Abstract
The vitamin A metabolite retinoic acid (RA) regulates intestinal immune responses through immunomodulatory actions on intestinal dendritic cells (DCs) and lymphocytes. Here, we show that retinoic acid also controls the generation of gut-tropic migratory DC precursors, referred to as pre-mucosal DCs (pre-μDCs). Pre-μDCs express the gut trafficking receptor α4β7 and home preferentially to the intestines. They develop in the bone marrow, can differentiate into CCR9+ plasmacytoid DCs as well as conventional DCs (cDCs), but preferentially give rise to CD103+ intestinal cDCs. Generation of pre-μDCs in vivo in the bone marrow or in vitro is regulated by RA and retinoic acid receptor α signaling. The frequency of pre-μDCs is reduced in vitamin A-deficient animals and in animals treated with retinoic acid receptor inhibitors. The results define a novel vitamin A-dependent, retinoic-acid-regulated developmental sequence for dendritic cells and identify a targeted precursor for CD103+ cDCs in the gut.
INTRODUCTION
The maintenance of steady-state tolerance to commensal flora and the ability to rapidly clear pathogens in case of gut wall disruption require flexibility and sophistication in the mucosal immune system. Specialized antigen-presenting dendritic cells (DCs) in the gut wall and gut-associated lymphoid tissues (GALT) control the balance between intestinal immunity and inflammation1–9. It is now clear that vitamin A and its metabolite retinoic acid (RA) play critical roles in the local differentiation and function of intestinal DCs, especially the migratory CD103+ populations10. RA programs CD103+ DCs to upregulate retinaldehyde dehydrogenase (RALDH), the rate-limiting enzyme for conversion of vitamin A precursors into retinoic acid10. These mucosal DCs migrate to the draining mesenteric lymph nodes where they present RA along with processed antigen to T cells2,4. RA imprints responding T cells with gut homing properties11 and, in the absence of danger signals, favors the induction of tolerogenic regulatory T cells8 by suppressing memory/effector T cell mediated inhibition of Treg conversion from naïve T cells12. Thus RA plays a critical local role in intestinal dendritic cell function and immune regulation, but its involvement in the origin of intestinal DC precursors has not been studied.
Here we describe a targeted gut homing DC precursor, designated pre-mucosal DCs (pre-μDCs), whose development in the bone marrow is regulated by retinoic acid. Pre-μDCs are identifiable phenotypically as lineage−CD11cintB220+CCR9−cells that express the intestinal homing receptor α4β7. They can arise in vitro from CD11cintB220+ bone marrow precursors that lack both CCR9 and α4β7. Pre-μDCs give rise to CCR9+ plasmacytoid DCs (pDCs) and to conventional DCs (cDCs), and home preferentially to the intestines in vivo. They are particularly efficient at generating CD103+ cDCs in vitro and replenishing intestinal CD103+ cDCs in vivo. RA signaling through retinoic acid receptor α (RARα) drives pre-μDC differentiation from bone marrow progenitors, and the frequency of pre-μDCs was reduced in vitamin A-deficient animals and in animals treated with an inhibitor of RAR signaling. Retinoic acid thus plays a unifying role in intestinal DC development and function, regulating both the generation of gut-homing precursors and the specialized functions of DC within the gut environment.
RESULTS
Identification of a phenotypically unique α4β7+ DC subset in vivo
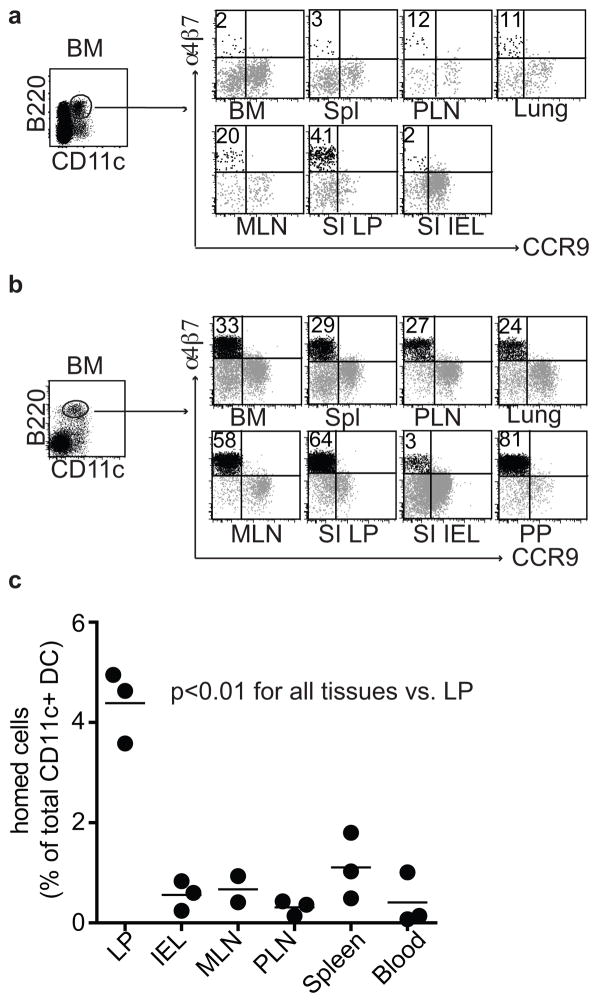
In studies of DC subsets expressing B220, we discovered a subset of B220+CD11cint DCs that expresses the α4β7 integrin, a homing receptor that mediates lymphocyte recruitment into the intestinal wall through interaction with the mucosal vascular addressin MAdCAM113(Figs. 1a and S1a). α4β7+B220+ DCs are found in secondary lymphoid tissues and the bone marrow (BM), and are particularly prominent among the B220+ DCs in the gut-associated mesenteric lymph nodes (MLN) and in the small intestine (SI) lamina propia (LP) (Figs. 1a and S1a). These cells are phenotypically distinct from previously described peripheral or BM DC populations or precursors, including splenic cDCs, BM CCR9+ pDCs, common DC progenitors (CDPs), and pre-cDCs (Figs. S1b and S1c). We took advantage of an Fms-like tyrosine kinase 3 ligand (Flt3L)-expressing B16 melanoma to expand DC subsets in vivo with minimal alterations in their phenotypic or functional capabilities for homing and adoptive transfer studies14. The α4β7+B220+ DCs were dramatically expanded in Flt3L-treated mice, suggesting a proliferative or progenitor potential (Fig. 1b). We refer to them hereafter as pre-μDCs, short for pre-mucosal DCs.
Figure 1. Identification of a phenotypically unique α4β7 expressing, gut-homing DC subset in vivo.
a) Surface expression of α4β7 and CCR9 on live lin− CD11c+B220+ DCs from lymphoid and non-lymphoid tissues from 2–3 week old C57Bl/6 mice; lin indicates CD3, CD19, and NK1.1. Data are representative of seven independent experiments. b) Surface expression of α4β7 and CCR9 on live lin−CD11c+B220+ DCs from lymphoid and non-lymphoid tissues taken from Flt3L-treated mice. Data are representative of at least three independent experiments. c) Three million purified pre-μDCs, sorted from peripheral lymphoid tissues from Flt3L-treated B6.CD45.2 mice, were transferred intravenously into B6.CD45.1 recipients. Tissues were harvested on day 3 after transfer and donor-derived cells were quantified. Data are presented as the percentage of total CD11c+ host cells. Each dot represents 1 individual animal, n=3 from two independent experiments. p<0.01 for LP vs. all other tissues by Student’s t-test.
We hypothesized that, by virtue of their expression of α4β7, pre-μDCs might home efficiently from the blood into the gut wall. Pre-μDCs were sorted from lymphoid tissues of Flt3L-treated B6.CD45.2 mice. Localization in vivo was assessed 3 days after intravenous transfer into congenic B6.CD45.1 recipients. Pre-μDCs preferentially homed to the SI LP (Fig. 1c). Preferential homing of pre-μDCs to the SI LP and colon was also apparent in shorter-term (12-hour) homing studies (data not shown).
