Abstract
The four types of platelet-derived growth factors (PDGFs) and the two types of PDGF receptors (PDGFRs, which belong to class III receptor tyrosine kinases) have important functions in the development of connective tissue cells. Recent structural studies have revealed novel mechanisms of PDGFs in propeptide loading and receptor recognition/activation. The detailed structural understanding of PDGF-PDGFR signaling has provided a template that can aid therapeutic intervention to counteract the aberrant signaling of this normally silent pathway, especially in proliferative diseases such as cancer. This review summarizes the advances in the PDGF system with a focus on relating the structural and functional understandings, and discusses the basic aspects of PDGFs and PDGFRs, the mechanisms of activation, and the insights into the therapeutic antagonism of PDGFRs.
Keywords: growth factor, signal transduction, receptor tyrosine kinase, receptor activation, propeptide recognition
1. Introduction
Platelet-Derived Growth Factors (PDGFs) are a family of four cystine-knot-type growth factors (PDGF-A, -B, -C and -D) which control the growth of connective tissue cells such as fibroblasts and smooth muscle cells [1, 2]. By acting on these mesenchymal cells, PDGFs critically regulate embryonic development, especially the formation of vessels and organs (reviewed in [3]). There are two types of receptors for PDGFs, PDGFRα and PDGFRβ, which belong to the class III receptor tyrosine kinases (RTKs), and have different expression patterns and physiological roles. PDGFRα signaling controls gastrulation and the development of several organs such as lung, intestine, skin, testis, kidney, bones, and neuroprotective tissues. PDGFRβ signaling is better recognized as an essential regulator of early hematopoiesis and blood vessel formation [3]. While PDGF-PDGFR signaling plays important roles during developmental stages, the expression of both PDGFs and PDGFRs are tightly controlled in adulthood. Enhanced PDGF-PDGFR signaling, except when happening briefly during wound repair, is generally considered abnormal, and is an important feature in a number of diseases involving proliferation, including many types of cancers, inflammation, pulmonary fibrosis and restenosis, and notably atherosclerosis [4].
PDGFs signaling through PDGFRs utilizes the general strategy for RTKs, which involves ligand-induced receptor dimerization, and the subsequent receptor conformational changes that are coupled to the activation of intracellular tyrosine kinase domain (reviewed in [5]). The activation of PDGFR signaling pathways is built on structural platforms of both the ligands and the receptors, which are predicted to be conserved family-wise based on the sequence similarities between different PDGF and PDGFR subtypes. However, for decades, except for a crystal structure of the PDGF-B ligand [6], detailed mechanistic and structural understanding of PDGFR recognition and activation had been hampered until the recent elucidation of the PDGF-B:PDGFRβ complex structure. Inhibiting PDGF-PDGFR signaling, especially by selectively blocking the extracellular assembly through antibodies, ligand decoy or receptor decoys, is actively pursued in anti-cancer drug development, as these ligands and receptors serve in multiple aspects of tumor progression, such as mediating tumor growth in an autocrine fashion, recruiting fibroblast-rich tumor stroma, and regulating tumor vasculature [4]. The therapeutic efforts targeting PDGF/PDGFR are well compatible with strategies that can be derived from structural templates of both ligands and receptors. This review will summarize some current structural and functional understandings of the four PDGF ligands and the two PDGFRs.
2. The four types of PDGFs
PDGF(s) were discovered in 1970s as a platelet-dependent serum factor that stimulates the proliferation of fibroblasts, arterial smooth muscle cells, and glial cells [1, 2, 7]. There are four types of PDGF polypeptide chains as encoded by four genes. Of these four genes, PDGF-B was first characterized by amino acid sequencing to reveal its surprisingly close homology to the simian sarcoma virus oncogene v-sis [8, 9]. The cDNA of PDGF-A was subsequently cloned and its chromosomal localization was identified [10]. The PDGF-A and PDGF-B dimeric proteins, together with the biochemically observed PDGF-AB heterodimer, constitute the classical PDGF proteins. In early 2000s, two additional members of the family, PDGF-C and PDGF-D, were identified as new, protease-activated ligands for PDGFRs using genetic and biochemical approaches [11–13].
All PDGFs have a conserved growth factor domain in the cystine-knot fold that is related to vascular endothelial growth factors (VEGFs), which is the only part of PDGFs and VEGFs that is functionally and evolutionarily conserved, and is primarily responsible for recruiting receptors [14]. Outside the growth factor domain, there are significant sequence and domain variations between PDGFs (Fig. 1). As paracrine factors that exert functions in the extracellular space, all PDGFs possess secretion signals sequences with 18–22 amino acids in length that share little homology between different types. Following the signal peptides, both PDGF-A and PDGF-B contain pro-peptide sequences that are ~60 amino acids in length, and are cleaved from the mature growth factors by furin or other proprotein convertases in the intracellular secretion pathway (reviewed in [15]). This sequence in fact is also present in PDGF-C and PDGF-D, as revealed by structure-guided sequence analysis [16]. In addition to this short sequence that should have a role in binding to the mature growth factor domain, PDGF-C and PDGF-D also have a large CUB (Complement subcomponents C1r/C1s, Urchin EGF-like protein, and Bone marrow protein 1) domain at the N-terminal part of the long pro-sequences. At the C-terminus of the growth factor domain, the PDGF-A and PDGF-B gene contain ~30 amino acids and ~60 amino acids tails respectively, which are highly polar in amino acid composition and likely adopt flexible structures. Most of the tail of PDGF-A is missing in an alternatively spliced, short isoform [17]. The tails of PDGF-A and PDGF-B are both rich in positively charged amino acids such as arginine and lysine, and are involved in retention and distribution by binding to heparin and heparin sulfate proteoglycans (HSPGs) [18, 19]. PDGF-C and PDGF-D lack the tail sequences following the growth factor domain for retention, but their CUB domains may regulate the extracellular distribution of latent forms by interacting with other proteins or carbohydrates [20].
Fig. 1.
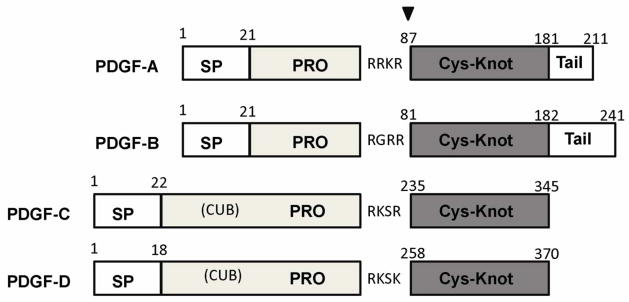
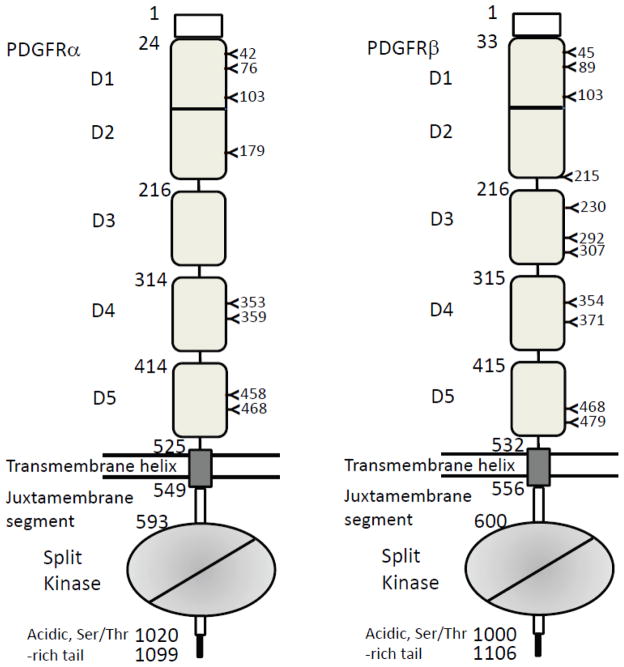
The four types PDGFs and their domain compositions. The starting numbers of specific domains/segments in the coding sequences are marked above the boundaries. Shown are all numbers for human PDGFs. The proprotein convertase-recognition sequences are indicated and positions of cleavage are marked with a black triangle. Abbreviations used: SP: signal peptide; PRO: the pro-sequence preceding the growth factor domain; Cys-Knot, the cysteine-knot growth factor domain that is responsible for receptor recognition; CUB, the Complement subcomponents C1r/C1s, Urchin EGF-like protein, and Bone marrow protein 1-like domain, which can be considered as part of the pro-sequence in PDGF-C and PDGF-D, but PDGF-C and PDGF-D also have sequences homologous to the PDGF-A and PDGF-B pro-sequences.
PDGF-A and PDGF-B are believed to be proteolytically processed already before being secreted (reviewed in [21]), which is consistent with their protease-recognition sequences (RRKR in human PDGF-A, which is ideal for furin specificity [22]; RGRR in human PDGF-B, less specific for furin but is compatible with furin and other furin-like proprotein convertases inside ER [23]). Rather than being processed intracellularly, PDGF-C and PDGF-D are thought to be secreted in latent forms and cleaved in the extracellular space for activation (reviewed in [20]). The responsible extracellular proteases are demonstrated to be plasmin [11, 24] and tPA (tissue plasminogen activator) [25] for PDGF-C, and plasmin [12] and uPA (urokinase plasminogen activator ) [26] for PDGF-D. However, the protease-recognition sequences of PDGF-C and PDGF-D (RKSR for human PDGF-C and RKSK for human PDGF-D, Fig. 1) are well compatible with intracellular ER-residing proprotein convertases such as furin [22], and it is conceivable that significant amount of PDGF-C and PDGF-D secreted are already cleaved inside the cell. Indeed, recent recombinant expression experiments on PDGF-D showing that a high percentage of PDGF-D is already cleaved when secreted, despite that the expression level driven by recombinant cassettes is much higher than the physiological level [16]. For physiologically expressed amounts of PDGF-C and PDGF-D, a larger percentage of secreted proteins should be cleaved by ER-resident proteases because of the lower load. It is possible that, for the already cleaved PDGF-C and PDGF-D, the extracellular proteases modulate their activation by displacing the pro-sequences from the signaling-active growth factor domains.
