Abstract
Absence of petals, or being apetalous, is usually one of the most important features that characterizes a group of flowering plants at high taxonomic ranks (i.e., family and above). The apetalous condition, however, appears to be the result of parallel or convergent evolution with unknown genetic causes. Here we show that within the buttercup family (Ranunculaceae), apetalous genera in at least seven different lineages were all derived from petalous ancestors, indicative of parallel petal losses. We also show that independent petal losses within this family were strongly associated with decreased or eliminated expression of a single floral organ identity gene, APETALA3-3 (AP3-3), apparently owing to species-specific molecular lesions. In an apetalous mutant of Nigella, insertion of a transposable element into the second intron has led to silencing of the gene and transformation of petals into sepals. In several naturally occurring apetalous genera, such as Thalictrum, Beesia, and Enemion, the gene has either been lost altogether or disrupted by deletions in coding or regulatory regions. In Clematis, a large genus in which petalous species evolved secondarily from apetalous ones, the gene exhibits hallmarks of a pseudogene. These results suggest that, as a petal identity gene, AP3-3 has been silenced or down-regulated by different mechanisms in different evolutionary lineages. This also suggests that petal identity did not evolve many times independently across the Ranunculaceae but was lost in numerous instances. The genetic mechanisms underlying the independent petal losses, however, may be complex, with disruption of AP3-3 being either cause or effect.
Keywords: ABC model, development, flower, parallel evolution
Petals are sterile floral organs that lie between the outer sepals and the inner reproductive parts (i.e., stamens and carpels) of a flower with a dimorphic perianth. Compared with sepals, which are the outermost parts of the flower, petals are usually brightly colored and/or unusually shaped to attract pollinators. Petals show tremendous diversity in number, size, shape, structure, arrangement, orientation, and coloration, and are believed to be results of both divergent and parallel/convergent evolution (1, 2). Traditional morphological and anatomical studies have proposed that petals originated multiple times in different lineages, either from stamens (andropetals) or from sepals (bractopetals) (3, 4). Recent studies, however, tend to suggest that although andropetals and bractopetals are morphologically and anatomically distinguishable, they share a common developmental program (i.e., process homology) (5–7). According to the “ABC”, “ABCE”, and “Quartet” models for flower development, this program requires combinational activities of A-, B-, and E-class genes, the representatives in the model plant Arabidopsis thaliana (hereafter called Arabidopsis) being APETALA1 (AP1), APETALA3 (AP3) and PISTILLATA (PI), and SEPALLATA1-4 (SEP1-4), respectively (8–12). The fact that counterparts (or orthologs) of these genes are expressed in petaloid organs across flowering plants (including the basal-most lineage, Amborella) (13–16), raises the possibility that the genetic program for petal development was established before the diversification of extant angiosperms. Then, during evolution, this program has been modified in different directions and to varying degrees, giving rise to the production of a huge variety of petals and petaloid structures (6, 17). The petal identity program may have also been inactivated multiple times, leading to the independent origins of apetalous groups in different evolutionary lineages (2, 4, 18, 19). Interestingly, some of the apetalous groups, such as the piper (Piperaceae), sandalwood (Santalaceae), mulberry (Moraceae), elm (Ulmaceae), nettle (Urticaceae), birch (Betulaceae), oak (Fagaceae), and walnut (Juglandaceae) families, as well as several genera within the buttercup family (Ranunculaceae), are very rich in species diversity, suggesting that loss of petals is not necessarily disadvantageous.