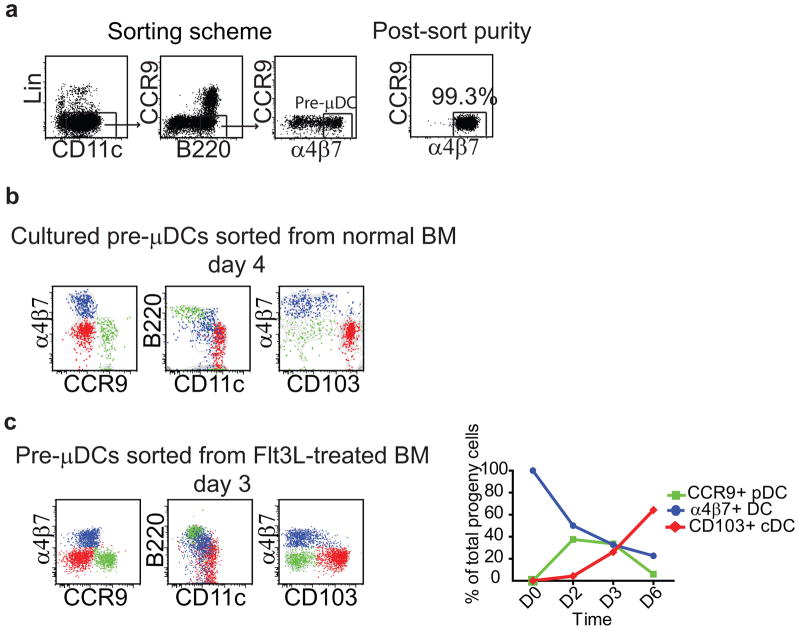
Pre-μDCs give rise to CCR9+ pDCs and to CD103+ cDCs in vitro
To assess the progenitor potential of pre-μDCs, we sorted them from BM and cultured them in vitro with total BM cells taken from CD45 allotype congenic mice as feeder cells (Fig. 2a). In some experiments, we also used pre-μDCs sorted from the BM of Flt3L-treated mice; these cells are phenotypically similar to normal BM pre-μDCs, the classical CCR9+ pDC markers PDCA1, Siglec H, and Ly6c are down-regulated, not unlike the surface phenotype of pre-μDCs in normal spleen (data not shown). Cells were cultured with recombinant Flt3L and their progeny were analyzed by flow cytometry after 3–6 days. By day 3–4, the cultures contained three prominent and phenotypically distinctive pre-μDC-derived populations (Figs. 2b and 2c): CCR9+ pDCs, which retained high levels of B220 and intermediate expression of CD11c; CD103+ DCs that were α4β7−CCR9−B220− and CD11c+, essentially a cDC phenotype; and a population of α4β7+ pre-μDC-like cells whose phenotype (lower B220 and slightly higher CD11c levels than starting pre-μDCs) appeared transitional between that of BM pre-μDCs and cDCs. In fact, a fraction of the α4β7+ DC had upregulated CD103. By day 6 pre-μDC-derived progeny were mostly CD103+ cDC (70–80%, N>3) (Fig. 2c). However, the absolute numbers of all three subsets peaked on day 3 and decreased by day 6, suggesting limited life span of pre-μDC and their progeny in vitro (Fig. S2a). Similar results were seen whether pre-μDCs were from normal or Flt3L-treated mouse BM (Figs. 2b and 2c). Consistent with prior studies of Flt3L-driven in vitro BM-derived cDC, pre-μDC progeny in vitro did not express surface CD8α (data not shown). The results demonstrate that pre-μDCs can give rise rapidly to both classical CCR9+ pDCs and to CD103+ cDCs. We were unsuccessful in efforts to evaluate pre-μDC development at the clonal level and therefore cannot exclude the possibility that distinct pDC and cDC precursors exist within the gut-tropic pre-μDC pool.
Figure 2. Pre-μDCs give rise to CCR9+ pDCs and to CD103+ cDCs in vitro.
a) Sorting scheme and purity analysis for pre-μDCs. Lin indicates CD3, CD19, and NK1.1. b) Pre-μDCs were sorted from BM of B6.CD45.2 mice and cultured with total BM taken from congenic B6.CD45.1 mice in complete RPMI supplemented with 100 ng/ml rFlt3L. Cultured cells were analyzed by flow cytometry on day 4. Pre-μDC-derived cells on day 4 included three distinct populations: CCR9+ DCs (green), CD103+ DCs (red) and α4β7+ DCs (blue). Grey contour plots show all pre-μDC-derived cells. Data represent one of three independent experiments with similar results. c) Pre-μDCs were sorted from Flt3L-treated mouse BM and cultured in the conditions described in panel b legend. Pre-μDC derived cells on day 3 consist of three populations similar to those from untreated mice. Line graphs show that by day 6, most pre-μDC-derived CCR9+ pDCs disappeared and the cultures consisted mostly of CD103+ cDCs. Data are representative of three independent experiments with similar results.
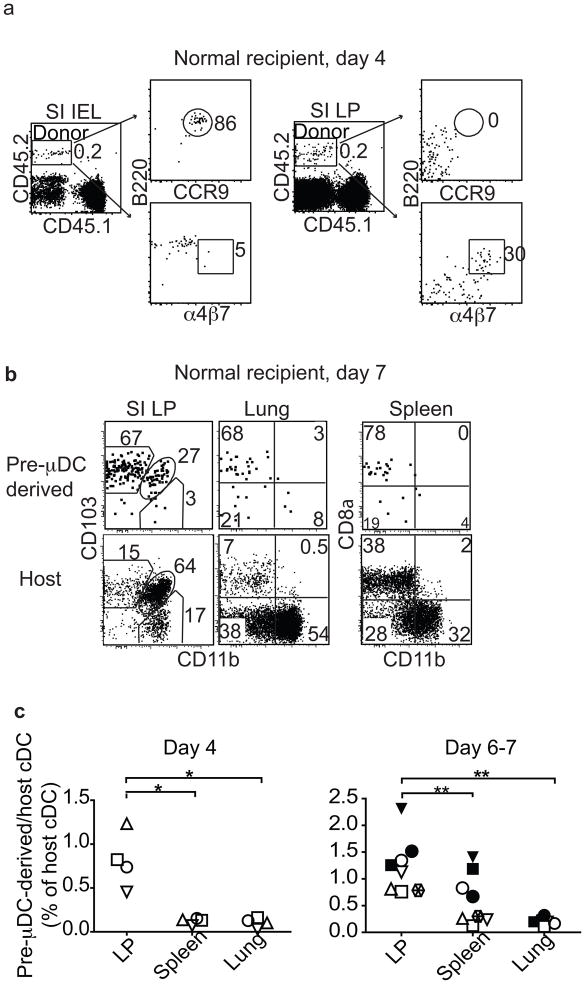
Pre-μDCs give rise to CCR9+ pDCs and to CD103+ cDCs in vivo
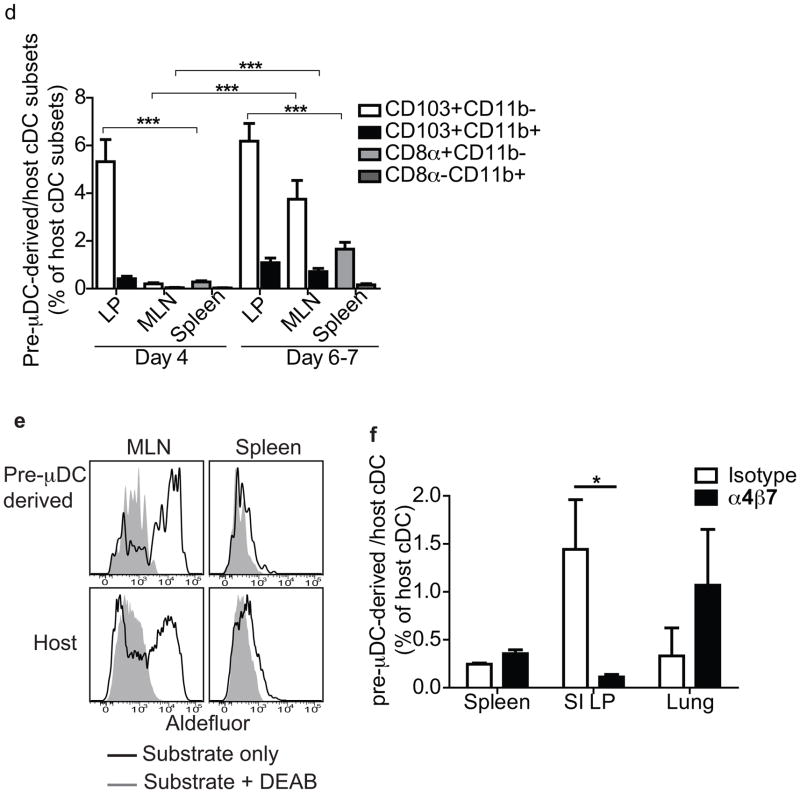
We next assessed the development of pre-μDC in vivo in a homeostatic setting. We sorted pre-μDCs from Flt3L-treated B6.CD45.2 mice and injected them into congenic B6.CD45.1 recipients. Recipient mice were sacrificed on day 4 or 7. On day 4 after transfer, both cDCs and CCR9+ pDCs were observed among CD45.2+ pre-μDC-derived progeny cells in the spleen, small intestine, and lung. In the intraepithelial lymphocytes (IEL), CCR9+ pDCs (CD11cintB220+CCR9+) predominated among pre-μDC-derived cells (Fig. 3a, left). In the SI LP, a subset of transferred cells maintained the pre-μDC phenotype (α4β7+, CD103−), with some showing reduced B220 expression (Fig. 3a, right). Pre-μDC progeny in the SI LP and in other tissues analyzed were mainly CD103+ cDCs (mucosal tissues) or CD8α+ cDC (in the spleen) on both day 4 and 7 (Fig. 3b; and data not shown). By day 7, most pre-μDC-derived CCR9+ pDCs and pre-μDC themselves had disappeared from all tissues, suggesting rapid turnover of pre-μDC-derived CCR9+ pDCs and a limited self-renewal capacity of pre-μDCs. Consistent with their preferential homing to the SI LP, a higher percentage of cDCs in the SI LP than in the spleen or the lung originated from pre-μDCs (Fig. 3c). Similar results were seen