Abstract
Red seaweeds are key components of coastal ecosystems and are economically important as food and as a source of gelling agents, but their genes and genomes have received little attention. Here we report the sequencing of the 105-Mbp genome of the florideophyte Chondrus crispus (Irish moss) and the annotation of the 9,606 genes. The genome features an unusual structure characterized by gene-dense regions surrounded by repeat-rich regions dominated by transposable elements. Despite its fairly large size, this genome shows features typical of compact genomes, e.g., on average only 0.3 introns per gene, short introns, low median distance between genes, small gene families, and no indication of large-scale genome duplication. The genome also gives insights into the metabolism of marine red algae and adaptations to the marine environment, including genes related to halogen metabolism, oxylipins, and multicellularity (microRNA processing and transcription factors). Particularly interesting are features related to carbohydrate metabolism, which include a minimalistic gene set for starch biosynthesis, the presence of cellulose synthases acquired before the primary endosymbiosis showing the polyphyly of cellulose synthesis in Archaeplastida, and cellulases absent in terrestrial plants as well as the occurrence of a mannosylglycerate synthase potentially originating from a marine bacterium. To explain the observations on genome structure and gene content, we propose an evolutionary scenario involving an ancestral red alga that was driven by early ecological forces to lose genes, introns, and intergenetic DNA; this loss was followed by an expansion of genome size as a consequence of activity of transposable elements.
The red algae, together with the glaucophytes and the Chloroplastida, are members of the Archaeplastida, the phylogenetic group formed during the primary endosymbiosis event that gave rise to the first photosynthetic eukaryote. Red algal genomes, both plastid and nuclear, also contributed, via secondary endosymbiosis, to several other eukaryotic lineages, including stramenopiles, alveolates, cryptophytes, and haptophytes (1), and thus genes of red algal origin are spread widely among the eukaryotes. Knowledge about red algal genes and genomes therefore is crucial for understanding eukaryote evolution. The red macroalgal fossil record stretches back 1.2 billion years, providing the oldest evidence of morphologically advanced, multicellular, sexually reproducing eukaryotes (2). Ecologically, red algae represent the most species-rich group of marine macrophytes with more than 6,000 described species (www.algaebase.org). They are important components of many marine ecosystems, including rocky intertidal shores and coral reefs, and also are present in fresh water (3). Red algae also show some unusual physiological traits. Their photosynthetic antennae are built with phycobiliproteins, the thylakoids are unstacked, and they totally lack flagella and centrioles. In contrast to Chloroplastida, which produce starch in their chloroplasts, red algae store carbon as starch granules in their cytosol (floridean starch) (3). Their cell wall is a complex assemblage of cellulose, various hemicelluloses, and unique sulfated galactans (agars and carrageenans) (4). Economically, red macroalgae are important for their polysaccharide content. For example, carrageenans, the main sulfate-containing compounds in many red algae, are used as texturing agents and had a market value of more than US$500 million in 2010 (5). Red algae, especially nori (Pyropia and Porphyra species), also are used directly for human consumption with a market value of ∼US$1,300 million/y (6).
A number of transcriptomic studies are available on red algae, including the genera Porphyra, Chondrus, and Gracilaria (see ref. 7 and references therein), which investigate developmental processes and physiological responses and establish the contribution of red algae to diverse evolutionary lineages via secondary endosymbiosis events. However, red macroalgae have been the last group of complex multicellular organisms lacking a high-quality reference genome sequence. The closest fully sequenced relative of the red macroalgae is the unicellular extremophile Cyanidioschyzon merolae, which has a reduced genome (8) and belongs to the Cyanidiales, a group that diverged from other red algae about 1.4 billion years ago (1).
In the present study, we analyze the genome of Chondrus crispus Stackhouse (Gigartinales), or Irish moss, an intertidal red seaweed, up to 20 cm long, found on rock shores in the northern Atlantic Ocean. Chondrus is a member of the florideophytes, the largest group of extant red algae, representing 95% of known species (3). It is a common seaweed with a typical red algal triphasic life history with easy access to all three life cycle phases: the haploid female and male gametophytes, the diploid tetrasporophyte, and the diploid carposporophyte (present on the female gametophyte). The cell wall contains carrageenan, typically with ι- and κ-carrageenan in the gametophyte and λ-carrageenan in the sporophyte. In contrast to most other red algae, important scientific background knowledge exists for Chondrus, including studies on the mitochondrial genome (9), transcriptomics (10, 11), interactions with pathogens (12), effects of UV radiation (13), stress metabolism (14), and population ecology (15, 16). Thus, the availability of the C. crispus genome should help promote this organism as a model species for florideophyte algae and shed light on key aspects of eukaryotic evolution.
Results and Discussion
Reduced Genome with Exceptionally Compact Clustered Genes.