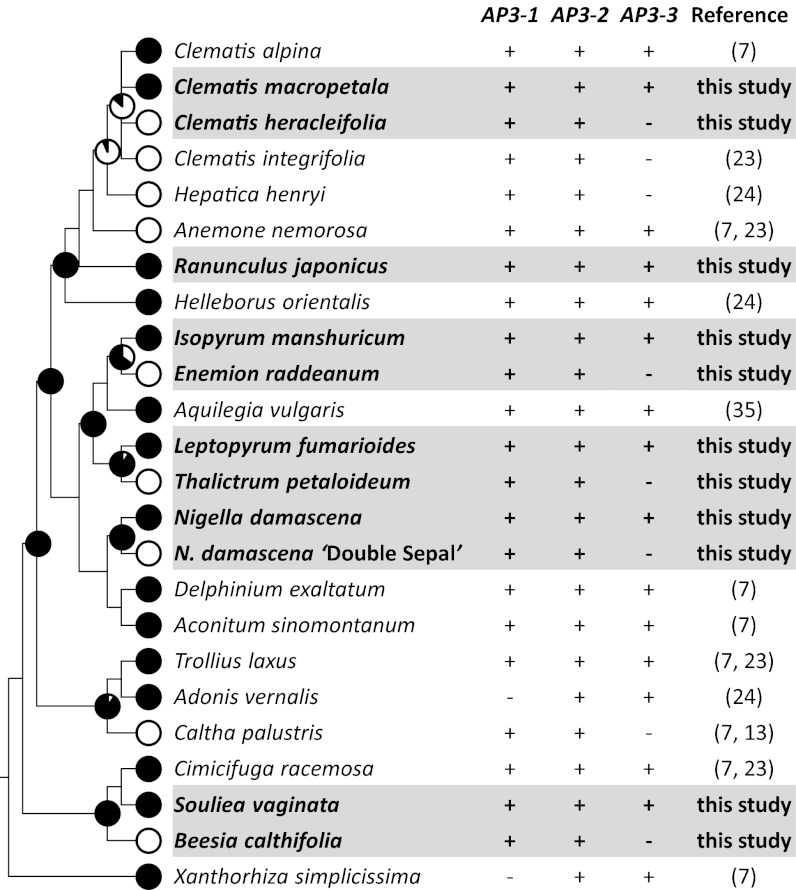
Ranunculaceae is a large, globally distributed family that consists of around 1,800 species in about 50 genera (20). The family has been the focus of centuries of morphological, anatomical, systematic, evolutionary, and phytochemical studies owing to its unusual position in the angiosperm phylogeny, enormous diversity in reproductive and vegetative structures, and wide ornamental and medical uses. Petals in this family are of particular interest because they show tremendous diversity in both appearance and function (20). In many species, petals are laminar, peltate, tubular, labiate, or cup-shaped and possess spurs or scale-/pocket-like structures that secrete nectar. In others, petals are equally showy, attractive, and specialized but do not produce nectar, suggestive of shifts in function. There are also genera in which petals are entirely missing and the attractive function is fulfilled mostly by sepals, which are commonly showy, attractive, and petaloid across the family, while stamens occupy the positions where petals are expected to be. Based on comparative morphological and anatomical studies, it has been proposed that petals in this and related families (such as Berberidaceae, Menispermaceae, and Lardizabalaceae) are all andropetals that resulted from independent modifications of stamens (1, 2). Recent phylogenetic analyses, however, tend to support the idea that having petals, or being petalous, is an ancestral character state whereas being apetalous is a derived one (21, 22). Indeed, when the two states of this character were mapped onto the phylogenetic tree of the family, it became evident that in at least seven lineages apetalous genera have had petalous ancestors (Fig. 1 and Fig. S1). This suggests that, rather than being independently gained many times, petals have been lost in parallel in different lineages during the evolution of the Ranunculaceae.
Fig. 1.
Correlation between loss of petals and expression of AP3-3 across the Ranunculaceae. Filled and open circles on the phylogenetic tree indicate the presence and absence of petals, respectively. The probability of having petals in ancestral taxa is indicated for each interior node (Fig. S1 gives details). The symbols + and − denote whether the AP3 paralogs can or cannot be isolated using the conventional RT-PCR technique, respectively.
The mechanism underlying the parallel petal losses within the Ranunculaceae is still unclear. However, recent studies indicated that absences of petals in this and related families are correlated with lack of the expression of a floral organ identity gene, AP3-3 (7, 23, 24). AP3-3 is an AP3-like gene, which, along with its two closest paralogs, AP3-1 and AP3-2, was generated through two successive gene duplication events before the divergence of the Ranunculaceae from its allies (7, 24). In the species that possess petals, AP3-3 orthologs are specifically expressed in petal primordia and developing petals, with their transcripts being detectable by using the conventional reverse transcription-polymerase chain reaction (RT-PCR) and in situ hybridization techniques. In the species lacking petals, however, AP3-3 expression could not be detected, despite considerable efforts. Based on these observations, it has been postulated that the independent petal losses in different Ranunculaceous lineages were caused by parallel inactivation of the AP3-3 orthologs (7, 24). However, because only a few species have been investigated extensively, it is still unclear whether petal losses are indeed correlated with AP3-3 expression, how AP3-3 orthologs could have been silenced or down-regulated, and whether silencing/down-regulation of AP3-3 was the only cause for parallel petal losses.
Results
AP3-3 Orthologs Are Generally Not Expressed in Apetalous Taxa.
