Abstract
Contagious cancers that pass between individuals as an infectious cell line are highly unusual pathogens. Devil facial tumor disease (DFTD) is one such contagious cancer that emerged 16 y ago and is driving the Tasmanian devil to extinction. As both a pathogen and an allograft, DFTD cells should be rejected by the host–immune response, yet DFTD causes 100% mortality among infected devils with no apparent rejection of tumor cells. Why DFTD cells are not rejected has been a question of considerable confusion. Here, we show that DFTD cells do not express cell surface MHC molecules in vitro or in vivo, due to down-regulation of genes essential to the antigen-processing pathway, such as β2-microglobulin and transporters associated with antigen processing. Loss of gene expression is not due to structural mutations, but to regulatory changes including epigenetic deacetylation of histones. Consequently, MHC class I molecules can be restored to the surface of DFTD cells in vitro by using recombinant devil IFN-γ, which is associated with up-regulation of the MHC class II transactivator, a key transcription factor with deacetylase activity. Further, expression of MHC class I molecules by DFTD cells can occur in vivo during lymphocyte infiltration. These results explain why T cells do not target DFTD cells. We propose that MHC-positive or epigenetically modified DFTD cells may provide a vaccine to DFTD. In addition, we suggest that down-regulation of MHC molecules using regulatory mechanisms allows evolvability of transmissible cancers and could affect the evolutionary trajectory of DFTD.
Keywords: canine transmissible venereal tumor, immune evasion, transplantation, primary tumour
The adaptive immune system should prevent cancer cells passing from one individual to another. However, contagious cancers can emerge in nature and provide a unique opportunity to study mechanisms of immune escape and tumor evolution in cancers that are continually passaged between individuals. Devil facial tumor disease (DFTD) is one such contagious cancer that emerged 16 y ago in the Tasmanian devil, a marsupial carnivore found only on the island of Tasmania (1).
The emergence of DFTD triggered immediate comparisons to the only other naturally occurring contagious cancer, canine transmissible venereal tumor (CTVT) (1, 2). CTVT is a sexually transmitted tumor in dogs that has existed as a parasitic cell line for thousands of years (2, 3). Despite their shared ability to pass between individuals, DFTD and CTVT have contrasting relationships with their respective hosts. Although DFTD causes complete mortality among infected devils (4), CTVT does not usually kill host dogs (5). This contrasting impact on the host species must be linked to how DFTD and CTVT interact with the immune system of their respective hosts. Although there is some understanding as to how CTVT avoids the immune response and progresses in its host (6, 7), very little is understood about how DFTD evades the immune response of host devils.
Recognition of allograft cells and malignant cells by the adaptive immune system depends on the interaction of host T cells with highly polymorphic MHC class I and class II molecules (8). In the endoplasmic reticulum (ER), MHC class I heavy chains bind peptide and β2-microglobulin (β2m), a single domain protein essential for stabilizing the heavy chain, resulting in a stable trimeric complex trafficked to the cell surface where peptide is presented to CD8+ T cells (9). Other proteins are required for successful peptide binding, including the transporter associated with antigen processing (TAP; a heterodimer of TAP1 and TAP2) that pumps peptides from the cytoplasm into the ER, and tapasin, which facilitates peptide binding (9). MHC class II molecules present peptides to CD4+ T cells, and are heterodimers encoded by A and B genes, also requiring other proteins to facilitate peptide binding, including the chaperone DMB (10).
Regulation of MHC expression occurs through a combination of cytokines, transcription factors, and epigenetic modifications (11, 12). Both MHC class I and class II molecules can be up-regulated by cytokines such as IFN-gamma (IFN-γ), released by lymphocytes upon stimulation (12). The response to IFN-γ results in expression of the MHC class II transactivator (CIITA), a transcription factor that binds to the SXY site within the promoter elements of MHC class I, MHC class II, and β2m genes, inducing or up-regulating expression (13). In addition to transcription factors and cytokines, MHC gene expression is influenced by the physical state of chromatin within the promoter regions of relevant genes. Acetylated histones and demethylated DNA within promoter elements are generally associated with relaxation of chromatin structure, binding of transcription factors to DNA, and transcription (14). MHC class II expression is tightly regulated, and these molecules are only found on the surface of antigen presenting cells, whereas MHC class I molecules are expressed on the surface of nearly all cells.
The success of CTVT as a contagious cancer depends on a balance between tumor growth and the ability of the immune system to control tumor growth, which is intricately linked to MHC expression. After transmission of CTVT, tumors appear within 2 mo and cells undergo an initial growth stage where the tumor cells lack expression of class I and class II molecules, and lymphocytes fail to infiltrate the tumor (6, 15). This period of tumor growth does not continue indefinitely, and after 3 to 9 mo, tumor growth either stabilizes or begins to regress, which is associated with a significant increase in MHC class I and class II expression on the surface of the CTVT cells and infiltration of lymphocytes into the tumor mass (6, 15). Outside the laboratory setting, CTVT tumors often enter a stationary phase in which the tumor neither grows nor regresses (5).
In contrast to CTVT, the mechanisms that allow DFTD to transmit successfully have been a subject of confusion. It has been proposed that DFTD successfully passes as an allograft due to low genetic diversity at MHC genes (16). This view has been supported by evidence for expression of MHC genes in tumor cell lines and biopsies (16, 17), intact antigen presentation genes in the tumor genome (18), and a lack of tumor infiltrating lymphocytes (19). However, devils are not monomorphic at MHC (20, 21), so reduced MHC genetic diversity cannot explain the sustained lack of immune response to the tumor. Thus, how DFTD cells move between individuals without immune rejection has remained unknown.
It is predicted that the rapid spread of DFTD will cause extinction of Tasmanian devils in the wild (22). DFTD arose in a Tasmanian devil in northeastern Tasmania but has since spread to all devil populations in eastern and central Tasmania (1, 22). DFTD cells are thought to pass between animals when they bite each other during social interactions and, once transmitted, large tumors form around the face and neck that cause 100% mortality (1). The spread of DFTD has caused the population to decline rapidly (23, 24) and to be classified as endangered (www.iucnredlist.org). Without an accurate understanding of how DFTD escapes the host–immune response, any hope of conserving the devil in the wild appears slim.