in lethally irradiated recipients (Fig. S2b). Intestinal CD103+ cDCs comprise 2 distinct subsets: CD11b+ and CD11b−. Pre-μDCs gave rise to both CD103+CD11b− and CD103+CD11b+ intestinal cDCs in the SI LP (Figs. 3b and d). The intestinal CD103+CD11b− cDCs are developmentally related to the splenic CD8a+CD11b− cDCs, as their development depends on common transcription factors15,16. No developmental equivalent of the intestinal CD103+CD11b+ cDCs have been identified in the spleen. Most of the pre-μDC-derived CD8α+CD11b− cDCs in the spleen expressed a low level of CD103 (data not shown). Bogunovic M. et al have shown that migratory pre-cDC (Lineage−CD11c+MHCII−CD135+Sirpαlow) can also give rise to CD103+ cDC in the SI LP2. When co-transferred with pre-cDC, pre-μDC are 7–10 fold better than pre-cDC at giving rise to cDC in the SI LP on a per cell basis (Figure S2c). This is not surprising because unlike pre-μDC, pre-cDC do not express gut-specific homing receptors and thus are not primed to repopulate the intestines (see Figure S1c).
Figure 3. Pre-μDCs give rise to CD103+ cDCs and to CCR9+ pDC in vivo and preferentially reconstitute the small intestine.
One to 2.5 million FACS-sorted pre-μDCs (CD45.2) were injected into CD45.1 recipients and analyzed on day 4 and day 6 or 7. Day 4: n=4 from two independent experiments. Day 6 or 7: n=8 from five independent experiments. * p<0.05, ** p<0.01, *** p<0.001 by Student’s t-test. a) CCR9, α4β7 and B220 expression on total pre-μDC-derived cells (CD45.2) in SI IEL (left) and LP (right) on day 4 after transfer. Numbers indicate percentage cells. Data shown are from one of four recipients with similar results. b) CD103 and CD11b on pre-μDC-derived CD11c+ cells and host CD11c+ cells on day 7. The plot shows data from one of eight recipients with similar results. c) Pre-μDC-derived CD11c+ cells (per million input cells) as percentage of host CD11c+ cells in different tissues on day 4 and day 6 or 7. Symbols of the same shape and color represent tissues from the same animal. d) Pre-μDC-derived CD103+CD11b− and CD103+CD11b+ cDCs from SI LP or MLN and CD8α+CD11b− and CD8α−CD11b+ cDCs from spleen are shown as percentage of the corresponding subset from the host on day 4 and day 6 or 7 after transfer. e) Aldefluor staining of pre-μDC-derived and host CD11c+ cDCs from MLN and spleen. Negative control staining in the presence of ALDH inhibitor DEAB is shown for comparison. Data are representative of three independent experiments with similar results. f) Pre-μDCs were sorted from Flt3L-treated mice and pre-incubated with 250 μg of either α4β7-blocking antibody (DATK32) or isotype control for 10 minutes at room temperature before transfer into congenic recipients. Recipient mice received 250 μg of α4β7-blocking antibody or isotype control every 12 hours and were sacrificed on day 4. Pre-μDC-derived total CD11c+ cells were calculated as percentage of host CD11c+ cells. Data show combined results from analyses of three mice from two independent experiments. Error bars show SEM. * p<0.05 by Student’s t-test.
Few pre-μDC-derived cells were seen in the MLN 4 days after transfer but by day 7, significant fractions of CD103+CD11b− and CD103+CD11b+ cDCs in the MLN were derived from pre-μDCs (Fig. 3d). Given preferential homing of pre-μDC to the LP rather than the MLN, pre-μDCs likely differentiate into CD103+ mucosal DCs in the gut wall and subsequently migrate to the MLN. This is consistent with the known migratory behavior of small intestine CD103+ cDCs 17. Importantly, pre-μDC-derived cDCs (CD11c+MHCII+B220−CD103+CD11b+ or CD11b−) in GALT displayed the characteristic aldehyde dehydrogenase (ALDH) activity of endogenous CD103+ gut cDCs: ~50% of pre-μDC-derived cDCs in the MLN stained strongly with Aldefluor™ (Fig. 3e), a fluorogenic substrate used to detect ALDHs important in the metabolism of retinal to retinoic acid 18. In contrast, pre-μDCs in the BM (data not shown) and pre-μDC-derived splenic cDCs did not display ALDH activity (Fig. 3e). Thus, pre-μDCs give rise to both CD11b+ and CD11b− subsets of CD103+ cDCs in the SI LP and acquire mucosal characteristics in response to the intestinal or GALT environment. Furthermore, antibody blockade of α4β7 during pre-μDC transfer drastically reduced the number of pre-μDC-derived cDC in the LP, but not in the spleen or lung (Fig. 3f), consistent with an important role for α4β7-mediated homing in the pre-μDC contribution to the intestinal DC pool.