To determine whether petal loss is indeed correlated with AP3-3 expression, we first sought to isolate AP3-3 orthologs from representative apetalous species. Four apetalous species (Thalictrum petaloideum, Beesia calthifolia, Enemion raddeanum, and Clematis heracleifolia; hereafter called Thalictrum, Beesia, Enemion, and apetalous Clematis, respectively) were sampled, each of which represents a separate lineage (Fig. 1 and Fig. S1). Because these apetalous species are only distantly related to each other, we also included four petalous species (Leptopyrum fumarioides, Souliea vaginata, Isopyrum manshuricum, and Clematis macropetala; hereafter called Leptopyrum, Souliea, Isopyrum, and petalous Clematis, respectively) as references. In addition, as has been pointed out by several authors, petals of the Clematis species may have been derived from stamens secondarily and recently, and as such are no longer homologous to those in other petalous genera (20, 25); our own ancestral character state reconstruction also supports this viewpoint (Fig. S1). For this reason, we included Ranunculus japonicus (hereafter called Ranunculus) as a second petalous reference to the apetalous Clematis. More interestingly, during the study, we encountered a “Double Sepal” mutant of Nigella damascena (hereafter called Nigella) in which petals are transformed into sepals. Because this mutant has apparently experienced a recent petal-loss event, we also added it into the analyses. By using the conventional RT-PCR technique, we obtained AP3-3 orthologs from floral buds of Leptopyrum, Souliea, Isopyrum, Ranunculus, the petalous Clematis, and the wild-type Nigella, but not from those of Thalictrum, Beesia, Enemion, the apetalous Clematis or the mutant Nigella (Fig. 1). Using each transition for presence/absence of petals and AP3-3 expression (Fig. 1), we conducted a 2x2 conditional binomial exact test and found a significant evolutionary correlation between petal loss and non-expression of AP3-3 (one-sided, P = 0.0146). The fact that the other two AP3-like genes, AP3-1 and AP3-2, could be obtained from all these species, however, suggests that the failure to isolate AP3-3 from apetalous species was not due to technical problems.
AP3-3 Orthologs Are Highly and Specifically Expressed in Petals.
To understand the roles of AP3-3 orthologs in flower development, we investigated their expression patterns in detail. Our quantitative real-time RT-PCR (qRT-PCR) analyses indicated that in five of the six petalous taxa (Leptopyrum, Souliea, Isopyrum, Ranunculus, and the wild-type Nigella, but not the petalous Clematis), the AP3-3 orthologs are all specifically expressed in petals, and the expression levels are always exceptionally high, at least compared with AP3-1 and AP3-2 (Figs. 2 and 3). In situ hybridization analyses further revealed that the Nigella, Leptopyrum, and Souliea AP3-3 genes are initially expressed in petal primordia and developing petals and then restricted to the inner parts of the near-mature petals, from where nectary tissues will be formed (Fig. 2 and see Figs. S3 and S4). Therefore, in petalous species (except for Clematis), AP3-3 may function to specify petal identity at early stages of flower development and control nectary formation at late stages. In the petalous Clematis, however, the role of AP3-3 is still unclear, because its expression level is very low, even in petals (Fig. 3). Presumably, petals in Clematis are controlled by a different developmental program, to which the contribution of AP3-3 is negligible.
Fig. 2.
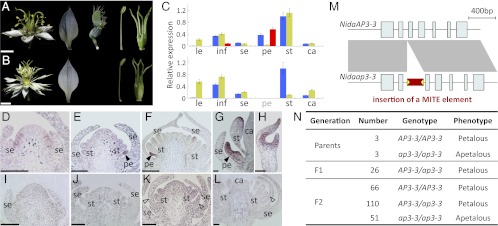
Genetic basis of the apetalous mutant of Nigella. (A and B) Flower and floral organs of the wild-type (A) and mutant (B) plants. (Scale bars, 0.5 cm.) Floral organs from left to right are sepals, petals, stamens, and carpels. Note that petals are absent in the mutant plants. (C) Relative expression of AP3-1 (blue bars), AP3-2 (yellow bars), and AP3-3 (red bars) in tissues from the wild-type (Upper) and mutant (Lower) plants measured using qRT-PCR. Tissues assayed are leaf (le), inflorescence (inf), sepal (se), petal (pe), stamen (st), and carpel (ca). (D–L) Expression pattern of AP3-3 in the wild-type (D–H) and mutant (I–L) flowers as revealed by in situ hybridization analyses. (Scale bars, 50 μm.) (M) Exon–intron structures of the functional (NidaAP3-3; Upper) and nonfunctional (Nidaap3-3; Lower) alleles. The red block in the second intron of Nidaap3-3 indicates the predicated Gypsy/Ty3-like MITE element. (N) Segregation pattern of the two alleles. The nonfunctional allele, which carries the MITE insertion, cosegregates with the apetalous phenotype.
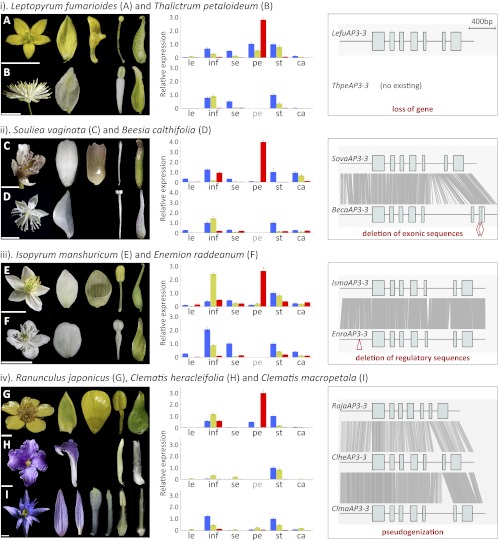
Fig. 3.