Here, we report our investigation of MHC expression and regulation in DFTD cells. We find that DFTD cells do not express cell surface MHC molecules, due to a down-regulation of genes essential to the antigen-processing pathway. Loss of expression is not due to structural mutations, but regulation including epigenetic modification of histones. We show that MHC class I molecules can be restored to the surface of DFTD cells by using recombinant devil IFN-γ. These results demonstrate how DFTD passes as an allograft, revealing characteristics that may be important in the emergence and evolutionary success of contagious cancers more generally. Further, these results have implications for the development of a vaccine against DFTD.
Results
DFTD Cells Lack Cell Surface MHC Class I Molecules.
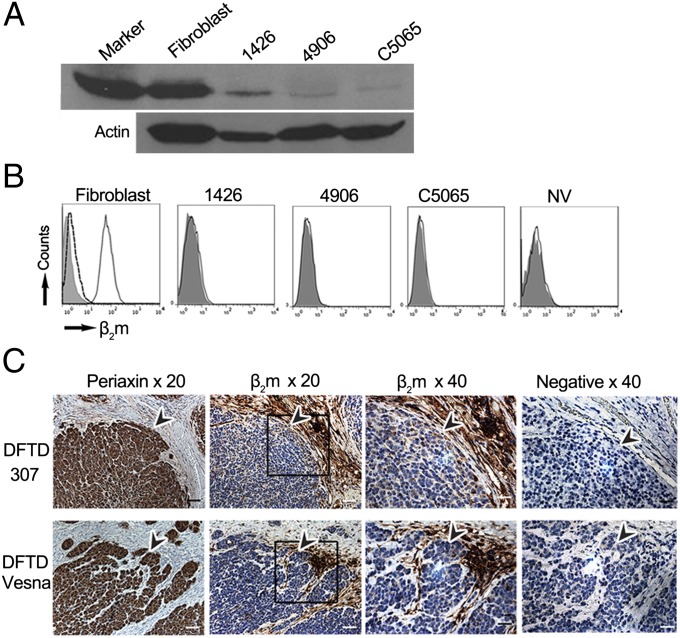
We developed a monoclonal antibody against Tasmanian devil MHC class I heavy chain (MHCI-mAb; raised against the cytoplasmic tail of the class I heavy chain) and a polyclonal antibody against devil β2m (β2m-Ab; raised against native β2m). Using these antibodies, we examined MHC class I and β2m expression in four DFTD cell lines (1426, C5065, 4906, and NV) and a devil fibroblast cell line. Cell lines 1426, 4906, and C5065 have been kept in culture since 2005, whereas cell line NV was in culture for only 3 wk before analysis. Only a small amount of class I protein could be detected by Western blot of DFTD cells compared with fibroblast cells (Fig. 1A). No β2m protein was detected on the surface of DFTD cells stained with β2m-Ab and analyzed by using flow cytometry (Fig. 1B). Therefore, functional MHC class I molecules, associated with β2m and peptide, are not present at the surface of DFTD cells in vitro.
Fig. 1.
DFTD cells have low levels of intracellular MHC class I and no surface expression of β2m in vitro and in vivo. (A) Western blot of fibroblast and DFTD whole-cell protein probed with MHCI-mAb, with the MHC class I band shown at 40 kDa for a 20-min exposure to X-ray film. (A Lower) A loading control probed with an antibody to β-actin. By Bradford assay for protein, 35 µg from fibroblast lysate and 50 µg from each DFTD lysate were loaded on the gel. (B) Flow cytometry of fibroblast and DFTD cells, with fluorescence intensity on x axis and number of cells (counts) on y axis. Shaded area, stained with the preimmune serum; solid black line, stained with β2m-Ab; dashed black line, stained with β2m-Ab blocked with native devil β2m protein. (C) IHC on serial sections of primary DFTD biopsies from wild devils stained with an antibody to periaxin (a marker specific for DFTD cells, enabling DFTD cells to be distinguished from host devil cells), devil β2m-Ab, and the preimmune rat serum as a negative control for the devil β2m-Ab. Boxes indicate areas shown at 40× magnification, and arrowheads indicate similar positions in the serial sections, pointing toward DFTD cells as defined by periaxin staining. Positive cells for each marker are stained brown; nuclei are stained blue. (Scale bars: 20× magnification, 50 μm; 40× magnification, 20 μm.)
Immunohistochemistry (IHC) was used to examine β2m protein expression in DFTD cells in vivo. Primary DFTD biopsies stained with periaxin, a marker specific for Schwann cells and DFTD cells (17), highlight the presence of host connective tissue within the tumor mass of all samples, demarcating the tumor cells in each section (Fig. 1C and SI Appendix, Fig. S1). Serial sections show that the DFTD tumor cells have low levels of β2m staining, whereas host cells within the connective tissue stain strongly for β2m (Fig. 1C and SI Appendix, Fig. S1). Thus, DFTD cells in vivo have low levels of β2m protein and, consequently, few if any MHC class I molecules.
Loss of MHC Class I Molecules Is Due to Down-Regulation of Genes Essential for Antigen Loading.