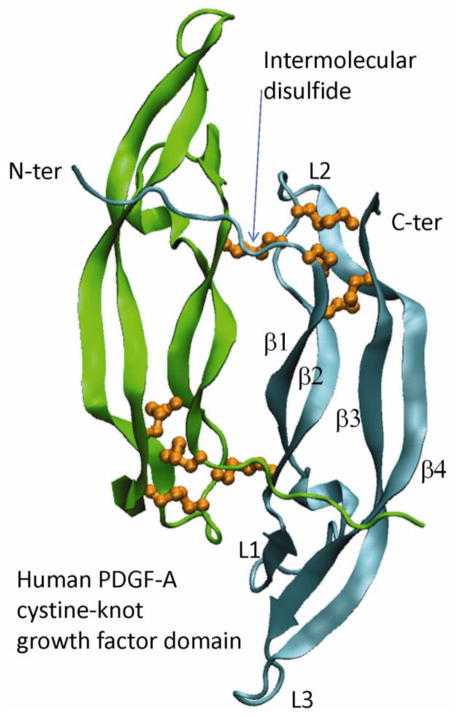
The growth factor domains of PDGFs belong to the cystine-knot fold, which is a set of anti-parallel β-strands linked by at least three disulfide bridges. The defining feature of this fold is a rotaxane substructure: the polypeptide between two disulfide bridges forms a loop through which a third disulfide bridge passes. This fold offers substantial structural stability and promotes the formation of homodimers or heterodimers; it is not only adopted by PDGFs, but also by glycoprotein hormones [27], the TGFβ family [28], and the nerve growth factor family [29] (reviewed in [30, 31]). In PDGFs, a pair of additional inter-molecular disulfide bridges further crosslink the two protomers in the dimer (Fig. 2). In PDGF-C and PDGF-D, additional intramolecular disulfide bridge(s) likely exist(s) due to the presence of additional cysteines in the sequences. It should be noted that the growth-factor domains, or the mature forms of PDGF-AA and PDGF-BB are commercially available by a number of companies, which are produced from bacterial fermentation and refolded from misfolded inclusion bodies. The PDGF-BB crystal structure has been determined [6] two decades ago. Although the free PDGF-AA structure has not yet been determined, a propeptide-bound PDGF-A crystal structure is available [16]. In both PDGF-A and PDGF-B, the four anti-parallel-β-strands, β1-β4, from the center of the dimer to the outside of the dimer, form three inter-strand loops (L1, L2, and L3). The sizes of the three loops rank in the order of L1>L3>L2. Because the β-sheets of the two protomers contact each other with their inner β1 strands, they form a super-sheet-like structure (β4-β3-β2-β1-β1-β2-β3-β4), which is flat in nature and necessitates the involvement of the N-terminal segment preceding the β1 strand. The N-terminal segment of each protomer extends across the top of the other protomer, serving as a stabilizer that prevents the twisting of the β-sheets. The vertical interaction between the N-terminal segment of each PDGF protomer and the β-strands of the other PDGF protomer is highly hydrophobic.
Fig. 2.
The cystine-knot growth-factor domain of PDGFs. Shown is the ribbon model of a PDGF-A dimer, as selected from PDB ID 3MJK. The two protomers in the dimer are colored in green and cyan respectively. The disulfide bridges are depicted as orange balls and sticks. Note that three intermolecular disulfide bridges are concentrated at one end of each protomer, and inter-molecular disulfide bridges are located at the dimerization interface. The N-terminal segment of each protomer, depicted as a coil, runs vertical to the β-strands of the other protomer.
3. The Propeptides of PDGFs and their association with the growth factor domains
As mentioned above, all PDGFs have pro-sequences, but PDGF-A and PDGF-B’s pro-sequences contain only ~60 amino acids, whereas PDGF-C and PDGF-D have over 200 amino acids preceding the cysteine-knot growth factor domains. Of the long pro-sequences of PDGF-C and PDGF-D, the N-terminal part is a CUB domain which spans ~110 residues. A three-dimension structures of the PDGF-C/-D CUB domain is not yet available, but by analogy to known CUB domain structures [32], they should fold in a β-sandwich of two 5-stranded anti-parallel β-sheets. This leaves a long sequence between the CUB domain and the cystine-knot growth factor domain undefined, which was previously named as the hinge region [20]. The recent structure of the PDGF-A/propeptide complex enables a sequence alignment of the hinge regions of PDGF-C and PDGF-D to the pro-peptides of PDGF-A and PDGF-B [16], and suggests that these hinge regions play a similar structural and functional role as the propeptides.
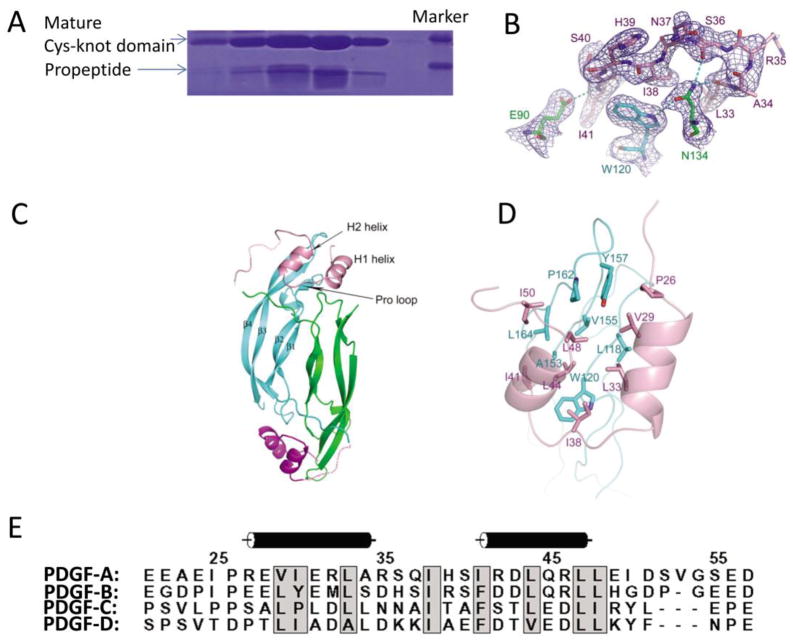
In PDGF-A, in contrary to the common belief that the propeptides of growth factors are released after cleavage, the propeptide can remain tightly bound to the mature domain [16]. The propeptide in fact is needed for the expression of most PDGFs, including PDGF-A, PDGF-B, and PDGF-D, in recombinant mammalian expression systems, with the only exception being PDGF-C [16]. Proteolytic cleavage take place during secretion of all three PDGFs that are dependent on pro-peptide for expression. It was unlikely that the cleavage occurs in post-secretion exposure to proteases, because the mammalian expression system used is a non-lytic system [16]. PDGF-A is cleaved the most efficiently because its cleavage sequence RRKR is the most optimal for furin, the most widely expressed proprotein convertase in the ER [22]. In gel filtration, cleaved PDGF-A elutes as a monodispersed peak, and SDS-PAGE analysis of the peak shows that the cleaved PDGF-A consists of two stoichiometric bands, corresponding in size to the pro-peptide and the mature cystine-knot domain, suggesting they are tightly associated with each other (Fig. 3A). The propeptide consists of two short helices (H1 and H2) linked by a short loop (Fig. 3B, 3C and 3D). The N-terminal segment preceding the H1 helix is well-ordered, but the segment after the H2 helix preceding the proprotein convertase-recognition site is mostly disordered, facilitating the presentation of the substrate sequence (e.g., RRKR) to the catalytic site. Both helices are amphipathic, with a hydrophobic side and a hydrophilic side. The hydrophobic side is used to interact with the large hydrophobic area on mature PDGF-A, and the hydrophilic side is exposed (Fig. 3B, 3C and 3D). The propeptide binds at the dimeric seam of the cystine-knot domain around the tips consisting of the three loops, L1 and L3 from one protomer and L2 from the other protomer. The interaction between the propeptide and mature PDGF-A is mostly hydrophobic, except Asn134 of the mature PDGF-A serving as a hub for hydrogen bonding at the interface (Fig. 3B). Asn134 would be a potential N-linked glycosylation site as its surrounding sequence fits the consensus sequence recognized by N-glycosyltransferases (Asn134-Thr135-Ser136). Yet the involvement of such extensive hydrogen bonding suggests that this site is not glycosylated, consistent with earlier findings that naturally purified PDGF proteins are non-glycosylated proteins. Of particular importance at the propeptide:PDGF-A interface is Trp120 which, with its large hydrophobic side chain, provides a nucleation site for a large number of hydrophobic interactions (Fig. 3B).
Fig. 3.
The association between propeptide and mature growth factor domain in PDGF-A. (A) The pro-peptide of PDGF-A is cleaved from mature PDGF-A, but remains tightly associated in gel filtration. The fractions from the elution peak are shown. (B) Two residues in mature PDGF-A, W120 and N134, play organizing roles in the association of the propeptide with the mature domain. (C) Ribbon model of the propeptide-loaded PDGF-A dimer, with the mature domains colored in green and cyan, whereas the propeptides colored in pink and magenta. (D) The hydrophobic interaction pattern between PDGF-A propeptide and mature domain. The backbone of the propeptide is shown as ribbons, and the backbone of the mature domains is shown as coils. The interacting sidechains are shown as sticks, colored according to their backbones. (E) Sequence comparison between all four types of PDGFs shows that the two helices used in the interactions with mature growth factors domains are a common feature, and the hydrophobic residues for interactions are preserved.
