Abstract
AIM: To investigate the relationship between low molecular polypeptide-7 (LMP-7) gene polymorphism and response to interferon (IFN) therapy in chronic hepatitis C virus (HCV) patients.
METHODS: LMP-7 polymorphism at codon 49 with nucleotide substitution from A to C was amplified in 104 chronic HCV patients of genotype 4. The amplicons were digested with restriction endonuclease BsmI and the produced restriction fragment length polymorphism was analyzed. Patients received IFN + regional blood volume therapy for 48 wk and the frequency of this single nucleotide polymorphism (SNP) was statistically correlated with treatment response. The exclusion criteria for these patients were stated by the national health program for treating viral hepatitis. Main exclusion criteria included co-infection with hepatitis B virus or schistosomiasis, thyroid dysfunction, uncontrolled diabetes mellitus, history of long term drug or alcohol intake and autoimmune hepatitis. Multivariate analyses were done to correlate LMP-7 SNP plus several factors such as age, gender, weight, serum alpha-fetoprotein (AFP) and alanine aminotransferase levels, liver activity, fibrosis score and viral load with response to therapy.
RESULTS: The data presented in this study clearly demonstrated statistically significant differences between sustained virological response (SVR) (defined as the absence of HCV RNA levels in the patient’s sera at least 6 mo after discontinuation of treatment) and non-response (NR) (where HCV RNA levels in the patient’s sera never become undetectable for 6 mo during or after treatment). Variables were described as odds ratio with 95%CI. The data were considered significant if P values were ≤ 0.05; highly significant if P < 0.01 and very highly significant if P < 0.001. Current data showed that 91.7% of patients carrying LMP-7 C/C allele were associated with SVR, while the other two genotypes C/A and A/A were associated with NR patients, 83.3% and 64.3% respectively, showing that genotype CC was strongly associated with response to interferon (95%CI: 12.0719-134.6572, P = 0.0001). The majority of parameters recorded in SVR and NR patients included higher values of mean age (P = 0.004), alanine aminotransferase (P = 0.001), AFP (P = 0.001), body weight (P = 0.025), viral load (P = 0.025), higher fibrosis and histological activity index indices among NR vs SVR patients. Also, the multivariate statistical analysis of the different factors of fibrosis score, liver activity grade, genotypes and alleles of LMP-7 gene polymorphism in responders and NRs of HCV patients in this study showed that HCV patients with A allele had a very highly significant association with the NRs, high fibrosis and higher liver activity, while the C allele had a very highly significant association with the responders, low fibrosis and lower liver activity (95%CI: 3.5800-13.2519, P = 0.0001).
CONCLUSION: LMP-7 SNP is a candidate gene that should be considered when designing a mathematical model for predicting response to therapy and disease progression in HCV patients.
Keywords: Hepatitis C virus, Interferon therapy, Low molecular mass polypeptide, Host gene, Single nucleotide polymorphism
INTRODUCTION
More than 170 million people are currently affected by chronic hepatitis C virus (HCV) infection worldwide, with the highest prevalence in Africa and Asia[1]. HCV cirrhosis, fibrosis and hepatocellular carcinoma are major sequelae of HCV chronic infection. The latter is one of the leading causes of mortality, morbidity and liver transplantation worldwide. Pegylated interferon (IFN) and ribavirin combined therapy is the standard approved treatment for HCV and is only effective in around 50% of patients[2]. Both virus and host-related elements have been reported as factors correlated to therapeutic effects of combination therapy, such as viral load, genotype and viral kinetics gender, age, ethnicity, obesity, degree of liver fibrosis and genetic polymorphisms[3]. The role of host genetic factors encoding gene products involved in the immune response following HCV infection are likely to influence the disease susceptibility and progression. As it has been shown that different host gene’s single nucleotide polymorphisms (SNPs) were mediators of the antiviral and antiproliferative effects of IFN[4], it is plausible that host genetic factors control the immune responses and play a critical role in determining the outcome of HCV infection. Viral infection of host cells by HCV leads to the release of the INFs (α, β, γ) which induce the formation of immunoproteosomes [catalytic core that consists of low molecular polypeptide-2 (LMP-2), LMP-7 and LMP-10] that produce immunogenic epitopes that bind to major histocompatibility complex (MHC) I molecules and when presented to CD8+ CTLs, enhances antigen presentation and triggers an antiviral response in the infected organism[5]. LMP-7 gene is a subunit of proteosomes encoded in the class II region of MHC on chromosome No. 6 adjacent to the TAP transporter genes[6]. The antigen recognition by cytotoxic CD8+T cells is dependent upon a number of crucial steps in antigen processing, including LMP-7 that can alter the pool of peptides available for class I antigen presentation, improving the CD8+T cells in response to viral antigens[7,8].