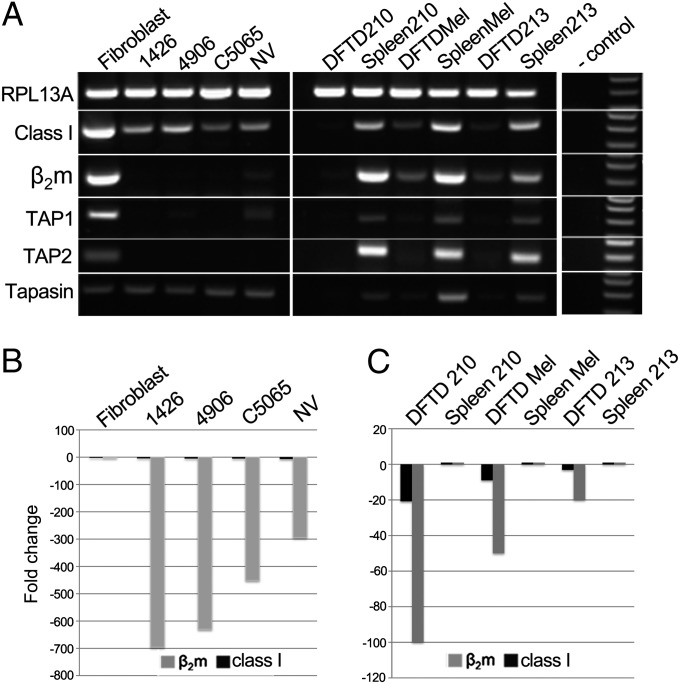
Using RT-PCR and quantitative RT-PCR (RT-qPCR), we examined the expression of genes required for functional class I and class II molecule expression in the same four DFTD and fibroblast cell lines described above. Consistent with a previous report (16), we found that DFTD cells express class I heavy chain (Fig. 2A) and class II B transcripts at the RNA level (SI Appendix, Fig. S2). Sequencing of PCR products (SI Appendix, Fig. S3) confirmed the amplification of the recently identified polymorphic class I and class II B loci (25). In addition, tapasin levels are equivalent between DFTD cells and fibroblast cells. However, other genes essential to MHC antigen processing are expressed at low levels or not at all in the four DFTD cell lines. β2m transcripts in the DFTD cell lines were 450- to 695-fold lower than in devil fibroblast cells (Fig. 2B). No TAP1 transcripts could be detected in cell lines 1426 and C5065 by RT-PCR, and only trace levels were present in cell line 4906 and NV. No TAP2 could be detected in all four DFTD cell lines. Thus, functional class I molecules should not be expressed in DFTD cells, as we found by using antibody staining. Similarly, few if any class II molecules would be expressed in the DFTD cells, based on detection of no DMB and only trace levels of class II A transcripts compared with devil spleen cells (SI Appendix, Fig. S2).
Fig. 2.
DFTD cells down-regulate mRNA of genes essential for antigen loading and presentation. (A) RT-PCR amplification of RPL13A (ribosomal protein L13A), MHC class I, β2m, TAP1, TAP2, and tapasin from RNA of a fibroblast cell line and DFTD cell lines in Left, DFTD biopsies and matched host spleen samples in Center, and no-cDNA negative control and markers in Right. Amplicons are between 100 and 300 bp. (B and C) RT-qPCR of β2m (gray) and MHC class I (black) gene expression normalized against RPL13A as a housekeeping gene for tumor lines relative to fibroblast cell line (B), and each DFTD biopsy relative to the matched host spleen sample (C). All samples were tested in triplicate.
We also examined gene expression in three primary tumor biopsies compared with spleen samples from the same hosts. MHC class I, β2m, TAP1, and TAP2 RNA levels were consistently lower in tumor biopsies than in the matched host spleen (Fig. 2 A and C). DFTD biopsies had 20- to 100-fold less β2m and 3- to 20-fold less MHC class I transcripts than matched spleen (Fig. 2C). Four additional primary tumor biopsies, two of which (DFTD_Vesna and DFTD_307) were also used in IHC experiments (Fig. 1C), were examined by RT-PCR, and also had reduced β2m, TAP1, and TAP2 RNA levels compared with fibroblast cells (SI Appendix, Fig. S4). β2m expression was variable between biopsies and higher than in the cell lines, which is due to the presence of host connective tissue strongly staining for β2m protein in these samples, as illustrated in the IHC experiments (Fig. 1C and SI Appendix, Fig. S1).
Loss of MHC Class I Expression Is Not Due to Structural Mutations.
Next we sought to determine the mechanism for down-regulation of genes involved in antigen presentation in DFTD cells. The loss of β2m, TAP1, TAP2, DMB, and MHC class II A chain genes from the DFTD genome was excluded by amplifying these genes from the DNA of fibroblast and DFTD cell lines (SI Appendix, Fig. S5). Moreover, full length class I and β2m transcripts isolated from DFTD and fibroblast cells have all of the features indicating that they are capable of translation and of forming functional molecules (SI Appendix, Figs. S6 and S7). The sequences of the β2m promoter from fibroblast and DFTD cells are virtually identical, with an intact IFN response element, S/W, X/X2, Y motifs and TATA box. Finally, the promoter sequences of the TAP1 and TAP2 genes are identical between fibroblast and DFTD cells (SI Appendix, Fig. S8). Therefore, there is no evidence for structural mutations in the DFTD β2m, TAP1, TAP2, or class I transcripts or promoters.