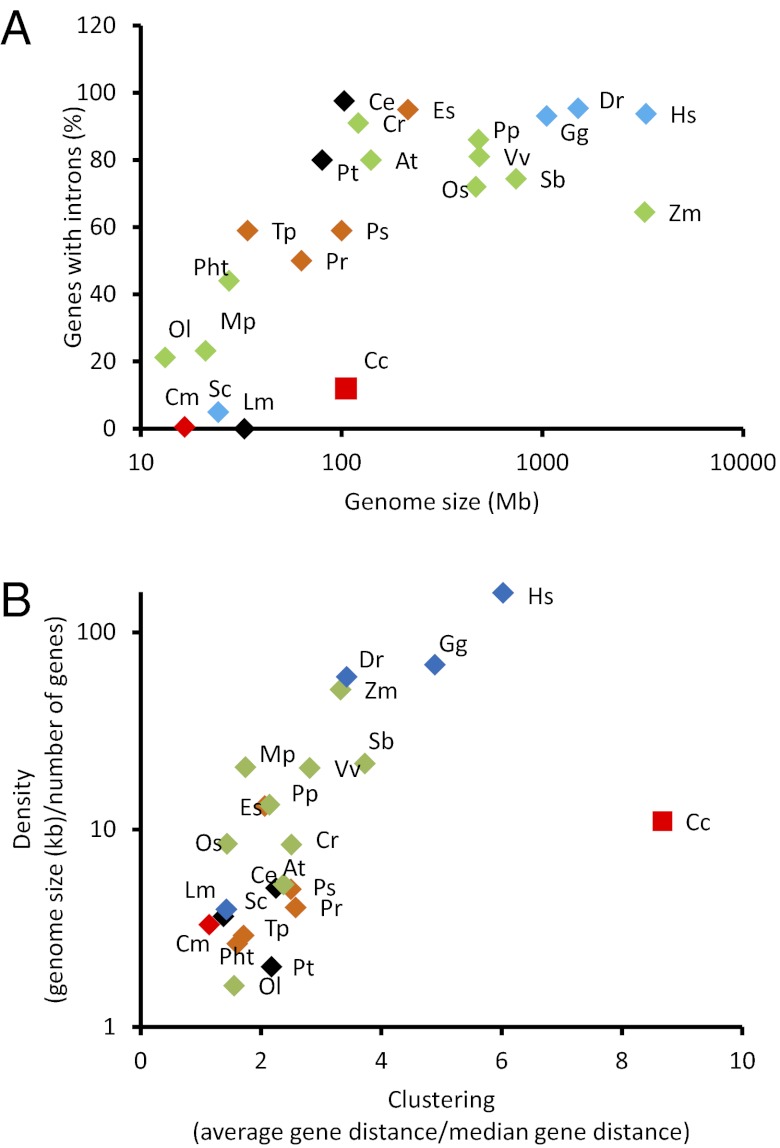
The genome sequence was obtained using DNA purified from a clonally growing unialgal culture of a gametophyte of Chondrus crispus and was sequenced using the Sanger technology. The assembled nuclear genome of Chondrus contains 1,266 scaffolds totaling 105 Mbp. A combination of expert and automatic annotation predicts 9,606 genes. The results of the annotation are described in detail in the SI Appendix. Genes are remarkably compact, containing only 1.32 exons on average (i.e., many fewer than other organisms of similar genome size), and most genes (88%) are monoexonic (Fig. 1A). The sparse introns are small, with an average length of 182 nucleotides (Table 1). The intron content of Chondrus and its distant relative C. merolae, as well as the limited data available on the gene structure of other red algae (17), suggest that compact genes are typical for this group and thus possibly are an ancestral trait. It is worth noting that the nucleomorphs of red algal origin in cryptomonads also have low intron content (18). Although we cannot exclude the possibility that a massive loss of introns could have occurred after the secondary endosymbiotic event, this observation suggests that the ancestral endosymbiotic red alga, which gave rise to these nucleomorphs, also had few introns. There is increasing evidence that the last eukaryotic common ancestor was intron rich and that there have been both intron losses and intron gains in the evolution of eukaryotes (19). The low number of introns in red algae thus would be a secondary feature that arose after the split between the green and red lineages about 1.5 billion years ago (1). The few introns that are present in Chondrus possibly have a regulatory function because, on average, transcripts for intron-containing genes accumulated to higher levels than those of monoexonic genes (SI Appendix, Fig. S1.1B). This result is in line with previous observations in other eukaryotes (20, 21).
Fig. 1.
Structural features of the Chondrus crispus (Cc) genome. (A) Percentage of genes with introns as a function of genome size in selected eukaryotes. (B) Gene density as a function of clustering in selected eukaryotes. Species: Ostreococcus lucimarinus (Ol), Cyanidioschyzon merolae (Cm), Micromonas pusilla (Mp), Saccharomyces cerevisiae (Sc), Phaeodactylum tricornutum (Pht), Leishmania major (Lm), Thalassiosira pseudonana (Tp), Phytophthora ramorum (Pr), Paramecium tetraurelia (Pt), Phytophthora sojae (Ps), Caenorhabditis elegans (Ce), Chlamydomonas reinhardtii (Cr), Arabidopsis thaliana (At), Ectocarpus siliculosus (Es), Oryza sativa (Os), Physcomitrella patens (Pp), Vitis vinifera (Vv), Sorghum bicolor (Sb), Gallus gallus (Gg), Danio rerio (Dr), Zea mays (Zm), Homo sapiens (Hs). Green symbols indicate chloroplastides; red, rhodophytes; blue, opisthokonts; brown, stramenopiles; and black, others.
Table 1.
Genome statistics from C. crispus and selected photosynthetic species
| Species | Genome size (Mbp) | Protein-coding loci | % coding | Introns per gene | Average intron length (bp) |
| C. crispus | 105 | 9,606 | 8 | 0.32 | 123* |
| Cyanidioschyzon merolae | 16.5 | 5,331 | 50 | 0.005 | 248* |
| Arabidopsis thaliana | 140.1 | 27,416 | 24 | 4.4 | 55* |
| Physcomitrella patens | 480 | 35,938 | 9 | 3.9 | 311* |
| Chlamydomonas reinhardtii | 121 | 14,516 | 16 | 7.4 | 174* |
| Ectocarpus siliculosus | 214 | 16,281 | 12 | 7.0 | 704† |
| Thalassiosira pseudonana | 34 | 11,242 | 32 | 1.4 | 132† |
| Ostreococcus lucimarinus | 13.2 | 7,551 | 71 | 0.27 | 187* |
*Median.
