Abstract
In Gram-negative bacteria, integral outer membrane β-barrel proteins (OMPs) are assembled by the beta-barrel assembly machine (Bam) complex. The essential components of this complex are the OMP BamA [which contains a carboxyl-terminal β-barrel and an amino-terminal periplasmic module composed of five polypeptide transport associated (POTRA) domains] and the lipoprotein BamD. In Escherichia coli, the Bam complex also contains three nonessential lipoproteins (BamBCE), all of which require the barrel-proximal POTRA domain (P5) for stable interactions with BamA. We have previously reported that the BamA β-barrel assumes two different conformations. A method for conformation-specific labeling of BamA described here reveals that these conformers reflect the degree of surface exposure of the conserved sixth extracellular loop (L6). L6 is surface accessible in one conformation but not in the other, likely because it occupies the lumen of the BamA β-barrel in the latter case. A gain-of-function mutation that promotes Bam activity (bamDR197L) and a loss-of-function mutation that decreases the activity of Bam (ΔbamE) both favor surface exposure of BamA L6, suggesting that BamD and BamE normally act to control L6 exposure through opposing functions. These results, along with the synthetic lethality of the bamDR197L ΔbamE double mutant, imply a cyclic mechanism in which the Bam lipoproteins regulate the conformation of BamA during the OMP assembly reaction. Our results further suggest that BamDE controls L6 exposure via conformational signals transmitted through P5 to L6.
Keywords: genetics, OM biogenesis, Omp85, membrane protein folding, conformational dynamics
How proteins are successfully integrated into lipid membranes is a fundamental biological problem. In Gram-negative bacteria, this problem is compounded by the need to transport and assemble a diverse array of proteins into two distinct membrane compartments. The transmembrane domains of inner membrane (IM) proteins are α-helical in nature, whereas proteins that span the outer membrane (OM) are instead rich in β-structure that permits the formation of integral β-barrels. Periplasmic proteins modified with N-terminal lipid moieties, which can be found tethered to one or the other membrane, represent an additional class of membrane-associated protein. In Escherichia coli, dedicated transport pathways enable targeting and assembly of envelope proteins into the correct membranous compartment (1). Proteins that span the OM are assembled locally by the beta-barrel assembly machine (Bam) complex (2, 3).
The process of assembling OM β-barrel proteins (OMPs) is an intensely studied phenomenon, and much has been learned since the initial discovery of the Bam complex. In E. coli, this complex is composed of the OMP BamA and the four associated lipoproteins BamBCDE (4, 5). BamA and BamD are essential for growth, and both are highly conserved among Gram-negative species (4, 6). Individually deleting the genes encoding BamB, BamC, and BamE does not affect viability; however, there are modest OMP assembly defects of varying degrees associated with mutation of these genes (4–7). Synthetic phenotypes are observed when these mutations are combined, with the ΔbamB ΔbamE double mutant being the most impaired (8, 9).
Bam components are not equally conserved across Gram-negative bacteria. The two essential components, BamA and BamD, are the most highly conserved, followed by the nonessential lipoproteins BamE and BamB. BamC is apparently found only in β- and γ-proteobacteria (10); it is perhaps unsurprising, then, that bamC mutations do not cause detectable OMP assembly defects in E. coli (5, 6). However, all five Bam proteins must be present to assemble OMPs with maximal efficiency in vitro (11). Biochemical and genetic experiments have demonstrated that the E. coli Bam holocomplex can be separated into two essential subcomplexes: BamAB and BamCDE (11, 12).
BamA is a bipartite OMP that contains a C-terminal β-barrel domain and an N-terminal periplasmic module that can be further subdivided into five polypeptide transport associated (POTRA) domains. The β-propeller protein BamB and the tetratricopeptide-repeat protein BamD associate directly with one or more of the POTRA domains (13), and BamCE dock onto BamA indirectly through stable interactions with BamD (5, 6). Deletion of any POTRA domain except POTRA 1 (P1) prevents formation of BamAB; however, P5 alone is sufficient for the BamA–BamCDE interaction (13). Thus, P5 is essential for the interaction between BamA and BamD, which is apparently stabilized by BamCE (5).
Recent genetic and biochemical analyses further suggest that P5 is critical for conformational signaling between BamA and BamD. We have described a substitution in P5 (E373K) that abrogates the stable interaction between BamA and BamD (12). A strong suppressor of bamAE373K was isolated in bamD, but this mutation (bamDR197L) does not lead to restoration of the BamA–BamD interaction. Rather, bamDR197L is a gain-of-function allele that renders BamD active even in the absence of a stable interaction between the Bam subcomplexes. Genetic evidence suggests that bamDR197L bypasses the need for activation of BamD by BamA, implying that BamD is a dynamic component of the Bam complex.
Concurrent findings shed light on the functional relationship between BamD, BamE, and BamA. We have shown that BamE functions uniquely among the nonessential Bam lipoproteins; specifically, BamE modulates the conformation of the BamA β-barrel domain (8). Because BamE is not thought to directly interact with BamA, this activity is presumably mediated through BamD. The change in BamA conformation was demonstrated by increased protease sensitivity of BamA in a ΔbamE mutant. These findings suggest that the accessory Bam lipoproteins can influence the state of the BamA β-barrel.
In this report we describe assays that distinguish the different conformations of the BamA β-barrel domain, and we probe the mechanism by which BamDE control the conformation of BamA. We show that a highly conserved extracellular loop of BamA [sixth extracellular loop (L6)] is accessible to a specific chemical labeling reagent and that accessibility to this reagent increases in bamD and bamE mutant strains. Increased labeling of L6 correlates with increased sensitivity of BamA to protease. Additionally, we present genetic evidence showing that BamE modulates the conformation of BamA through direct control of BamD activity. Taken together, these findings suggest that surface accessibility of L6 is diagnostic for a conformational change in the BamA β-barrel that is controlled by BamDE. Our genetic analysis of bamA mutants and their suppressors supports the idea that the conformation of the BamA β-barrel is modulated by the Bam lipoproteins via P5 and implies a regulated reaction cycle that underlies the OMP assembly process in E. coli.