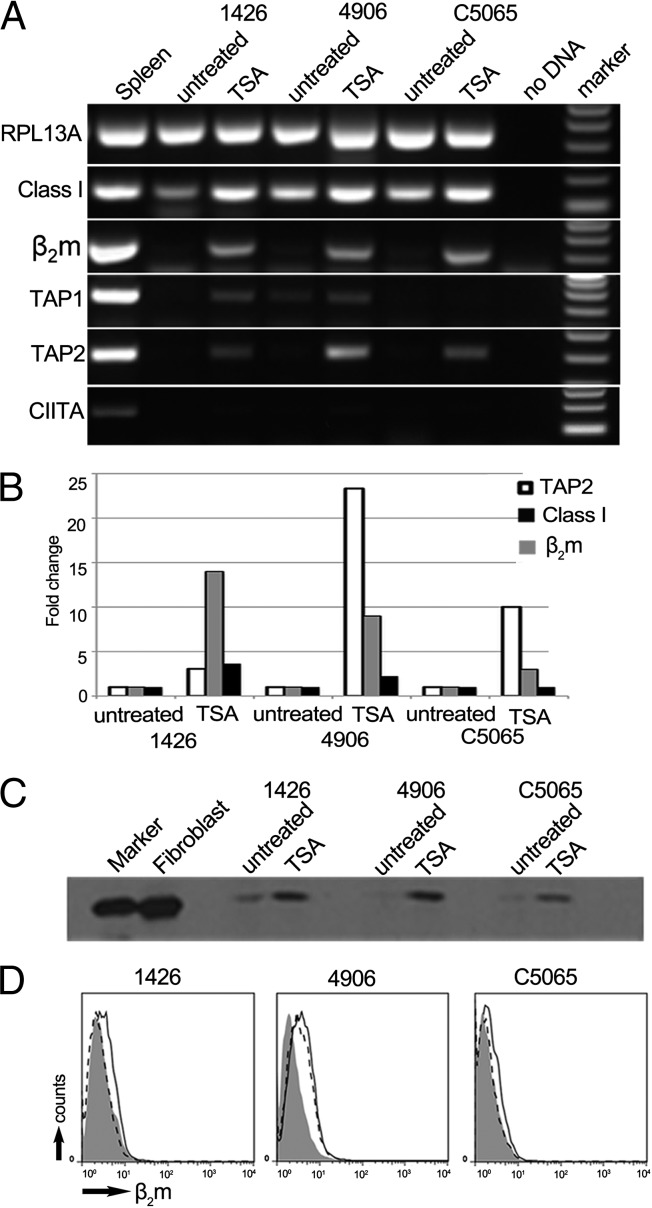
The combined loss of β2m and TAP in tumor cells suggested a common regulatory mechanism suppressing the transcription of these genes. Bisulphite sequencing across CpG islands within the β2m and TAP1 promoters showed no significant increase in methylation of DFTD promoters compared with fibroblast promoters (SI Appendix, Figs. S9 and S10). However, treatment of DFTD cells with the histone deacetylase inhibitor trichostatin A (TSA) resulted in increased expression of class I, β2m, TAP1, and TAP2 transcripts, but not the transcription factor CIITA (Fig. 3 A and B). Treatment with TSA resulted in an increase of MHC class I protein detectable by Western blot (Fig. 3C), but restored, at best, trace amounts of β2m to the surface of DFTD cells (Fig. 3D). These results show that down-regulation of MHC genes by DFTD is due to neither structural mutations in the coding region or promoter regions of these genes nor to hypomethylation of the β2m and TAP1 promoters. However, the up-regulation of class I, β2m, and TAP genes after treatment with TSA indicates that suppression of these genes is related to the acetylation state of histones and, thus, chromatin structure within regulatory regions.
Fig. 3.
MHC class I protein is up-regulated in DFTD cells after treatment with the deacetylation inhibitor, Trichostatin A (TSA). (A) RT-PCR amplification of RPL13A, MHC class I, β2m, TAP1, TAP2, and CIITA from RNA of TSA-treated and untreated cells. Amplicons are between 100 and 300 bp. (B) RT-qPCR of TAP2 (white), β2m (gray), and MHC class I (black) in the tumor lines after treatment with TSA. Fold change is relative to the untreated control cells for each cell line and normalized against RPL13A as a housekeeping gene. A standard curve was constructed from fibroblast expression. DFTD samples were tested in triplicate. (C) Western blot of whole-cell protein from treated and untreated DFTD cells, probed with MHCI-mAb, showing MHC class I at 40 kDa. By Bradford assay for protein, 24 µg from lysate of fibroblasts, 25 µg from lysate of untreated DFTD cells, and 20 µg from lysate of treated DFTD cells were loaded on the gel. (D) Flow cytometry to test for surface expression of β2m on TSA-treated DFTD, with fluorescence intensity on x axis and number of cells (counts) on y axis. Shaded area, stained with serum from a preimmunized rat; dashed line, untreated cells stained with β2m-Ab; solid black line, TSA-treated cells stained with β2m-Ab.
MHC Class I Expression Can Be Restored on DFTD Cells both in Vitro and in Vivo.
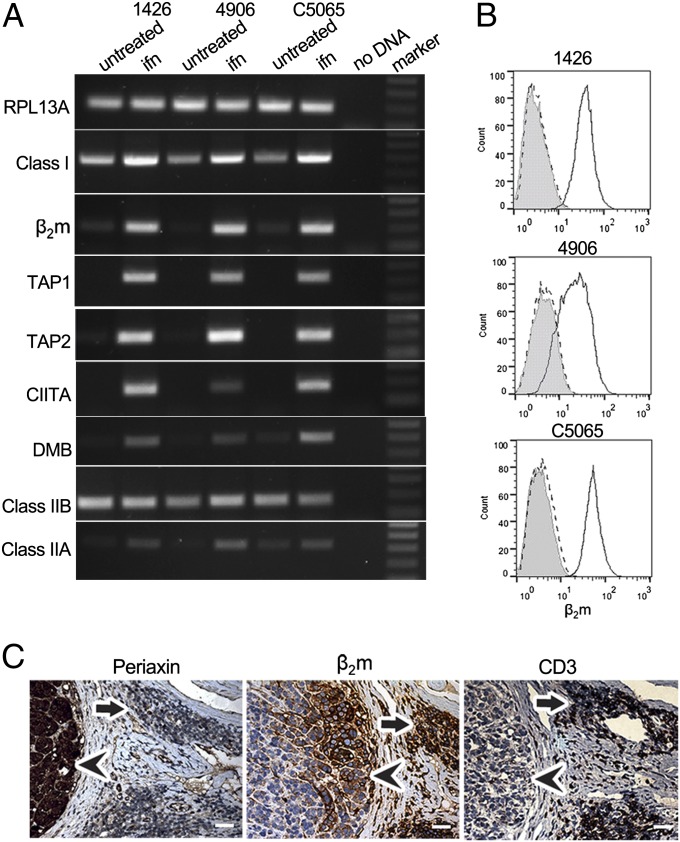
To determine whether MHC molecules could be expressed on the surface of DFTD cells, we cloned and expressed recombinant devil IFN-γ (SI Appendix, Fig. S11). Treatment with IFN-γ caused an enormous increase of β2m on the surface of DFTD cells in culture, as assessed by flow cytometry (Fig. 4B). This significant increase in surface MHC class I expression is associated with up-regulation of RNA from MHC class I, β2m, TAP1, and TAP2 genes along with the transcription factor CIITA. CIITA is required for MHC class II expression, and we found that DMB and class II B and A transcripts are also up-regulated after IFN-γ treatment (Fig. 4A).