Relative expression and exon–intron structures of the AP3-3 orthologs from petalous and apetalous Ranunculaceous species. (Left) Flower and floral organs of the sampled species. (Scale bars, 0.5 cm.) (Center) Expression of AP3-1 (blue bars), AP3-2 (yellow bars), and AP3-3 (red bars) as revealed by qRT-PCR. Tissues assayed are leaf (le), inflorescence (inf), sepal (se), petal (pe), stamen (st), and carpel (ca). (Right) Exon–intron structures of the AP3-3 orthologs. Alignable regions between orthologs are shown with gray lines. Two red diamonds in BecaAP3-3 and one red triangle in EnraAP3-3 represent the deletions in the coding and regulatory regions, respectively.
Apetalous Nigella Mutant Was Caused by Insertion of a Transposable Element.
To understand why mRNAs of AP3-3 could not be isolated from the apetalous Nigella mutant, we conducted additional expression analyses using the qRT-PCR and in situ hybridization techniques. We found that AP3-3 is simply not expressed in the mutant, whereas AP3-1 and AP3-2 show normal expression patterns (Fig. 2 and Fig. S2). This suggests that AP3-3 has been completely inactivated in the mutant. We then obtained its genomic sequence and found that the mutant sequence is completely identical to the wild-type one except in the length of the second intron (Fig. 2 and Fig. S2). Close inspection of this intron further revealed that the mutant contains a 253-bp fragment that has the characteristics of a transposable element and can form stem-loop structures (Fig. S2). Specifically, this fragment resembles a Gypsy/Ty3-like miniature inverted-repeat transposable element (MITE) (26). Numerous studies have indicated that when a MITE was inserted into a gene, the gene became inactivated immediately because the formation of one or more stem-loop structures can interrupt the transcription and splicing processes (27, 28). Therefore, in the newly formed apetalous Nigella mutant, petal loss via transformation into sepals may have been caused by the insertion of the MITE element and the disruption of the AP3-3 expression. To search for more evidence supporting this idea, we examined the genotypes of the Nigella plants. We found that whereas all apetalous plants are homozygous for the insertion allele (i.e., ap3-3/ap3-3), the petalous ones are either AP3-3/AP3-3 or AP3-3/ap3-3. This, together with the segregation data obtained from crossing AP3-3/AP3-3 and ap3-3/ap3-3 individuals (Fig. 2), confirmed that loss of petals in the Nigella mutant was indeed caused by the MITE insertion.
AP3-3 Ortholog in Thalictrum No Longer Exists.
To understand whether AP3-3 has been down-regulated, silenced, or lost in Thalictrum, we conducted qRT-PCR experiments. However, because no AP3-3 ortholog has ever been obtained from this species, we used instead the primers designed for its ortholog in Leptopyrum. Meanwhile, to ensure the validity of the experiments, we included AP3-1 and AP3-2 as positive controls because, theoretically, if this interspecific amplification strategy works for one gene, it should work for others. We found that whereas analyses of AP3-1 and AP3-2 gave expected results, no signal could be obtained for AP3-3 (Fig. 3), suggesting that the failure to capture AP3-3 transcripts from Thalictrum was not due to its extremely low level of expression. Rather, it may have been caused by degeneration or deletion of the primer regions, or inactivation or loss of the gene. For this reason, we then performed interspecific in situ hybridization analyses using the probes designed for the Leptopyrum AP3-1, AP3-2, and AP3-3 genes. Again, hybridization of the first two genes gave reasonable results, whereas no signal was detected for the last one (Fig. S3). This suggests that the lack of qRT-PCR product for AP3-3 was not due to degeneration or deletion of the primer regions but, rather, lack of expression. We therefore attempted to amplify its genomic sequences, but no product could be obtained, even after extensive efforts (i.e., multiple primer combinations, high numbers of PCR cycles, and the use of an additional, congeneric species, Thalictrum aquilegifolium, as DNA template). Finally, we performed Southern hybridization using a probe derived from the Leptopyrum AP3-3. We found that while no signal could be detected for Thalictrum, the probe cross-reacted with Leptopyrum and a more distantly related species, Aquilegia coerulea (hereafter called Aquilegia) (Fig. S3). This, together with the positive Southern results for AP3-1 and AP3-2, strongly suggests that the AP3-3 ortholog has been lost altogether from the Thalictrum genome.
AP3-3 Ortholog in Beesia Is Down-Regulated by Two Deletions in Coding Regions.