The association between propeptide and mature peptide is likely conserved for all PDGFs, as has been biochemically observed for PDGF-A, -B and–D [16]. Sequence alignment shows that the hydrophobic residues involved in the interactions between PDGF-A propeptide and the mature peptide are similarly conserved in PDGF-B/C/D (Fig. 3E). Functionally, the 2-helix pro-sequence is important for the folding of at least PDGF-A, -B, and–D, probably also because the hydrophobic nature of the propeptide: mature domain associations. Although the growth factor domain of PDGF-C was not dependent on pro-sequence in recombinant expression, the association between the pro-sequence and mature sequence was not tested [16], and most likely it would still contribute to the stability of the mature domain when the pro-sequence was included in expression, whether recombinantly or physiologically. During the secretion of PDGF-A and PDGF-B, the propeptide is retained after cleavage, most likely to protect the hydrophobic surface of the mature proteins. They can be released after receptor binding [16], although physiologically the exact time point of releasing the propeptides remain to be clarified. For PDGF-C and PDGF-D, it has been suggested that the CUB domains prevent the cystine-knot domains from binding to receptors and keeping them in the latent form [20]. But from the PDGF-A example, it would appear more likely that the tightly-bound 2-helix regions (or the hinge regions) next to the CUB domains, are responsible for keeping the mature domain in the latent, inactive form, regardless whether they are cleaved or not by the extracellular proteases, since the cleaved pro-peptides can remain tightly bound to the mature domains. The CUB domains of PDGF-C and PDGF-D also possibly associate with the hinge region or the growth factor domain by domain-domain interactions independent of peptide-linkers, yet the idea remains to be tested.
4. Two types of PDGFRs
The two receptors for PDGFs, PDGFRα and PDGFRβ, belong to the class III receptor tyrosine kinases (RTKs), a clan of five members including PDGFRα and PDGFRβ, KIT, FMS and FLT3 [5]. Like all RTKs, the PDGFR family of receptors have a modular architecture that utilize the extracellular domain to recognize ligands, a single transmembrane helix to pass structural/informational input from outside the cell, and an effector tyrosine kinase domain that respond to the extracellular signals, and undergoes phosphorylation to induce downstream recruiting/signaling events. Compared to other RTKs, PDGFRs and related KIT, FMS, and FLT3 feature five immunoglobulin-like (Ig) domains in the extracellular segment (D1-D5), and a split kinases domain that contains an insert of variable length between the N-terminal and C-terminal halves [33, 34]. KIT, FMS and FLT3 are considered a distinct subgroup from PDGFRα and PDGFRβ in class III RTKs because they bind ligands of the four-helix bundle fold instead of the PDGF-like cystine-knot fold [35–40].
The coding sequences of PDGFRα and PDGFRβ both start with a signal peptides of different length (23 amino acids for PDGFRα and 32 amino acids for PDGFRβ), leading to the Ig domains. Among the five Ig domains, D1, D2, and D3 belong to the I-set in the immunoglobulin-fold, and D4 and D5 have not been characterized, but likely are also I-set Ig domains. These domains all contain two layers of anti-parallel β-sheets, and the strands are arranged in a Greek-key fashion. D1 and D2 domains are linked by an unusually long linker, but this linker does not generate inter-domain flexibility; rather, it length enables the D1 domain to lean toward D2, forming a large hydrophobic interaction area between D1 and D2. D1 and D2 therefore are an integral structural module [16]. The role of such bent conformation of D1 is not entirely clear in PDGFRs, but for related KIT, the bent D1 allows it to contact its 4-helix bundle ligand stem cell factor (SCF). In PDGFRs, D1 is not used in ligand binding, but it could serve as a hydrophilic hat for the protection of the ligand-binding region, particular the adjacent D2 which uses a large hydrophobic area to capture its ligand. Indeed, D1 is heavily coated with N-linked glycans, but D2 carries no or low amount of glycosylation (Fig. 4). The N-linked glycans are not only highly hydrophilic, and also provide steric hindrance to potential non-specific hydrophobic interactions, therefore is beneficial to the stability of PDGFRs, likely in both the quiescent state and the ligand-bound state. D2 and D3 are linked by a short linker accommodative to hinge movement. The exact orientation between D2 and D3 is most likely within a small rotational range, and can be fixed upon ligand binding at both domains. This inter-D2-D3 flexibility has been also observed for the related class III RTKs, such as FMS, the receptor for another 4-helxi bundle ligand Macrophage colony-stimulating factor (M-CSF or CSF-1) [35, 39–41]. The D3-D4 and D4-D5 hinges are likely rigid because of their needs to convey the ligand-binding signal that can only reach the D3 level. Both PDGFRα and PDGFRβ are heavily glycosylated in these Ig domains (8 in PDGFRα and 11 in PDGFRβ), probably reflecting the need of the receptors to balance the hydrophobic area at the ligand-binding surface. In fact, their ligands, as discussed earlier, are also hydrophobic, especially in its mature, processed forms. It appears that this signaling system has evolved coping strategies at both the ligand and the receptor levels, using either amphipathic propeptides or a heavy glycan coat for the protection of the key recognition areas.
Fig. 4.
The two types of PDGFRs and their domain compositions. The starting numbers of the specific domains/segments in the coding sequences are marked left to the boundaries. Shown are all numbers for human PDGFRs. The positions of N-linked glycosylations are also marked. The lipid bilayer is represented by two straight lines. Note that D1 and D2 are an integral module, and the intracellular kinase domain is a split domains with an insert between N-terminal and C-terminal lobes.
The single transmembrane helix in each PDGFR likely serves as a simple tether, to pass on the positioning information from the D4 and D5 domains to the intracellular part. The linkers between D5 and the transmembrane helix are only 3–4 amino acids in both PDGFRα and PDGFRβ, suggesting that two transmembrane helices from the dimerized PDGFRs are geometrically unlikely to form inter-helix interactions due to that fact that D5 domains cannot be too close to clash into each other. A possibility is for the two transmembrane helices to be tilted for inter-helical interactions. But considering that the PDGFR transmembrane helices are of average length (25 amino acids), severely tilting in the lipid membrane is unlikely for PDGFR transmembrane helices to be thermodynamically stable. Nevertheless, a recently study has observed strong self-association behavior of the PDGFRβ transmembrane helix, and its implication in the full length receptor and in the physiological state remains to be characterized [42].
PDGFRs contains a polypeptide of ~40 amino acids in length between the transmembrane helix and the kinase domain, often described as the juxtamembrane (JM) segment, which plays a role in maintaining the kinase domain in an auto-inhibited state until activation by ligand binding. In related class III RTKs such as KIT, FMS and FLT3, the JM segments adopt a similar conformation to auto-inhibit the kinase domains by occupying the cleft near the catalytic site between the N-terminal and C-terminal lobes [43–45]. Several tyrosines in the JM segment, which are conserved among all class III RTKs, are responsible for anchoring the segment to the kinase domain. By analogy, it is likely that PDGFRs are also auto-inhibited by the JM segment in a similar fashion. Importantly, phosphorylation of these conserved tyrosines renders PDGFRβ constitutively active [46]. Similar results have been observed for KIT, FMS and FLT3 [43, 47, 48]. It is likely that the phosphate group added to the tyrosines prevents the binding of the JM segment to the kinase groove. The same tyrosines may also be involved in the ligand-induced PDGFR activation. As observed for KIT, auto-phosphorylation of these tyrosines in trans could be responsible for the activation of the kinase domain [49].
The tyrosine kinase domain is the effector domain in PDGFRs. In addition to the phosphorylated tyrosines in the JM region, the kinase domain of PDGFRs also carries two major tyrosine auto-phosphorylation sites, and one of these sites, Y751 in PDGFRβ, is specifically located in the insert region within the kinase [50]. The phosphorylation of these tyrosines provides docking sites for downstream signaling molecules [51, 52]. At the C-terminus of the encoding sequences, both PDGFRα and PDGFRβ have a highly acidic region which is also rich in serine and threonine. These sequences are involved in ubiquitination and receptor down-regulation [53].
5. PDGF:PDGFR recognition
The PDGF dimer, as mentioned above, has a flat shape, with all β-strands forming a super-sheet, leaving the inter-strand loops at the ends of these strands. These loops are not only used for propeptide binding, but also for receptor binding. The binding of receptor to PDGFs is sterically incompatible with the simultaneous binding of propeptides to PDGFs. When the PDGF-A/propeptide complex and the PDGF-B/PDGFRβ complex are superimposed with the backbones of the growth factor domains overlaid, it is apparent that these two binding events are mutually exclusive: the same hydrophobic residues important for propeptide association are also used for receptor binding. Consequently, receptor-binding can displace the propeptide that is bound at the same site [16]. The loops at each end of the super-sheet, L1 and L3 from one protomer, and L2 from the other protomer, bind PDGFR in a clamp-like fashion (Fig. 5A). The L1/L3 arm of the clamp is more protruding, whereas the L2 arm of the clamp is more receding. The two arms of the ligand clamp PDGFR perpendicularly near the receptor’s D2-D3 boundary. For PDGFRβ, the D2-D3 linker uses an extended conformation to open up a large cleft for contacting PDGF-B. The overall shape of the PDGF-B:PDGFRβ recognition complex resembles other class III RTKs such as KIT and FMS [35–37, 39, 40], but not FLT3 [38]. The positions of the D3 domains are similar in the SCF/KIT complex, the M-CSF/FMS complex, and the PDGF-B/PDGFRβ complex, despite that the positions of the D1 and D2 domains are dramatically different.