Previously, the relationship between LMP-7 genetic variation and some diseases has been reported. It has also been shown that LMP-7 might play an important role in the immunological reaction to HCV infection[7-9]. Thus, the aim of the current study is to determine the frequency of SNP at codon 49 of LMP-7 gene in chronic HCV patients receiving IFN therapy and to correlate the SNP frequency with clinical status of patients and the response to therapy.
MATERIALS AND METHODS
Subjects
Patients: The current study comprises of 104 patients (85 males and 19 females with an age range of 7-64 years old) infected with chronic HCV genotype 4. Only those patients fulfilling the criteria for being covered by the national health program for treating viral hepatitis were included. The criteria include absence of co-infection with hepatitis B virus (HBV), human immunodeficiency virus or schistosomiasis, patients with co morbid disorders such as thyroid dysfunction, uncontrolled diabetes mellitus, history of long term drug or alcohol intake and autoimmune hepatitis were excluded. Liver biopsy was carried out for histological examination and assessment of the stages of fibrosis and activity. Liver fibrosis score was divided into 5 stages from F0 to F4 and liver activity grade divided into 4 stages from A0 to A3. The number of patients belonging to each stage was detected in the two studied groups [responders and non-responders (NR)]. In NR, patient HCV viral loads persisted at or more than 2 logs of pre-treatment levels. Among those patients with an initial response to treatment, up to 50% will relapse after treatment is discontinued and are known as relapsed responders, whereas the rest will achieve a sustained virological response (SVR), as determined by the absence of detectable viremia six months after the end of treatment response. Patients received weekly sc injection of pegylated IFN α + daily oral ribavirin at a dose of 1000-1200 mg, depending on body weight, for 48 wk.
Controls: Healthy subjects (n = 33) with no history of acute or chronic disease and absence of any other viruses, diseases or disorders served as controls for LMP-7 typing.
Methods
HCV RNA tests: These include qualitative HCV nested reverse transcription-polymerase chain reaction (RT-PCR), quantitative real-time PCR determination and genotyping of HCV RNA genome. Methods used for these assays were previously described as nested RT-PCR[10], quantitative test of HCV RNA (Copus Amplicore, HCV monitor test roche, United States) and HCV genotyping[11].
DNA extraction: A salting out technique was used to extract DNA from whole blood[12].
Genotyping of LMP-7 gene: The second exon of LMP-7 at codon 49 spanning the tested SNP was amplified by PCR using sequence specific primers as follows forward primer (F): 5’-CGGACAGATCTCTGGGTGCT-3’ and reverse primer (R): 5’-TCTCCGGGACTGAAGGCT-3’. Reaction mix (50 μL) contained 2 units Taq polymerase (finnzyme, Finland), 5 μL of 10 × PCR buffer (supplied with the enzyme), 0.2 mmol/L dNTPs (Promega, Madison WI, United States), 1.5 mmol/L MgCl2, 5 μmol/L from each primer, 3 μL of DNA and DDW to 50 μL. Thermal cycling protocol comprised of initial denaturation at 94 °C for 5 min, followed by 35 cycles, each includes 94 °C for 1 min, 57 °C for 1 min and 72 °C for 1 min and a final extension step at 72 °C for 10 min. Amplification product was resolved on 2% agarose gel electrophoresis. A fragment of 304 bp indicates successful amplification (Figure 1). Amplified products were subjected to digestion with 10 units of BsmI restriction endonuclease (Amersham Pharmacia-Biotech, St Albans, United Kingdom) at 37 °C overnight. Restricted fragments were resolved on 2% agarose gel electrophoresis parallel with a DNA size marker (Amersham Pharmacia-Biotech) where C/C homozygotes appear as 2 bands at 174 and 130 bp, C/A heterozygotes appear as 3 bands of 174, 130 and 304 bp and uncut fragment at 304 bp represents A/A genotype (Figure 1).
Figure 1.
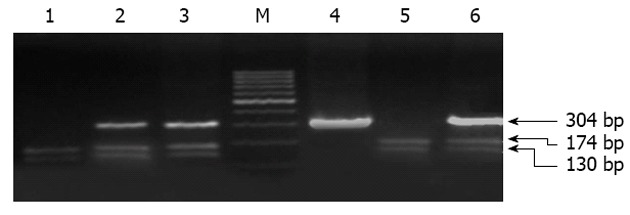
Agrose gel electrophoresis of digested low molecular polypeptide-7 amplicons. Purified genomic DNA from each patient was subjected to polymerase chain reaction amplification using specific primers bracketing the low molecular polypeptide-7 gene followed by BsmI digestion and the products were resolved on 2% agarose gel. Lanes 2, 3 and 6 represent heterozygous C/A; Lanes 1 and 5 show C/C genotype; Lane 4 shows A/A genotype; M indicated molecular weight marker.
