 4). In abetalipoproteinemia patients, microsomal triglyceride transfer protein gene (MTTP) is mutated leading to abnormal lipid accumulation in enterocytes, a phenotype resembling that of the Dicer knockout mice. We found that microsomal triglyceride transfer protein (Mttp) was downregulated 11.4-fold in the small intestine of Dicer gene-deleted mice (Figure 5, A, B). The major function of Mttp is to package triglycerides into pre-chylomicron that is exported out of enterocytes into the lymphatic system and transported to peripheral tissues (e.g., adipose tissue, skeletal muscle, and liver) for storage or catabolism. Dihydroxyacetone kinase (Dak) was downregulated 32.3-fold in the small intestine of the Dicer knockout mice. Dak phosphorylates dihydroxyacetone, to generate dihydroxyacetone phosphate (DHAP), an intermediate metabolite in glycolysis. Downregulation of Dak can lead to the accumulation of dihydroxyacetone, which in turn may prevent glycerol (a precursor for triglyceride synthesis) from entering the glycolytic pathway, potentially channeling the glycerol substrate toward the triglyceride synthesis pathway.
Table 3.
Regulation of lipid metabolism-associated proteins in mouse small intestine by Dicer knockout
| Gene Symbol |
Protein | mRNA (Knockout /Control) |
Protein (Knockout /Control) |
Potential regulating miRNA (Knockout /Control) |
|---|---|---|---|---|
| Acadvl | Very-long-chain fatty-acyl-CoA dehydrogenase, mitochondrial precursor | 0.50 | 0.60 | NA |
| Acsl5 | Long-chain fatty-acyl-CoA ligase 5 | 0.26 | 0.22 | NA |
| Adh6a | Alcohol dehydrogenase 6A (class V) | NA | 0.06 | NA |
| Aldh2 | Aldehyde dehydrogenase, mitochondrial precursor | 0.82 | 0.33 | NA |
| Aldob | Fructose-bisphosphate aldolase B | 0.10 | 0.02 | mmu-miR-23a(1.80),-23b(1.18),-26a(1.02),-26b(1.13),-146a(1.83),-146b(2.95),-486(1.23) |
| Dak | Bifunctional ATP-dependent dihydroxyacetone kinase/FAD-AMP lyase (cyclizing) | 0.18 | 0.03 | mmu-miR-23a(1.80),-23b(1.18),-139-5p(1.11),-143(1.67),-193b(1.19),-382(1.01),-494(1.14),-541(1.11),-709(1.22), |
| Fabp1 | Fatty acid-binding protein-1 | 0.02 | 0.03 | NA |
| Mttp | Microsomal triglyceride transfer protein large subunit isoform 1 | 0.11 | 0.09 | NA |
NA: not available
Figure 5.
MS and MS/MS spectra of peptides from representative proteins involved in lipid metabolism or ribosome biogenesis. Peptides from microsomal triglyceride transfer protein (A: MS; B: MS/MS) and fatty acid binding protein (C: MS; D: MS/MS) were lower in abundance in Dicer knockout mice (light versions of the peptides) than in control mice (heavy peptides). Conversely, nucleolin-derived peptide (E: MS; F: MS/MS) was upregulated in samples from Dicer knockout. The peptide sequence in each case in indicated in the MS/MS spectrum.
Other downregulated enzymes included alcohol dehydrogenase 6A (Adh6a, 16.1-fold) and aldehyde dehydrogenase (Aldh2, 3.0-fold), fatty acid binding protein-1 (Fabp1, NP_059095.1, 31.3-fold), long-chain fatty-acyl-CoA ligase 5 (Acsl5, NP_082252.1, 4.6-fold), and very-long-chain fatty-acyl-CoA dehydrogenase (Acadvl, NP_059062.1, 1.7-fold). Fabp1 is a lipid chaperone, playing an important role in facilitating the entry of long-chain fatty acids into smooth endoplasmic reticulum. In Dicer knockout mice, Fabp1 protein levels reduced 31-fold compared to control animals (Figure 5, C, D). Acadvl plays an important role in mitochondria β-oxidation of long-chain fatty acids. Reducing Acadvl protein levels promotes accumulation of long-chain fatty acids. Overall, our quantitative proteomic analysis suggests that proteins involved in lipid metabolism are significantly altered in the small intestine of Dicer knockout mice, providing a possible mechanism accounting for abnormal lipid droplet accumulation in enterocytes of these animals (Figure 6). Our global transcriptomic analysis also revealed a general concordance between changes in mRNA level and protein levels for these molecules involved in metabolism (Figure 4B, the correlation coefficient=0.68). Reduction in the protein levels of Mttp, Acsl5, Fabp1 and Dak was mirrored by the downregulation of their corresponding mRNA levels (9.4-fold, 3.8-fold, 49.9-fold and 5.5-fold, respectively). In addition, we have also identified two downregulated proteins normally involved in glucose metabolism—fructose bisphosphate aldolase B (Aldob, NP_659152.1, 47.6-fold) and fructose bisphosphatase 1 (Fbp1, NP_062268.1, 18.2-fold).