Fig. 5.
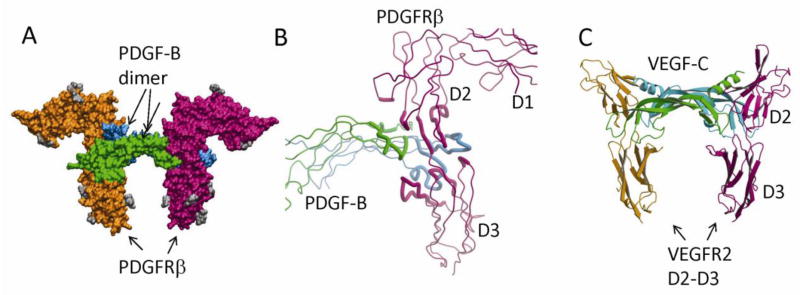
PDGF:PDGFR recognition. (A) The surface model of the PDGF-B in complex with the D1-D3 domains of PDGFRβ. PDGF-B protomers are colored in green and cyan, and PDGFRβ is colored in magenta and orange. The N-linked glycans are colored in gray. (B) The recognition involves the dimeric seam of PDGF-B, extending two arms clamping the D2-D3 boundary of PDGFRβ. Both PDGF-B and PDGFRβ are shown as tubes, and the interacting parts are shown as thicker tubes than the rest. (C) PDGF:PDGFR recognition is reminiscent of VEGF:VEGFR recognition. Shown is the ribbon model of VEGF-C in complex with the D2-D3 domains of VEGFR2. Despite roughly equivalent structural elements involved, there are major differences including the interface chemistry, the domain orientations, and the length of the L1 loops.
The PDGF-B:PDGFRβ interface is large, burying a 2870 Å2 solvent-accessible surface area contributed roughly two thirds by the protruding L1/L3 arm, and one third by the receding L2 arm (Fig. 5B). In addition, the N-terminal segment of PDGF-B protomer on the L2 arm side also contributes some small contacts with the receptor. On the receptor side, the interface is contributed nearly equally by the D2 and D3 domains. The D2 domain of PDGFRβ uses its A′-, G-, and F-strands on one of the β-sheets to occupy the upper side of the PDGF-B clamp. In addition, its CD loop also contacts PDGF-B’s N-terminal segment at the edge of the interface. The D3 domain of PDGFRβ occupies the lower side of the PDGF-B clamp with its BC, DE and FG loops extending near the D2-D3 boundary.
The large PDGF-B/PDGFRβ interface can be conveniently divided into two subinterfaces, based on the involvement of either D2 or D3 domain of PDGFRβ, using the L1-L3 plane of the PDGF-B clamp as the divider. Above this plane is the upper subinterface, and below the plane is the lower subinterface. The two subinterfaces are chemically different in nature. The upper subinterface mainly consists of an aromatic-rich hydrophobic cluster, with Tyr205, Tyr207, Phe136, Phe138 from PDGFRβ-D2 and Trp40, Leu38, Ile75, Ile77, Pro82, Phe84 from the PDGF-B. Of the hydrophobic residues of PDGF-B at this sub-interface, Trp40 plays an organizing role, as surrounded by other residues Tyr205, Tyr207, and Phe136. At this sub-interface, the protruding PDGF-B protomer (L1/L3) is more involved than the receding protomer (L2), which plays an auxiliary role by forming hydrophilic interactions with PDGFRβ.
The lower sub-interface involves mostly the long L1 loop of the protruding PDGF-B protomer, and the inter-strand loops of PDGFRβ D3. The L1 loop of PDGF-B drops from the L2-L3 dividing line of the clamp, reaching down to the waistline of the PDGFRβ-D3 βsandwich. The interaction at the lower sub-interface also has a significant hydrophobic composition. However, this is dependent on the conformation of the PDGF-B L1 loop, which is likely induced by the binding of the receptor. Interestingly, the conformation of this loop is the same in the propeptide-loaded PDGF-A and in the receptor-loaded PDGF-B. Therefore, either propeptide or receptor can aid the formation of such a conformation in the L1 loop, but without theses aids, the loop tends to be disordered as shown in the structure of the mature PDGF-B structure [6]. A structural determinant of the conformation of this loop is the buried terminal guanidine group of Arg27 of PDGF-B, which organizes a network of hydrogen bonds that maintain the backbone conformation of PDGF-B. Arg27 and Ile30 are two major determinants for PDGF-B:PDGFRβ interaction as demonstrated by mutagenesis data [54]. Both residues are involved in maintaining the kinked conformation of the PDGF-B L1 loop, and preparing it for receptor contact. Interestingly, the same study also mutated many other residues that can be mapped to the PDGFB:PDGFRβ interface and most of these mutations only slightly compromised receptor/ligand binding [54]. It appears that point-to-point contacts at the interface are less important in PDGF-B:PDGFRβ binding than the global conformation of the tertiary structures. This is consistent with the hydrophobic nature of the PDGFB:PDGFRβ interactions, which tend to be less sensitive to the loss of additive contact points.
The L1 loop is the longest loop in PDGFs, and was disordered in the free, mature PDGF-BB structure [6]. A structural organization is needed for this loop to be able to recognize the receptor, which can also be viewed as a receptor-induced PDGF conformational change. The conformation of the L1 loop is supported by interactions on two sides: on one side with PDGFRβ-D3, and on the other side by hydrophobic interactions with the rest of PDGF-B. It is likely this loop exists in equilibrium between the folded and unfolded states, with the folded state preferentially selected by receptor binding.
Because PDGF:PDGFR interactions are built on a conserved scaffold, most of these structural features in recognition between PDGF-B and PDGFRβ should be shared by all PDGF:PDGFR interactions. The binding between mature PDGF-C, the only PDGF that can be recombinantly expressed from a eukaryotic source, and PDGFRα, has been examined, and the reaction is enthalpically favorable but entropically unfavorable [16]. Enthalpically dominant interactions typically indicate primarily hydrophilic interactions, whereas entropically dominant interactions typically indicate primarily hydrophobic interactions that exclude ordered water molecules. Because PDGFRα contains the hydrophobic residues implicated in PDGF binding as PDGFRβ does, a large buried hydrophobic area should be buried between PDGF-C and PDGFRα, even if the receptor-binding surface on PDGF-C is hydrophilic (this is highly unlikely by inspecting the sequences). The large entropic decrease of PDGF-C:PDGFRα binding cannot be explained by a hydrophilic interaction mode, but can be explained by the ordering of large structural elements, such as the long inter-strand loops, as was observed in the PDGF-B:PDGFRβ structure. Since the L1 loop of PDGF-C is 3 amino acids shorter than PDGF-A and PDGF-B, it could be ordered even in its free form. A better candidate for conformational reorganization is the L3 loop in PDGF-C, which is 4 amino acids longer than in PDGF-A/PDGF-B (in PDGF-D it is 7amino acids longer, even more prone to flexibility in unbound state). Therefore, it may be the long L3 loop, rather than the shorter L1, in PDGF-C and PDGF-D, that undergoes structural organization in response to receptor binding.
In comparison with PDGFs, the related VEGFs also have a cystine-knot growth factor domain for receptor recognition, and their receptors are also composed of Ig domains, except that VEGFRs have 7 Ig domains whereas PDGFRs have 5 Ig domains. VEGFRs also recognize ligands with the second and third Ig domains [55, 56], as also revealed by the electron microcopy (EM) analysis of the VEGF-A/VEGFR2 complex [57]. The recent crystal structure of VEGF-C and VEGFR2-D2-D3 complex showed some similarities but significant differences from the PDGF/PDGFR complex [58]. In VEGF-C/VEGFR2 interaction, the N-terminal segment (a helix in VEGF versus a linear segment in PDGFs) of VEGF-C forms more significant contacts with the CD loop of VEGFR2-D2. The D2 subinterface of VEGF-C/VEGFR2 is primarily hydrophilic, in contrast with the extreme hydrophobic nature of the PDGF-B/PDGFRβ D2 subinterface. The L1 loop of VEGF-C, much shorter than the PDGF L1 loops, contact the D3 of VEGFR2 with much smaller area, albeit this interaction can still make 20–1000 fold differences in VEGF-VEGFR binding affinities [57, 59]. The major structural differences between VEGF/VEGFR and PDGF/PDGFR may be underlined by their strategies to cope with different functional needs. In particular, paracrine ligands like PDGFs and Wnt molecules [60] often utilizes their hydrophobicity to confine their distribution. In comparison, because VEGFs can serve as endocrine ligands, they need to be hydrophilic enough to be soluble in the plasma in order to reach distant target organs [61]. Consequently, VEGFs and VEGFRs have evolved to be primarily hydrophilic in their recognition mode while PDGF and PDGFRs have evolved to by primarily hydrophobic in their recognition mode, even though they evolve from a common ancestor [62]. With much hydrophobic areas to protect on both the ligand and the receptor sides, PDGFs have evolved to be dependent on amphipathic pro-peptides for cover-up, whereas PDGFR D2 domains have become more dependent on the nearby D1 domains to balance its overall physiochemical surface properties. The VEGF and PDGF ligands have co-evolved with their respective receptors. Despite the major differences in recognition, it has been reported that VEGF could signal through PDGFR in bone-marrow-derived mesenchymal cells [63], although this was not supported by direct binding experiments using recombinant VEGF and PDGFRs [16].