Results
Perturbations to the BamCDE Subcomplex Influence the Conformation of BamA.
In previous studies, we uncovered evidence that both BamA and BamD can exist in different states. In contrast to the protease-resistant BamA form found in wild-type cells, BamA exists primarily in an alternate, protease-sensitive conformation in the absence of BamE (8). Additionally, we isolated a gain-of-function mutation in bamD (bamDR197L) that was proposed to stabilize the activated state of BamD (12). Given that BamA and BamD normally interact with each other directly and that this interaction is critical for efficient OMP assembly, we wondered if the activation state of BamD could influence the conformational state of BamA.
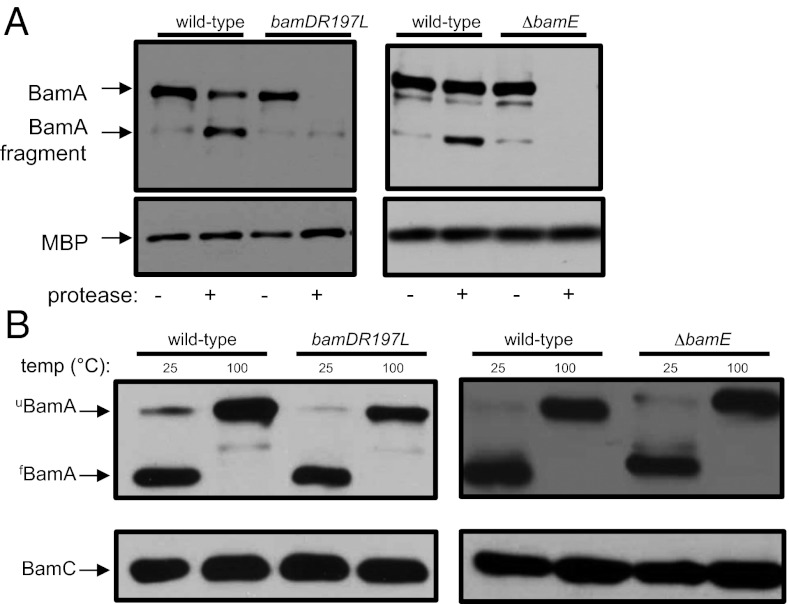
To test this, we first assayed the protease sensitivity of BamA in a bamDR197L background. As shown in Fig. 1A, the protease sensitivity of BamA in a bamDR197L strain is comparable to that observed in a ΔbamE mutant strain. This confirms our suspicion that activation of BamD (i.e., bamDR197L) and deactivation of BamE (i.e., ∆bamE) lead to the same net result—an increase in the ratio of protease-sensitive (BamAS) to protease-resistant (BamAR) species of BamA.
Fig. 1.
BamA exists in two conformations. (A) Cell pellets from wild-type, bamDR197L, and ΔbamE strains were treated with proteinase K where indicated. The protease was inactivated by treatment with PMSF, and samples were analyzed by SDS/PAGE and immunoblot using anti-BamA and anti-MBP antibodies. MBP serves as a loading control to ensure that protease does not enter the periplasm. (B) Wild-type, bamDR197L, and ΔbamE cells were processed by gentle lysis and treated at the indicated temperatures. Samples were analyzed by SDS/PAGE and immunoblot using anti-BamA and anti-BamC antibodies.
Although the protease assay is useful for distinguishing BamAS from BamAR, it does not allow us to interrogate the conformational dynamics of BamA that impact protease susceptibility. For example, our observations may suggest that the β-barrel domain is destabilized in the BamAS form in a way that renders it highly susceptible to proteolysis. To test this possibility, we took advantage of the intrinsic stability of β-barrel proteins. The β-barrel domain of BamA is, like most OM β-barrels, naturally resistant to denaturation by SDS. However, the β-barrel domain is sensitive to heat denaturation. Thus, when protein extracts are analyzed by SDS/PAGE following gentle cell lysis at room temperature, BamA remains fully folded and migrates faster than heat-denatured BamA, a property known as heat modifiability (14). However, the electrophoretic mobility of BamA is retarded in the presence of mutations that destabilize or impair folding of the β-barrel domain (9). We subjected wild-type, bamDR197L, and ΔbamE strains to gentle lysis and examined BamA folding by SDS/PAGE and immunoblot (Fig. 1B). As expected, BamA migrated at the predicted molecular weight in samples incubated at 100 °C. In samples incubated at room temperature, BamA remained folded as evidenced by its migration at a lower apparent molecular weight. Using this assay, we found that folding of BamA was indistinguishable among the three strains. This indicates that the protease assay reports on a specific structural feature of BamA and does not reflect β-barrel instability. To identify this feature, we first sought to predict the topology and structure of the BamA β-barrel domain.
Method for Selective Surface Labeling of BamA Loop 6.
Although the structure of the periplasmic domain of BamA has been determined in various conformations by multiple groups (13, 15–18), the structure of the β-barrel domain has not been reported. We investigated the topology of the β-barrel domain using the PRED-TMBB algorithm (19). BamA is predicted to form a β-barrel with 16 transmembrane β-strands and 8 extracellular loops. Of these loops, the sixth loop (L6) is significantly longer and better conserved than the others; a portion of this loop constitutes a motif common to all Omp85 family proteins (20). The full-length structure of the related Omp85 transporter FhaC revealed L6 in an extended conformation folded into the lumen of the β-barrel domain such that the tip of the loop reaches the periplasmic space (21, 22). Moreover, FhaC L6 is known to be accessible to surface proteases only in the presence of FHA, the substrate of the transporter (23), suggesting that the loop is flexible. Accordingly, we wondered whether the protease sensitivity of BamA observed in bamDR197L and ∆bamE backgrounds might reflect the surface exposure status of L6. To test this possibility, we developed an assay to monitor surface exposure of L6 specifically.