Figure 6.
Downregulation of enzymes in lipid metabolic pathways by Dicer knockout. Major categories of processes/pathways involved in the metabolism of lipids are shown as boxes along with the major enzymes that were observed to be altered in their expression levels.
Because changes in miRNAs can affect mRNA and protein abundance, we sought to correlate these parameters for the above-mentioned proteins involved in lipid metabolism. TargetScanMouse (Release 6.0) analysis of potential targets of upregulated miRNAs highlighted mmu-miR-23a and -23b, which were upregulated 1.8- and 1.2-fold, respectively, in Dicer knockout mice with their potential cognate targets being Aldob and Dak. mRNA levels of these two proteins were decreased 10- and 5.6-fold, respectively, in the transcriptomic analysis. Given their relatively long 3’ untranslated regions, Aldob (1086 nt) and Dak (1580 nt) can be targeted by multiple miRNAs including mmu-miR-23a and -23b, both of which were upregulated in the small intestine of Dicer knockout mice (Table 3).
Validation of Dicer-regulated changes in proteins involved in lipid metabolism through MRM
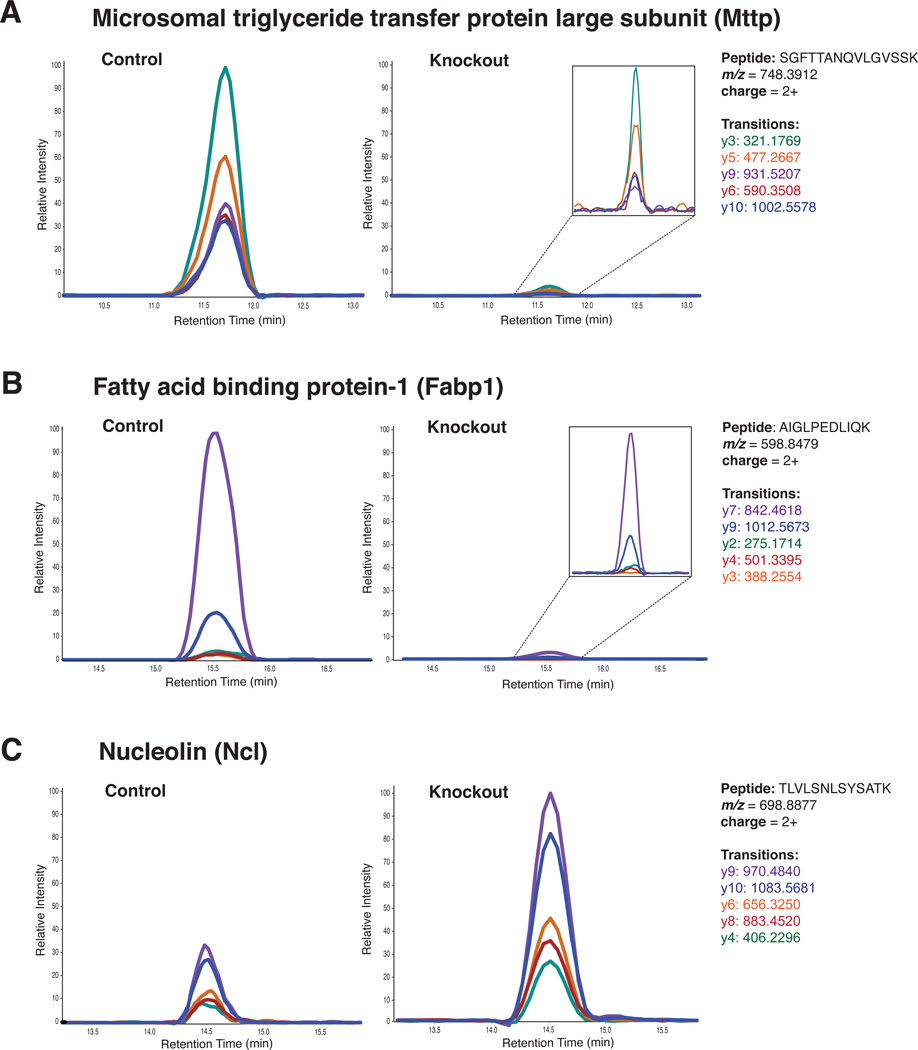
To validate the altered levels of proteins identified from SILAC mice, we developed targeted MRM-based assays to quantify changes induced by Dicer gene deletion in a different cohort of unlabeled mice. We carried out biological experiments in quadruplicate for this purpose. Equal amounts of protein lysates were subjected to basic RPLC fractionation and analyzed on a triple quadrupole mass spectrometer. The list of all candidate peptides selected for MRM-based validation and the corresponding fold-change values obtained from MRM assays are provided in Supplementary Table 5. We monitored five transitions for each peptide. Of the downregulated proteins, we selected candidate peptides from Mttp and Dak for validation. We targeted three peptides from Mttp: SGFTTANQVLGVSSK (charge: +2, monoisotopic m/z: 748.3921) (Figure 7A), AFALNFQQTIAGK (charge: +2, monoisotopic m/z: 704.8828), and EFYSYENEPVGIENLK (charge: +2, monoisotopic m/z: 965.96118). The ratios of ion count between Dicer knockout and control for these three peptides were 0.09, 0.09, and 0.10; the ratio of this protein obtained from the SILAC mice was 0.09.
Figure 7.
Multiple reaction monitoring (MRM) analysis for validation of differentially expressed proteins. Five transitions were chosen for each proteotypic peptide and are listed in descending order of their intensities. In the case of knockouts in panels A and B, the insets depict a zoomed-in view of the transitions.