†Mean
Genes in Chondrus are clustered in gene-dense regions interspersed with sequences containing numerous repetitive elements. As a result, we observed a low median distance (0.8 kbp) between genes compared with the average distance (6.9 kbp). The ratio between average and median intergenic distances in different eukaryotes makes it clear that Chondrus presents an exceptionally low gene density and a high degree of clustering (Fig. 1B). The proximity of coding ORFs is enhanced by short untranslated regions (on average 142 bp). Although different in size, the C. merolae and Chondrus genomes are similar in that they are regionally compact with few introns and a limited number of genes compared with other eukaryotic species (Table 1). Therefore it is possible that red macroalgae (and other non-Cyanidiales red algae) share with C. merolae a common ancestor that had a reduced genome and that the expansion of the size of the macroalgal genome [red macroalgal genome sizes are 80–1,200 Mbp (22)] occurred after the separation from Cyanidiales.
Recent Genome Expansion Resulting from Transposable Element Invasion.
Repeated sequences constitute 73% of the Chondrus genome. The most abundant transposable elements are class I LTR retrotransposons, representing 58 Mbp; non-LTR retroelements were found also. Twenty-one families of terminal inverted repeat elements (class II elements), representing 13 Mbp of the genome, were found, as was one active helitron family. The retrotransposon component of the genome is extremely complex, not only because of the enormous number of recently transposed elements but also because each family has members that have diverged significantly. The analysis showed evidence for an ongoing burst of transposition activity that is responsible for at least 18 Mbp of the genome. The histogram of the similarity between LTRs shows a unimodal distribution, indicating that the transposition of all elements occurred concomitantly and is very recent (SI Appendix, Fig. S2.3). The mean similarity is 98%, with well over 100 elements exhibiting identical LTRs. The sizes of the copia and gypsy reference elements also are remarkably similar, suggesting a low rate of occurrence of insertions/deletions. Together these results indicate that LTR retroelements have been a major driving force in shaping the genome of Chondrus and that their proliferation has increased the genome size significantly in the last 300,000 y (SI Appendix, Fig. S2.3).
Reduced Gene Content.
In agreement with the compact structure of its genome, there are many examples of reduced gene diversity in Chondrus. For example, we did not find typical and widespread eukaryotic genes such as those for selenoproteins, the machinery for DNA methylation, sulfatases, core components of the endocytic machinery (Rab5 GTPase, AP-2 adaptor complex, endocytic Qc-SNARE), heterotrimeric G proteins, or flagella-specific genes (red algae lack flagella in all life cycle phases). In addition, and surprisingly for a photosynthetic organism, only one photoreceptor was found, a cryptochrome, and Chondrus therefore seems to lack most of the photoreceptor types known to date, including aureochromes, phytochromes, rhodopsin, or phototropins. Furthermore, most gene families are small, with few paralogs involved in a given functional process. For example, Chondrus encodes 82 genes for cytoplasmic ribosomal proteins, compared with 349 in Arabidopsis thaliana, even though nearly all ribosomal protein types are present in Chondrus (SI Appendix, Table S4.8). Starch metabolism is another example of the use of a minimum set of genes for a function (see below). The number of transcription factors and transcriptional regulators encoded also is small: 193 proteins, compared with 161 in the unicellular red alga C. merolae, 401 in the multicellular brown alga Ectocarpus siliculosus, which has similarly complex morphology, and more than 1,500 in the morphologically more complex embryophyte A. thaliana (23). Even though the number of transcription factors is limited, it is worth noting that both Dicer and Argonaute, genes, which are involved in small RNA processing (24), are found in the genome. Argonaute genes have not been described in unicellular red algae, glaucophytes, or most prasinophytes, and Dicer cannot be detected in any other red, green, or unicellular heterokont algae (23). This observation suggests a complex regulation by miRNAs in Chondrus, comparable to that found in multicellular plants and animals.
Taken together, these findings prove that pathway simplification, along with gene and intron losses, is ancestral to rhodophytes and not derived in Cyanidiales and other unicellular red algal lineages.
Large Unexplored Gene Diversity.
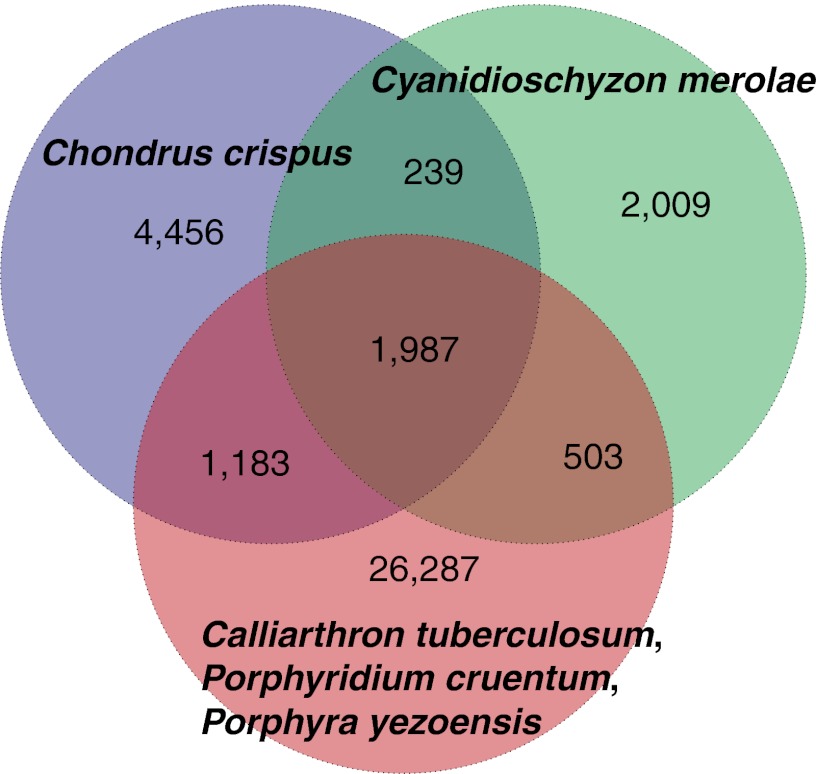
This study provides an insight into the large number of hitherto unknown genes found in Chondrus, i.e., the 52% of genes that had no counterpart (blastp e-value >10−5) in GenBank. The predicted proteins in the Chondrus genome were compared with the 5,064 proteins from C. merolae (25), the 23,961 predicted proteins of Calliarthron tuberculosum (26), and the 839 proteins of Pyropia (Porphyra) yezoensis present in GenBank. This set of proteins was completed with 22,431 ESTs of P. yezoensis and 36,167 ESTs of Porphyridium cruentum (26) (for details, see SI Appendix). As shown in Fig. 2, 57% of Chondrus orthology groups were not found in other red algae, demonstrating large gene diversity even within this lineage.