Statistical analysis
Data were expressed as mean ± SD of percentage of each genotype (C/C, A/A or C/A) among the subject population whether SVR or NR. Comparison between mean values of different variables among SVR and NR patients was performed using unpaired student t test. Comparison between categorical data [n (%)] was done using χ2 test. Multinomial logistic regression with forward stepwise variable selection was used to identify predictors associated with SVR rates. Variables were described as odds ratio with 95%CI. The data were considered significant if P values were ≤ 0.05; highly significant if P < 0.01 and very highly significant if P < 0.001[13,14].
RESULTS
Demographic data
The data presented in Table 1 clearly demonstrate statistically significant differences between SVR and NR patients in the majority of parameters recorded, including higher values of mean age, alanine aminotransferase (ALT), alpha-fetoprotein (AFP), body weight, viral load, higher fibrosis and histological activity index (HAI) indices among NR vs SVR patients, whereas low fibrosis and HAI indices were statistically elevated in SVR patients compared with NR patients. All CHC patients in this study (n = 104) had genotype 4a of HCV RNA, using the method described previously[11].
Table 1.
The demographic data of chronic hepatitis C virus patients infected with hepatitis C virus type 4a (sustained virological response vs non-response) after treatment
| NR (n = 48) | SVR (n = 56) | P value | |
| Age (yr) | 0.004 | ||
| min-max | 12-64 | 7-58 | |
| mean ± SD | 44.17 ± 10.97 | 37.91 ± 10.91 | |
| Gender | 0.9061 | ||
| Female | 9 (18.75) | 10 (17.86) | |
| Male | 39 (81.25) | 46 (82.14) | |
| Weight (kg) | 85.05 ± 13.98 | 78.54 ± 15.10 | 0.025 |
| ALT | 89.15 ± 62.70 | 48.39 ± 40.52 | 0.001 |
| AFP | 30.56 ± 22.58 | 11.28 ± 15.33 | 0.001 |
| Viral load | 686 150.38 ± 680 474.25 | 15 656.09 ± 452 782.09 | 0.001 |
| Low fibrosis (F0-F1) (n = 39) | 7 (17.95) | 32 (82.05) | 0.001 |
| High fibrosis (F2-F4) (n = 65) | 41 (63.08) | 24 (36.92) | 0.035 |
| A0-A1 (n = 71) | 24 (33.8) | 47 (66.2) | 0.006 |
| A2-A3 (n = 33) | 24 (72.73) | 9 (27.27) | 0.009 |
Data are expressed as mean ± SD or n (%). NR: Non-response; SVR: Sustained virological response; ALT: Alanine aminotransferase; AFP: Alpha-fetoprotein.
Not significant.
Frequencies of LMP-7 genotypes
Current data showed that 91.7% of patients carrying LMP7 C/C allele were associated with SVR, showing that genotype CC was strongly associated with response to interferon and these data were very highly significant (P = 0.001) (Tables 2 and 3). However, the LMP-7 C/C allele constituted 54.5% of the healthy population which is slightly higher than the CHC patients 46% (Table 2, Figure 2), suggesting a possible protective role of C/C genotype. On the other hand, the genotypes C/A and A/A were more associated with NR, since 69% and 57% of NR patients were associated with C/A and A/A respectively, while 14.3% and 7.3% of relapsers carried C/A and A/A genotypes respectively, thus making a total of 83.3% C/A and 64.3% A/A as NR+ relapsed patients (Tables 2 and 3, Figure 3).
Table 2.
The percentage of sustained virological response and non-response patients compared with control subjects in various genotypes of low molecular polypeptide-7 gene n (%)
| NR (n = 48) | SVR (n = 56) | All patients (n = 104) | Control (n = 33) | |
| AA (n = 14) | 9 (64.29) | 5 (35.71) | 14 (13.46) | 3 (9) |
| CA (n = 42) | 35 (83.33) | 7 (16.67) | 42 (40.38) | 12 (36.5) |
| CC (n = 48) | 4 (8.33) | 44 (91.67) | 48 (46.15) | 18 (54.5) |
NR: Non-response; SVR: Sustained virological response.
Table 3.