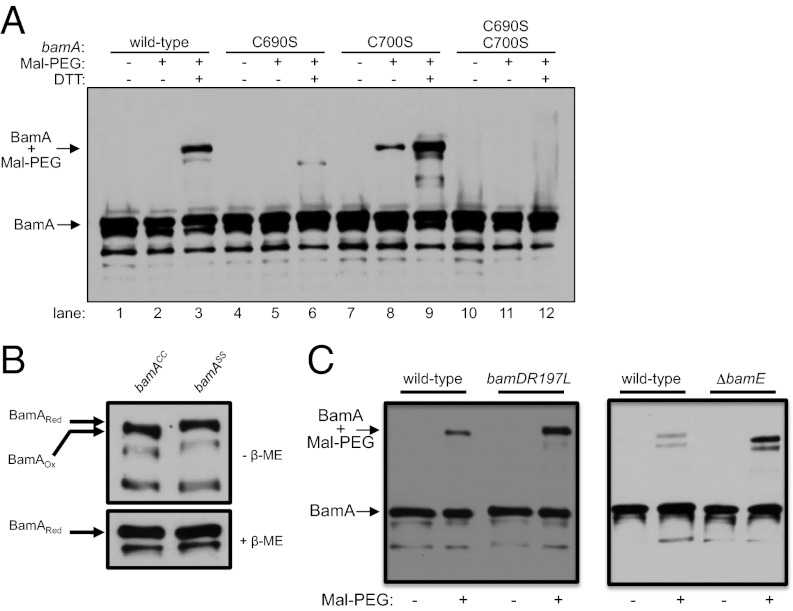
Precise monitoring of the exposure status of L6 requires a reagent that specifically labels this loop. To that end, we exploited the fact that there are two naturally occurring cysteine residues in BamA (Cys690 and Cys700), both of which are found in L6. By treating whole cells with methoxypolyethylene glycol-maleimide (Mal-PEG), an ∼20-kDa cysteine-reactive reagent that is too large to enter the periplasm, we can detect labeling of BamA by size shift on immunoblots. We find that wild-type BamA is efficiently labeled with Mal-PEG when cells are pretreated with the reducing agent DTT (Fig. 2A, lane 3). Mal-PEG labeling is specific to L6, as replacement of both cysteine residues (BamAC690S+C700S) with serine precludes labeling of BamA (Fig. 2A, lane 12). To confirm that Mal-PEG cannot penetrate the OM and label cysteines in the periplasm, we took advantage of a bamD allele that encodes a cysteine substitution at position 203 (bamDR197L+R203C). As expected, Mal-PEG labeling of BamDR197L+R203C occurs only when cells are permeabilized with SDS (Fig. S1), demonstrating the OM impermeability of the Mal-PEG reagent.
Fig. 2.
BamA L6 is surface-exposed and contains an intramolecular disulfide bond. (A) Cells from wild-type or bamA cysteine mutants were labeled with Mal-PEG as described in SI Materials and Methods either with or without DTT pretreatment. (B) Folding of BamA was assessed in wild-type (bamACC) and double mutant (bamASS) cells by performing SDS/PAGE under reducing (Lower) or nonreducing (Upper) conditions. β-Mercaptoethanol (β-ME) was used where indicated as the reducing agent. (C) Labeling as described in A was performed on wild-type, bamDR197L, or ΔbamE strains. Blots were probed with anti-BamA antibodies.
The fact that efficient labeling of BamA requires pretreatment with DTT indicates that the cysteine residues at positions 690 and 700 are typically oxidized, perhaps suggesting that their thiol groups are coupled in a disulfide bond. A number of OM β-barrel proteins are known to contain intramolecular disulfide bonds (24–26), and these proteins may exhibit altered mobility on SDS/PAGE when maintained in an oxidized state (25). To explore the possibility that the BamA β-barrel normally contains a disulfide bond, we prepared whole-cell lysates of cells expressing either wild-type BamA (BamACC) or a cysteine-less variant of BamA (BamAC690S+C700S or BamASS) in the presence or absence of reducing agent (β-mercaptoethanol) and compared the electrophoretic mobility of these variants by SDS/PAGE. We find that the oxidized species of BamA+ (BamAox) migrates faster than the reduced form (BamAred), yielding an apparent size shift; however, we do not detect any such mobility shift for the BamASS variant (Fig. 2B). This suggests that the observed shift in BamA+ is a consequence of disulfide bond formation between Cys690 and Cys700. In support of this result, we find that Mal-PEG labeling of BamAC700S (BamACS) is significantly higher than that of BamACC in the absence of DTT (Fig. 2A; compare lane 8 with lane 2), suggesting that residue Cys690 is normally inaccessible to Mal-PEG due to its involvement in the Cys690-Cys700 disulfide bond. Although some OMPs (e.g., the LPS insertase LptD) are known to contain essential intramolecular disulfide bonds (25), the Cys690-Cys700 disulfide must be dispensable for BamA folding and activity, as there are no differences between strains expressing BamACC and BamASS with respect to OM permeability, OMP levels, BamA levels, or BamA stability (Fig. S2). Additionally, a number of Omp85 family members and BamA homologs lack cysteines in the β-barrel domain, suggesting that this disulfide bond is not highly conserved.
We next determined whether BamA is more sensitive to Mal-PEG labeling in bamDR197L and ΔbamE mutant backgrounds (Fig. 2C). BamA was labeled more efficiently with Mal-PEG in both mutant strains than in wild-type E. coli. This finding confirms that the protease assay is diagnostic for the surface accessibility of L6 rather than the stability of the BamA β-barrel domain. It is worth noting that some proportion of wild-type BamA can be labeled with Mal-PEG. We take this to mean that both species of BamA (BamAR and BamAS) are normally present in wild-type cells, suggesting that BamA is capable of interconverting between the BamAR and BamAS conformers.
bamE Is Conditionally Essential in a bamDR197L Strain Background.