Fig. 4.
MHC class I surface expression can be restored on DFTD cell lines after treatment with recombinant devil IFN-γ. (A) RT-PCR amplification of RPL13A, MHC class I, β2m, TAP1, TAP2, CIITA, DMB, MHC class IIB, and MHC class IIA from three DFTD cell lines treated with recombinant devil IFN-γ (ifn) compared with untreated. A no-DNA control and a marker are included. Amplicons are between 100 and 300 bp. (B) Flow cytometry showing β2m expression on the surface of IFN-γ–treated DFTD cells. Shaded area, treated DFTD stained with the preimmune serum; dashed line, untreated DFTD cells stained with β2m-Ab; solid line, treated DFTD cells stained with β2m-Ab. (C) Serial sections of DFTD biopsy 307 stained for periaxin (Left), β2m (Center), and CD3ε (Right), all at 20× magnification. Arrowheads indicate tumor cells and arrows indicate CD3-positive infiltrating cells. (Scale bars: 20× magnification, 50 μm; 40× magnification, 20 μm.)
Significantly, we have found that in rare cases, DFTD cells can also express MHC molecules in vivo. In some tumor biopsies, lymphocytes that stain for CD3 [T cells or natural killer (NK) cells] are found within the connective tissue. The DFTD cells near these lymphocytes stain strongly for β2m protein, in stark contrast to DFTD cells farther within the tumor mass that remain β2m negative (Fig. 4C). This result suggests that lymphocytes can approach and respond to DFTD cells in vivo, secreting cytokines like IFN-γ to up-regulate MHC molecules on tumor cells nearby.
Discussion
Here we show that DFTD cells do not express functional MHC class I molecules in vitro and in vivo, explaining how DFTD escapes the T-cell response typical of allograft rejection. Further, we show that loss of MHC molecules from the cell surface of DFTD cells is due to coordinated down-regulation of genes essential to the antigen-processing pathway, and that this loss is by regulatory mechanisms including epigenetic modifications rather than structural mutations. Finally, we show that the class I-negative phenotype of DFTD cells can be reversed in vitro and in vivo.
DFTD cells lack MHC molecules due to down-regulation of multiple components of the antigen processing pathway. Without β2m and peptide (pumped by the TAPs), MHC class I heavy chain produced by DFTD cells will be retained in the ER and degraded (9). Although more β2m RNA was detected in the DFTD biopsy samples compared with the cell lines, IHC using the same biopsy samples demonstrates the presence of β2m in connective tissue derived from the host devil. These results explain the higher level of β2m RNA we detected in biopsy samples compared with DFTD cell lines, as well as a previous analysis (17) of a DFTD biopsy that did not detect any differences in expression levels of β2m, TAP1, and TAP2 RNA between DFTD and host tissue.
The lack of MHC class I molecules expressed on DFTD cells most likely represents a down-regulation of class I expression that occurred during the transformation of the ancestral DFTD cell to a malignant cell. DFTD cells are reported to be of Schwann cell origin (17), and MHC expression is known to be regulated in the nervous system to safeguard against hypersensitivity (26). Tasmanian devil Schwann cells have not been isolated. However, human and rodent Schwann cells constitutively express class I but not class II molecules, although both can be up-regulated with IFN-γ (27, 28). Thus, assuming that devil Schwann cells have a similar phenotype to those of placental mammals, modifications in the DFTD cell have resulted in loss of MHC class I expression, giving these cells the ability to move as an allograft. However, the absence of MHC class II RNA expression may represent the ancestral state of the DFTD cell.
Loss of MHC class I expression in DFTD cells is not caused by structural mutations, but by regulatory modifications including epigenetic changes. The β2m, TAP1, and TAP2 genes are structurally sound, and their expression can be increased in vitro by treatment with either TSA or recombinant devil IFN-γ. However, the inhibition of histone deactylases with TSA only partially restores gene expression and does not up-regulate CIITA expression. In contrast, treatment with IFN-γ results in expression of CIITA, a more pronounced expression of β2m and TAP genes, and cell surface expression of MHC class I molecules. As well as facilitating the binding of other transcription factors to MHC class I, class II, and β2m promoters (13, 29), CIITA also has intrinsic acetyltransferase activity and, upon binding, it relaxes chromatin structure, allowing transcription factors to access DNA (30). Thus, although inhibition of histone deacetylation results in a small increase in gene expression, CIITA may be required for full acetylation and gene expression. Alternatively, IFN-γ may control other transcription or epigenetic factors. Regardless, the down-regulation of MHC molecules via regulatory rather than structural mutations has implications for the interaction of the tumor with the devil immune system and may be exploited to design an effective vaccine against DFTD.
We propose that priming the devil immune system with MHC-positive and TSA-treated DFTD cells could provide an effective vaccine against DFTD. A whole-cell vaccine would expose devil T cells to antigenic peptides derived from the DFTD cells and presented by foreign MHC molecules. Upon subsequent challenge with wild-type DFTD cells, host cells should be activated against those antigens found even at low levels on the surface of DFTD cells and/or intracellular antigens released by DFTD cells during tumor growth. Once an immune response is initiated, the release of cytokines such as IFN-γ should stimulate wild-type DFTD cells to express MHC molecules, as we have shown, potentially leading to a more significant and protective immune response.
The regulation of MHC gene expression we describe for DFTD also has implications for the evolution of DFTD, perhaps representing an advantage for long-lived transmissible tumors. The only other naturally occurring contagious cancer, CTVT, has existed for as much as 2,500 y (2) and is rarely fatal to dogs. CTVT down-regulates MHC class I expression during tumor transmission and growth before up-regulating expression as it enters a stationary phase associated with lymphocyte infiltration and IFN-γ release (7). Like DFTD, CTVT has not switched off MHC expression permanently by structural mutations, but regulates MHC expression, although the mechanisms of regulation are not fully understood (2). Evolutionary pressure may have favored CTVT subclones that can subsist in the population through a balance between tumor growth and the host–immune response (2). It remains to be seen whether DFTD will evolve into a less aggressive cancer and ensure its own survival, but in any case, control of gene expression by regulatory, rather than structural mutations, gives DFTD cells the ability to adjust MHC expression in response to changing cellular environments.