CD11cintB220+ DCs were comprised of α4β7+ pre-μDCs, CCR9+ pDCs, and a subset of α4β7−CCR9− (double negative or DN) cells (Fig. 1, Fig. S3a). The DN DCs are particularly abundant relative to pre-μDCs in the bone marrow. When cultured in vitro with Flt3L, DN DCs gave rise rapidly (within 24 hours) to CCR9+ pDCs and to α4β7+ pre-μDCs (Fig. S3b). Although both DN DCs and pre-μDCs gave rise to CCR9+ pDCs in vitro, DN DCs did so more rapidly and resulted in a higher proportion of CCR9+ pDCs (compare Fig. S3b and Figs. 2b and 2c). In contrast, sorted CCR9+ pDCs retained their phenotype in culture, indicating that these cells were terminally differentiated (data not shown). Like pre-μDCs, upon longer culture DN DCs developed into cDCs, including CD103+ cDC, although cDC development from DN DCs was delayed relative to that from pre-μDCs (Fig. S3c). When transferred into normal or irradiated recipients, DN DCs were slightly less efficient than pre-μDC at generating cDCs in lymphoid tissues and did not preferentially contribute to intestinal vs. splenic cDC populations at the time points examined (day 5–7, Fig. S3d). These results suggest that pre-μDCs arise from DN DCs in the BM, but that pre-μDCs may be more developmentally primed to migrate and reconstitute peripheral tissues, especially the intestinal tract, than are DN DCs.
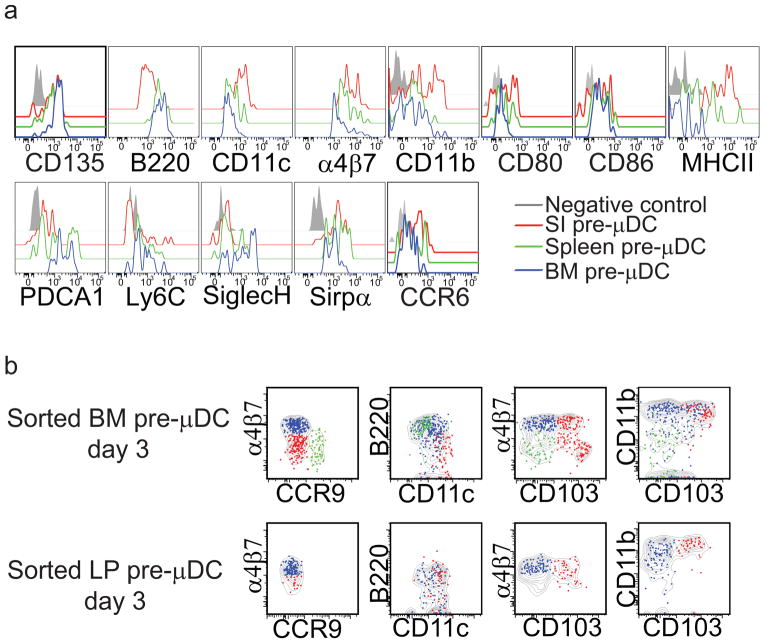
Intestinal pre-μDCs display a transitional phenotype but retain progenitor activity
Pre-μDCs are most highly represented within the GALT and SI LP of those tissues evaluated. Compared to BM pre-μDCs, pre-μDCs in the LP displayed higher expression of CD11c, but lower or no expression of pDC markers including B220, PDCA1, Ly6C, and Siglec H (Fig. 4a). LP pre-μDCs also appeared more mature than their BM counterparts, with higher MHCII, CD11b,α4β7, and CCR6 expression but similar CD80 and CD86 co-stimulatory molecule expression (Fig. 4a). Splenic pre-μDCs had an intermediate phenotype (Fig. 4a). When cultured with Flt3L and total BM cells, SI LP pre-μDCs retained the ability to generate CD103+ cDCs; but they displayed little or no potential to give rise to pDCs (Fig. 4b). Furthermore, ~ 30% of LP pre-μDC expressed proliferating cell nuclear antigen (PCNA) and transferred pre-μDC can still be detected 4 days after transfer, suggesting that pre-μDCs can proliferate locally in the SI LP (data not shown). We conclude that pre-μDCs in the LP retain cDC progenitor activity, but have a transitional phenotype likely reflecting their programmed development into CD103+ intestinal cDCs.
Figure 4. SI pre-μDCs are more differentiated than BM pre-μDCs and have lost potential to give rise to CCR9+ pDCs.
a) Surface antigen expression on pre-μDCs from the BM, spleen, and SI LP. b) pre-μDCs were FACS sorted from BM and SI LP and cultured with congenic total BM feeder cells in complete RPMI supplemented with 100 ng/ml rFlt3L. Cultures were analyzed by flow cytometry on day 3. Pre-μDC-derived subsets are coded as follows: green, CCR9+; blue, α4β7+; red, CD103+; grey contour plot indicates total pre-μDC-derived cells. Data are representative of two experiments with similar results.
Retinoic acid controls pre-μDC development in the BM
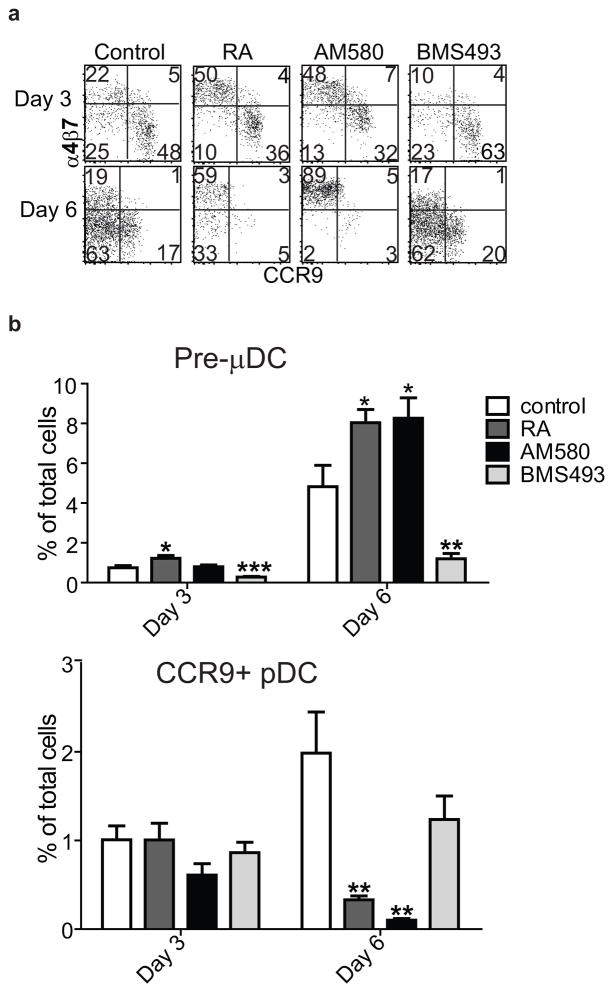
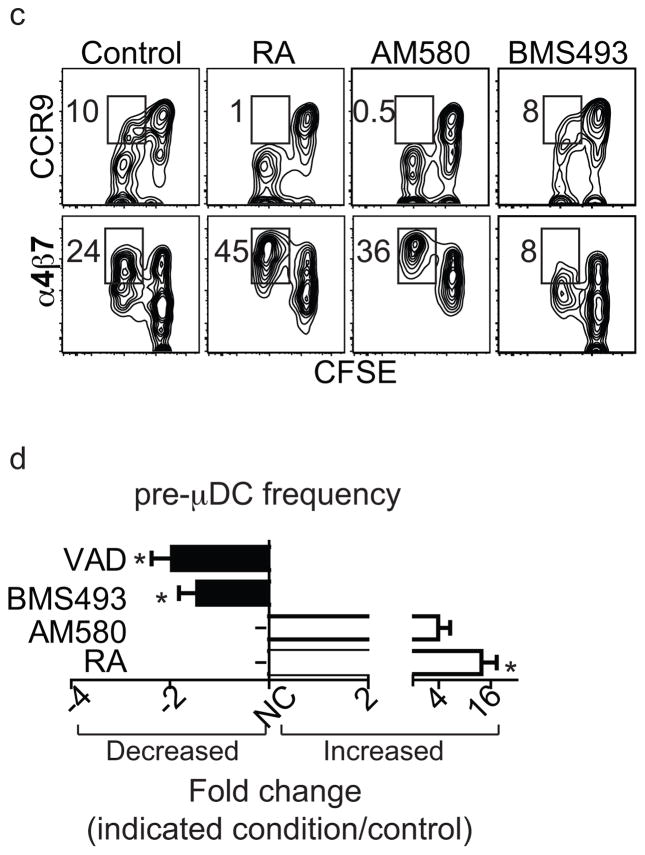
We next asked whether RA regulates the development of α4β7+ pre-μDCs from BM progenitors. Addition of RA to cultures of total BM dramatically increased the frequency of pre-μDCs among total progeny cells at the expense of CCR9+ pDCs and DN DCs (Figs. 5a and 5b and data not shown). Consistent with a requirement for RA signaling through its nuclear receptor RARα, pre-μDC generation was also enhanced by the selective RARα agonist AM580 (Figs. 5a and 5b, S4a). When cultured with delipidated serum, which is free of retinoids, fewer pre-μDC were generated. When RA or AM580 was added to the culture with delipidated serum, pre-μDC generation was enhanced (Fig. S4b). Furthermore, addition of the pan-RAR inverse agonist BMS493 or pan antagonist LE540 inhibited pre-μDC development in vitro (Figs. 5a and 5b and data not shown). Like pre-μDC found in the BM in vivo, pre-μDC generated in vitro with or without RAR signaling are mostly CD103− (Fig S4c). To assess the involvement of proliferation in these effects, we labeled BM with CFSE prior to culture. Under control conditions or with the RAR antagonist, a significant fraction of developing CCR9+ pDCs were derived from cells that had divided, but RA or the RARα agonist almost completely eliminated this population of previously divided CCR9+ pDCs. Given the limited self-proliferative potential of CCR9+ pDCs, it is likely that RAR signaling directly inhibits CCR9+ pDC development from progenitors in the BM. In contrast, RA signaling enhanced the percentage of α4β7+ pre-μDCs arising from dividing cells (Fig. 5c). Thus RA regulates the generation of pre-μDC.