Sustained virological response and non-response frequencies in various low molecular polypeptide-7 genotypes n (%)
| AA (n = 14) | CA (n = 42) | CC (n = 48) | P value | |
| NR (n = 48) | 9 (18.75) | 35 (72.92) | 4 (8.33) | 0.001 |
| SVR (n = 56) | 5 (8.93) | 7 (12.50) | 44 (78.57) | 0.001 |
Polymerase chain reaction-restriction fragment length polymorphism analysis were performed on chronic hepatitis C virus patients and the low molecular polypeptide-7 genotype frequencies were calculated as percentages of each group. NR: Non-response; SVR: Sustained virological response.
Figure 2.
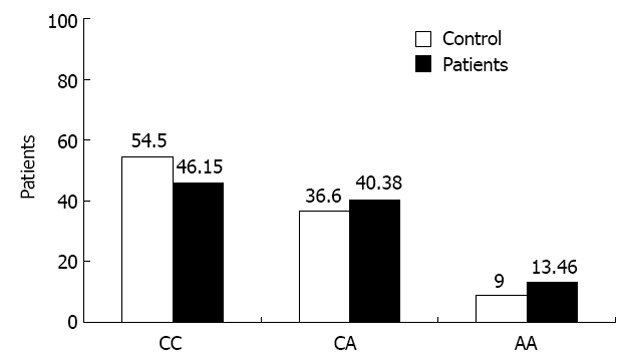
Frequencies of low molecular polypeptide-7 genotypes in control Egyptian subjects compared with chronic hepatitis C virus patients. Typing of low molecular polypeptide-7 (LMP-7) single nucleotide polymorphism was determined in 33 control subjects and 104 chronic hepatitis C virus patients. The frequency of each LMP-7 genotype in both patient groups was presented as columns.
Figure 3.

Rates of sustained virological response and non-response among the three genotypes of low molecular polypeptide-7 gene. Genotyping of low molecular polypeptide-7 (LMP-7) was determined in 56 sustained virological response (SVR) patients and in 48 non-response (NR) patients. Rates of SVR (black column) and NR (white column) were independently plotted as a function of each LMP-7 genotype, i.e., CC, CA and AA.
Genetic association of LMP-7 SNP with pathobiology of the liver in CHC patients
The strongest association between LMP-7 variations and pathological changes in the liver of chronic C patients was observed during stages of fibrosis where the percentage of C/C genotype was the highest in the lower fibrosis stage, i.e., F0 and F1 (70% and 62.07%) respectively and these data were highly significant (P = 0.045 and P = 0.004, respectively) and decreases gradually till it reaches 0% at the severe fibrosis stage (F4). Also, C/C genotype was the highest at lower grades of liver activity, i.e., A0-A1 (77.78% and 54.84% respectively) which was also highly significant (P = 0.018 and P = 0.001, respectively) and decreases gradually until it reaches its lowest value (16.67%) at (A3) (Table 4, Figures 4 and 5).
Table 4.
The relationship of the genotypes of low molecular polypeptide-7 gene polymorphism within different liver fibrosis and activity stages in sustained virological response and non-response groups after treatment n (%)
| AA (n = 14) | CA (n = 42) | CC (n = 48) | P value | |
| F0 (n = 10) | 1 (10.00) | 2 (20) | 7 (70) | 0.045 |
| F1 (n = 29) | 4 (13.79) | 7 (24.14) | 18 (62.07) | 0.004 |
| F2 (n = 36) | 4 (11.11) | 14 (38.89) | 18 (50.00) | 0.013 |
| F3 (n = 23) | 4 (17.39) | 14 (60.87) | 5 (21.74) | 0.019 |
| F4 (n = 6) | 1 (16.67) | 5 (83.33) | 0 (0) | 0.1021 |
| A0 (n = 10) | 1 (11.11) | 1 (11.11) | 7 (77.78) | 0.018 |
| A1 (n = 29) | 9 (14.52) | 19 (30.65) | 34 (54.84) | 0.001 |
| A2 (n = 36) | 3 (11.11) | 18 (66.67) | 6 (22.22) | 0.001 |
| A3 (n = 6) | 1 (16.67) | 4 (66.67) | 1 (16.67) | 0.023 |
Not significant.
Figure 4.

The fibrosis stages among low molecular polypeptide-7 genotypes polymorphism in patient groups (sustained virological response and non-response) after treatment. Frequencies of low molecular polypeptide-7 genotypes were determined in chronic hepatitis C virus patients after treatment with different fibrosis scores (F0-F4).
Figure 5.