Fig. 2.
Orthology groups within red algal protein-coding genes. The Venn diagram shows the ortholog groups identified within the genomes of C. crispus and Cyanidioschyzon merolae and within the available sequences of Calliarthron tuberculosum, P. cruentum, and Pyropia (Porphyra) yezoensis.
Unique Carbohydrate Metabolism.
The Chondrus genome contains 31 glycoside hydrolases (GH) and 65 glycosyltransferases (GT) belonging to 16 GH and 27 GT families, respectively (SI Appendix, Table S7.13). These enzymes are involved in cell-wall metabolism and in the synthesis of other polysaccharides as well as protein and lipid glycosylation. Chondrus features all the genes needed to synthesize and recycle starch (SI Appendix, Table S7.14) but with a surprisingly low redundancy. The finding of only 12 starch-related genes revolutionizes our understanding of the building of this important polymer. Indeed, until very recently it was assumed that the complexity of starch metabolism in the green lineage reflected the complexity of the structure of starch granules. The Chondrus genome clearly invalidates this hypothesis.
One gene homologous to family GT7 chondroitin synthase and nine genes similar to carbohydrate sulfotransferases (CSTs) were identified. These enzymes are involved in the biosynthesis of sulfated polysaccharides, glycosaminoglycans, in animals, suggesting that their Chondrus homologs are involved in carrageenan biosynthesis. CSTs also are conserved in brown algae but are absent in available genomes of terrestrial plants. The clustering of red algal CSTs with those of animals and brown algae (SI Appendix, Figs. S7.3–S7.5) confirms that the synthesis of sulfated polysaccharides is an ancient eukaryotic capacity which has been lost by plants during the conquest of land (27). In addition, Chondrus possesses 12 galactose-6-sulfurylases, which are responsible for the last step of carrageenan biosynthesis and are unique to red algae (28), and three GH16 enzymes related to κ-carrageenases from marine bacteria (29), which putatively are involved in cell-wall expansion and recycling.
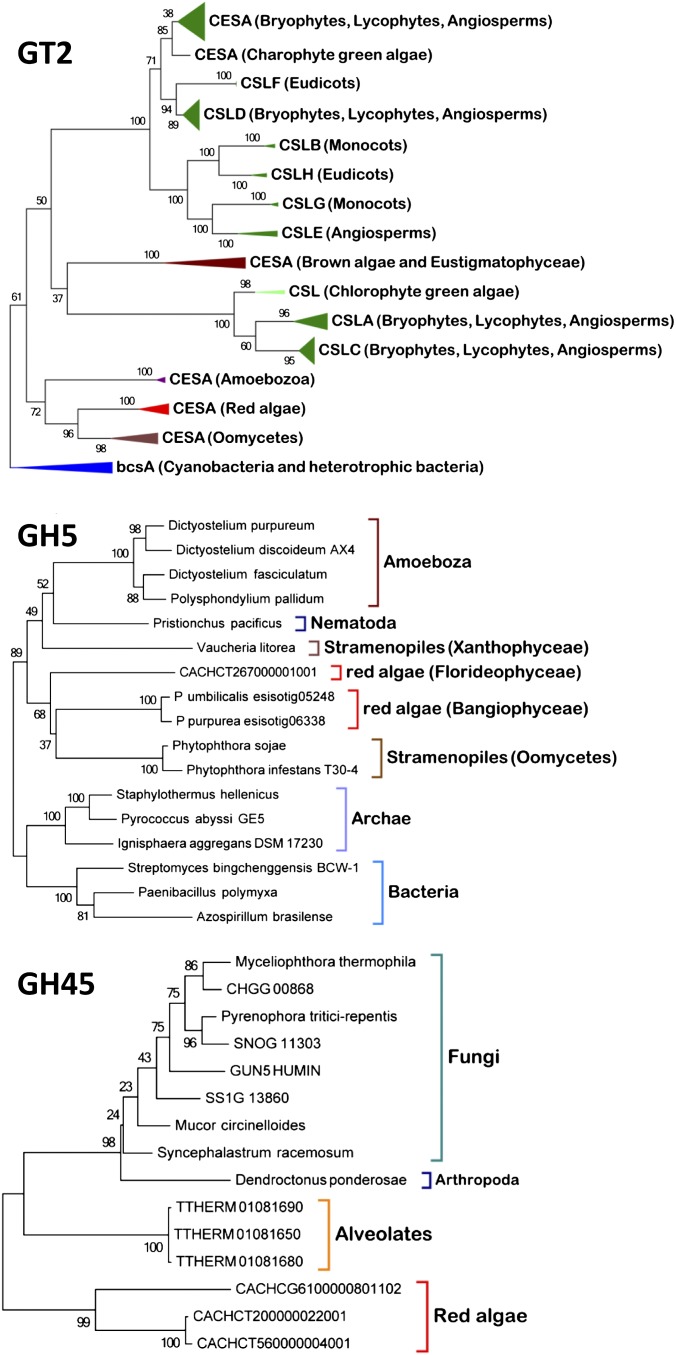
Chondrus contains two cellulose synthases (CESA) similar to those of the red algae Porphyra sp. (55% identity) and Griffithsia monilis (62% identity). Like the CESA from G. monilis, the Chondrus CESAs display a CBM48 in N terminus (30). In a Blast search against the NR database, the closest homologs of red algal sequences are CESA from Oomycetes (∼35% identity), from Dictyostelium spp. (∼30% identity), and from various bacteria (∼28% identity). In contrast, CESA from land plants are more distant (∼20% identity). A phylogenetic analysis with the bacterial CESAs as outgroup indicates that CESA and cellulose synthase-like proteins (CSL) from Chloroplastida diverge into two unrelated clades (Fig. 3). The first clade encompasses CESA and CLSB, D, E, F, G, and H and likely derived from the single cellulose synthase of charophytes. The second clade, which includes CSLA and CSLC, originates from a CSL from chlorophytes which is not found in the transcriptomes of charophytes (31). Red algal CESA emerge together with CESA from oomycetes in a distinct cluster rooted by CESA from Amoebozoa, confirming the tendency observed in blastp searches. Therefore, the CESAs from red algae and from green algae and embryophytes have different origins. Amoebozoa were not involved in the primary plastid endosymbiosis; thus acquisition of the bacterial cellulose synthase likely occurred before the primary endosymbiosis, and is not necessarily of cyanobacterial origin (32, 33). The nature of the different ancestral bacteria involved in horizontal gene transfer (HGT) with red algae and green algae is difficult to resolve, because all bacterial cellulose synthesis A (bCsA) genes tend to cluster together in an unrooted tree. Chondrus lacks family GH9 cellulases, which are found in land plants. In contrast, the genome