We also applied the same interspecific strategy to Beesia. As expected, analyses of AP3-1 and AP3-2 gave excellent results (Fig. 3 and Fig. S4), suggesting that the primers and probes derived from the Souliea genes worked well for Beesia. In the AP3-3 case, however, only very weak signals were detected in Beesia (Fig. 3 and Fig. S4), suggesting that the gene may have been expressed but the expression was too weak to be detected using the conventional RT-PCR technique. To gain more insight into the possible reasons for its down-regulation, we obtained the genomic sequence of the gene. We found that, compared with its ortholog in Souliea, the Beesia AP3-3 gene is defective in its exon–intron structure: Two deletions in the last exon, one 4 bp long and the other 12 bp long, have resulted in a shift in reading frame and the occurrence of a premature stop codon (Fig. 3 and Fig. S4). Because the 4-bp deletion is located at the 5′ end of the last exon, it may have also affected or even interrupted the transcription and/or splicing process. In eukaryotes, this kind of structural defect is usually a signature of imprecise transposon excision and can cause instability of unspliced mRNAs (27). Thus, in Beesia, it is possible that the two deletions are responsible for the down-regulation. To test this hypothesis, we examined the expression of the Beesia AP3-3 gene and its corrected version in Arabidopsis. We found that, compared with the corrected version, the uncorrected one was expressed at significantly lower levels (Fig. S4), suggesting that the deletions can indeed cause down-regulation.
AP3-3 Ortholog in Enemion Has a Deletion in the Promoter Region.
To determine why AP3-3 was not expressed in Enemion, we obtained its genomic sequence. Comparison of this gene with its orthologs in Isopyrum and other genera suggests that the gene is not defective in its exon–intron organization or coding regions (Fig. 3 and Fig. S5). However, qRT-PCR analyses using multiple primer combinations indicated that its expression level is negligibly low (Fig. 3 and Table S1). For this reason, we also inspected its regulatory regions and found a 12-bp deletion in an otherwise conserved promoter region upstream of a highly conserved CArG-box motif (Fig. S5). In eukaryotes, and plants in particular, CArG-box motifs are the DNA binding sites of MADS-box–containing transcription factors proteins. AP3-like genes are themselves MADS-box genes, whose expression are commonly regulated by AP3 and other MADS-box proteins (29–31). It has been shown that to ensure the occurrence and stability of the protein–DNA interaction, the flanking DNA sequences of the CArG-box motifs must bend to some extent to form a special secondary/tertiary structure (32–34). Therefore, the 12-bp deletion in this region may have affected or even prohibited the protein-DNA interaction and caused down-regulation or inactivation of the gene. Notably, another AP3-3 homolog, the Aquilegia AqAP3-3b, which appears to be a pseudogene and is expressed at very low levels, also has deletions in this region (Fig. S5), suggesting that this region may be critical for proper functioning of the AP3-3 genes. However, it is also possible that the deletion has destroyed a separate cis-regulatory element, or that additional transregulatory changes are responsible for the down-regulation.
AP3-3 Orthologs in Clematis Have the Characteristics of Pseudogenes.
To understand why the expression levels of the Clematis AP3-3 genes are so low, we obtained their genomic sequences. Comparison of these sequences with those of their orthologs in other species, however, failed to detect any obvious defect in coding or regulatory regions (Fig. 3 and Fig. S6). Nevertheless, we noticed that the coding regions of the two Clematis genes are quite divergent from each other and from their orthologs, suggestive of functional divergence or possible pseudogenization. We therefore performed molecular evolutionary studies. We found that while the AP3-3 orthologs from all other petalous species have evolved under strong purifying selection, the Clematis genes, as well as the Aquilegia AqAP3-3b, have evolved under relaxed, or nearly neutral, selection (Fig. S6). This suggests that the Clematis genes may have become pseudogenes, although the gene structures remain intact. The AP3-3 orthologs from two other apetalous genera (Beesia and Enemion), however, do not show clear signature of relaxed selection, suggesting that they may either still be functional or have become pseudogenes very recently.
Discussion
AP3-3–Based Petal Identity.
It is remarkable that parallel petal losses in different Ranunculaceous lineages were strongly correlated with silencing or down-regulation of a single gene, AP3-3. What intrigues us more is that the apparent cause for the change in AP3-3 expression varies from species to species. In Thalictrum, the gene has been lost altogether, whereas in Beesia, Enemion, and the apetalous Nigella mutant the gene was disrupted by deletions or insertion in the coding, promoter, or intronic regions. In Clematis, the AP3-3 orthologs have become pseudogenes, although the underlying mechanism for the possible pseudogenization or reduced expression is still unclear. This, together with the fact that knockdown of the AP3-3 gene in Aquilegia can result in the petal-to-sepal transformation (24), strongly suggests that AP3-3 is a petal identity gene, and that the specific role of AP3-3 in petal identity is broadly conserved across the Ranunculaceae. More importantly, because Aquilegia and Nigella are two deeply divergent members of the family, these results provide the most compelling argument yet that the AP3-3–based petal identity program did not evolve many times independently but rather was lost in numerous instances.