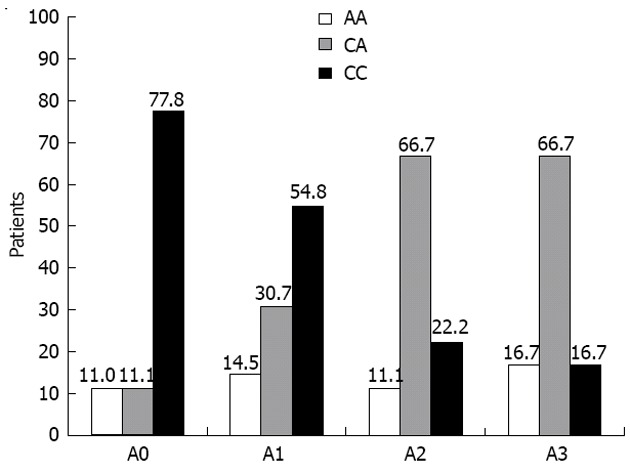
Distribution of low molecular polypeptide-7 genotypes in chronic hepatitis C virus patients after treatment with various hepatic activities (A0-A3). Frequencies of low molecular polypeptide-7 genotypes in chronic hepatitis C virus patients after treatment with various hepatic activities (A0-A3).
Multivariate analysis of different host factors, including LMP-7 variants in SVR and NR patients
The data presented in Table 5 summarize the role of several factors in achieving SVR to combined therapy. Patients with lower stages of fibrosis (F0-F1) are more associated with SVR than patients with advanced fibrosis (F2-F4) (95%CI: 0.0490-0.3346, P = 0.0001). Likewise, patients with higher liver activity (A2-A3) had lower chances of SVR compared to those with high activity (A0-A1) (95%CI: 0.0777-0.4759, P = 0.0004). Patients with higher levels of AFP (> 5 ng/mL) had less chance of SVR compared to those with AFP levels below 5 ng/mL (95%CI: 0.0871-0.5948, P = 0.0002). LMP-7 C/C genotype had significant association with SVR compared with C/A and A/A variants (95%CI: 12.0719-134.7572, P = 0.0001). In general, patients with A allele had significant association with NR, while C carriers had significant association with SVR (95%CI: 3.5800-13.2519, P = 0.0001).
Table 5.
Multivariate analysis of different clinical factors and alleles of low molecular polypeptide-7 gene polymorphism in responders and non-responders hepatitis C virus patients (%)
| Regression coefficient | SE | Odds ratio | 95%CI | P value | |
| F0-F1 vs F2-F4 | 2.0553 | 0.490 | 0.128 | 0.0490-0.3346 | 0.0001 |
| A0-A1 vs A2-A4 | -1.6529 | 0.465 | 0.192 | 0.0771-0.4759 | 0.0004 |
| AFP (< 5 ng/mL vs > 5 ng/mL) | -1.4800 | 0.490 | 0.228 | 0.0871-0.5948 | 0.00025 |
| CC vs CA + AA | 3.6972 | 0.616 | 40.333 | 12.0719-134.7572 | 0.0001 |
| C allele vs A allel | 1.9298 | 0.334 | 6.888 | 3.5800-13.2519 | 0.0001 |
DISCUSSION
Recently, it has been shown that there is a complex relationship between HCV and immune responses of the host. MHC is a crucial molecule which initiates or regulates the immune response by presenting foreign or self-antigens to T lymphocytes that play essential roles in the HCV infection or even viral clearance[15]. It has been shown that LMP-7 is important in the MHC class I antigen presentation pathway. LMP-7 is a subunit of a multifunctional proteosome that plays a pivotal role in selective degradation of endogenous proteins into peptides suited for binding to human MHC class I molecule[9]. LMP-7 polymorphisms were found to be related to a number of immune diseases and HBV infection, while actually few studies have examined the relationship between LMP-7 and HCV infection[8,16,17]. The aim of the present study is to investigate the possible association between LMP-7 polymorphism and HCV infection in response to IFN therapy. Data of the current study show that demographic and clinical factors influence the likelihood of HCV viral clearance. Age is considered one of the contributing factors to treatment outcome. This was supported by earlier reports[18,19] that younger patients responded better to current HCV treatments than older ones. This is due to a couple of reasons: the immune system of someone who is younger is more intact and better able to help with the task of fighting HCV. Here, the longer that one has hepatitis C, the more the virus can replicate and possibly cause damage to the liver. This is why some medical providers now believe that people should be treated early before any liver damage has a chance to occur. The present work found no significant difference in gender distribution, similar to an earlier study[17]. However, others reported that females seem to respond better than males which is likely to be related to the positive effects of estrogens on immune responses that are influenced by IFN therapy[20,21]. Our data confirm the relationship between a patient’s weight and response to interferon therapy, in agreement with other studies which showed that obesity is a predictor of disease progression in patients with chronic hepatitis C[22,23]. Serum ALT level is a marker of liver cell damage and the efficacy of antiviral treatment[24], as shown in our results where the mean value of ALT in SVR is much less than that in NR patients. Viral load is a key in predicting response to IFN. Many studies have shown that the lower the viral load, the more likely a patient is to eradicate the virus with IFN-based therapies[25,26]; these data support our results as the mean value in NR is much higher than in SVR patients. In general, lower viral loads are associated with less circulating viral quasispecies and also with a faster time towards HCV RNA negativity during treatment. A significant association was observed between LMP-7 polymorphism and viral clearance following IFN therapy. A strikingly higher frequency of the C/C genotype (91.67%) was observed in SVR vs 8.33% in NR cases (P = 0.001). Also, C allele was detected in 68.84% of SVR vs 31.16% in NR patients (P = 0.001). On the other hand, A allele frequency was 24.29% in SVR vs 75.71% in NR patients (P = 0.001). In addition to that, C/A genotype distribution was 16.67% in responders vs 83.33% in non-responders, while that of A/A was 35.71% in responders vs 64.29% in non-responders. Earlier studies reported conflicting data where some reported that CA or AA genotypes LMP7 could increase the risk of HCV persistence compared with C/C genotype[21], while others showed that LMP-7 A/A genotype is higher in SVR patients than in non-responders[27].