contains three other families of cellulases (GH5, GH6, and GH45), which are absent in Chloroplastida but are conserved in various bacteria and heterotrophic eukaryotes. Phylogenetic analyses confirm that the GH5 cellulases emerge in a clade encompassing cellulases from oomycetes, Amoebozoa, and Nematoda, whereas red algal GH45 cellulases are related to cellulases from fungi (Fig. 3). GH6 cellulases from Chondrus are conserved both in bacteria and fungi but seem closer to bacterial GH6 cellulases. Because at least the red algal GH5 and GH45 cellulases share common ancestors with cellulases from opisthokonts or Amobozoa, these proteins are ancient eukaryotic enzymes predating the primary plastid endosymbiosis. Thus, these ancestral cellulases probably were involved initially in the degradation of bacterial cellulose. After the acquisition of the cellulose biosynthetic pathway, these red algal enzymes likely evolved to participate in cell-wall remodeling.
Fig. 3.
Phylogenetic trees of the cellulose synthases CESA and cellulose synthase-like proteins CSL (family GT2) and of the cellulases of the GH5 and GH45 families. All phylogenetic trees were constructed using the maximum likelihood (ML) approach with the program MEGA 5.05 (www.megasoftware.net). Numbers indicate the bootstrap values in the ML analysis.
Unusual Metabolic Features.
Because of their evolutionary history and their habitat, red algae feature some uncommon enzymes related to primary and secondary metabolism. As an illustration, the Chondrus genome contains a gene similar to the mannosylglycerate synthase (MGS) from the marine bacterium Rhodothermus marinus (48% identity) and from some Archaea (∼29% identity). This family GT78 enzyme synthesizes mannosylglycerate, an osmolyte required for thermal adaptation in thermophilic microorganisms (34, 35). This rare compound is known in red algae as “digeneaside” and accumulates during photosynthesis (36). MGS are not found in the available genomes of glaucophytes, green algae, or land plants, with the exception of Physcomitrella patens and Selaginella moellendorffii. Nonetheless, we have identified GT78 homologs in transcriptomic data of five other red algae and five streptophyte algae. A phylogenetic analysis indicates that the GT78 sequences from red algae, streptophytes, mosses, and lycophytes constitute two distinct clades, rooted by the MGS from R. marinus (SI Appendix, Fig. S7.2). Thus, there was a lateral transfer between a common ancestor of green and red algae with a thermophilic marine bacterium. Most extant red algae retained this enzyme; in the green lineage this gene was lost early by chlorophytes, but it was conserved by streptophytes, mosses, and lycophytes. It finally was lost by land plants after the divergence from lycophytes.
Several sets of anabolic and catabolic reactions previously considered specific to plants or animals were found in Chondrus. This result raises intriguing questions about the biological roles and the regulation of the related genes, molecules, and metabolic pathways, in particular whether their functions and mechanisms of action are conserved across different lineages. Examples are the C18 (plant-like) and C20 (animal-like) oxylipins and related compounds that have been identified in Chondrus in the context of studies on biotic stress response (28, 29). Interestingly, only two genes encoding lipoxygenase (SI Appendix, Table S7.10) have been identified, a surprising result given the diversity of oxylipins observed in this alga. The presence of methyl jasmonate, a plant hormone involved in stress signaling, has been detected in vitro after incubation with linolenic acid (37, 38). However, no candidates for allene oxide synthase, allene oxide cyclase, or jasmonic acid carboxyl methyltransferase were found. This outcome indicates that methyl jasmonate and oxylipin synthesis in Chondrus may be carried out by enzymes other than the ones characterized so far.