Why AP3-3?
As one of the three AP3-like genes in the Ranunculaceae and related families, AP3-3 is unique in its expression pattern and functional properties. Unlike AP3-1 and AP3-2, whose expression patterns are rather broad and nonspecific, the AP3-3 orthologs are usually specifically expressed in petals, and the expression levels are generally exceptionally high (7, 35). Silencing of AP3-3 in Aquilegia and Nigella only resulted in homeotic transformations of petals into sepals, whereas all other floral organs remained largely unchanged (24). Up to now, it is still unclear whether AP3-3 evolved its current functions through changes in coding, regulatory, or both regions, yet the available data suggest that (i) it is a key regulator of petal development and (ii) it does not have pleiotropic roles. Being a key regulator of petal development implies that each time when having petals is no longer advantageous it will become useless and accumulate destructive mutations; otherwise, it evolves under stringent functional constraints, as is confirmed by a recent molecular evolutionary study (24). Meanwhile, because it is not pleiotropic, and because of the existence of two functionally redundant (at least in stamens) paralogs (AP3-1 and AP3-2), down-regulation, silencing, or loss of it will only affect the expression of a few downstream genes and, consequently, is unlikely to cause obvious phenotypic alterations outside petals. It may be because of these properties that AP3-3, rather than its paralogs, has the potential to be the key to understanding the underlying mechanisms for parallel petal losses.
Cause or Effect?
Despite the potential, it is still difficult to conclude that disruption of AP3-3 was the sole cause for petal losses, for two reasons. First, in the Nigella and Aquilegia ap3-3 mutants, petals were converted into sepals and thus the number of sepals increased (24). In the natural apetalous species, such as Thalictrum, Beesia, Enemion, and Clematis, however, the numbers of sepals did not increase, at least compared with their respective petalous relatives (36). This suggests that silencing of AP3-3 itself may not be sufficient for petal loss in natural apetalous species; other factors, such as shifts in the expression of C-function genes, may also be necessary. According to the ABC model for flower development (8), outward expansion of the C function into petals will lead to the homeotic transformations of petals into stamens, whereas the number of sepals remains unchanged. Therefore, if loss of petals was actually caused by petal-to-stamen transformation, outward expansion of the C function would be a more probable explanation. Second, although deletions in the coding and regulatory regions were very likely the causes for the silencing or down-regulation of the Beesia and Enemion AP3-3 orthologs, they can also be explained as degeneration following nonfunctionalization and relaxed selection. Because AP3-3 is specifically expressed in petals, if this identity program is silenced via a genetically upstream mechanism, expression of AP3-3 will be lost and the gene will evolve under relaxed selection, accumulating destructive mutations. In this case, defects in AP3-3 would be the effect of, rather than cause for, petal losses. More data are needed to elucidate the cause-or-effect relationship between the loss of petals and the disruption of AP3-3.
Loss of Petals Could Be Advantageous.
The parallel petal losses within the buttercup family and many other lineages not only reflect the existence of similar selective pressures in different spatial and temporal dimensions, but also highlight the astonishing ability of plants to adjust their phenotypes in response to a changing environment. In flowering plants, it has long been postulated that presence of petals is beneficial because the showy and attractive organs can facilitate pollination (1). Indeed, the reason why some groups of flowering plants (such as the Orchidaceae, Zingiberales, Lamiales, and so forth) are extremely rich in species diversity is that they have evolved conspicuous and specialized petals or petal-like structures (1, 2, 19). However, petal loss may have also been advantageous, at least in certain circumstances, as is evidenced by the occurrence of apetalous species in many lineages. In addition, in the Ranunculaceae and many other families, sepals or sometimes bracts are large, showy, and attractive, whereas petals are relatively small in size and inconspicuous in appearance (18, 20). Therefore, loss of petals would unlikely change the general display of the flower. Rather, because the formation of petals and nectaries requires energy, having petals may not be advantageous when resources become limited and pollinators are not specialized. Actually, many Ranunculaceous species have evolved rather generalized pollination systems, in which open flowers can use pollen as a reward (37). Therefore, except in a few extraordinary cases (such as Aquilegia, Aconitum, and Delphinium), the selective pressure to maintain nectar-secreting petals is not strong. In contrast, in many taxa, sepals, stamens, or even filaments have become showy, conspicuous, and attractive to insects, and thus petals are no longer a decisive factor for pollination. The fact that some Thalictrum species have evolved syndromes of wind pollination (37) further suggests that the negative effect caused by petal losses, if any, may be compensated by other factors.
Materials and Methods
Gene Isolation and Confirmation.