The results presented here explain why the adaptive immune system fails to reject DFTD cells and provide the basis for answering other important questions. For instance, down-regulation of class I molecules should make DFTD cells good targets for NK cells (31). Do DFTD cells use regulatory mechanisms to alter the balance of activating and inhibitory NK ligands as an additional mechanism of immune escape? In a more general sense, dogs control CTVT whereas devils do not control DFTD, although both tumors down-regulate MHC molecules and up-regulate them upon treatment with IFN-γ. What additional mechanisms of immune evasion (speed of replication, release of immunosuppressive cytokines, manipulation of the tumor environment) make DFTD tumor cells so difficult for the devil immune system to control? And finally, DFTD and CTVT are contagious cancers, but they share some features with trophoblasts in the fetus of placental mammals (32). To what extent are the regulatory mechanisms shared between transmissible tumors and normal cells in specialized situations? The mechanisms of immune escape used by DFTD have the potential to provide a greater understanding of the complex interaction of a tumor with its host, in addition to more general mechanisms of immune surveillance and regulation.
Materials and Methods
Cells and Cell Culture Conditions.
A devil fibroblast cell line (18) was used as a control. Three cell lines derived from DFTD primary tumors (1426, 4906 and C5065) are described (33, 34). Complete medium and cell culture conditions are described in SI Appendix. An additional DFTD cell line (DFTD_NV) was derived from a DFTD biopsy taken from a wild Tasmanian devil as follows. A fine needle aspiration from the DFTD tumor mass was placed into complete medium (but with kanamycin at 200 μg/mL). The cells were centrifuged (350 g for 10 min) and the resulting pellet was resuspended in complete medium before plating at ∼2 × 106/mL followed by incubation at 35 °C with 5% (vol/vol) CO2. Cell lines 1426, 4906, and C5065 have been kept in culture since 2005, whereas cell line NV was in culture for only 3 wk before analysis. All field procedures were carried out with approval from the University of Tasmania's Animal Ethics Committee (AEC Ref # A0011696).
Cell Treatments.
DFTD cells were treated with the histone deacetylase inhibitor trichostatin A (Sigma; T1952) at 10 ng/mL in culture for 72 h. A range of concentrations (5–20 ng/mL) and culture times (24, 48, and 72 h) were trialed to ensure minimal cell death during treatment.
Three DFTD cell lines (1426, 4906, and C5065) were treated with recombinant devil IFN-γ. Briefly, devil IFN-γ was identified in the Tasmanian devil genome sequence (ref. 16; www.ensembl.org/Sarcophilus_harrisii; Location: GL861606.1:1664620–1670021:1) and amplified by using the following primers (F – 5′ AGCGGATCCGCCATGAATTATTCAAGCTACCTCTTAGC 3′ and R - 5′ TATCTCTAGATTACTGTGTGATTTTTCCTTGGCTTTT 3′). The amplicon was cloned into the pcDNA3.1 expression vector (Invitrogen) by using standard molecular biology procedures. The resulting construct was sequenced in both directions to ensure no errors were introduced during amplification or cloning. The construct (pcDNA3-IFN-γ) was transiently transfected into Chinese hamster ovary (CHO) cells (cultured as described in SI Appendix) by using FuGENE transfection reagent (Promega). A construct-only control, transfection reagent-only control, and untreated cells were also included in the experiment. Cells were cultured for 30 h, before the supernatants from transfected and control cells were harvested and filtered by using a 0.45-μm filter (Millipore). DFTD cells were cultured for 48 h in 50% (vol/vol) culture supernatant from transfected or control Chinese hamster ovary cells, before cells were harvested for RT-PCR and flow cytometry analysis, as described below.
RT-PCR and DNA PCR.
RNA was extracted from cells by using the Nucleospin RNA II kit (Macherey-Nagel). One microgram of RNA was reverse transcribed to cDNA by using Verso cDNA synthesis kit (Thermo Scientific). DNA was extracted from cultured cells by using DNeasy blood kit (Qiagen). Primers were designed for RPL13A, TAP1, TAP2, β2m, MHC class I heavy chain, tapasin, CIITA, MHC class IIB, class II A, and DMB, and the promoters of β2m, TAP1, and TAP2 (500 base pairs upstream of the translation start sites), by aligning sequences from a range of species or where possible from the reference genome for the Tasmanian devil. All primers with the reactions and cycling conditions are in SI Appendix, Tables S1–S4). All gel products were purified (QiaQuick gel purification kit; Qiagen) and cloned into pJET plasmid (CloneJet; Fermentas). Six clones for each PCR were selected and sequenced in both directions by using T7 and bovine growth hormone primers, to determine whether the RT-PCR primers amplify all sequences from the devil MHC loci recently identified (25).
RT-qPCR.
RT-qPCR was carried out for RPL13A, MHC class I, and β2m genes on the Biorad iCycler (Biorad) with cDNA generated as described above, using the Absolute Blue Sybr Green Fluorescein qPCR mix (Thermo Scientific). Details of reaction conditions, housekeeping genes, and analysis can be found in SI Appendix.
RACE PCRs.
5′ and 3′ RACE cDNAs were constructed from total RNA of DFTD cells and fibroblast cells (isolated as described above) by using the GeneRacer kit according to the manufacturer’s instructions (GeneRacer; Invitrogen), with primers and PCR conditions described in SI Appendix, SI Methods. All gel products were cloned, and 12 clones were sequenced as described above.
Bisulphite Sequencing of β2m, TAP1, and TAP2 Promoters.