Despite the overall reduced genome, a number of gene families have remained diverse or were subject to recent diversification and expansion. One example of this diversity is the comparatively large set of genes related to halogen metabolism. Halogens play an important role in the metabolism of marine red algae (9), and transcriptomic data indicate that the corresponding genes are highly expressed (S1 Appendix, Table S8.1). For example, 20 genes encoding animal-like heme peroxidase homologs were identified (SI Appendix, Fig. S8.4). In mammals these genes play a major role during pathogen ingress, releasing hypohalous acids (39), but their function in red algae is unknown. To our knowledge, animals and marine bacteria are the only groups of organisms in which this type of protein has been found. Their occurrence in Chondrus provides additional evidence for the hypothesis that proteins from the peroxidase-cyclooxygenase superfamily (such as heme peroxidases) have an ancient origin (40). In addition, the Chondrus genome encodes 15 members of the phosphatidic acid phosphatase type 2-haloperoxidase family. Interestingly, it also harbors a group of haloalkane dehalogenase and haloacid dehalogenase enzymes, which remove halogens from alkanes. This group of enzymes previously has been found only in prokaryotes and in the brown seaweed E. siliculosus (41). The large size of these halogen-related gene families likely is a specific evolutionary adaptation to the marine environment, allowing brown and red macroalgae to take benefit of halide chemistry and to modulate finely halogen metabolism, which plays an important role in defense reactions, redox reactions, and the production of secondary metabolites. Supporting this hypothesis are the facts that E. siliculosus has a similarly rich repertoire of peroxidases and haloperoxidases, with ∼16 representatives (41), and that brown algae in general have an active halogen metabolism (42).
Evolutionary Scenario.
The Chondrus genome sheds lights on the early evolution of Archaeplastida. The presence of cellulase families GH5 and GH45 in Chondrus supports the notion that the ancestor of the Archaeplastida was a cellulolytic protist feeding on bacterial exopolysaccharides such as cellulose. This hypothesis is consistent with the ancient eukaryotic origin of family GH9 cellulases (43). After their divergence, red algae only kept GH5 and GH45 cellulases, and green algae and plants lost these genes and conserved GH9 cellulases. Repeated exposition to bacterial genomic DNA also could explain the HGTs of various bacterial origins found in the Chondrus genome (e.g., GT2, GT78). Cellulose biosynthesis was acquired independently in red algae and green algae; independent acquisition could partially explain the structural diversity of cellulose-synthesizing enzyme complexes and cellulose microfibrils in Archaeplastida (44).
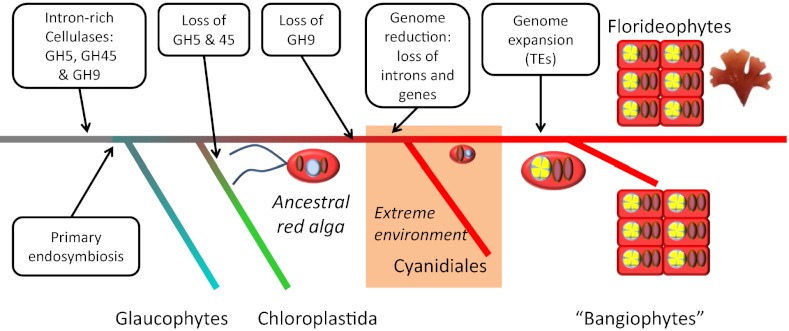
The compact structure of the nonrepetitive part of the Chondrus genome and genes also indicates that the red algal lineage went through an evolutionary bottleneck (Fig. 4). Early in the evolution of red algae, but after their divergence from green algae, selective pressure for small physical size or low nutrient requirements probably caused a reduction of the genome, with loss of introns and intergenetic material. This bottleneck also could explain the lack of flagella in all life-cycle stages in red algae, because the corresponding genes may have been lost during the genome compaction. It has been suggested previously (45) that, because of the limited low pH tolerance of cyanobacteria, early eukaryotic algae would have had less competition in acidic environments where fewer photosynthetic organisms were present. The extant red algae C. merolae or Galdieria sulphuraria live in an environment with high temperature and low pH, and even though it is not obvious why such conditions would reduce genome size, it is clear that these conditions favor compact genomes in red algae and may indicate that the ancestral red alga was an acido- and thermophilic organism. The evolutionary bottleneck also might explain the high number of orphan genes in the genome, because red algae were forced to reinvent gene functions that were lost during the genome reduction. If this hypothesis is correct, we predict that the ongoing red algal genome projects on Porphyra spp (6), P. cruentum (46), and C. tuberculosum (47) will show similar gene and genome organization.
Fig. 4.
Proposed scenario for the evolution of red algae. An ancestor with flagella and an intron-rich genome invaded an extreme environment, possibly acidic and high temperature, with a strong selection pressure toward a reduced genome, where a genome reduction took place. The red algae later recolonized the marine and freshwater environments and experienced an expansion of the genome through the activity of transposable elements. They now are represented by the florideophytes and the bangiophytes (red algae that are neither Cyanidiales nor florideophytes). Red ovals represent plastids; light blue circles, nucleus with ancestral genes; yellow, transposable elements.
In conclusion, this study presents a reference genome for a multicellular red alga and provides a number of unexpected insights into the origin and evolution of this ancestral plant lineage. It also provides fundamental data on the unique metabolic pathways of this large and economically important group of marine algae. In addition, because of its unique genome characteristics, C. crispus constitutes a novel model species for studying the complex evolutionary forces that shape eukaryotic genomes. Finally, as an archive of the gene content of ancestral marine plants, this genome will help comparatively delineate the innovations that were necessary for the emergence of land plants and their adaptation to the terrestrial environments.
Materials and Methods
A gametophyte of C. crispus Stackhouse (Gigartinales) was collected at Peggy’s Cove, Nova Scotia, Canada (44°29′31′′N, 63°55′11′′W) in 1985 by Juan Correa and since then has been growing vegetatively in unialgal culture.