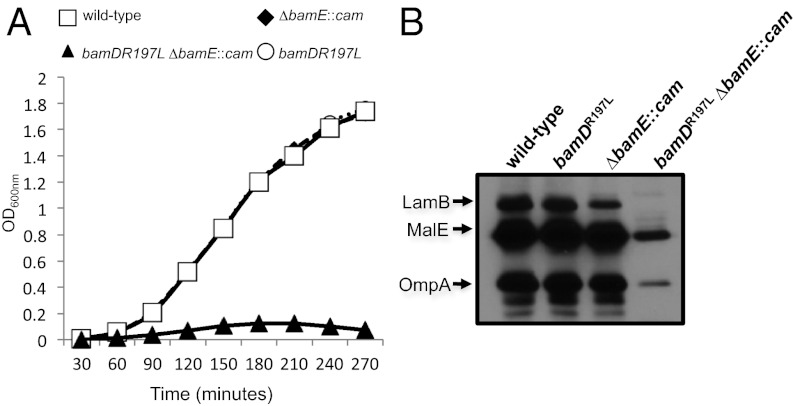
As described above, the consequence of bamDR197L and ΔbamE mutations is the same; in either case, BamA exists predominantly in the protease-sensitive, Mal-PEG–accessible conformation (BamAS). Although the equilibrium of the two forms of BamA appears to be influenced by the BamDE lipoproteins, there does not seem to be any significant impact on OMP assembly in either mutant described above. Despite this fact, we find that bamE is conditionally essential in a bamDR197L background (Fig. 3A). Null alleles of bamE can be transduced into a bamDR197L background at 30 °C, but the resulting strain exhibits profound growth and OMP biogenesis defects (Fig. 3B) and does not grow at elevated temperatures. In contrast, bamDR197L mutants are indistinguishable from wild-type E. coli by every measure, and deletion of bamE has no affect on growth and only mild effects on OM permeability and β-barrel assembly. The conditional lethality of bamDR197L ∆bamE, then, cannot be explained by invoking additive effects.
Fig. 3.
Genetic interactions between bamDR197L and ΔbamE. (A) Growth of wild-type (square), bamDR197L (circle), ΔbamE::cam (diamond), and bamDR197L ΔbamE::cam (triangle) strains at 37 °C was monitored by measuring OD600. All strains were grown to stationary phase at 30 °C and then diluted 1:1,000 in fresh LB and grown at 37 °C. (B) OMP levels were determined by immunoblot in cell extracts of wild-type, bamDR197L, ΔbamE::cam, and bamDR197L ΔbamE::cam strains grown at 30 °C.
Genetic Interaction Between POTRA 5 and Loop 6.
The results presented above show that BamA can exist in two distinct conformations that can be discriminated on the basis of the surface accessibility of L6. Both conformations are observed in wild-type cells, but the protease-sensitive, Mal-PEG–reactive conformation apparently accumulates in the presence of a gain-of-function bamD mutation or a loss-of-function bamE mutation, suggesting that BamD and BamE have opposing roles in regulation of BamA β-barrel dynamics. How might periplasmic lipoproteins (BamDE) modulate the conformation of an OM-integral β-barrel protein? Several observations led us to investigate the possibility that the barrel-proximal POTRA domain (P5) serves as an intermediary between the BamA β-barrel and the BamCDE subcomplex. First, BamCDE associate physically with BamA exclusively through P5 (13). Second, the activating bamDR197L mutation was isolated as a suppressor of a conditional lethal bamA allele (bamAE373K) that encodes a charge–change substitution in P5 (12). A third connection between P5 and L6, described below, was uncovered in the course of analyzing a nonlethal substitution at position 373.
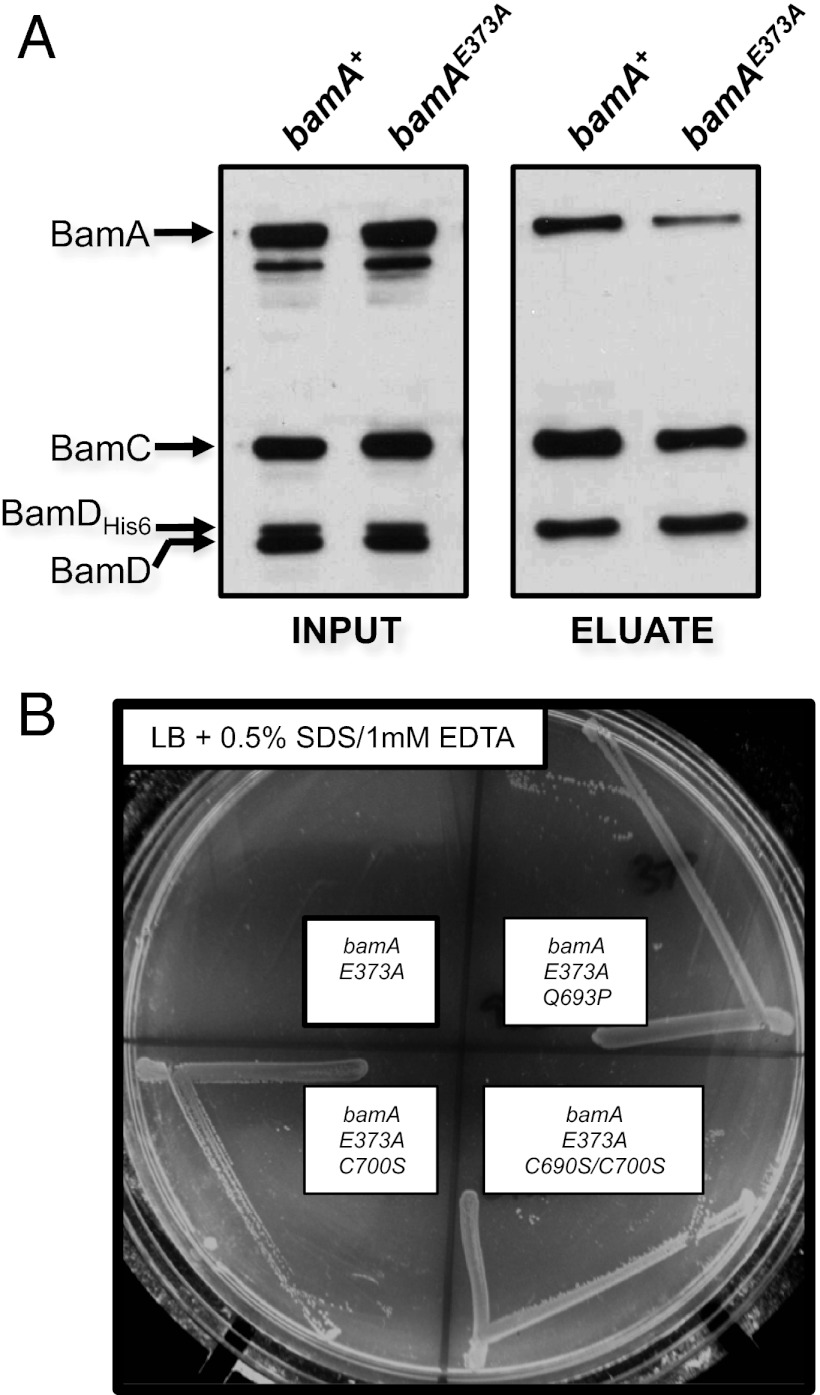
The bamAE373K mutant grows poorly even at the permissive temperature and is therefore difficult to manipulate experimentally. However, when we replace Glu373 with a nonpolar residue (E373A), we find the resulting strain to be viable, showing no OMP assembly defects but exhibiting increased sensitivity to detergent (Table 1). Additionally, we find that the amount of BamAE373A that copurifies with BamD-His6 is reduced compared with wild-type BamA (Fig. 4A), although the BamA–BamD interaction is not compromised to nearly the degree observed in the presence of BamAE373K (12). This mutation clearly causes defects that are relatively mild in comparison with bamAE373K, but the apparent destabilization of BamAD observed in a bamAE373A background implies that the E373A and E373K substitutions affect BamA function in a similar manner yet differ in severity.