For each species (details in Table S2), total RNA was extracted from floral buds at various developmental stages using PureLink Plant RNA Reagent (Invitrogen), and then reverse-transcribed into cDNA with the SuperScript III first-strand cDNA synthesis kit (Invitrogen). AP3-like genes were amplified by two rounds of conventional RT-PCR with degenerate forward primers and a reverse adaptor primer (Tables S1 and S3). Amplified fragments of expected lengths were then purified and cloned into pEASY-T3 cloning vector (TransGen). At least 20 positive clones were sequenced for each gene, and the sequences were confirmed by BLAST searches and phylogenetic analysis as described (16) (Fig. S7). To obtain the genomic DNA of the AP3-3 orthologs, gene-specific primers were designed, and a PCR-based genome walking method was performed (Takara). Full-length fragments of each gene were assembled by ContigExpress and confirmed by PCR-based sequencing.
qRT-PCR.
Total RNAs were isolated from leaves, inflorescences (including floral buds at different developmental stages), and individual floral organs at the nearly-mature stages. mRNAs were first purified from total RNAs with Oligotex mRNA kit (Qiagen) and then reverse-transcribed into cDNA as above described. Amplification efficiency of the primer combinations (Tables S1 and S3) for each gene was determined by comparing the standard curves, and the best combinations were used for further qRT-PCR, which was performed using PrimerScript RT reagent kit (Perfect Real Time)(Takara) in a Stratagene Mx3000P. All reactions were run with three biological replicates (except for the Leptopyrum petals, which are very tiny and difficult to collect) and three technical replicates. Relative gene expression values were first normalized a well-known house-keeping gene, ACTIN, and then renormalized to the expression of AP3-1 in stamens (38). Previous (7, 24, 39) and our own observations have found that the expression of the AP3-1 orthologs in stamens are generally comparable between species.
In Situ Hybridization.
Floral buds at various development stages were fixed in 4% (wt/vol) paraformaldehyde and embedded in Paraplast (Sigma). A fragment spanning the C-terminal end of the coding region and the 3′ UTR was used as template for synthesis of sense and antisense digoxigenin-labeled RNA probes with the DIG RNA labeling kit (Roche) (Tables S1 and S3). Treatments of the sections (8–10 µm) were performed as described (40), with several modifications depending on species. Images were captured with a Zeiss Axio imager microscope.
Supplementary Material
Acknowledgments
We thank Genlu Bai and You Zhou for assistance in collecting plant materials and Veronica Di Stilio, Xuejun Ge, Yanping Guo, Anmin Lu, Hong Ma, Jongmin Nam, Masatoshi Nei, Kaiyu Pan, Guangyuan Rao, and Ji Yang for valuable comments. This work was supported by National Basic Research Program of China Grant 2009CB941500 and National Natural Science Foundation of China Grants 31125005, 30870179, and 31170215.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219690110/-/DCSupplemental.
References
- 1.Endress PK. Diversity and Evolutionary Biology of Tropical Flowers. Cambridge, UK: Cambridge Univ Press; 1994. [Google Scholar]
- 2.Soltis DE, Soltis PS, Endress PK, Chase MW. Phylogeny and Evolution of Angiosperms. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- 3.Kozo-Poljanski BM. An Introduction to Phylogenetic Systematics of the Higher Plants. Voronezh, USSR: Nature and Culture; 1922. [Google Scholar]
- 4.Takhtajan A. Flowering Plants. Berlin: Springer; 2009. [Google Scholar]
- 5.Kramer EM, Irish VF. Evolution of genetic mechanisms controlling petal development. Nature. 1999;399(6732):144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- 6.Irish VF. Evolution of petal identity. J Exp Bot. 2009;60(9):2517–2527. doi: 10.1093/jxb/erp159. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen DA, Kramer EM, Zimmer EA. One size fits all? Molecular evidence for a commonly inherited petal identity program in Ranunculales. Am J Bot. 2009;96(1):96–109. doi: 10.3732/ajb.0800038. [DOI] [PubMed] [Google Scholar]
- 8.Coen ES, Meyerowitz EM. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353(6339):31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 9.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405(6783):200–203. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- 10.Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature. 2001;409(6819):525–529. doi: 10.1038/35054083. [DOI] [PubMed] [Google Scholar]
- 11.Theissen G, Saedler H. Plant biology: Floral quartets. Nature. 2001;409(6819):469–471. doi: 10.1038/35054172. [DOI] [PubMed] [Google Scholar]