6. Recognition specificity between PDGFs and PDGFRs
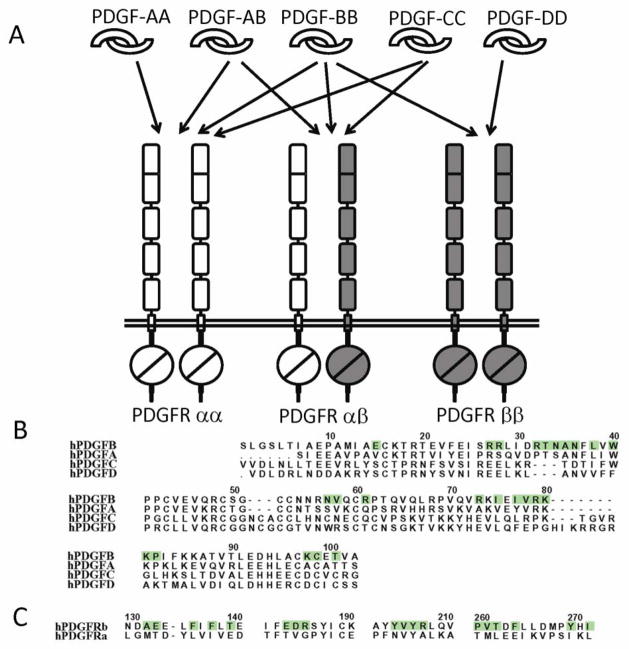
PDGF-A and PDGF-B are often expressed in the same types of cells (reviewed in [64]) and the assembly of PDGF-AB heterodimers has been observed [65–67]. A study even suggested that the PDGF-AB heterodimer is preferentially formed over the homodimers [68]. There have been, however, no evidence that PDGF-C and PDGF-D form heterodimers. Therefore, considering all homodimers and heterodimers, five types of PDGF dimer proteins currently exist (Fig. 6). Given that PDGFRs are dimerized by the ligands, depending on the expression pattern of the receptors, either PDGFRα or PDGFRβ, or both types of receptors, can be recruited to bind the ligands. Indeed, formation of PDGFR heterodimer has been observed in vivo by crosslinking studies [69]. Therefore, there exists a multitude of possible PDGF-PDGFR interactions. PDGFRα binds the PDGF-A, -B and –C chains all with high affinities, and PDGFRβ binds the PDGF-B and PDGF-D chains with high affinities. Consequently, PDGFRα are activated by homodimeric PDGF-AA, PDGF-BB, and PDGF-CC, and heterodimeric PDGF-AB. PDGFRβ can only be activated by PDGF-BB and PDGF-DD (reviewed in [21]). The PDGFRαβ heterodimerization can be induced by the PDGF-BB homodimer or PDGF-AB heterodimer. This interaction pattern indicates that PDGFRα is more promiscuous than PDGFRβ, and PDGF-B is more promiscuous than other PDGFs.
Fig. 6.
The specificity in PDGF:PDGFR recognition. (A) The biochemically defined interactions between PDGF homodimers/heterodimers and the PDGFR homodimers/heterodimers. Note that there is no proof for pre-associated PDGFR dimers, therefore the receptor dimers are just a result of ligand-driven clustering. (B) Sequence comparison of PDGFs with the residues involved in PDGF-B:PDGFRβ interaction highlighted. This comparison shows that the hydrophobic residues used for the core of the PDGF:PDGFR interface are preserved, but the hydrophilic residues at the periphery of the interface have significant variations. (D) Sequence comparison between the PDGFRs fragments used at the interface, with the PDGFRβ residues in ligand recognition highlighted. This comparison also shows that the hydrophobic nature of the ligand-recognition surface is preserved. (B) and (C) also show changes from aromatic residues to branched residues, or from larger to smaller residues, particularly between PDGF-A and PDGF-B, and between PDGFRα and PDGFRβ.
In comparison to PDGFRβ, the promiscuity of PDGFRα is largely attributed to the smaller, conformationally less specific residues at the ligand-receptor interface. In particular, PDGFRβ has a large number of aromatic residues at the ligand-binding surface; all but one (Tyr207) of these aromatic residues are replaced by non-aromatic, smaller residues (Tyr205Asn, Phe138Leu, Tyr270Ile, Phe245Leu, Phe264Ile) in PDGFRα (Fig. 6). In PDGFRβ, the bulky, rigid aromatic residues are difficult to rotate, providing extra requirements in shape complementarity and hydrogen-bonding networks, thereby raising the specificity of protein-protein interactions. The replacement of these aromatic residues with non-aromatic residues in PDGFRα makes the ligand recognition surface of PDGFRα more adaptive to a wider variety of PDGF ligand surfaces.
Rules governing PDGF selectivity and promiscuity can likewise be deduced from available crystal structures. The promiscuity of PDGF-B over PDGF-A is largely due to its presence of a large number of long-chain hydrophilic residues at the edge of the receptor-binding surface, which are substituted by smaller residues in PDGF-A (Glu15Val, Arg18Ser, Arg32Pro, Asn34Ser, Asn55Thr, Arg56Ser, Asn57Ser, Arg73Ala and Lys99Ala, using residues numbers for PDGF-B) (Fig. 6). These long chain hydrophilic residues in PDGF-B, being at the edge but not the center of the interface, can be flexible to reach interacting residues at the opposite side of the interface, hence are not as selective as the smaller, more rigid residues. The other two PDGFs, namely PDGF-C and PDGF-D, diverge from PDGF-A/B in sequence, and hence their interaction pattern with receptors cannot be accurately predicted based on sequence homology alone. In particular, for PDGF-C and PDGF-D, there are extra deletions and insertions in the loops (i.e., L1 and L3) involved in receptor recognition (Fig. 6). Their selectivities towards PDGFRs α and PDGFRβ remain to be elucidated. PDGF-C is the only member of the PDGF family that is not dependent on propeptide for the recombinant expression of the growth factor domain [16], and its receptor-binding surface may be more hydrophilic than other PDGFs. Therefore, PDGF-C could fit the ligand binding surface of PDGFRα better, which has less aromatic, hydrophobic residues compared to PDGFRβ.
7. Activation of PDGFRs
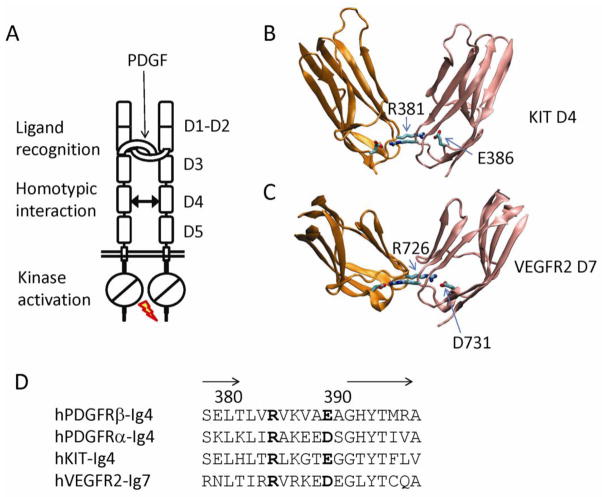
The activation of PDGFRs is not entirely understood mechanistically, but it likely requires receptor conformational changes in multiple steps. Initially, the bivalent nature of PDGF ligands brings two receptor protomers into proximity with one another. Importantly, activation further requires contacts between the receptor membrane proximal regions [71], so that the intracellular kinase domains can be brought together for transphosphorylation. Although the molecular details of the interactions of the PDGFR membrane proximal regions are not available, comparison with the related class III RTK KIT, the SCF receptor, and mutagenesis experiments have offered much insight. In KIT, stimulation by SCF not only brings two copies of KIT together, but also induces a swing of the membrane-proximal domain, as compared to the free, unliganded KIT structure [37]. Neighboring KIT D4 domains then contact each other through a pair of salt bridges mediated between E386 and R381 (Fig. 7). There are few other interactions besides these salt bridges, although there may be additional functionally important, less intimate interactions at the inner faces of the D4 and D5 domains. The same salt-bridge forming residues in PDGFRβ, E390 and R385, when mutated to alanine, compromised PDGF-induced PDGFRβ activation, and impaired a variety of PDGF-induced cellular responses, suggesting that the pairing of these salt bridges are essential for the precise positioning of the membrane proximal regions [71]. Using a chemical crosslinking approach, it was further shown that the Glu390 mutant of PDGFRβ can still be dimerized by PDGFs, but are inactive dimers and show reduced internalization and degradation [71].
Fig. 7.
The activation of PDGFRs is dependent on the homotypic interactions at the membrane-proximal region of the extracellular segment. (A) Schematic model of PDGFR activation by PDGF-B. The dimeric PDGF ligand binds the D2-D3 boundary of PDGFRs, enabling or forcing the interaction between PDGFR D4 domains. The precise positioning of D4-D5 domains lead to transmembrane and juxtamembrane changes, ultimately leading to the activations of the tyrosine kinase domain. (B) The PDGFR D4-D4 homotypic interaction is likely analogous to the KIT D4- D4 interaction. Shown is the part of the structure of stem cells factor (SCF) in complex with KIT D1-D5. The D4 domains from two copies of receptors are colored in orange and pink respectively. The inter-receptor salt bridges are shown as sticks. (C) PDGFR D4-D4 interaction is also likely to be analogous to that of the D7 domains of VEGFRs, the class of RTKs evolutionarily related to PDGFRs. (D) Sequence comparison of the segment that provides inter-domain interactions for PDGFRs, KIT, and VEGFR2. Note that arginine is always required at one position, but the other position can have either glutamate or aspartate, which are different in length and could result in the difference in the interaction strength depending on the contexts.