Table 1.
Growth phenotypes of bamA E373 mutants compared with wild type
| Growth condition |
||||
| LB |
||||
| bamA allele | 24 °C | 37 °C | Bile salts | SDS |
| bamA+ | +++ | +++ | +++ | +++ |
| bamAE373K | + | − | − | − |
| bamAE373A | +++ | +++ | +++ | − |
Growth of each strain on the indicated agar plates was scored after incubation overnight. Strains were scored on the following scale: +++, normal growth; +, weak growth; −, no growth. The results shown are from a representative experiment.
Fig. 4.
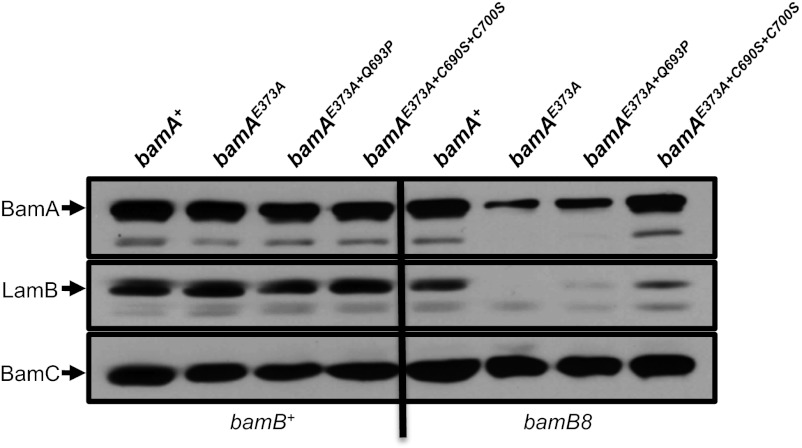
BamA L6 and BamDE communicate via POTRA5. (A) A His-tagged variant of BamD was expressed in bamA+ or bamA E373A strains to enable copurification of the Bam complex. Extracts of each strain were purified using a Ni-NTA affinity resin and eluted with imidazole. Input and eluted fractions were analyzed by SDS/PAGE and immunoblot with antibodies that recognize BamA, BamC, or BamD. (B) Strains carrying bamAE373A, bamAE373A+Q693P, bamAE373A+C700S, or bamAE373A+C690S+C700S mutations were streaked onto LB agar plates supplemented with 0.5% SDS and 1.0 mM EDTA. The image was captured after overnight growth at 37 °C.
We isolated detergent-resistant suppressors of bamAE373A, expecting that suppressor analysis might reveal residues implicated in either the BamA-CDE association or the activity of BamA. One suppressor that we identified carried a mutation in L6 that changed residue 693 from glutamine to proline (Q693P). This substitution does not affect the folding or stability of the BamA β-barrel, which retains the property of heat modifiability in the presence of the suppressor mutation (Fig. S3). However, the close proximity of this residue to C690 in the primary sequence of BamA and the propensity of proline to disrupt local secondary structure raised the possibility that the Q693P mutation might impact L6 structure in the segment containing the C690-C700 disulfide bond. We reasoned that destabilizing this segment of L6 by dissolving the C690-C700 disulfide bridge would also confer detergent resistance in a bamAE373A background. To test this, we mutated one or both L6 cysteine residues to serine and determined whether these mutations phenocopy the spontaneous Q693P suppressor. Consistent with our prediction, we find that both the C700S single mutation and the C690S/C700S double mutation restore detergent resistance to a bamAE373A mutant (Fig. 4B). Suppression must not require disruption of the disulfide bond, as BamAQ693P is apparently properly oxidized (Fig. S4). These data suggest that the suppressive effect of Q693P on the P5 mutation (E373A) is related to the conformation of L6 or a segment thereof. This result implies an association, perhaps direct, between P5 and L6, and further suggests that functional defects caused by mutation of E373 reflect some impairment in the activity or folding of the BamA β-barrel (particularly L6).