The main raw data are 14-fold coverage shotgun reads sequenced with Sanger sequencing produced from five libraries with various insert sizes (SI Appendix, Table S1.1). Their assembly with ARACHNE (48) generated a collection of 925 scaffolds, covering 104.8 Mbp. An automated annotation based partially on 300,000 cDNA reads was performed and was used as a basis for expert annotation. For details on the different analyses and available data, see SI Appendix. An outline of SI Appendix content is shown in Fig. 5.
Fig. 5.
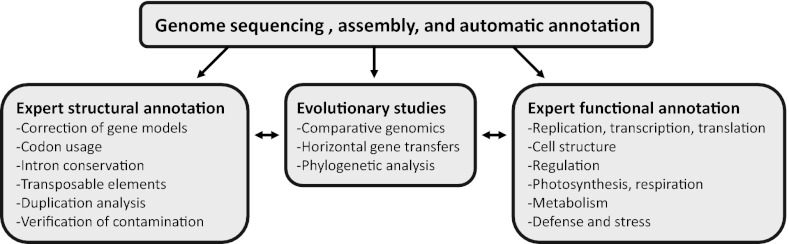
Snapshot of the C. crispus genome analysis and an outline of the contents of the SI Appendix.
Supplementary Material
Acknowledgments
We thank Pr. Juan Correa (Pontificia Universidad Católica de Chile) for providing the sequenced strain. This work was supported by funding from IDEALG Grants ANR-10-BTBR-04-02 and 04-04 “Investissements d’avenir, Biotechnologies-Bioressources,” Groupement d'Intérêt Scientifique Génomique Marine, NERC NE/J00460X/1, Network of Excellence Marine Genomics Europe (GOCE-CT-2004-505403), Conseil Regional de Bretagne, and the Czech Science Foundation. A.Z. was supported by a bursary grant from the Scottish Association for Marine Science, travel grants from the Seventh Framework Program Association of European Marine Biological Laboratories (ASSEMBLE), and a The Marine Alliance for Science and Technology for Scotland (MASTS) Visiting Researcher Fellowship. C.M.M.G. was supported by Grant Natural Environment Research Council NE/J00460X/1. This work was supported by funding from the Commissariat à l'Energie Atomique (CEA).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (sequence nos. CAKH01000001–CAKH01003241).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221259110/-/DCSupplemental.
References
- 1.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21(5):809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 2.Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26(3):386–404. [Google Scholar]
- 3.Woelkerling WJ. In: Biology of the Red Algae. Cole KM, Sheath RG, editors. Cambridge, UK: Cambridge Univ Press; 1990. pp. 1–6. [Google Scholar]
- 4.Popper ZA, et al. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu Rev Plant Biol. 2011;62:567–590. doi: 10.1146/annurev-arplant-042110-103809. [DOI] [PubMed] [Google Scholar]
- 5.Bixler HJ, Porse H. A decade of change in the seaweed hydrocolloids industry. J Appl Phycol. 2010;23(3):321–335. [Google Scholar]
- 6.Blouin NA, Brodie JA, Grossman AC, Xu P, Brawley SH. Porphyra: A marine crop shaped by stress. Trends Plant Sci. 2011;16(1):29–37. doi: 10.1016/j.tplants.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Chan C, et al. Porphyra (Bangiophyceae) transcriptomes provide insights into red algal development and metabolism. J Phycol. 2012;48(6):1328–1342. doi: 10.1111/j.1529-8817.2012.01229.x. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428(6983):653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 9.Leblanc C, et al. Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales). Gene content and genome organization. J Mol Biol. 1995;250(4):484–495. doi: 10.1006/jmbi.1995.0392. [DOI] [PubMed] [Google Scholar]
- 10.Collén J, Hervé C, Guisle-Marsollier I, Léger JJ, Boyen C. Expression profiling of Chondrus crispus (Rhodophyta) after exposure to methyl jasmonate. J Exp Bot. 2006;57(14):3869–3881. doi: 10.1093/jxb/erl171. [DOI] [PubMed] [Google Scholar]
- 11.Collén J, Guisle-Marsollier I, Léger JJ, Boyen C. Response of the transcriptome of the intertidal red seaweed Chondrus crispus to controlled and natural stresses. New Phytol. 2007;176(1):45–55. doi: 10.1111/j.1469-8137.2007.02152.x. [DOI] [PubMed] [Google Scholar]
- 12.Bouarab K, Potin P, Correa J, Kloareg B. Sulfated oligosaccharides mediate the interaction between a marine red alga and its green algal pathogenic endophyte. Plant Cell. 1999;11(9):1635–1650. doi: 10.1105/tpc.11.9.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kräbs G, Watanabe M, Wiencke C. A monochromatic action spectrum for the photoinduction of the UV-absorbing mycosporine-like amino acid shinorine in the red alga Chondrus crispus. Photochem Photobiol. 2004;79(6):515–519. doi: 10.1562/2003-12-14-ra.1. [DOI] [PubMed] [Google Scholar]
- 14.Collén J, Davison I. Stress tolerance and reactive oxygen metabolism in the intertidal red seaweeds Mastocarpus stellatus and Chondrus crispus. Plant Cell Environ. 1999;22(9):1143–1151. [Google Scholar]
- 15.Krueger-Hadfield SA, Collén J, Daguin-Thiébaut C, Valero M. Genetic population structure and mating system in Chondrus crispus (Rhodophyta) J Phycol. 2011;47(3):440–450. doi: 10.1111/j.1529-8817.2011.00995.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. Inter-simple sequence repeat (ISSR) analysis of genetic variation of Chondrus crispus populations from North Atlantic. Aquat Bot. 2008;88(2):154–159. [Google Scholar]
- 17.Hoef-Emden K, et al. Actin phylogeny and intron distribution in bangiophyte red algae(rhodoplantae) J Mol Evol. 2005;61(3):360–371. doi: 10.1007/s00239-004-0366-3. [DOI] [PubMed] [Google Scholar]