The proteotypic peptides targeted in Fabp1 were AIGLPEDLIQK (charge: +2, monoisotopic m/z: 598.8479) (Figure 7B) and NEFTLGEECELETMTGEK (charge: +2, monoisotopic m/z: 1058.9561). The ratios of ion counts between Dicer knockout and control were 0.04 and 0.03; the value obtained from SILAC mice result was 0.03. These two downregulated proteins in Dicer knockout mice are responsible for the packaging and transport for triglycerides. Among the upregulated proteins, we targeted two proteotypic peptides in nucleolin, TLVLSNLSYSATK (charge: +2, monoisotopic m/z: 698.8877) (Figure 7C) and GLSEDTTEETLK (charge: +2, monoisotopic m/z: 661.8197). Again, the results from MRM quantification corroborated results obtained from SILAC mice, which were 2.7 and 3.0 vs. 2.2. Overall, the quantification values from MRM validation and SILAC mice appeared robust and consistent, especially for those proteins with marked changes caused by Dicer gene deletion.
CONCLUSIONS
In this study, we have elucidated an important but previously unappreciated role of Dicer in maintaining small intestinal tissue homeostasis. Through inducible disruption of the Dicer gene in adult mice, we uncovered a Dicer-dependent mechanism regulating lipid transport and catabolism in enterocytes. In our mouse model, deletion of Dicer also reduced cellularity within the bone marrow niche. It is interesting that intestine and bone marrow exhibited pronounced phenotypes when Dicer was deleted because of high rates of proliferation and turnover that are common to both of these organs. Overall, our data clearly highlight the role of Dicer beyond embryonic development and reveal its critical role in maintaining tissue homeostasis in adult animals.
Our quantitative proteomic analysis provided mechanistic insights into metabolic processes regulated by Dicer, the absence of which resulted in abnormal lipid accumulation in enterocytes similar to abetalipoproteinemia in humans. Several enzymes involved in lipid transport (e.g. Mttp and Fabp1) and catabolism (e.g. Acadvl, Acsl5, Adh6a, Aldh2, and Dak) were strikingly downregulated. Downregulation of lipid-metabolizing enzymes in Dicer knockout mice occurred despite upregulation of ribosomal proteins and protein constituents of the translational machinery. In a simplified model, Dicer generates mature miRNAs, which suppress the translation of cognate mRNA targets or destabilize the transcripts. In this model, Dicer knockout is predicted to result in upregulation of proteins. However, we identified proteins that are downregulated as well as those that are upregulated. Since multiple functions of Dicer outside of microRNA biogenesis have been described, further studies are required to dissect additional roles of Dicer.