Deletion of Accessory bam Genes Causes Lethality in a bamAE373A Background.
Single bam mutations that cause subtle defects often exhibit synergistic effects when combined (5, 8). As these phenotypes have the potential to both unmask critical roles for the various components in OMP assembly and reveal the step(s) at which they act, we sought to determine whether such interactions are observed when accessory Bam components are deleted in a bamAE373A mutant background. Although deletion of bamC did not affect the growth or viability of a bamAE373A mutant, we found that introduction of a loss-of-function bamB mutation (bamB8) (7) precludes growth of bamAE373A at 37 °C and that bamE cannot be deleted in a bamAE373A background under any growth condition (Table 2). Thus, a bamAE373A bamB8 double mutant is conditionally lethal and bamE is essential for growth in a bamAE373A background.
Table 2.
Genetic interactions between bamA, bamB, bamC, and bamE mutants
|
bamB8 |
ΔbamC |
ΔbamE::cam |
||||
| bamA allele | 30 °C | 37 °C | 30 °C | 37 °C | 30 °C | 37 °C |
| bamA+ | +++ | +++ | +++ | +++ | +++ | +++ |
| bamAE373A | ++ | − | +++ | +++ | − | − |
| bamAE373A+Q693P | ++ | ++ | ND | ND | − | − |
| bamAE373A+C690S+C700S | +++ | +++ | ND | ND | − | − |
Growth of each strain on LB agar plates at the indicated temperature was scored after incubation overnight. Strains were scored on the following scale: +++, normal growth; ++, intermediate growth; +, weak growth; −, no growth; ND, not determined. The results shown are from a representative experiment.
In an attempt to understand the basis of these synthetic effects, we determined whether the L6 mutations described above (Q693P, C690S, C700S) could restore growth to a bamAE373A mutant lacking either BamB or BamE. None of the mutations above suppresses the synthetic lethality of a bamAE373A ΔbamE double mutant. However, these L6 mutations individually restore growth of the bamAE373A bamB8 double mutant at the nonpermissive temperature (Table 2). Levels of BamAE373A and the model OMP LamB, which are significantly reduced in a bamAE373A bamB8 background, are increased by these suppressor mutations to varying degrees, with the disulfide-disrupting mutations exhibiting the most significant effect (Fig. 5).
Fig. 5.
Mutations in BamA L6 can suppress the synthetically lethal bamAE373A bamB8 double mutant. Immunoblot analysis of extracts from the indicated strains were prepared as described in Materials and Methods. Extracts were probed with anti-BamA, anti-BamC, and anti-LamB antibodies.
Discussion
We have shown that BamA exists in two distinct conformations that reflect the degree of surface exposure of L6. In one conformation, L6 is largely protected from Mal-PEG and externally added protease, most likely for reasons noted below, because this loop is folded back inside the β-barrel where it could interact with POTRA 5 (BamAR); in the other conformation, L6 is surface-exposed and accessible to both reagents (BamAS). Because both conformations can be detected in wild-type cells, we suggest that both are physiologically relevant and present at equilibrium.
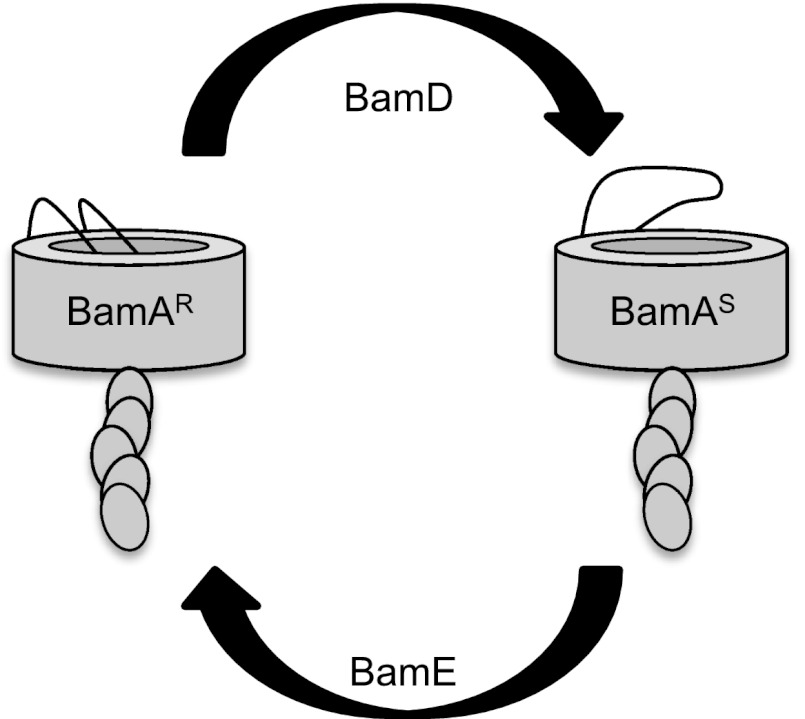
The bamDR197L and ΔbamE mutations alter this equilibrium so as to favor BamAS. Our genetic and biochemical analyses indicate that, despite this apparent similarity, BamD and BamE must have opposing mechanistic roles in modulating the conformational state of BamA. This must be so because bamDR197L and ΔbamE oppositely affect the phenotype of a bamA101 mutant. Recall that the bamA101 mutation, which alters the bamA promoter but not the ORF, causes a growth defect secondary to a 10-fold reduction in BamA levels (27). Whereas the gain-of-function mutation bamDR197L is a suppressor of bamA101 growth defects (12), the loss-of-function ΔbamE mutation is synthetically lethal with bamA101 (8). This suggests that bamDR197L increases the activity of Bam. In contrast, Bam must be less active in the absence of BamE because a significant reduction in cellular BamA levels is tolerated only when BamE is present. To account for these facts, we propose that BamD activity shifts the conformational equilibrium toward BamAS, whereas BamE activity shifts the equilibrium toward BamAR as depicted in Fig. 6.
Fig. 6.