Figure 5. Retinoic acid regulates pre-μDC development in the BM.
a–b) Total BM cells were cultured in complete media supplemented with 100 ng/ml of Flt3L in the absence (control) or presence of 1 nM RA, 10 nM AM580 (an RARα agonist), or 100 nM BMS498 (a pan RAR inverse agonist). Cells were harvested on days 3 and 6 and stained for pre-μDC and CCR9+ pDC markers. a) Cells were gated on Lin−CD11c+B220+ (Lin indicates CD3, CD19, DX-5, Ter-119, Ly6G). Numbers in each quadrant indicate percent cells. b) Percent of pre-μDCs (top) and CCR9+ pDCs (bottom) on days 3 and 6 in total BM cultures, grown under the conditions described in panel a legend. Data show combined results from four independent experiments with n=6–8 for each condition. Error bars show SEM. * p<0.05, **p<0.01, ***p<0.001 compared to control by Student’s t-test. c) Total BM cells were labeled with CFSE and cultured as described in the panel a legend. Cultures were harvested on day 3 and cells were gated on Lin−CD11c+B220+. Numbers indicate percentage cells. d) Fold change of pre-μDC frequency in BM from mice treated with each indicated condition compared to control. VAD: vitamin A deficient mice, n=10, control: n=8; BMS493/AM580/RA: mice received BMS493 or vehicle (for 7–18 days, n=4 each), AM580 or vehicle (4–6 days, n=2) or RA or vehicle (4–6 days, n=3). Pre-μDC frequency (among total BM cells) was determined. The fold change in frequency in treated vs. control mice in each paired comparison was calculated, and is shown with SEM or (for AM580) range. * p<0.05 by Mann Whitney test between treated and control mice.
Manipulation of the RAR signaling pathway had similar effects on pre-μDC development in vivo. Mice treated with RA and AM580 displayed an increase in pre-μDC frequency in the BM, consistent with our in vitro observations. In contrast, treatment with BMS493 reduced the frequency of pre-μDCs (Figs. 5d). More importantly, pre-μDCs were also significantly reduced in the BM of mice fed a vitamin A-deficient diet compared to mice fed a control diet (Fig. 5d). We conclude that vitamin A and its metabolite RA regulate the development of a targeted precursor for intestinal dendritic cells.
DISCUSSION
We have identified a migratory common DC precursor that preferentially homes to the gut wall and gives rise to intestinal DCs, especially CD103+ subsets. Pre-μDCs have an immature phenotype and share phenotypic features with CCR9+ pDCs, including expression of pDC markers such as PDCA1, Siglec H, Ly6C, and B220, but are distinguished from CCR9+ pDCs by differential trafficking receptor expression and by their developmental potential. Pre-μDCs expressα4β7 but not CCR9 and proliferate and differentiate into cDCs and CCR9+ pDCs in vitro and in vivo, whereas conventional CCR9+ pDCs are α4β7− and are terminally differentiated. Bone marrow pre-μDCs are closely related to and can arise from CD11cintB220+CCR9−α4β7− DN DCs.
Development of pre-μDCs in the bone marrow is regulated by retinoic acid. Treatment of BM cultures with RAR agonists increased and RAR antagonists decreased the frequency of pre-μDCs. All trans-RA, a pan-RAR agonist, and AM580, an RARα specific agonist, had similar effects on pre-μDC development, suggesting that modulation of RAR signaling in the BM is primarily through RARα. RAR signaling had a similar modulating effect on pre-μDC development in vivo. Vitamin A deficiency, which leads to reduced circulating retinol11, reduced the number and frequency of pre-μDCs in the BM. Similarly, systemic treatment of mice with the pan RAR inverse agonist BMS493 reduced pre-μDC, whereas intraperitoneal administration of all trans-RA or the RARα agonist AM580 increased the frequency and number of pre-μDCs dramatically. RA regulates hematopoietic progenitor development, and modulation of RARa/RXR signaling regulates hematopoietic stem cell (HSC) differentiation, proliferation, and self-renewal19–21. Furthermore, RA modulates neutrophil differentiation from myeloid progenitors under steady state conditions22. While BM pre-μDCs lack ALDH activity (as indicated by Aldefluor assay; data not shown), subsets of BM hematopoietic cells, including HSCs, express retinaldehyde dehydrogenases21,23, providing a potential source for RA generation from retinol within the local bone marrow environment.
Earlier studies showed that retinoic acid regulates cDC function within the intestinal LP and GALT. RA induces the RALDH gene Aldh1a2 in intestinal DCs10. RALDH allows intestinal cDCs to convert retinal to retinoic acid, creating a positive feedback loop and also allowing GALT DCs to present RA along with antigen to T cells. RA in turn induces T cell expression of the gut homing receptors α4β7 and CCR9. In this manner, T cells responding to gut antigens are programmed to traffic back to the intestines11. Interestingly, although RA also induces α4β7 expression on developing pre-μDCs, it does not induce CCR9 expression. Indeed, expression of α4β7 and CCR9 is mutually exclusive among B220+CD11cint DCs, and RA actually suppresses the development of CCR9-expressing pDCs in our in vitro and in vivo models. Thus the signaling pathways that control expression of these trafficking receptors must differ in DCs and lymphocytes. BM pre-μDCs do not display intrinsic aldehyde dehydrogenase activity, but we show that their CD103-expressing cDC progeny upregulate ALDH activity within the gut environment. Furthermore, pre-μDC derived splenic cDC and CD103+CD11b− intestinal cDC express CD8α in vivo but pre-μDC derived CD103+ cDC do not express CD8α in vitro. This suggests that the in vivo tissue environments of the spleen or LP must comprise additional factors or stimuli not present in our in vitro culture system.