DNA from three DFTD cell lines (1426, 4906, and C5065) and a fibroblast cell line was treated with bisulphite to convert cytosines to thymines by using the Epitect Kit according to the manufacturer’s instructions (Qiagen). Primers were designed to amplify the CpG islands across the promoter sequences of β2m and TAP1, using amplification conditions in SI Appendix, Tables S3 and S4. Amplified sequences were cloned, and 12 clones for each sample were sequenced as described above. The promoter of TAP2 was not amplified as no CpG dinucleotides were detected.
Development of Antibodies.
Mice were immunized s.c. three times with 25 μg of a peptide representing the cytoplasmic region of devil MHC class I [GGKGGDYVPAAGN: based on Saha*01, a previously published full-length class I transcript (National Center for Biotechnology Information accession no. EF591089)] coupled to diphtheria toxoid by using glutaraldehyde. The antigen was adsorbed to Al(OH)3 and mixed in 1:1 ratio with incomplete Freund’s adjuvant. Four days before the fusion, the mice received an i.v. injection with 25 μg of antigen administered with adrenalin. Spleen cells and SP2/0-AG14 myeloma cells were used for fusion. Positive clones were selected by screening against the peptide coupled to ovalbumin in ELISA, with specificity of the MHCI-mAb clone TD50 illustrated in SI Appendix, Fig. S12.
Full-length devil β2m was amplified from cDNA derived from fibroblast cells with primer B2mF (5′ TTGCCATATGGTCACAAGTCCTCCCAGAGTTC 3′) and B2mR (5′ GCACCAAGTTCTGTTCTGGATCCCATTTAATTAC 3′). The subsequent amplicon was cloned into the pET22b+ vector (Novagen) and transformed into Rosetta pLysS cells (Novagen) according to the manufacturer’s instructions. Details of the expression induction and protein purification can be found in SI Appendix.
Rats of the Sprague–Dawley strain were immunized s.c. at 2- to 3-wk intervals by using ∼30 μg recombinant devil β2m in 100 μL of PBS mixed with 100 μL of the GERBU 10 adjuvant (Gerbu Biotechnik). Two weeks after each immunization, the rats were bled from the tail vein and antibodies were recovered as EDTA plasma. Specificity of the β2m-Ab is shown in Fig. 1 by preincubation of the antibodies with purified recombinant devil β2m.
Flow Cytometry.
Cells were incubated on ice with protein-G purified β2m-Ab (3 μg/mL) or protein-G purified preimmune rat serum for 20 min, followed by secondary antibody (goat anti-rat IgG conjugated to FITC; Sigma, F6258) for 20 min. In addition to secondary antibody-only and no-antibody controls, the specificity of the antibody was determined by adding 1 mg of devil β2m protein to the β2m-Ab and incubating on ice for 30 min before incubation with cells. Cells were analyzed on the FACScan cytometer (BD Biosciences), with data analyzed by using FlowJo software.
Western Blots.
Cells were detached by using PBS with 2 mM EDTA, cell pellets were lysed on ice for 30 min in a lysis buffer [100 mM TrisCl, 150 mM NaCl, 1 mM MgCl2, 0.5 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride, and 1% digitonin], and the lysates were clarified by centrifugation to give 108 cells per mL, as described (35). The total protein in lysates was measured by using Bradford Reagent (Sigma) following the manufacturer’s instructions. Electrophoresis and blotting was performed as described (35) with the primary antibody, a tissue culture supernatant containing mAb TD50, and incubated overnight at 4 °C. To ensure equal loading of all samples gels were stained with Commassie Brilliant Blue posttransfer and membranes were stripped (Restore Stripping Buffer; Thermo Scientific) and blotted with a primary actin antibody (clone AC-15; Sigma) according to the manufacturer’s instructions.
Immunohistochemistry.
DFTD primary tumors and metastases were fixed in 10% (mass/vol) PBS-buffered formalin solution for 2 to 4 d. Tissues were processed and embedded in paraffin blocks, which were cut onto 3-aminotriethoxysilane–coated slides at 3-µm thickness. Sections were deparaffinized in xylene and rehydrated through graded alcohol solutions to water and antigen epitopes were retrieved by using heat treatment with citrate buffer solution (pH 6) for 15 min. Endogenous peroxidase and nonspecific protein binding were blocked by incubation of the slides with 3% (mass/mass) hydrogen peroxide (Analar) and serum-free block solution (Dako). Sections were then incubated with protein G-purified anti-devil β2m-Ab (1.5 mg/mL), protein G-purified preimmune rat serum, anti-periaxin (Sigma; diluted 1:300), or anti-CD3ε (Sigma; A0452) (list of antibodies in SI Appendix, Table S5), all diluted in antibody diluent (Dako) and incubated overnight at 4 °C. Primary antibody binding was detected with peroxidase-coupled secondary antibody (Envision kit; Dako). Sections were counterstained with hematoxylin for 40 s, dehydrated through graded alcohol solutions to xylene and cover-slipped. Sections were visually analyzed by using a Leica DM 2500 microscope, and selected micrographs were obtained with a Leica FireCam DFC320 camera.
Supplementary Material
Acknowledgments
We thank Narelle Phillips for preparing tissue sections; Stephan Beck, Clive Tregaskes, and Ashley Moffett for valuable discussion; and Anne Cooke, Gillian Griffiths, and Clive Tregaskes for critically reading the manuscript. H.V.S. was supported by a National Health and Medical Research Council Overseas Postdoctoral Fellowship and is currently supported by a European Molecular Biology Organisation long-term fellowship. A.K. is supported by an Australian Research Council-linkage grant. The experiments in this paper were funded by a University of Tasmania Dr. Eric Guiler Tasmanian Devil Research Grant (to H.V.S.) and by Wellcome Trust programme Grant 089305 (to J.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219920110/-/DCSupplemental.