Conformational dynamics of the Bam complex. BamA cycles between two different conformations that involve movement of the conserved L6. Interconversion between the protease-resistant, Mal-PEG inaccessible form (BamAR) and the protease-sensitive, Mal-PEG accessible form (BamAS) is regulated by BamDE. In strains carrying gain-of-function bamDR197L and loss-of-function ΔbamE mutations, the equilibrium is shifted to the right.
It is not immediately clear how and why conformational changes in the BamA β-barrel influence its function. One interpretation is that these conformers reflect “active” and “inactive” states of the Bam complex and that the BamA forms that we can distinguish biochemically represent these two states. However, because the bamDR197L and ∆bamE mutations oppositely affect BamA activity but equivalently affect the ratio of BamAS to BamAR, it is unlikely that BamAS represents an active (“ON”) conformation of BamA. BamAS, furthermore, cannot represent an inactive molecule because this form is stabilized in the presence of the BamA-activating bamDR197L mutation. Consequently, we disfavor this model.
An alternative model, which we currently favor, suggests that OMP assembly in E. coli occurs via a reaction cycle involving the regulated, cyclic interconversion between an initial and a final state of the Bam complex. Our findings are consistent with a two-step model in which BamA is induced by BamD to assume the BamAS conformation while BamE promotes completion of the reaction cycle by indirectly restoring the initial BamAR state through regulation of BamD. In wild-type cells, an unfolded substrate would interact with and alter the conformation of BamA in a BamD-dependent manner, causing L6 to become surface-exposed. After a round of assembly is completed, the Bam lipoproteins act to restore the ground state, thereby priming the machine to accept and assemble another incoming substrate.
Two lines of evidence support this “reaction cycle” model. First, this model provides a satisfying explanation for the seemingly paradoxical synthetic lethality of bamDR197L ∆bamE. In the presence of BamDR197L, the conversion from the initial to the final state is accelerated, whereas in the absence of BamE, restoration of the initial state is substantially slowed. When the two mutations are combined, the equilibrium is shifted strongly in favor of the final state, which accumulates at the expense of the initial state. The overall effect is to reduce the efficiency of OMP assembly enough to inhibit cell growth by slowing the kinetics of Bam recycling. This reduction in efficiency is lethal at elevated temperatures when growth is fastest and demand on the Bam machine is greatest.
Second, there is precedent for such a reaction cycle that involves similar changes in β-barrel conformation. BamA is related phylogenetically to FhaC, an Omp85-type transporter that mediates secretion of a single substrate, FHA, across the OM of Bordetella pertussis. The FhaC β-barrel also contains a sixth extracellular loop that exhibits two conformational states. L6 of FhaC is susceptible to surface proteolysis only in the presence of the substrate FHA, which suggests that the conformation of the barrel is modulated only when the substrate engages the transporter (23). Jacob-Dubuisson and colleagues have proposed that FhaC L6 normally occupies the lumen of the β-barrel, as evidenced by the crystal structure, but is displaced when the transporter is activated (21, 28). We extend this principle to BamA by proposing that activation of the Bam machine by substrate involves a conformational change in the BamA β-barrel that is evidenced by surface exposure of L6 and that the Bam lipoproteins regulate OMP assembly by modulating this conformational change.
Our genetic analysis suggests that residue E373 defines a critical region in POTRA 5 (P5) that is necessary for communication between BamA and its associated lipoproteins. The conditional-lethal mutation bamAE373K disrupts the interaction between BamA and BamD, separating the heteropentameric Bam complex into two stable subcomplexes, BamAB and BamCDE (12). A less deleterious substitution at the same residue, E373A, which partially destabilizes the BamA–BamD interaction and renders BamB function essential, can be suppressed by mutations in L6 that likely affect the region containing the intramolecular disulfide bond identified here. Although the importance of the BamA disulfide remains to be determined, our results clearly demonstrate a genetic link between L6 and the periplasmic domain of BamA. Because the BamD-binding site on BamA is in P5 (13), we suggest that BamDE control the conformation of BamA via signals transmitted indirectly through P5 to L6. Our results also show that the function of BamB becomes critical when this communication network is compromised.
How structural information is transduced from P5 to L6 remains to be determined, although it is conceivable that L6 directly contacts the periplasmic domain of BamA as its length is more than sufficient to traverse the barrel lumen and extend into the periplasmic space (28, 29). Notably, suppressor mutations in L6 restore viability to strains lacking BamB, -C, and -E, corroborating our assertion that a functional relationship exists between the BamA β-barrel and the Bam lipoproteins (9).
Although the similarities between BamA and FhaC with respect to L6 surface exposure imply a conserved function common to all Omp85 family members, any comparison is limited by the fact that FhaC promotes protein translocation and BamA promotes membrane protein assembly. However, a role for Bam in protein secretion has been established (30–32), and it has been proposed that the hydrophilic extracellular loops of OMPs are translocated across the OM through the pore of the BamA β-barrel (14, 28). This raises the possibility that the two conformations of BamA are each involved in the assembly and/or secretion of different types of substrates. Clearly, more work is required to clarify the functional relevance of the BamA conformations identified here. However, we note in closing that SecYEG, the IM protein translocase involved in secretion and membrane assembly of envelope proteins, has a central channel gated with a plug that is expelled to accommodate both secreted proteins and integral IM proteins (33). Moreover, gain-of-function mutations (the prl mutations) that either destabilize the closed form or stabilize the open form can activate Sec, allowing secretion of substrates with defective signal sequences (34). Further elucidation of Bam conformational dynamics will likely provide significant mechanistic insight into the molecular underpinnings of protein integration into and translocation across the OM.
Materials and Methods
Strains and Media.
All strains and plasmids used in this study are listed in Table S1. All strains were grown in LB medium at 37 °C unless noted otherwise. Antibiotics were included in the culture medium where appropriate at the following concentrations: ampicillin (125 μg/mL), chloramphenicol (20 μg/mL), kanamycin (25 μg/mL), and tetracycline (25 μg/mL). Mutant strains were constructed by P1 transduction as described (35).