We focused primarily on cDC progeny in our analyses, even though pre-μDCs rapidly differentiated into CCR9+ pDCs as well both in vitro and in vivo. Pre-μDC-derived pDCs were especially abundant relative to pre-μDC-derived cDCs in the BM and the IEL compartments of the gut wall of adoptive recipients. This distribution parallels the characteristic homeostatic representation of DC subsets in these sites, where classical CCR9+ pDCs significantly outnumber cDCs (whereas cDCs predominate in most other tissues). Although pre-μDC-derived pDCs generated in the BM or other tissues can likely home to the gut after upregulation of CCR9, pre-μDCs may also be able to generate pDCs locally in the gut after entry from the blood. Importantly, in the gut, pre-μDC-derived pDCs appeared rapidly and then disappeared from the recipient IEL compartment within 5–6 days, whereas donor-derived intestinal cDCs remained readily detectable for at least 10 days after pre-μDC transfer. Overall, the tissue- and time-dependent representation of pre-μDC-derived pDCs and cDCs likely reflects multiple factors including environmental effects on pre-μDC differentiation into pDCs vs. cDCs, intrinsic and/or environment-specific differences in the survival and proliferation of progeny pDCs vs. cDCs, and a limited self-renewal potential of pre-μDCs. Another potential factor is the apparent progressive loss of pDC potential after pre-μDCs enter peripheral tissues, as evidenced by the inability of pre-μDCs from the SI LP to generate pDCs in culture.
In the BM, pre-μDCs are a minor subset of the B220+CD11cint DCs: The majority of B220+ DCs are CCR9+ pDCs and CCR9−α4β7− DN DCs. Importantly, DN DCs depleted of pre-μDCs gave rise to pre-μDCs in culture, and thus contain precursors for both pre-μDC and CCR9+ pDC. DN DC conversion into CCR9+ pDC and α4β7+ pre-μDC phenotypes occurred rapidly, within 2 days, and did not require cell division (data not shown). DN DCs also gave rise to cDCs, including CD103+ cDCs, in longer-term culture. These findings are consistent with the reported ability of splenic and BM CD11cintB220+CCR9− DCs to give rise to cDCs24,25 (and see next paragraph). Moreover, culture of classical CD11c− CDPs with Flt3L also led to production of DN DCs and pre-μDCs, as well as to pDCs and cDCs (data not shown). In the simplest interpretation, our data suggest a linear model in which bone marrow progenitors such as the common DC progenitor (CDP) and possibly early progenitors such as the macrophage, dendritic cell progenitor (MDP) give rise to CD11cintB220+ DN DCs, which in turn differentiate into pre-μDCs that rapidly exit the marrow and home to the gut to replenish intestinal DCs (Figure S5). However, given the emerging appreciation of nonlinear developmental sequences in hematopoiesis and of the malleability of progenitors in response to environmental signals25–27, we cannot exclude other pathways to pre-μDC development.
The gut homing ability of pre-μDC, their capacity to differentiate into pDC as well as cDC, and their preferential contribution to intestinal DC distinguishes them from other migratory DC precursors. BM pre-cDCs (lineage−B220−CD11c+MHCII− CD135+Sirpαlow cells) described by Liu et al. 28 and splenic pre-cDC (CD11cint MHCII−CD45RAlow CD43int Sirpαint) described by Naik SH et al.29 have a more restricted developmental potential, contributing to peripheral cDCs but not pDCs. Moreover, although pre-cDCs give rise to CD103+ intestinal DCs as well as other peripheral DC populations, they lack specialized gut homing ability (they are α4β7 and CCR9 negative). In fact, in our co-transfer experiments, pre-μDC are 7–10 fold better than pre-cDC at contributing to intestinal cDC on a per cell basis. Pre-μDC are more closely related phenotypically to recently identified “CCR9−pDC-like precursors”, a common DC precursor which (like pre-μDC and the DN DC population studied here) is B220+ and expresses intermediate levels of CD11c: Schlitzer et al25,30 showed elegantly that although these CCR9− pDC precursors can be induced to differentiate into cDC by GM-CSF or microenvironmental influences, they preferentially give rise to pDCs. This contrasts with the preferential cDC development displayed by pre-μDCs. Phenotypically, CCR9- pDC precursors are defined as SiglecHhi PDCA1hi CCR9− CD11c+ and B220+ 25,30; as such they comprise a small subset of pre-μDC (the PDCA1hiSiglecHhi fraction), as well as a large fraction of DN DCs enriched for high levels of expression of the pDC differentiation antigens PDCA1 and SiglecH. We have shown that, in contrast to pre-μDC but similar to Schlitzer’s pDC precursors, CCR9−α4β7− DN DCs preferentially give rise to pDCs at early time points. We propose therefore that the Lin−B220+C11cint CCR9− population of bone marrow cells comprises a diverse precursor pool for peripheral DC subsets, in which functional precursors with a preference towards pDC development are enriched in the PDCA1hi SiglecH hiα4β7−CCR9− pDC-like subset; whereas precursors biased towards development into intestinal CD103+ DCs are enriched in the gut homing α4β7hi pre-μDC population. Indeed, pre-μDC themselves may well be heterogeneous in potential: we cannot exclude the possibility that some pre-μDC are clonally committed to pDC vs. cDC development, or indeed that B220+C11cint BM DC, including gut tropic pre-μDC, are composed of distinct subsets of committed pDC vs. cDC precursors. This pool of B220+ DC precursors was not recognized in earlier studies that included B220 as a marker of exclusion in a “lineage negative” sort gate28. In the BM, pre-μDCs are a minor subset of this pool, which may reflect programming for rapid exit from the marrow to seed the gut wall. In addition to these CD11c+ migratory precursors, earlier progenitors, including CD11c− CDPs and even pluripotent hematopoietic stem cells, may be able to seed peripheral tissues and contribute to tissue DC populations31,32. The rates of generation and exit of each of these distinctive DC precursors, their relative contributions to CD103+CD11b− vs. CD103+CD11b+ intestinal DC and to other peripheral DC populations, and their relative importance in homeostatic vs. inflammatory settings remain to be determined.
The recruitment of blood leukocytes requires adhesion and chemoattractant receptors to mediate endothelial adhesion, arrest, and subsequent diapedesis33. Pre-μDCs express α4β7, an integrin receptor for the mucosal vascular addressin MAdCAM1, on gut postcapillary venules, and blockade of α4β7 prevents pre-μDCs from contributing to intestinal DC populations. It is unclear which chemoattractant receptors are involved in this process. CCR9 is an important chemokine receptor for T and B lymphocytes and in CCR9+ pDC homing to the SI. However, pre-μDCs do not express CCR9. Moreover blocking antibody to the CCR9 ligand CCL25 had no effect on pre-μDC-derived cDC in the SI, and pre-μDCs from CCR9-knockout mice homed to and gave rise to CD103+ cDCs in the SI as efficiently as wild-type cells (data not shown). Although BM pre-μDCs express minimal CCR6, CCR6 is upregulated on pre-μDC in the SI LP, thus CCR6 might contribute to pre-μDC homing. Pre-μDCs also express CCR5 and CXCR3 that could participate in gut homing. On the other hand, pre-μDC lack the lymphoid organ trafficking receptor CCR7, consistent with their inability to home well to mesenteric lymph nodes in spite of α4β7 expression.
In conclusion, we identify a gut homing common DC precursor whose development in BM is regulated by retinoic acid and impaired in vitamin A deficiency. Pre-μDCs can generate cDC and CCR9+ pDC, but more efficiently give rise to CD103+ cDC populations in the small intestines. Gut-specific homing of DC precursors may allow targeted replacement of intestinal dendritic cells as a function of ongoing tissue-specific immune requirements.