The mechanism by which LMP-7 influences response to IFN remains unclear. Several reports have indicated that patients who display successful clearance of HCV either spontaneously or after IFN α therapy express strong T-cell reaction and increased IFN γ production in response to HCV antigens[28,29]. Since the expression of LMP-7 is up-regulated by IFN γ, the differences of LMP-7 phenotypes could alter the proteosome function of patients treated with IFN α. Moreover, an amino acid substitution [LMP-7-Q (Gln) of genotype (CC) and LMP-7-K (Lys)] of genotype (AA) alters its electric charge (from non-charged polar to charged polar) with possible functional consequences. This confirms the phenotypic effects of LMP-7 on IFN response in patients with chronic hepatitis[27,30].
The present work also shows a relationship between LMP-7 gene polymorphisms with different fibrosis stages and indicates that frequency of C/C genotype was the highest in lower fibrosis stage F0 and F1 (70% and 62.07%, respectively, P = 0.045 and P = 0.004). C/C genotype frequency decreases gradually until it reaches 0% during the severe fibrosis stage (F4). On the other hand, C/A genotype frequency was the highest in advanced fibrosis stages F3 and F4 (60.87% and 83.33%, respectively, P = 0.019) and decreases gradually until it reaches its lowest value (20%) at F0. In agreement with our data, other studies reported that LMP-7 activity influences the rate of progression of liver fibrosis and the increased risk of the clinical course of HBV and HCV infection[17,21,27]. Even further, impairment of LMP function may enhance human tumorigenesis[8,31].
In conclusion, the present findings implicate genetic variation of LMP-7 gene polymorphisms in the outcome of HCV infection and have a great association with response to IFN-therapy and could be used as a diagnostic marker for disease progression and prediction of drug response.
COMMENTS
Background
Hepatitis C virus (HCV) is a global health problem that infects more than 170 million people worldwide. HCV cirrhosis, fibrosis and hepatocellular carcinoma are major sequelae of HCV chronic infection and are the leading causes of liver transplantation, mortality and morbidity worldwide. Pegylated interferon (IFN) and ribavirin combined therapy is the standard approved treatment for HCV and is only effective in around 50% of patients. Host genetic markers are required to predict drug induced viral clearance.
Research frontiers
HCV results in chronic hepatitis in more than 70% of infected patients, while 20%-30% of patients recover spontaneously. This indicates the role of the host genetic factors that may influence the ability to clear the virus after infection and in response to antiviral therapy. In this study, the authors investigated the association between low molecular polypeptide-7 (LMP-7) single nucleotide polymorphism (SNP) and drug induced clearance of HCV as a possible host gene for pre treatment prognosis.
Innovations and breakthroughs
Predictors of response are very important for HCV treatment. There are various genetic predictors of response. In this study, LMP-7 is presented as a new novel SNP which predicts sustained virological response (SVR) in patients with chronic HCV (CHC). Indeed, the data of the current study indicate that LMP-7 gene polymorphism is a better prognostic marker for response to therapy than interleukin-28B (IL28B) SNP since both markers were examined in the same patient cohort with 91.7% in LMP-7 vs 67% in IL28B association of the protective allele with SVR.
Applications
The authors hereby introduce a novel predictor for response to therapy in CHC patients. This marker will be a candidate for designing a model for viral clearance, either drug induced or spontaneous, as well as for progression of chronic HCV disease.