Mal-PEG Labeling Assay.
Cells from exponential-phase cultures were harvested and washed in reaction buffer as described above for the protease assay. Where indicated, DTT was added to samples at a final concentration of 10 mM followed by incubation at room temperature for 10 min. The cells were then harvested by centrifugation and washed twice in an equal volume of reaction buffer. Where appropriate, Mal-PEG (Laysan Bio) was added to the resuspended cells and incubated at room temperature for 10 min. The cells were washed twice as before, resuspended in an equal volume of SDS/PAGE sample buffer, and then analyzed by SDS/PAGE and immunoblot.
Supplementary Material
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant GM34821 (to T.J.S.) and by National Institutes of Health Postdoctoral Fellowship GM093768 (to N.W.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302662110/-/DCSupplemental.
References
- 1.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricci DP, Silhavy TJ. The Bam machine: A molecular cooper. Biochim Biophys Acta. 2012;1818(4):1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rigel NW, Silhavy TJ. Making a beta-barrel: Assembly of outer membrane proteins in Gram-negative bacteria. Curr Opin Microbiol. 2012;15(2):189–193. doi: 10.1016/j.mib.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121(2):235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104(15):6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61(1):151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 7.Ruiz N, Falcone B, Kahne D, Silhavy TJ. Chemical conditionality: A genetic strategy to probe organelle assembly. Cell. 2005;121(2):307–317. doi: 10.1016/j.cell.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Rigel NW, Schwalm J, Ricci DP, Silhavy TJ. BamE modulates the Escherichia coli beta-barrel assembly machine component BamA. J Bacteriol. 2012;194(5):1002–1008. doi: 10.1128/JB.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tellez R, Jr, Misra R. Substitutions in the BamA β-barrel domain overcome the conditional lethal phenotype of a ΔbamB ΔbamE strain of Escherichia coli. J Bacteriol. 2012;194(2):317–324. doi: 10.1128/JB.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb CT, Heinz E, Lithgow T. Evolution of the β-barrel assembly machinery. Trends Microbiol. 2012;20(12):612–620. doi: 10.1016/j.tim.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328(5980):890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci DP, Hagan CL, Kahne D, Silhavy TJ. Activation of the Escherichia coli β-barrel assembly machine (Bam) is required for essential components to interact properly with substrate. Proc Natl Acad Sci USA. 2012;109(9):3487–3491. doi: 10.1073/pnas.1201362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317(5840):961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 14.Stegmeier JF, Andersen C. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J Biochem. 2006;140(2):275–283. doi: 10.1093/jb/mvj147. [DOI] [PubMed] [Google Scholar]
- 15.Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: Conformational flexibility and substrate recognition. Structure. 2008;16(12):1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: The protein insertion machine of the outer membrane. Structure. 2010;18(11):1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knowles TJ, et al. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68(5):1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- 18.Ward R, et al. The orientation of a tandem POTRA domain pair, of the beta-barrel assembly protein BamA, determined by PELDOR spectroscopy. Structure. 2009;17(9):1187–1194. doi: 10.1016/j.str.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: A web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32(Web Server issue):W400–W404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moslavac S, et al. Conserved pore-forming regions in polypeptide-transporting proteins. FEBS J. 2005;272(6):1367–1378. doi: 10.1111/j.1742-4658.2005.04569.x. [DOI] [PubMed] [Google Scholar]
- 21.Clantin B, et al. Structure of the membrane protein FhaC: A member of the Omp85-TpsB transporter superfamily. Science. 2007;317(5840):957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- 22.Delattre A-S, et al. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 2010;277(22):4755–4765. doi: 10.1111/j.1742-4658.2010.07881.x. [DOI] [PubMed] [Google Scholar]
- 23.Guédin S, et al. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J Biol Chem. 2000;275(39):30202–30210. doi: 10.1074/jbc.M005515200. [DOI] [PubMed] [Google Scholar]
- 24.Schirmer T, Keller TA, Wang YF, Rosenbusch JP. Structural basis for sugar translocation through maltoporin channels at 3.1 A resolution. Science. 1995;267(5197):512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 25.Ruiz N, Chng S-S, Hiniker A, Kahne D, Silhavy TJ. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc Natl Acad Sci USA. 2010;107(27):12245–12250. doi: 10.1073/pnas.1007319107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugawara E, Nagano K, Nikaido H. Factors affecting the folding of Pseudomonas aeruginosa OprF porin into the one-domain open conformer. MBio. 2010;1(4):e00228–10. doi: 10.1128/mBio.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoki SK, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol. 2008;70(2):323–340. doi: 10.1111/j.1365-2958.2008.06404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob-Dubuisson F, Villeret V, Clantin B, Delattre A-S, Saint N. First structural insights into the TpsB/Omp85 superfamily. Biol Chem. 2009;390(8):675–684. doi: 10.1515/BC.2009.099. [DOI] [PubMed] [Google Scholar]
- 29.Leonard-Rivera M, Misra R. Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol. 2012;194(17):4662–4668. doi: 10.1128/JB.00825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the beta-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67(1):188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 31.Ieva R, Tian P, Peterson JH, Bernstein HD. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc Natl Acad Sci USA. 2011;108(31):E383–E391. doi: 10.1073/pnas.1103827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106(45):19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalbey RE, Kuhn A. Protein traffic in Gram-negative bacteria: How exported and secreted proteins find their way. FEMS Microbiol Rev. 2012;36(6):1023–1045. doi: 10.1111/j.1574-6976.2012.00327.x. [DOI] [PubMed] [Google Scholar]
- 34.Smith MA, Clemons WM, Jr, DeMars CJ, Flower AM. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J Bacteriol. 2005;187(18):6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhavy TJ, Berman ML, Enquist LW. Experiments with Gene Fusions. NY: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1984. [Google Scholar]
- 36.Misra R, Peterson A, Ferenci T, Silhavy TJ. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J Biol Chem. 1991;266(21):13592–13597. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.