- 12.Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol. 2004;14(21):1935–1940. doi: 10.1016/j.cub.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Kramer EM, Dorit RL, Irish VF. Molecular evolution of genes controlling petal and stamen development: Duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics. 1998;149(2):765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, et al. Expression of floral MADS-box genes in basal angiosperms: Implications for the evolution of floral regulators. Plant J. 2005;43(5):724–744. doi: 10.1111/j.1365-313X.2005.02487.x. [DOI] [PubMed] [Google Scholar]
- 15.Zahn LM, et al. The evolution of the SEPALLATA subfamily of MADS-box genes: A preangiosperm origin with multiple duplications throughout angiosperm history. Genetics. 2005;169(4):2209–2223. doi: 10.1534/genetics.104.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan H, et al. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes. Mol Phylogenet Evol. 2007;44(1):26–41. doi: 10.1016/j.ympev.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Baum DA, Whitlock BA. Plant development: Genetic clues to petal evolution. Curr Biol. 1999;9(14):R525–R527. doi: 10.1016/s0960-9822(99)80327-3. [DOI] [PubMed] [Google Scholar]
- 18.Heywood VH, Brummitt RK, Culham A, Seberg O. Flowering Plant Families of the World. Richmond Hill, ON, Canada: Firefly Books; 2007. [Google Scholar]
- 19.Judd WS, et al. Plant Systematics: A Phylogenetics Approach. Sunderland, MA: Sinauer Associates; 2008. [Google Scholar]
- 20.Tamura M. ANGIOSPERMAE: Ordnung Ranunculales. Fam. Ranunculaceae. Berlin: Duncker & Humblot; 1995. [Google Scholar]
- 21.Damerval C, Nadot S. Evolution of perianth and stamen characteristics with respect to floral symmetry in Ranunculales. Ann Bot (Lond) 2007;100(3):631–640. doi: 10.1093/aob/mcm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W, et al. Phylogeny and classification of Ranunculales: evidence from four molecular loci and morphological data. Perspect Plant Ecol Evol Syst. 2009;11:81–110. [Google Scholar]
- 23.Kramer EM, Di Stilio VS, Jaramillo MA. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int J Plant Sci. 2003;164:1–11. [Google Scholar]
- 24.Sharma B, Guo C, Kong H, Kramer EM. Petal-specific subfunctionalization of an APETALA3 paralog in the Ranunculales and its implications for petal evolution. New Phytol. 2011;191(3):870–883. doi: 10.1111/j.1469-8137.2011.03744.x. [DOI] [PubMed] [Google Scholar]
- 25.Miikeda O, Kita K, Handa T, Yukawa T. Phylogenetic relationships of Clematis (Ranunculaceae) based on chloroplast and nuclear DNA sequences. Bot J Linn Soc. 2006;152:153–168. [Google Scholar]
- 26.Lloréns C, Futami R, Bezemer D, Moya A. The Gypsy Database (GyDB) of mobile genetic elements. Nucleic Acids Res. 2008;36(Database issue):D38–D46. doi: 10.1093/nar/gkm697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wessler SR. Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci USA. 2006;103(47):17600–17601. doi: 10.1073/pnas.0607612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9(5):397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riechmann JL, Wang M, Meyerowitz EM. DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996;24(16):3134–3141. doi: 10.1093/nar/24.16.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development. 1998;125(9):1647–1657. doi: 10.1242/dev.125.9.1647. [DOI] [PubMed] [Google Scholar]
- 31.Wuesta SE, et al. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA. 2012;109:13453–13457. doi: 10.1073/pnas.1207075109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AG, Sharrocks AD. The role of DNA-bending in MADS-box transcription factor function. Biochem Soc Trans. 1997;25(4):S639. doi: 10.1042/bst025s639. [DOI] [PubMed] [Google Scholar]
- 33.West AG, Shore P, Sharrocks AD. DNA binding by MADS-box transcription factors: A molecular mechanism for differential DNA bending. Mol Cell Biol. 1997;17(5):2876–2887. doi: 10.1128/mcb.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Folter S, Angenent GC. trans meets cis in MADS science. Trends Plant Sci. 2006;11(5):224–231. doi: 10.1016/j.tplants.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Kramer EM, et al. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia. Plant Cell. 2007;19(3):750–766. doi: 10.1105/tpc.107.050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker SC, Hodges SA. Floral ontogeny of Aquilegia, Semiaquilegia, and Enemion (Ranunculaceae) Int J Plant Sci. 2005;166:557–574. [Google Scholar]
- 37.Soza VL, Brunet J, Liston A, Smith PS, Di Stilio VS. Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae) Mol Phylogenet Evol. 2012;63(1):180–192. doi: 10.1016/j.ympev.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Sharma B, Kramer E. Sub- and neo-functionalization of APETALA3 paralogs have contributed to the evolution of novel floral organ identity in Aquilegia (columbine, Ranunculaceae) New Phytol. 2013;197(3):949–957. doi: 10.1111/nph.12078. [DOI] [PubMed] [Google Scholar]
- 40.Kramer EM. Methods for studying the evolution of plant reproductive structures: Comparative gene expression techniques. Methods Enzymol. 2005;395:617–636. doi: 10.1016/S0076-6879(05)95032-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.