MATERIALS AND METHODS
Mice
C57Bl/6.CD45.2 (B6.CD45.2) and C57Bl/6.CD45.1 (B6.CD45.1) mice were originally purchased from Jackson Laboratory and were maintained and bred in specific pathogen-free conditions in the animal facility in Veterans Affairs Palo Alto Health Care Systems (VAPAHCS). To generate B16/Flt3L-injected mice, 5 million B16/Flt3L cells were injected subcutaneously near the neck and animals were sacrificed 11–14 days later. All animals were used in accordance with guidelines set forth by the animal committee of VAPAHCS. Vitamin A deficient and control mice were generated as described11. In some experiment, mice were i.p. injected with BMS493 (25mM in DMSO) at 1μl per gram of weight in 25 μl of olive oil. Control animals received the same amount of DMSO in olive oil. All trans-retinoic acid was made into suspension in olive oil at 25mg/ml and injected i.p. at 125μg per gram of weight, control animals received olive oil. AM580 was dissolved in DMSO at a concentration of 40mg/ml and injected i.p. at 1μl per gram of weight with 100ml of olive oil; control animals received the same amount of DMSO in olive oil.
Flow cytometry
Samples (single-cell suspensions) were first blocked with FACS buffer (HBSS with 2% FCS) containing 100X dilution of antibody against mouse FcγIII/II receptor (BD Bioscience) and rat serum to prevent non-specific binding of monoclonal antibodies. The following antibodies were used for staining: CD3-PECy7/CD3-Biotin/CD3-PerCpCy5.5 (145-2C11), CD19-PECy7/CD19-biotin/CD19-PerCPCy5.5 (ID3), NK1.1-PECy7/NK1.1-biotin/NK1.1-PerCPCy5.5 (PK136), CD49b-Biotin(DX-5), Ly6G-Biotin (1A8), Ter-119(Ter-119), B220-PECy7/B220-Biotin/B220-PerCPCy5.5 (RA3-6B2), MHCII-AF700(M5/114.15.2)/MHCII-Biotin (2G9), CD11c-PB (N418),α4β7-APC/α4β7-PE (DATK32), CCR9-APC/CCR9-PE/CCR9-FITC (242503), CCR9-PECy7 (CW1.1), CD103-PE(M290), CD11b-AF700 (M1/70), CD8α-PE (53-6.7), CD45.1-PerCPCy5.5/CD45.1-APC, CD45.2-FITC/CD45.2-PerCPCy5.5 (RA3-6B2), CD135-PE/CD135-PECy5 (A2F10), CD115-APC/CD115-FITC (AFS98), CD117-PECy7 (2B8), PDCA1-FITC/PDCA1-PerCP-eFluor710(927), SiglecH-FITC (eBio440C), Ly6C-FITC(AL-21), CD4-AF700(RM4-5), CD9-AF647 (MZ3), Sirpα-FITC/Sirpα-PerCP-eFluor710 (P84), α4-PE (9C10, BD), β1-PECy7 (HMb1-1, eBioscience), αV-PE (RMV-7, BD Bioscience), β3-PE(2C9.G2, BD Bioscience), CD62L-FITC (MEL14), CXCR3-APC (CXCR3-173, Biolegend), CCR2-APC (475301, R&D), CCR5-PE (2D7/CCR5, BD Bioscience), CCR7-APC(4B12, eBioscience), CXCR5-PE (BD, 2G8) and CCR6-PE (140706, R&D).
Cell isolation from tissues
Femurs and tibias were collected and crushed using a 5ml syringe. Bone fragments were removed by filtering through a wire mesh to isolate bone marrow cells.
Spleens, inguinal, auxiliary and brachial lymph nodes (LNs) were isolated and digested with RPMI media containing 5% FCS, 500 unit/ml of collagenase IV (Worthington Biochemical)and 1 unit/ml of DNaseI(Sigma) for 30 minutes at 37°C and made into single cell suspension.
Lungs were perfused with 25 ml of PBS and cut into small pieces (~1–2mm) and digested as were spleen and LNs.
Full-length SI (with Peyer’s Patches removed) was cut open longitudinally and rinsed twice in HBSS (without Ca2+ and Mg2+) with 2% FCS and cut into small pieces. To isolate IEL, the cut SI was incubated twice in 10 ml of 2% HBSS containing 15 mg/100ml dithiothreitol (DTT) at 37 °C for 20 minute, twice, and supernatant containing IEL was collected after each incubation. Residual tissues were digested in 10 ml RPMI containing 5% FCS and 0.5 mg/ml of collagenase IV (Sigma C2139) at 37°C for 30 minutes, three times. Supernatant containing LP cells was collected. Mononucleated cells from the IEL and LP supernatant were isolated by gradient separation with 40% and 75% percoll solutions (GE, 17-0891-01) at 2000 RPMI for 20 minutes at room temperature. Cells of interest were located at the interface. When young mice (10–16 days of age) were used, Peyer’s Patches were not removed because they are not clearly visible at this age.
Cell sorting
To sort pre-μDCs and DN DCs from normal and Flt3L-treated mice, BM cells were isolated as described above and enriched by magnetic activated cell sorting (MACS) using the pan-DC kits from Miltenyi. The cells were then sorted on Aria II or III (BD) for Lineage (CD3, CD19, NK1.1)-CD11cintB220+α4β7+CCR9− pre-μDC and Lineage− CD11cintB220+α4β7−CCR9− DN DC.
Adoptive transfer
For transfer into lethally irradiated recipients, B6.CD45.1/45.2 F1 mice were given full body irradiation with a lethal dose of 900 rads total from a 131 Cs source at two doses (4 hours apart, 450 rads each). Irradiated mice were given total BM (CD45.1) and sorted pre-μDCs (CD45.2 or vice versa) at a ratio of 10:1 (5×106 total BM and 0.5×106 sorted pre-μDC). Mice were kept on antibiotic water until they were sacrificed on day 4,7,10 or 14. Adoptive transfer into normal recipients was performed the same way except recipient mice were not irradiated and received only sorted pre-μDC (1–3×106). For homing experiments, 3×106 sorted pre-μDCs were transferred into each congenic recipient.
Blocking experiments
Sorted pre-μDC from Flt3L-treated mice were pre-blocked with 250 μg of anti-α4β7 antibody made in house (DATK32) or isotype control (Rat IgG2a) in 200 ml PBS for 10 minute at room temperature before injection. Subsequently recipients received blocking antibody or isotype control every 12 hours via intraperitoneal injection until mice were sacrificed. All cell transfers were done via retro-orbital injection.
In vitro culture
Pre-μDCs and DN DCs were sorted from BM (or SI LP for pre-μDC in some experiments) from normal or Flt3L-treated mice and cultured with congenic total BM cells at a density of 3–5 million cells/ml, 200μl per well in flat-bottom 96-well plate in complete RPMI media (10% FCS, 1X penicillin/streptomycin) supplemented with 100ng/ml recombinant Flt3L. Media were changed on day 3. Total BM cells were cultured in complete media with normal FCS or delipidated FCS with 100ng/ml of rFlt3L in the presence of 1nM of ATRA, 10nM of AM580 or 100nM of BMS493. Media were changed on day 3.
CFSE labeling: Total BM cells were incubated with 1μM of CFSE at a density of 10E6 cells/ml in PBS for 10 min at 37°C and washed by incubating in complete media for 5 min at 37°C.
Analysis of ALDH activity
Aldehyde dehydrogenase (ALDH) activity in each cell was analyzed using Aldefluor Staining Kit (StemCell Technologies) per the manufacturer’s protocol with modifications as previously described18. Briefly, cells were suspended in Aldefluor assay buffer containing activated substrate (150 nM) with or without the ALDH inhibitor DEAB (100μM) at a density of 10E6 cells/ml and incubated in 37°C water bath for 30 minutes. Subsequent surface antigen staining was performed in Aldefluor assay buffer. Cells were immediately analyzed on LSRII analyzer (BD) without fixing.