Terminology
Host genes were shown to play important roles in the pathogenesis of chronic hepatitis C and serve to guide improvements to IFN-based therapy and the development of novel anti-HCV therapeutics. This led the authors to study one of these genetic variants as LMP-7 in the chronically infected to clarify the association of these variants with the spontaneous viral elimination, stage of fibrosis and response to antiviral therapy.
Peer review
This is a good study in which the authors investigated the relationship between LMP-7 gene polymorphism and response to IFN therapy in CHC for the importance of host genetic factors in either spontaneous or drug induced viral clearance. It revealed a novel SNP which predicts SVR in patients with CHC and could be used in the future as a useful marker for prediction of response to therapy.
Footnotes
P- Reviewer Ahn SH S- Editor Song XX L- Editor Roemmele A E- Editor Li JY
References
- 1.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 2.Ferenci P. Treatment of chronic viral hepatitis. Best Pract Res Clin Gastroenterol. 2004;18 Suppl:113–120. doi: 10.1016/j.bpg.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 3.Shirakawa H, Matsumoto A, Joshita S, Komatsu M, Tanaka N, Umemura T, Ichijo T, Yoshizawa K, Kiyosawa K, Tanaka E. Pretreatment prediction of virological response to peginterferon plus ribavirin therapy in chronic hepatitis C patients using viral and host factors. Hepatology. 2008;48:1753–1760. doi: 10.1002/hep.22543. [DOI] [PubMed] [Google Scholar]
- 4.Mascheretti S, Hinrichsen H, Ross S, Buggisch P, Hampe J, Foelsch UR, Schreiber S. Genetic variants in the CCR gene cluster and spontaneous viral elimination in hepatitis C-infected patients. Clin Exp Immunol. 2004;136:328–333. doi: 10.1111/j.1365-2249.2004.02444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzushita N, Hayashi N, Kanto T, Takehara T, Tatsumi T, Katayama K, Ohkawa K, Ito A, Kasahara A, Moribe T, et al. Involvement of transporter associated with antigen processing 2 (TAP2) gene polymorphisms in hepatitis C virus infection. Gastroenterology. 1999;116:1149–1154. doi: 10.1016/s0016-5085(99)70018-1. [DOI] [PubMed] [Google Scholar]
- 6.Trowsdale J, Hanson I, Mockridge I, Beck S, Townsend A, Kelly A. Sequences encoded in the class II region of the MHC related to the “ABC” superfamiliy of transporters. Nature. 1990;348:741–744. doi: 10.1038/348741a0. [DOI] [PubMed] [Google Scholar]
- 7.Prahalad S, Kingsbury DJ, Griffin TA, Cooper BL, Glass DN, Maksymowych WP, Colbert RA. Polymorphism in the MHC-encoded LMP7 gene: association with JRA without functional significance for immunoproteasome assembly. J Rheumatol. 2001;28:2320–2325. [PubMed] [Google Scholar]
- 8.Cao B, Tian X, Li Y, Jiang P, Ning T, Xing H, Zhao Y, Zhang C, Shi X, Chen D, et al. LMP7/TAP2 gene polymorphisms and HPV infection in esophageal carcinoma patients from a high incidence area in China. Carcinogenesis. 2005;26:1280–1284. doi: 10.1093/carcin/bgi071. [DOI] [PubMed] [Google Scholar]
- 9.Vargas-Alarcón G, Gamboa R, Zuñiga J, Fragoso JM, Hernández-Pacheco G, Londoño J, Pacheco-Tena C, Cardiel MH, Granados J, Burgos-Vargas R. Association study of LMP gene polymorphisms in Mexican patients with spondyloarthritis. Hum Immunol. 2004;65:1437–1442. doi: 10.1016/j.humimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 10.El-Awady MK, Youssef SS, Omran MH, Tabll AA, El Garf WT, Salem AM. Soluble egg antigen of Schistosoma Haematobium induces HCV replication in PBMC from patients with chronic HCV infection. BMC Infect Dis. 2006;6:91. doi: 10.1186/1471-2334-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, Mukaide M, Williams R, Lau JY. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–207. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thakeb F, El-Serafy M, Zakaria S, Monir B, Lashin S, Marzaban R, El-Awady M. Evaluation of liver tissue by polymerase chain reaction for hepatitis B virus in patients with negative viremia. World J Gastroenterol. 2005;11:6853–6857. doi: 10.3748/wjg.v11.i43.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armitage P, Berry G. Statistical methods in medical research. 3rd ed. Science. 2000. [Google Scholar]