References
- 1.Pearse AM, Swift K. Allograft theory: Transmission of devil facial-tumour disease. Nature. 2006;439(7076):549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 2.Murgia C, Pritchard JK, Kim SY, Fassati A, Weiss RA. Clonal origin and evolution of a transmissible cancer. Cell. 2006;126(3):477–487. doi: 10.1016/j.cell.2006.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebbeck CA, Thomas R, Breen M, Leroi AM, Burt A. Origins and evolution of a transmissible cancer. Evolution. 2009;63(9):2340–2349. doi: 10.1111/j.1558-5646.2009.00724.x. [DOI] [PubMed] [Google Scholar]
- 4.Woods GM, et al. The immune response of the Tasmanian devil (sarcophilus harrisii) and devil facial tumour disease. EcoHealth. 2007;4(3):388–345. [Google Scholar]
- 5.Cohen D. The biological behaviour of the transmissible venereal tumour in immunosuppressed dogs. Eur J Cancer. 1973;9(4):253–258. doi: 10.1016/0014-2964(73)90090-x. [DOI] [PubMed] [Google Scholar]
- 6.Pérez J, Day MJ, Mozos E. Immunohistochemical study of the local inflammatory infiltrate in spontaneous canine transmissible venereal tumour at different stages of growth. Vet Immunol Immunopathol. 1998;64(2):133–147. doi: 10.1016/s0165-2427(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 7.Hsiao YW, Liao KW, Hung SW, Chu RM. Tumor-infiltrating lymphocyte secretion of IL-6 antagonizes tumor-derived TGF-beta 1 and restores the lymphokine-activated killing activity. J Immunol. 2004;172(3):1508–1514. doi: 10.4049/jimmunol.172.3.1508. [DOI] [PubMed] [Google Scholar]
- 8.Doherty PC, Zinkernagel RM. A biological role for the major histocompatibility antigens. Lancet. 1975;1(7922):1406–1409. doi: 10.1016/s0140-6736(75)92610-0. [DOI] [PubMed] [Google Scholar]
- 9.Lehner PJ, Cresswell P. Processing and delivery of peptides presented by MHC class I molecules. Curr Opin Immunol. 1996;8(1):59–67. doi: 10.1016/s0952-7915(96)80106-3. [DOI] [PubMed] [Google Scholar]
- 10.Villadangos JA. Presentation of antigens by MHC class II molecules: Getting the most out of them. Mol Immunol. 2001;38(5):329–346. doi: 10.1016/s0161-5890(01)00069-4. [DOI] [PubMed] [Google Scholar]
- 11.van den Elsen PJ, Holling TM, Kuipers HF, van der Stoep N. Transcriptional regulation of antigen presentation. Curr Opin Immunol. 2004;16(1):67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Ting JP, Baldwin AS. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5(1):8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 13.Martin BK, et al. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. 1997;6(5):591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- 14.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao YW, et al. Interactions of host IL-6 and IFN-gamma and cancer-derived TGF-beta1 on MHC molecule expression during tumor spontaneous regression. Cancer Immunol Immunother. 2008;57(7):1091–1104. doi: 10.1007/s00262-007-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siddle HV, et al. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Natl Acad Sci USA. 2007;104(41):16221–16226. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murchison EP, et al. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science. 2010;327(5961):84–87. doi: 10.1126/science.1180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murchison EP, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148(4):780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh R, et al. The immunohistochemical characterization of devil facial tumor disease (DFTD) in the Tasmanian Devil (Sarcophilus harrisii) Vet Pathol. 2006;43(6):896–903. doi: 10.1354/vp.43-6-896. [DOI] [PubMed] [Google Scholar]
- 20.Lane A, et al. New insights into the role of MHC diversity in devil facial tumour disease. PLoS ONE. 2012;7(6):e36955. doi: 10.1371/journal.pone.0036955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreiss A, et al. Allorecognition in the Tasmanian devil (Sarcophilus harrisii), an endangered marsupial species with limited genetic diversity. PLoS ONE. 2011;6(7):e22402. doi: 10.1371/journal.pone.0022402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCallum H, et al. Distribution and impacts of Tasmanian devil facial tumor disease. EcoHealth. 2007;4(3):318–325. [Google Scholar]
- 23.Lachish S, Jones M, McCallum H. The impact of disease on the survival and population growth rate of the Tasmanian devil. J Anim Ecol. 2007;76(5):926–936. doi: 10.1111/j.1365-2656.2007.01272.x. [DOI] [PubMed] [Google Scholar]
- 24.McCallum H, Jones M. To lose both would look like carelessness: Tasmanian devil facial tumour disease. PLoS Biol. 2006;4(10):e342. doi: 10.1371/journal.pbio.0040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, et al. Antigen-presenting genes and genomic copy number variations in the Tasmanian devil MHC. BMC Genomics. 2012;13:87. doi: 10.1186/1471-2164-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampson LA. MHC regulation in neural cells. Distribution of peripheral and internal beta 2-microglobulin and class I molecules in human neuroblastoma cell lines. J Immunol. 1990;144(2):512–520. [PubMed] [Google Scholar]
- 27.Meyer zu Hörste G, et al. Mouse Schwann cells activate MHC class I and II restricted T-cell responses, but require external peptide processing for MHC class II presentation. Neurobiol Dis. 2010;37(2):483–490. doi: 10.1016/j.nbd.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Armati PJ, Pollard JD, Gatenby P. Rat and human Schwann cells in vitro can synthesize and express MHC molecules. Muscle Nerve. 1990;13(2):106–116. doi: 10.1002/mus.880130204. [DOI] [PubMed] [Google Scholar]
- 29.Gobin SJ, et al. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta2-microglobulin genes. Immunity. 1998;9(4):531–541. doi: 10.1016/s1074-7613(00)80636-6. [DOI] [PubMed] [Google Scholar]
- 30.Raval A, et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol Cell. 2001;7(1):105–115. doi: 10.1016/s1097-2765(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 31.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11(7):237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 32.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6(8):584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 33.Pyecroft SB, et al. Towards a case definition for devil facial tumor disease: What is it? EcoHealth. 2007;4(3):346–351. [Google Scholar]
- 34.Deakin JE, et al. Genomic restructuring in the Tasmanian devil facial tumour: Chromosome painting and gene mapping provide clues to evolution of a transmissible tumour. PLoS Genet. 2012;8(2):e1002483. doi: 10.1371/journal.pgen.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker BA, et al. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci USA. 2011;108(20):8396–8401. doi: 10.1073/pnas.1019496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.