In addition to the discovery phase of our study, we developed MRM assays to target peptides identified in SILAC mice. Results from these MRM assays helped confirm that downregulation of lipid transport and metabolizing enzymes likely accounts for the abnormal lipid droplet accumulation in enterocytes of Dicer knockout mice. The use of these two quantitative methods in tandem provides a promising platform for discovering and confirming diverse roles of proteins in biological processes. Because of the regulatory role of miRNAs in protein abundance, we envisage that the approach presented here can be adopted for discovery and validation of miRNAs regulating proteins in metabolism research.
Supplementary Material
Figure S1. Bone marrow failure in mice lacking Dicer. The cellularity of bone marrow in Dicer knockout mice was drastically decreased, especially in the myeloid lineage, as compared to controls.
Figure S2. Validation of miRNA microarray experiments by qRT-PCR. TaqMan microRNA assays were used to quantitate miRNA expression levels for the indicated miRNAs. The red bars represent qRT-PCR results while the blue bars depict microarray results.
Figure S3. Validation of DNA microarray results by qRT-PCR. SYBR-Green assays were used to quantitate mRNA expression levels for the indicated transcripts. The green bars represent qRT-PCR results while the blue bars depict microarray results. A: genes observed to be upregulated in Dicer knockout mice; B: genes observed to be downregulated in Dicer knockout mice.
ACKNOWLEDGEMENTS
This study was supported in part by an NIH roadmap grant for Technology Centers of Networks and Pathways (U54RR020839) and a contract (HHSN268201000032C) from the National Heart, Lung and Blood Institute. G.W.W is supported by grants from the American Heart Association (SDG2260721) and the National Institute of Health (RO1 DK084171). J.M.P is supported by the NIH National Research Service Award (F32DK084607).
REFERENCES
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Knight SW, Bass BL. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Tainsky MA. Cancer Res. 2011;71:255–265. doi: 10.1158/0008-5472.CAN-10-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 6.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 7.Hebert SS, Papadopoulou AS, Smith P, Galas MC, Planel E, Silahtaroglu AN, Sergeant N, Buee L, De Strooper B. Hum Mol Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- 8.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS. Proc Natl Acad Sci U S A. 2009;106:10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Gastroenterology. 2009;136:2304–2315. doi: 10.1053/j.gastro.2009.02.067. e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Science. 2010;328:327–334. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaerkady R, Pandey A. Proteomics Clin Appl. 2007;1:1080–1089. doi: 10.1002/prca.200700284. [DOI] [PubMed] [Google Scholar]
- 17.Manes NP, Dong L, Zhou W, Du X, Reghu N, Kool AC, Choi D, Bailey CL, Petricoin EFr, Liotta LA, Popov SG. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.000927. M110.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Othman E-D, Issaq HJ, Hornung D, Al-Hendy A, Veenstra TD. Electrophoresis. 2008;29:2706–2713. doi: 10.1002/elps.200700837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondarenko PV, Chelius D, Shaler TA. Anal Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Sadygov RG, Yates JRr. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 21.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 24.Kim MS, Kandasamy K, Chaerkady R, Pandey A. J Am Soc Mass Spectrom. 2010;21:1606–1611. doi: 10.1016/j.jasms.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woolcock KJ, Gaidatzis D, Punga T, Buhler M. Nat Struct Mol Biol. 2011;18:94–99. doi: 10.1038/nsmb.1935. [DOI] [PubMed] [Google Scholar]
- 26.Giles KE, Ghirlando R, Felsenfeld G. Nat Cell Biol. 2010;12:94–99. doi: 10.1038/ncb2010. sup pp 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Bone marrow failure in mice lacking Dicer. The cellularity of bone marrow in Dicer knockout mice was drastically decreased, especially in the myeloid lineage, as compared to controls.
Figure S2. Validation of miRNA microarray experiments by qRT-PCR. TaqMan microRNA assays were used to quantitate miRNA expression levels for the indicated miRNAs. The red bars represent qRT-PCR results while the blue bars depict microarray results.
Figure S3. Validation of DNA microarray results by qRT-PCR. SYBR-Green assays were used to quantitate mRNA expression levels for the indicated transcripts. The green bars represent qRT-PCR results while the blue bars depict microarray results. A: genes observed to be upregulated in Dicer knockout mice; B: genes observed to be downregulated in Dicer knockout mice.