- 14.Riffenburgh RH. Statistics in Medicine. 2nd ed. United States: Elsevier Academic press; 2006. [Google Scholar]
- 15.Sun J, Li K, Shata MT, Chan TS. The immunologic basis for hepatitis C infection. Curr Opin Gastroenterol. 2004;20:598–602. doi: 10.1097/00001574-200411000-00016. [DOI] [PubMed] [Google Scholar]
- 16.Dai Y, Ning T, Li K, Qi SX, Jiang MW, Chai QB, Gai YH, Wang X. Association between LMP2/LMP7 gene polymorphism and the infection of hepatitis B virus. Beijing Daxue Xuebao. 2005;37:508–512. [PubMed] [Google Scholar]
- 17.Xu C, Qi S, Gao L, Cui H, Liu M, Yang H, Li K, Cao B. Genetic polymorphisms of LMP/TAP gene and hepatitis B virus infection risk in the Chinese population. J Clin Immunol. 2007;27:534–541. doi: 10.1007/s10875-007-9095-x. [DOI] [PubMed] [Google Scholar]
- 18.Poynard T, McHutchison J, Davis GL, Esteban-Mur R, Goodman Z, Bedossa P, Albrecht J. Impact of interferon alfa-2b and ribavirin on progression of liver fibrosis in patients with chronic hepatitis C. Hepatology. 2000;32:1131–1137. doi: 10.1053/jhep.2000.19347. [DOI] [PubMed] [Google Scholar]
- 19.Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A, et al. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124–134. doi: 10.1056/NEJMoa066403. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Y, Shimizu I, Shen M, Aoyagi E, Takenaka H, Itagaki T, Urata M, Sannomiya K, Kohno N, Tamaki K, et al. Effects of estradiol and progesterone on the proinflammatory cytokine production by mononuclear cells from patients with chronic hepatitis C. World J Gastroenterol. 2008;14:2200–2207. doi: 10.3748/wjg.14.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui Q, Zhang Y, Su J, Shi C, Lei N, Ding K, Li J. The Association between the genetic polymorphisms of LMP2/LMP7 and the Outcomes of HCV infection among drug users. Shengwu Yixve Yanjiu Zazhi. 2010;24:374–380. doi: 10.1016/S1674-8301(10)60050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ortiz V, Berenguer M, Rayón JM, Carrasco D, Berenguer J. Contribution of obesity to hepatitis C-related fibrosis progression. Am J Gastroenterol. 2002;97:2408–2414. doi: 10.1111/j.1572-0241.2002.05995.x. [DOI] [PubMed] [Google Scholar]
- 23.Charlton MR, Pockros PJ, Harrison SA. Impact of obesity on treatment of chronic hepatitis C. Hepatology. 2006;43:1177–1186. doi: 10.1002/hep.21239. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay KL. Introduction to therapy of hepatitis C. Hepatology. 2002;36:S114–S120. doi: 10.1053/jhep.2002.36226. [DOI] [PubMed] [Google Scholar]
- 25.Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, et al. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT) Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 26.McHutchison JG, Poynard T. Combination therapy with interferon plus ribavirin for the initial treatment of chronic hepatitis C. Semin Liver Dis. 1999;19 Suppl 1:57–65. [PubMed] [Google Scholar]
- 27.Sugimoto Y, Kuzushita N, Takehara T, Kanto T, Tatsumi T, Miyagi T, Jinushi M, Ohkawa K, Horimoto M, Kasahara A, et al. A single nucleotide polymorphism of the low molecular mass polypeptide 7 gene influences the interferon response in patients with chronic hepatitis C". J Viral Hepat. 2002;9:377–384. doi: 10.1046/j.1365-2893.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- 28.Missale G, Bertoni R, lamonaca V. Different clinical behaviours of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell mediated immune response. J Clin Invest. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cramp ME, Rossol S, Chokshi S, Carucci P, Williams R, Naoumov NV. Hepatitis C virus-specific T-cell reactivity during interferon and ribavirin treatment in chronic hepatitis C. Gastroenterology. 2000;118:346–355. doi: 10.1016/s0016-5085(00)70217-4. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka K, Tanahashi N, Tsurumi C, Yokota KY, Shimbara N. Proteasomes and antigen processing. Adv Immunol. 1997;64:1–38. doi: 10.1016/s0065-2776(08)60885-8. [DOI] [PubMed] [Google Scholar]
- 31.Seliger B, Bock M, Ritz U, Huber C. High frequency of a non-functional TAP1/LMP2 promoter polymorphism in human tumors. Int J Oncol. 2002;20:349–353. [PubMed] [Google Scholar]