Similar homotypic contacts that are required for receptor activation and cell signaling have also been observed for VEGFRs, the close relative of PDGFRs [72]. Of the 7 Ig domains of VEGFRs, D7, but not D4, was identified to be the functional equivalence of the D4 domains of class III RTKs [72]. These domains have an “L/Ix ΦxxxD/ExG “sequence motif that is located at EF loop. It should be noted that in the EM images of the VEGF-VEGFR2 complex, the D4 domains were observed to form inter-receptor contact as well, but the significance of this contact remains to be characterized [57]. In VEGFR2, when D731 and R726, the residues corresponding to PDGFRβ E390 and R385, were mutated to alanine, VEGF-stimulated receptor autophosphorylation was compromised. The D7-D7 association is weak by itself, and the isolated D7 domain remained monomeric even when its concentration reaches 0.1mM. However, the D7-D7 interaction was able to be recaptured in the crystal lattice, where the local concentration becomes extremely high. The crystal structure of VEGFR2 D7 showed that D731 and R726 indeed form a pair of anti-parallel salt bridges in the same fashion as the KIT-D4 salt bridges (Fig. 7) [72]. It is likely that when D7 in present in the full-length receptor, these salt bridges are much readier to form than in solution due to the confinement of both ligand binding and the membrane tethering. The weak affinity between these domains, and also between PDGFR D4 domains, is probably a necessity to prevent accidental, spontaneous reception activation in the absence of ligands.
Interestingly, in PDGFRα, the D4-D4 homotypic interaction is not simply weak, but is also a negative contributor to the association between PDGF-C and the entire ectodomain [16]. Using calorimetry, it was shown that PDGF-C binds PDGFRα D1-D3 domains with higher affinity than with PDGFRα D1-D5 domains. Since addition of D4-D5 compromises ligand-receptor binding, one can then imagine that in the intact, full-length receptor, the D4-D4 contact, the likely contact at the membrane-proximal region, is unfavorable and is forced by ligand-receptor binding at the D2-D3 position. This particularly weak association of PDGFRα-D4 may be a result of having aspartate for the salt bridge, whereas other class III RTKs such as PDGFRβ and KIT utilize glutamate at the position. Because aspartate is shorter than glutamate, PDGFRα D4 may need to move closer to each other than in other class III RTKs, which may be energetically unfavorable. It should be notated that among class III RTKs, FMS have the strongest D4-D4 interactions, as the addition of D4-D5 domain boosts the binding between M-CSF and FMS by 50-fold [35]. It is possible that FMS-D4 engages further favorable interactions in addition to the dual salt bridges.
The precise pairing of the D4-D5 regions of PDGFRs is coupled to the activation of the intracellular effector tyrosine kinase domain. How the coupling is configured is not entirely clear, but it may involve several steps. The contact of D4 domains may lead to transmembrane helix interactions, as suggested by the observation of self-association behavior of the PDGFRβ transmembrane helix [42]. However, in isolated state, two hydrophobic helices may interact non-specifically, not necessarily reflecting an interaction that provides specific geometry to support appropriate intracellular positions of the JM and kinase domains. It is likely that only transient or no interaction of the transmembrane helices are needed; rather, the mere proximity of the transmembrane helices could suffice to shift the equilibrium of the JM-kinase domain interactions from association towards dissociation as reflected in the phosphorylation/dephosphorylation balance of the key tyrosines in the JM region. Once the kinase domain is activated, it would be able to transphosphorylate specific tyrosines in the kinase regions, making it suitable for interacting with substrates. For example, binding to the SH2 domain of Grb2 leads to the activation of Raf-1 and MAPK pathways (reviewed in [73, 74]). In addition to adaptors like Grb2, the kinase domain can also activate downstream enzymes, notably PI3K and PLCγ [73, 75]. Furthermore, PDGFRs can also bind other types of signaling molecules including transcription factors [76].
8, perspectives in PDGFR targeting
PDGF-PDGFR signaling plays essential roles in development, but once adulthood is reached, its function is usually detrimental rather than constructive in human physiology. An exception is in tissue repair and wound healing. PDGF-BB and PDGFRβ appear to be involved in the formational of new vessels and collagen production [77]. For this reason recombinant human PDGF-B has been used in clinical studies. However, the general need is to block PDGF-PDGFR signaling to stop or reverse proliferative diseases. PDGFs and PDGFRs are validated therapeutic targets in a variety of diseases, especially cancer [78]. PDGFRs’ implications in cancer are multi-faceted, including but not limited to angiogenesis, a fact that is consistent with the mitogenic effect of PDGFs on mesenchymal cells that influence tumor growth [4].
PDGFs and PDGFRs can be pharmacologically inhibited in multiple ways, and the most direct way is to inhibit the intracellular kinase domain using small molecule inhibitors. Tyrosine kinases inhibitors (TKIs) generally operate by two fashions: either binding to the active site to block the enzymatic activity directly, or binding to an allosteric site outside the active site to affect the kinase’s activity by a conformational change [79]. Several TKIs have been developed against PDGFRs; yet they are usually nonspecific to PDGFRs, as they also act on KIT and FLT3 [78]. The lack of specificity offers advantages and disadvantages. On the one hand, these small molecule inhibitors can target all at once multiple class III RTKs implicated in cancers. On the other hand, the side effects due to the broad spectrum of specificity must also be considered (reviewed in [80]).
For high specificity the extracellular inhibition of PDGFRs is often considered. For inhibiting angiogenesis, blockade of the related VEGFR2 (e.g., by avastin/bevacizumab, a humanized monoclonal antibody) has thus far proven to be successful in clinical trials. Neutralizing antibodies have also been developed for PDGFs and PDGFRs (e.g., [81]), so is oligonucleic acid aptamers specific for PDGF-B [82]. The PDGF-B:PDGF-Rβ structure suggests a few additional directions besides antibodies and aptamers that may offer both high affinity and high specificity. For instance, receptor decoys derived from PDGFR-D2-D3 could be used for PDGF inhibition, analogous to the VEGFR-trap consisting of VEGFR D2-D3 domains [83]. Because there is a hydrophobic interface between D1 and D2, the D2-D3 decoy must carry hydrophilic mutations at the D1-D2 interface. The back of D2 that is not used for ligand recognition can be engineered to carry N-linked glycans to balance the hydrophobic nature of the PDGF-recognizing surface. Doing so would reduce nonspecific adhesion and the deposition of the PDGFR decoy at the drug administration site and improve the drug’s phamacokinetic profile. Another possibility is to design higher-affinity, broader-spectrum PDGF inhibitors by combining PDGFRα and PDGFRβ structural features, especially by replacing the aromatic residues with smaller aliphatic residues at the ligand-recognizing surface, based on the specificity determinants at the PDGF-PDGFR interfaces. The D2-D3 junction of PDGFRs can also be engineered to reduce hinge flexibility, rendering an entropic advantage in PDGF recognition, thereby increasing affinity. The simple 2-helix structure of the PDGF pro-peptide can be used as a template to design peptide mimics that bind the PDGF ligands in a tight, non-dissociable fashion. To do so, affinity maturation through surface display of altered PDGF propeptides could be implemented to increase the affinity of such peptides to PDGFs to a substantially higher level. In addition to interfering at the ligand-receptor interaction regions, antibodies that bind the membrane-proximal domains (D4 or D5) may also prevent theses domains to pair with each other for receptor activation.
Other than inhibition of the kinase domains or the extracellular domain, it is conceivable to antagonize the transmembrane segment or the JM segment through small molecules. However, these regions have more dynamic conformations, and achieving a therapeutic-level affinity level is intrinsically difficult. Another approach is to inhibit PDGF signaling at the transcription level in addition to the inhibition at the protein level. A recent study suggested the possibility of using the small molecule TMPyP4 to bind the promoter region of PDGF-A [84].
9. Conclusions
In the last a few years we have gained significant insight into the recognition and activation mechanisms of PDGFRs, a class of import receptor tyrosine kinase that is involved not only in physiological functions such as organogenesis and vessel formation, but also in several widespread diseases such as cancer and atherosclerosis. The significant roles of PDGFRs in cancer and vascular diseases underline the significant efforts in both academia and industry in search for specific, high-affinity, and easy-to deliver drugs for PDGF or PDGFR inhibition. The detailed structural understanding resulted from recent studies should certainly aid these efforts, although a more complete and in-depth picture about the full-length PDGFRs in the transmembrane format would greatly advance the PDGFR biology as well as therapeutic pursuits. The homotypic interaction regions of PDGFRs have not been thoroughly characterized, and any therapeutic design against the membrane proximal domains may have the advantage of not only bypass ligand-receptor interactions, but also bear the potential of limiting intracellular geometry in a state impermissible to kinase domain activation, even if the PDGFR oncogenic mutations were intracellular but not extracellular. The specific roles of the CUB domains in the newly-discovered PDGFs, PDGF-C and PDGF-D, also awaits further studies. As one of the earliest discovered classes of growth factors and growth factors receptors, PDGFs and PDGFRs have revealed many first principles in receptor signaling biology, yet more revelations in this important signaling system are yet to come, especially in its involvement and mechanism in the development of proliferative and chronic diseases, and in the therapeutic modulation of it in clinical studies.
Highlights.