- 18.Moore CE, Archibald JM. Nucleomorph genomes. Annu Rev Genet. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- 19.Csuros M, Rogozin IB, Koonin EV. A detailed history of intron-rich eukaryotic ancestors inferred from a global survey of 100 complete genomes. PLOS Comput Biol. 2011;7(9):e1002150. doi: 10.1371/journal.pcbi.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanier W, Moustafa A, Bhattacharya D, Comeron JM. EST analysis of Ostreococcus lucimarinus, the most compact eukaryotic genome, shows an excess of introns in highly expressed genes. PLoS ONE. 2008;3(5):e2171. doi: 10.1371/journal.pone.0002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shabalina SA, et al. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol. 2010;27(8):1745–1749. doi: 10.1093/molbev/msq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapraun DF. Nuclear DNA content estimates in multicellular green, red and brown algae: Phylogenetic considerations. Ann Bot (Lond) 2005;95(1):7–44. doi: 10.1093/aob/mci002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang D, et al. Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: A timeline of loss, gain, expansion, and correlation with complexity. Genome Biol Evol. 2010;2:488–503. doi: 10.1093/gbe/evq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457(7228):405–412. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 25.Nozaki H, et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan CX, et al. Red and green algal monophyly and extensive gene sharing found in a rich repertoire of red algal genes. Curr Biol. 2011;21(4):328–333. doi: 10.1016/j.cub.2011.01.037. [DOI] [PubMed] [Google Scholar]
- 27.Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010;188(1):82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- 28.Genicot-Joncour S, et al. The cyclization of the 3,6-anhydro-galactose ring of iota-carrageenan is catalyzed by two D-galactose-2,6-sulfurylases in the red alga Chondrus crispus. Plant Physiol. 2009;151(3):1609–1616. doi: 10.1104/pp.109.144329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel G, et al. The kappa-carrageenase of P. carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of Clan-B glycoside hydrolases. Structure. 2001;9(6):513–525. doi: 10.1016/s0969-2126(01)00612-8. [DOI] [PubMed] [Google Scholar]
- 30.Matthews PR, Schindler M, Howles P, Arioli T, Williamson RE. A CESA from Griffithsia monilis (Rhodophyta, Florideophyceae) has a family 48 carbohydrate-binding module. J Exp Bot. 2010;61(15):4461–4468. doi: 10.1093/jxb/erq254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE. 2012;7(1):e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobles DR, Romanovicz DK, Brown RM., Jr Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 2001;127(2):529–542. [PMC free article] [PubMed] [Google Scholar]
- 33.Nobles DR, Brown RM. The pivotal role of cyanobacteria in the evolution of cellulose synthases and cellulose synthase-like proteins. Cellulose. 2004;11(3–4):437–448. [Google Scholar]
- 34.Martins LO, et al. Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic characterization of a mannosylglycerate synthase. J Biol Chem. 1999;274(50):35407–35414. doi: 10.1074/jbc.274.50.35407. [DOI] [PubMed] [Google Scholar]
- 35.Empadinhas N, da Costa MS. Diversity, biological roles and biosynthetic pathways for sugar-glycerate containing compatible solutes in bacteria and archaea. Environ Microbiol. 2011;13(8):2056–2077. doi: 10.1111/j.1462-2920.2010.02390.x. [DOI] [PubMed] [Google Scholar]
- 36.Kremer B. Taxonomic implications of algal photoassimilate patterns. Br Phycol J. 1980;15(4):399–409. [Google Scholar]
- 37.Bouarab K, et al. The innate immunity of a marine red alga involves oxylipins from both the eicosanoid and octadecanoid pathways. Plant Physiol. 2004;135(3):1838–1848. doi: 10.1104/pp.103.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaquerel E, et al. Evidence for oxylipin synthesis and induction of a new polyunsaturated fatty acid hydroxylase activity in Chondrus crispus in response to methyljasmonate. Biochim Biophys Acta. 2007;1771(5):565–575. doi: 10.1016/j.bbalip.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 39.Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: From molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10(7):1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 40.Bernroitner M, Zamocky M, Furtmüller PG, Peschek GA, Obinger C. Occurrence, phylogeny, structure, and function of catalases and peroxidases in cyanobacteria. J Exp Bot. 2009;60(2):423–440. doi: 10.1093/jxb/ern309. [DOI] [PubMed] [Google Scholar]
- 41.Cock JM, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465(7298):617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- 42.La Barre S, Potin P, Leblanc C, Delage L. The halogenated metabolism of brown algae (Phaeophyta), its biological importance and its environmental significance. Mar Drugs. 2010;8(4):988–1010. doi: 10.3390/md8040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davison A, Blaxter M. Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol Biol Evol. 2005;22(5):1273–1284. doi: 10.1093/molbev/msi107. [DOI] [PubMed] [Google Scholar]
- 44.Tsekos I. The sites of cellulose synthesis in algae: Diversity and evolution of cellulose-synthesizing enzyme complexes. J Phycol. 2002;655(4):635–655. [Google Scholar]
- 45.Brock TD. Lower pH limit for the existence of blue-green algae: Evolutionary and ecological implications. Science. 1973;179(4072):480–483. doi: 10.1126/science.179.4072.480. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharya D, Price D, Yoon HS, Rajah V, Zaeuner S. Sequencing and analysis of the Porphyridium cruentum genome. J Phycol. 2012;48(s1):S5. [Google Scholar]
- 47.Chan C, Martone P. Recent advances in the calliarthron genome: Climate responses and cell wall evolution. J Phycol. 2012;48(s1):S1. [Google Scholar]
- 48.Batzoglou S, et al. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12(1):177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.