Statistical Analysis
The statistical significance of differences between two sets of data was assessed by Student’s t-test unless stated otherwise.
Supplementary Material
Acknowledgments
We thank L. Rott for assistance with flow cytometry and cell sorting and WW Agace for critical comments on early studies. The B16 melanoma cell line stably transfected with murine Flt3L was a kind gift from G. Dranoff, Dana-Farber Cancer Institute, Boston, MA. R.Z. is a recipient of the National Science Scholarship awarded by the Agency for Science, Technology And Research, Singapore; C.O. was supported by the Wenner-Gren Foundation, and by a Research Fellowship Award (#2791) from the Crohn’s & Colitis Foundation of America; R.Y. is supported by the Immunology training grant (5 T32 AI07290, Molecular and Cellular Immunobiology); M.L. is a recipient of a senior fellowship from the Crohn’s and Colitis Foundation, and was a fellow under the NIH Training Grant AI07290; H.H. is a recipient of an Investigator Career Award from the Arthritis Foundation and was a fellow under the NIH Training Grant AI07290; A.H. was supported by NIH grant DK085426. The work was supported in part by NIH grants AI047822, AI093981 and DK084647 to E.C.B., the FACS Core facility of the Stanford Digestive Disease Center under DK056339, and a Merit Award from the Department of Veterans Affairs.
Footnotes
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
R.Z. designed and performed experiments and wrote the manuscript; C.O carried out early phenotypic and other studies that led to the definition of pre-μDC, and performed the homing studies; R.Y. and M.L. performed experiments; H.H. was involved in the initiation of the project, including initial studies of pre-μDC development, and provided advice; A.H assisted with the homing study and provided advice; E.C.B directed the study and wrote the manuscript. All authors reviewed and commented on the manuscript.
References
- 1.Annacker O, Coombes JL, Malmstrom V, et al. Essential role for CD103 in the T cell-mediated regulation of experimental colitis. J Exp Med. 2005 Oct 17;202(8):1051–1061. doi: 10.1084/jem.20040662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogunovic M, Ginhoux F, Helft J, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009 Sep 18;31(3):513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki A. Mucosal dendritic cells. Annu Rev Immunol. 2007;25:381–418. doi: 10.1146/annurev.immunol.25.022106.141634. [DOI] [PubMed] [Google Scholar]
- 4.Johansson-Lindbom B, Svensson M, Pabst O, et al. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005 Oct 17;202(8):1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva MA. Intestinal dendritic cells and epithelial barrier dysfunction in Crohn’s disease. Inflamm Bowel Dis. 2009 Mar;15(3):436–453. doi: 10.1002/ibd.20660. [DOI] [PubMed] [Google Scholar]
- 6.Salazar-Gonzalez RM, Niess JH, Zammit DJ, et al. CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity. 2006 May;24(5):623–632. doi: 10.1016/j.immuni.2006.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varol C, Vallon-Eberhard A, Elinav E, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009 Sep 18;31(3):502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007 Aug 6;204(8):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. Feb 5;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaensson-Gyllenback E, Kotarsky K, Zapata F, et al. Bile retinoids imprint intestinal CD103+ dendritic cells with the ability to generate gut-tropic T cells. Mucosal immunology. 2011 Jul;4(4):438–447. doi: 10.1038/mi.2010.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004 Oct;21(4):527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 12.Hill JA, Hall JA, Sun CM, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008 Nov 14;29(5):758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berlin C, Berg EL, Briskin MJ, et al. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993 Jul 16;74(1):185–195. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 14.Mach N, Gillessen S, Wilson SB, Sheehan C, Mihm M, Dranoff G. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000 Jun 15;60(12):3239–3246. [PubMed] [Google Scholar]
- 15.Edelson BT, Kc W, Juang R, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. The Journal of experimental medicine. 2010 Apr 12;207(4):823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agace WW, Persson EK. How vitamin A metabolizing dendritic cells are generated in the gut mucosa. Trends in immunology. 2012 Jan;33(1):42–48. doi: 10.1016/j.it.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Schulz O, Jaensson E, Persson EK, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009 Dec 21;206(13):3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokota A, Takeuchi H, Maeda N, et al. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. International immunology. 2009 Apr;21(4):361–377. doi: 10.1093/intimm/dxp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chute JP, Ross JR, McDonnell DP. Minireview: Nuclear receptors, hematopoiesis, and stem cells. Mol Endocrinol. 2010 Jan;24(1):1–10. doi: 10.1210/me.2009-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Safi R, Muramoto GG, Salter AB, et al. Pharmacological manipulation of the RAR/RXR signaling pathway maintains the repopulating capacity of hematopoietic stem cells in culture. Mol Endocrinol. 2009 Feb;23(2):188–201. doi: 10.1210/me.2008-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chute JP, Muramoto GG, Whitesides J, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2006 Aug 1;103(31):11707–11712. doi: 10.1073/pnas.0603806103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastner P, Chan S. Function of RARalpha during the maturation of neutrophils. Oncogene. 2001 Oct 29;20(49):7178–7185. doi: 10.1038/sj.onc.1204757. [DOI] [PubMed] [Google Scholar]
- 23.Hess DA, Meyerrose TE, Wirthlin L, et al. Functional characterization of highly purified human hematopoietic repopulating cells isolated according to aldehyde dehydrogenase activity. Blood. 2004 Sep 15;104(6):1648–1655. doi: 10.1182/blood-2004-02-0448. [DOI] [PubMed] [Google Scholar]
- 24.Segura E, Wong J, Villadangos JA. Cutting edge: B220+CCR9-dendritic cells are not plasmacytoid dendritic cells but are precursors of conventional dendritic cells. J Immunol. 2009 Aug 1;183(3):1514–1517. doi: 10.4049/jimmunol.0901524. [DOI] [PubMed] [Google Scholar]
- 25.Schlitzer A, Loschko J, Mair K, et al. Identification of CCR9-murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood. 2011 Jun 16;117(24):6562–6570. doi: 10.1182/blood-2010-12-326678. [DOI] [PubMed] [Google Scholar]
- 26.Watowich SS, Liu YJ. Mechanisms regulating dendritic cell specification and development. Immunological reviews. 2010 Nov;238(1):76–92. doi: 10.1111/j.1600-065X.2010.00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franco CB, Chen CC, Drukker M, Weissman IL, Galli SJ. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell Stem Cell. 2010 Apr 2;6(4):361–368. doi: 10.1016/j.stem.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu K, Victora GD, Schwickert TA, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009 Apr 17;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naik SH, Metcalf D, van Nieuwenhuijze A, et al. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nature immunology. 2006 Jun;7(6):663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- 30.Schlitzer A, Heiseke AF, Einwachter H, et al. Tissue-specific differentiation of a circulating CCR9-pDC-like common dendritic cell precursor. Blood. 2012 Jun 21;119(25):6063–6071. doi: 10.1182/blood-2012-03-418400. [DOI] [PubMed] [Google Scholar]
- 31.Schmid MA, Takizawa H, Baumjohann DR, Saito Y, Manz MG. Bone marrow dendritic cell progenitors in mice sense pathogens via Toll-like receptors and subsequently migrate to inflamed lymph nodes. Blood. 2011 Sep 9; doi: 10.1182/blood-2011-03-344960. [DOI] [PubMed] [Google Scholar]
- 32.Massberg S, Schaerli P, Knezevic-Maramica I, et al. Immunosurveillance by hematopoietic progenitor cells trafficking through blood, lymph, and peripheral tissues. Cell. 2007 Nov 30;131(5):994–1008. doi: 10.1016/j.cell.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butcher EC. Leukocyte-endothelial cell recognition: three (or more) steps to specificity and diversity. Cell. 1991 Dec 20;67(6):1033–1036. doi: 10.1016/0092-8674(91)90279-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.