Recent advances on PDGFs and their receptors PDGFRs
Dissection of PDGF structural/functional understandings
Mechanistic insights into PDGF:propeptide recognition, receptor recognition and activation
Perspectives on therapeutic modulation of PDGFR signaling
Acknowledgments
The authors wish to thank Heli Liu and Ann Shim for comments and help in the preparation of some figures. This work in the authors’ laboratory is supported by the NIH grant 5R01GM098259.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kohler N, Lipton A. Platelets as a source of fibroblast growth-promoting activity. Exp Cell Res. 1974;87:297–301. doi: 10.1016/0014-4827(74)90484-4. [DOI] [PubMed] [Google Scholar]
- 2.Ross R, Glomset J, Kariya B, Harker L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc Natl Acad Sci U S A. 1974;71:1207–1210. doi: 10.1073/pnas.71.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostman A. PDGF receptors-mediators of autocrine tumor growth and regulators of tumor vasculature and stroma. Cytokine Growth Factor Rev. 2004;15:275–286. doi: 10.1016/j.cytogfr.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11:3921–3926. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westermark B, Wasteson A. A platelet factor stimulating human normal glial cells. Exp Cell Res. 1976;98:170–174. doi: 10.1016/0014-4827(76)90476-6. [DOI] [PubMed] [Google Scholar]
- 8.Waterfield MD, Scrace GT, Whittle N, Stroobant P, Johnsson A, Wasteson A, Westermark B, Heldin CH, Huang JS, Deuel TF. Platelet-derived growth factor is structurally related to the putative transforming protein p28sis of simian sarcoma virus. Nature. 1983;304:35–39. doi: 10.1038/304035a0. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle RF, Hunkapiller MW, Hood LE, Devare SG, Robbins KC, Aaronson SA, Antoniades HN. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983;221:275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- 10.Betsholtz C, Johnsson A, Heldin CH, Westermark B, Lind P, Urdea MS, Eddy R, Shows TB, Philpott K, Mellor AL, Knott TJ, Scott J. cDNA sequence and chromosomal localization of human platelet-derived growth factor A-chain and its expression in tumour cell lines. Nature. 1986;320:695–699. doi: 10.1038/320695a0. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, Soriano P, Betsholtz C, Heldin CH, Alitalo K, Ostman A, Eriksson U. PDGF-C is a new protease-activated ligand for the PDGF alpha-receptor. Nature cell biology. 2000;2:302–309. doi: 10.1038/35010579. [DOI] [PubMed] [Google Scholar]
- 12.Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U. PDGF-D is a specific, protease-activated ligand for the PDGF beta-receptor. Nature cell biology. 2001;3:512–516. doi: 10.1038/35074588. [DOI] [PubMed] [Google Scholar]
- 13.LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichenstein HS. PDGF-D, a new protease-activated growth factor. Nature cell biology. 2001;3:517–521. doi: 10.1038/35074593. [DOI] [PubMed] [Google Scholar]
- 14.McDonald NQ, Hendrickson WA. A structural superfamily of growth factors containing a cystine knot motif. Cell. 1993;73:421–424. doi: 10.1016/0092-8674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- 15.Fredriksson L, Li H, Eriksson U. The PDGF family: four gene products form five dimeric isoforms. Cytokine Growth Factor Rev. 2004;15:197–204. doi: 10.1016/j.cytogfr.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci U S A. 2010;107:11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rorsman F, Bywater M, Knott TJ, Scott J, Betsholtz C. Structural characterization of the human platelet-derived growth factor A-chain cDNA and gene: alternative exon usage predicts two different precursor proteins. Molecular and cellular biology. 1988;8:571–577. doi: 10.1128/mcb.8.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abramsson A, Kurup S, Busse M, Yamada S, Lindblom P, Schallmeiner E, Stenzel D, Sauvaget D, Ledin J, Ringvall M, Landegren U, Kjellen L, Bondjers G, Li JP, Lindahl U, Spillmann D, Betsholtz C, Gerhardt H. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes Dev. 2007;21:316–331. doi: 10.1101/gad.398207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindblom P, Gerhardt H, Liebner S, Abramsson A, Enge M, Hellstrom M, Backstrom G, Fredriksson S, Landegren U, Nystrom HC, Bergstrom G, Dejana E, Ostman A, Lindahl P, Betsholtz C. Endothelial PDGF-B retention is required for proper investment of pericytes in the microvessel wall. Genes Dev. 2003;17:1835–1840. doi: 10.1101/gad.266803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reigstad LJ, Varhaug JE, Lillehaug JR. Structural and functional specificities of PDGF-C and PDGF-D, the novel members of the platelet-derived growth factors family. FEBS J. 2005;272:5723–5741. doi: 10.1111/j.1742-4658.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 21.Heldin CH, Westermark B. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol Rev. 1999;79:1283–1316. doi: 10.1152/physrev.1999.79.4.1283. [DOI] [PubMed] [Google Scholar]
- 22.Izidoro MA, Gouvea IE, Santos JA, Assis DM, Oliveira V, Judice WA, Juliano MA, Lindberg I, Juliano L. A study of human furin specificity using synthetic peptides derived from natural substrates, and effects of potassium ions. Archives of biochemistry and biophysics. 2009;487:105–114. doi: 10.1016/j.abb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11:367–383. doi: 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 24.Gilbertson DG, Duff ME, West JW, Kelly JD, Sheppard PO, Hofstrand PD, Gao Z, Shoemaker K, Bukowski TR, Moore M, Feldhaus AL, Humes JM, Palmer TE, Hart CE. Platelet-derived growth factor C (PDGF-C), a novel growth factor that binds to PDGF alpha and beta receptor. J Biol Chem. 2001;276:27406–27414. doi: 10.1074/jbc.M101056200. [DOI] [PubMed] [Google Scholar]
- 25.Fredriksson L, Li H, Fieber C, Li X, Eriksson U. Tissue plasminogen activator is a potent activator of PDGF-CC. EMBO J. 2004;23:3793–3802. doi: 10.1038/sj.emboj.7600397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ustach CV, Kim HR. Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Molecular and cellular biology. 2005;25:6279–6288. doi: 10.1128/MCB.25.14.6279-6288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang X, Liu H, Chen X, Chen PH, Fischer D, Sriraman V, Yu HN, Arkinstall S, He X. Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. Proc Natl Acad Sci U S A. 2012;109:12491–12496. doi: 10.1073/pnas.1206643109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray-Rust J, McDonald NQ, Blundell TL, Hosang M, Oefner C, Winkler F, Bradshaw RA. Topological similarities in TGF-beta 2, PDGF-BB and NGF define a superfamily of polypeptide growth factors. Structure. 1993;1:153–159. doi: 10.1016/0969-2126(93)90029-g. [DOI] [PubMed] [Google Scholar]
- 29.He XL, Garcia KC. Structure of nerve growth factor complexed with the shared neurotrophin receptor p75. Science. 2004;304:870–875. doi: 10.1126/science.1095190. [DOI] [PubMed] [Google Scholar]
- 30.Sun PD, Davies DR. The cystine-knot growth-factor superfamily. Annual review of biophysics and biomolecular structure. 1995;24:269–291. doi: 10.1146/annurev.bb.24.060195.001413. [DOI] [PubMed] [Google Scholar]
- 31.Vitt UA, Hsu SY, Hsueh AJ. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 2001;15:681–694. doi: 10.1210/mend.15.5.0639. [DOI] [PubMed] [Google Scholar]
- 32.Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. Journal of molecular biology. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 33.Coussens L, Van Beveren C, Smith D, Chen E, Mitchell RL, Isacke CM, Verma IM, Ullrich A. Structural alteration of viral homologue of receptor proto-oncogene fms at carboxyl terminus. Nature. 1986;320:277–280. doi: 10.1038/320277a0. [DOI] [PubMed] [Google Scholar]
- 34.Rosnet O, Marchetto S, deLapeyriere O, Birnbaum D. Murine Flt3, a gene encoding a novel tyrosine kinase receptor of the PDGFR/CSF1R family. Oncogene. 1991;6:1641–1650. [PubMed] [Google Scholar]
- 35.Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 38.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, Savvides SN. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 39.Ma X, Lin WY, Chen Y, Stawicki S, Mukhyala K, Wu Y, Martin F, Bazan JF, Starovasnik MA. Structural basis for the dual recognition of helical cytokines IL-34 and CSF-1 by CSF-1R. Structure. 2012;20:676–687. doi: 10.1016/j.str.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Leo C, Chen X, Wong BR, Williams LT, Lin H, He X. The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochimica et biophysica acta. 2012;1824:938–945. doi: 10.1016/j.bbapap.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elegheert J, Desfosses A, Shkumatov AV, Wu X, Bracke N, Verstraete K, Van Craenenbroeck K, Brooks BR, Svergun DI, Vergauwen B, Gutsche I, Savvides SN. Extracellular complexes of the hematopoietic human and mouse CSF-1 receptor are driven by common assembly principles. Structure. 2011;19:1762–1772. doi: 10.1016/j.str.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oates J, King G, Dixon AM. Strong oligomerization behavior of PDGFbeta receptor transmembrane domain and its regulation by the juxtamembrane regions. Biochimica et biophysica acta. 2010;1798:605–615. doi: 10.1016/j.bbamem.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 43.Chan PM, Ilangumaran S, La Rose J, Chakrabartty A, Rottapel R. Autoinhibition of the kit receptor tyrosine kinase by the cytosolic juxtamembrane region. Molecular and cellular biology. 2003;23:3067–3078. doi: 10.1128/MCB.23.9.3067-3078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter M, Lucet IS, Patel O, Broughton SE, Bamert R, Williams NK, Fantino E, Wilks AF, Rossjohn J. The 2.7 A crystal structure of the autoinhibited human c-Fms kinase domain. Journal of molecular biology. 2007;367:839–847. doi: 10.1016/j.jmb.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 45.Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Molecular cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 46.Mori S, Ronnstrand L, Yokote K, Engstrom A, Courtneidge SA, Claesson-Welsh L, Heldin CH. Identification of two juxtamembrane autophosphorylation sites in the PDGF beta-receptor; involvement in the interaction with Src family tyrosine kinases. EMBO J. 1993;12:2257–2264. doi: 10.1002/j.1460-2075.1993.tb05879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohde CM, Schrum J, Lee AW. A juxtamembrane tyrosine in the colony stimulating factor-1 receptor regulates ligand-induced Src association, receptor kinase function, and down-regulation. J Biol Chem. 2004;279:43448–43461. doi: 10.1074/jbc.M314170200. [DOI] [PubMed] [Google Scholar]
- 48.Vempati S, Reindl C, Wolf U, Kern R, Petropoulos K, Naidu VM, Buske C, Hiddemann W, Kohl TM, Spiekermann K. Transformation by oncogenic mutants and ligand-dependent activation of FLT3 wild-type requires the tyrosine residues 589 and 591. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14:4437–4445. doi: 10.1158/1078-0432.CCR-07-1873. [DOI] [PubMed] [Google Scholar]
- 49.Mol CD, Lim KB, Sridhar V, Zou H, Chien EY, Sang BC, Nowakowski J, Kassel DB, Cronin CN, McRee DE. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- 50.Kazlauskas A, Cooper JA. Autophosphorylation of the PDGF receptor in the kinase insert region regulates interactions with cell proteins. Cell. 1989;58:1121–1133. doi: 10.1016/0092-8674(89)90510-2. [DOI] [PubMed] [Google Scholar]
- 51.Kazlauskas A, Cooper JA. Phosphorylation of the PDGF receptor beta subunit creates a tight binding site for phosphatidylinositol 3 kinase. EMBO J. 1990;9:3279–3286. doi: 10.1002/j.1460-2075.1990.tb07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Escobedo JA, Kaplan DR, Kavanaugh WM, Turck CW, Williams LT. A phosphatidylinositol-3 kinase binds to platelet-derived growth factor receptors through a specific receptor sequence containing phosphotyrosine. Molecular and cellular biology. 1991;11:1125–1132. doi: 10.1128/mcb.11.2.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lennartsson J, Wardega P, Engstrom U, Hellman U, Heldin CH. Alix facilitates the interaction between c-Cbl and platelet-derived growth factor beta-receptor and thereby modulates receptor down-regulation. J Biol Chem. 2006;281:39152–39158. doi: 10.1074/jbc.M608489200. [DOI] [PubMed] [Google Scholar]
- 54.Clements JM, Bawden LJ, Bloxidge RE, Catlin G, Cook AL, Craig S, Drummond AH, Edwards RM, Fallon A, Green DR, et al. Two PDGF-B chain residues, arginine 27 and isoleucine 30, mediate receptor binding and activation. EMBO J. 1991;10:4113–4120. doi: 10.1002/j.1460-2075.1991.tb04988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis-Smyth T, Presta LG, Ferrara N. Mapping the charged residues in the second immunoglobulin-like domain of the vascular endothelial growth factor/placenta growth factor receptor Flt-1 required for binding and structural stability. J Biol Chem. 1998;273:3216–3222. doi: 10.1074/jbc.273.6.3216. [DOI] [PubMed] [Google Scholar]
- 56.Fuh G, Li B, Crowley C, Cunningham B, Wells JA. Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J Biol Chem. 1998;273:11197–11204. doi: 10.1074/jbc.273.18.11197. [DOI] [PubMed] [Google Scholar]
- 57.Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14:249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- 58.Leppanen VM, Prota AE, Jeltsch M, Anisimov A, Kalkkinen N, Strandin T, Lankinen H, Goldman A, Ballmer-Hofer K, Alitalo K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci U S A. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 60.Janda CY, Waghray D, Levin AM, Thomas C, Garcia KC. Structural basis of Wnt recognition by Frizzled. Science. 2012;337:59–64. doi: 10.1126/science.1222879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tammela T, Enholm B, Alitalo K, Paavonen K. The biology of vascular endothelial growth factors. Cardiovascular research. 2005;65:550–563. doi: 10.1016/j.cardiores.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 62.Grassot J, Gouy M, Perriere G, Mouchiroud G. Origin and molecular evolution of receptor tyrosine kinases with immunoglobulin-like domains. Molecular biology and evolution. 2006;23:1232–1241. doi: 10.1093/molbev/msk007. [DOI] [PubMed] [Google Scholar]
- 63.Ball SG, Shuttleworth CA, Kielty CM. Vascular endothelial growth factor can signal through platelet-derived growth factor receptors. J Cell Biol. 2007;177:489–500. doi: 10.1083/jcb.200608093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dirks RP, Bloemers HP. Signals controlling the expression of PDGF. Molecular biology reports. 1995;22:1–24. doi: 10.1007/BF00996300. [DOI] [PubMed] [Google Scholar]
- 65.Hammacher A, Hellman U, Johnsson A, Ostman A, Gunnarsson K, Westermark B, Wasteson A, Heldin CH. A major part of platelet-derived growth factor purified from human platelets is a heterodimer of one A and one B chain. J Biol Chem. 1988;263:16493–16498. [PubMed] [Google Scholar]
- 66.Hammacher A, Nister M, Westermark B, Heldin CH. A human glioma cell line secretes three structurally and functionally different dimeric forms of platelet-derived growth factor. European journal of biochemistry / FEBS. 1988;176:179–186. doi: 10.1111/j.1432-1033.1988.tb14266.x. [DOI] [PubMed] [Google Scholar]
- 67.Hart CE, Bailey M, Curtis DA, Osborn S, Raines E, Ross R, Forstrom JW. Purification of PDGF-AB and PDGF-BB from human platelet extracts and identification of all three PDGF dimers in human platelets. Biochemistry. 1990;29:166–172. doi: 10.1021/bi00453a022. [DOI] [PubMed] [Google Scholar]
- 68.Hoppe J, Weich HA, Eichner W, Tatje D. Preparation of biologically active platelet-derived growth factor isoforms AA and AB. Preferential formation of AB heterodimers. European journal of biochemistry / FEBS. 1990;187:207–214. doi: 10.1111/j.1432-1033.1990.tb15296.x. [DOI] [PubMed] [Google Scholar]
- 69.Klinghoffer RA, Hamilton TG, Hoch R, Soriano P. An allelic series at the PDGFalphaR locus indicates unequal contributions of distinct signaling pathways during development. Developmental cell. 2002;2:103–113. doi: 10.1016/s1534-5807(01)00103-4. [DOI] [PubMed] [Google Scholar]
- 70.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. Journal of molecular biology. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 71.Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci U S A. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tallquist M, Kazlauskas A. PDGF signaling in cells and mice. Cytokine Growth Factor Rev. 2004;15:205–213. doi: 10.1016/j.cytogfr.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 74.Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 75.Hu Q, Klippel A, Muslin AJ, Fantl WJ, Williams LT. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science. 1995;268:100–102. doi: 10.1126/science.7701328. [DOI] [PubMed] [Google Scholar]
- 76.Erpel T, Courtneidge SA. Src family protein tyrosine kinases and cellular signal transduction pathways. Current opinion in cell biology. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 77.Pierce GF, Tarpley JE, Tseng J, Bready J, Chang D, Kenney WC, Rudolph R, Robson MC, Vande Berg J, Reid P, et al. Detection of platelet-derived growth factor (PDGF)-AA in actively healing human wounds treated with recombinant PDGF-BB and absence of PDGF in chronic nonhealing wounds. J Clin Invest. 1995;96:1336–1350. doi: 10.1172/JCI118169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levitzki A. PDGF receptor kinase inhibitors for the treatment of PDGF driven diseases. Cytokine Growth Factor Rev. 2004;15:229–235. doi: 10.1016/j.cytogfr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 79.Posner I, Engel M, Gazit A, Levitzki A. Kinetics of inhibition by tyrphostins of the tyrosine kinase activity of the epidermal growth factor receptor and analysis by a new computer program. Molecular pharmacology. 1994;45:673–683. [PubMed] [Google Scholar]
- 80.Homsi J, Daud AI. Spectrum of activity and mechanism of action of VEGF/PDGF inhibitors. Cancer control : journal of the Moffitt Cancer Center. 2007;14:285–294. doi: 10.1177/107327480701400312. [DOI] [PubMed] [Google Scholar]
- 81.Hou X, Kumar A, Lee C, Wang B, Arjunan P, Dong L, Maminishkis A, Tang Z, Li Y, Zhang F, Zhang SZ, Wardega P, Chakrabarty S, Liu B, Wu Z, Colosi P, Fariss RN, Lennartsson J, Nussenblatt R, Gutkind JS, Cao Y, Li X. PDGF-CC blockade inhibits pathological angiogenesis by acting on multiple cellular and molecular targets. Proc Natl Acad Sci U S A. 2010;107:12216–12221. doi: 10.1073/pnas.1004143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Akiyama H, Kachi S, Silva RL, Umeda N, Hackett SF, McCauley D, McCauley T, Zoltoski A, Epstein DM, Campochiaro PA. Intraocular injection of an aptamer that binds PDGF-B: a potential treatment for proliferative retinopathies. Journal of cellular physiology. 2006;207:407–412. doi: 10.1002/jcp.20583. [DOI] [PubMed] [Google Scholar]
- 83.Holash J, Davis S, Papadopoulos N, Croll SD, Ho L, Russell M, Boland P, Leidich R, Hylton D, Burova E, Ioffe E, Huang T, Radziejewski C, Bailey K, Fandl JP, Daly T, Wiegand SJ, Yancopoulos GD, Rudge JS. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci U S A. 2002;99:11393–11398. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]